The score is a number that reflects the conservation at a position.
position_aggregator: mean [default]
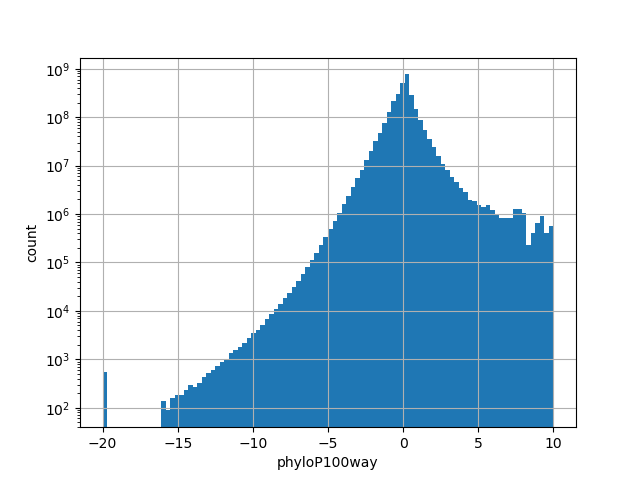
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
| Id: | pipeline/GPF-SFARI_annotation |
| Type: | annotation_pipeline |
| Version: | 0 |
| Summary: |
GPF-SFARI Production Annotation Pipeline |
| Description: |
This is the pipeline used in the GPF-SFARI instance. |
| Labels: |
| Summary | GPF-SFARI Production Annotation Pipeline |
|---|---|
| Description | This is the pipeline used in the GPF-SFARI instance. |
| Input reference genome | hg38/genomes/GRCh38-hg38 |
Worst effect accross all transcripts.
comma separated list of genes with worst effect.
<gene_1>:<effect_1>|... A gene can be repeated.
Effect details for each affected transcript. Format: < transcript 1 >:<gene 1>:<effect 1>:<details 1>|...
List of all genes
Annotator to identify the effect of the variant on protein coding.
The score is a number that reflects the conservation at a position.
position_aggregator: mean [default]

Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
The score is a number that reflects the conservation at a position.
position_aggregator: mean [default]
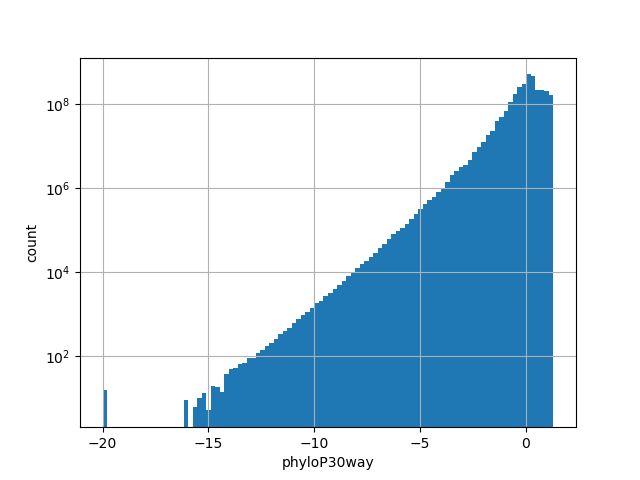
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
The score is a number that reflects the conservation at a position.
position_aggregator: mean [default]
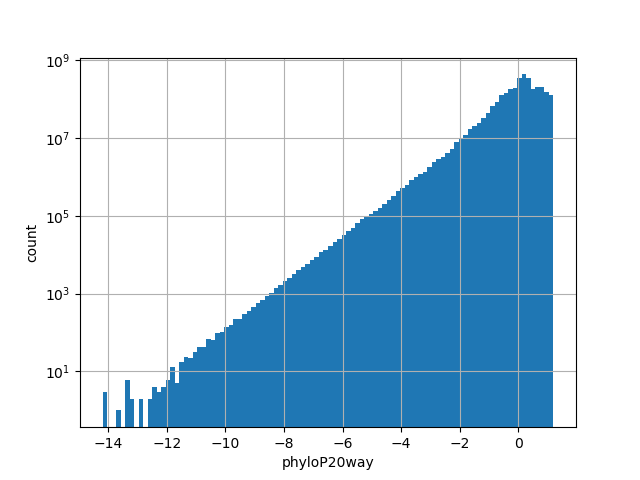
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
The score is a number that reflects the conservation at a position.
position_aggregator: mean [default]
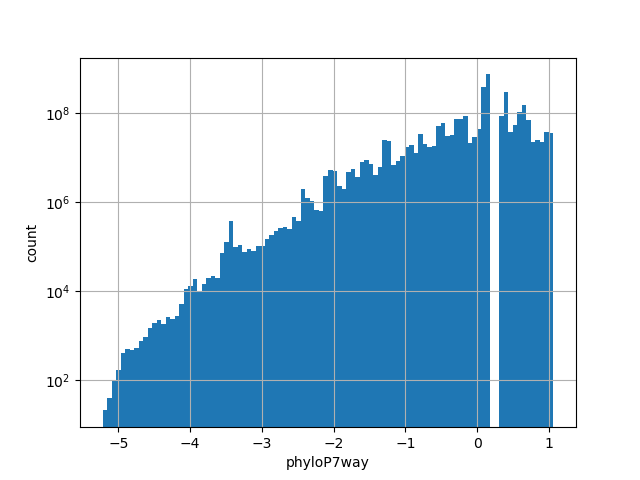
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
The score is a number that reflects the conservation at a position.
position_aggregator: mean [default]
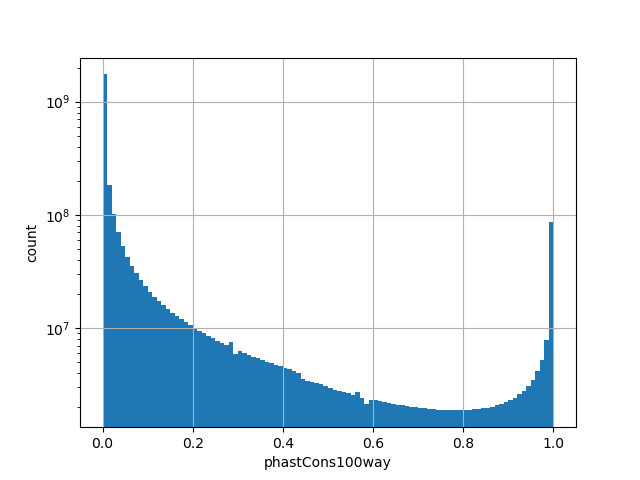
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
The score is a number that reflects the conservation at a position.
position_aggregator: mean [default]
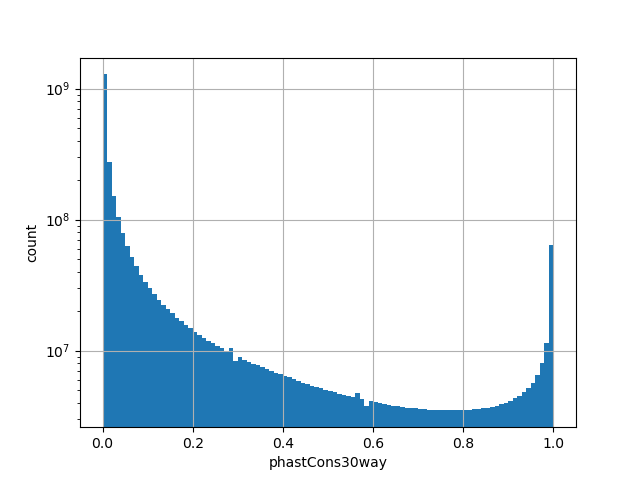
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
The score is a number that reflects the conservation at a position.
position_aggregator: mean [default]
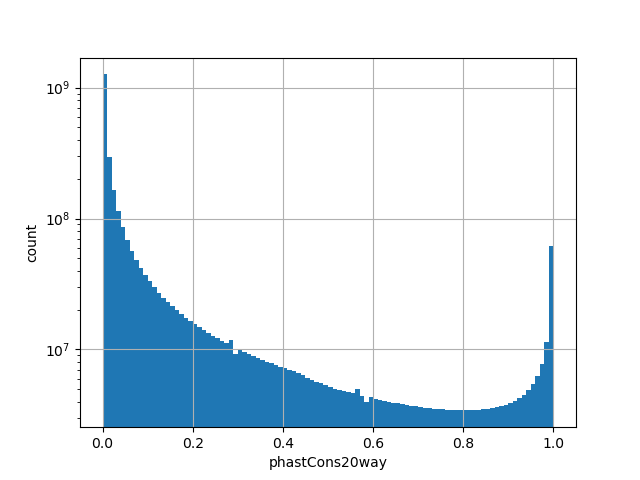
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
The score is a number that reflects the conservation at a position.
position_aggregator: mean [default]
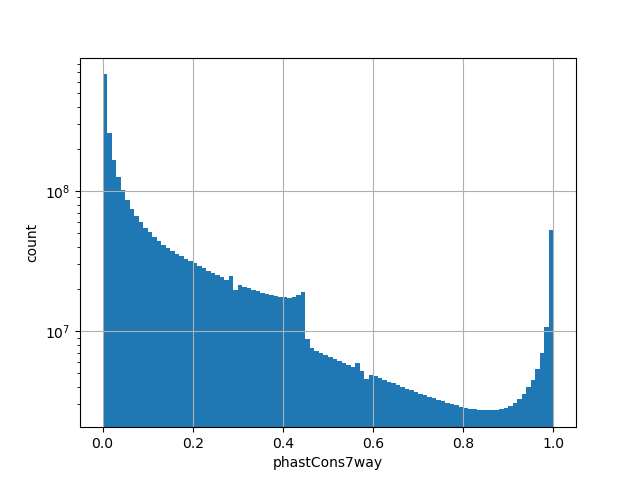
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
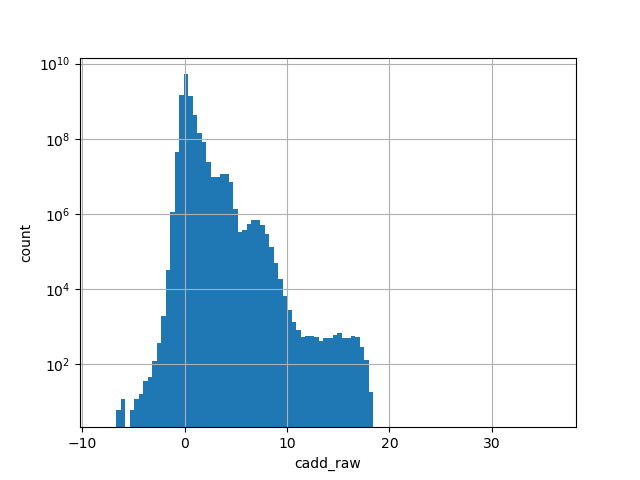
CADD raw score for functional prediction of a SNP. The larger the score the more likely the SNP has damaging effect
position_aggregator: mean [default]
allele_aggregator: max [default]
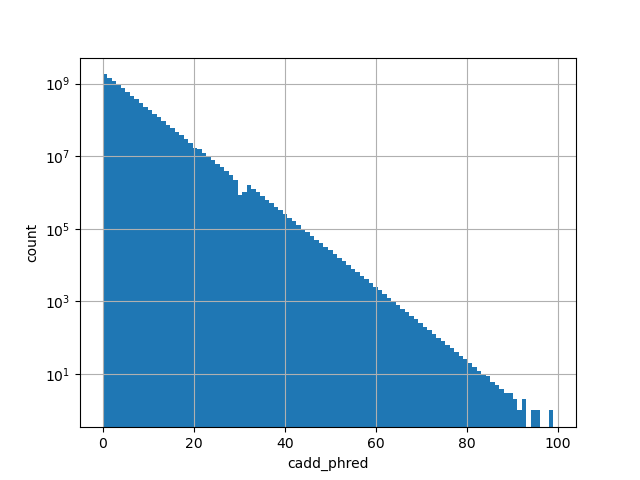
CADD phred-like score. This is phred-like rank score based on whole genome CADD raw scores. The larger the score the more likely the SNP has damaging effect.
position_aggregator: mean [default]
allele_aggregator: max [default]
Annotator to use with scores that depend on allele like variant frequencies, etc.
The lifted over annotatable
Annotator to lift over a variant from one reference genome to another.
probability that a point mutation at each position in a genome will influence fitness
position_aggregator: mean [default]
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
hg19_annotatableThe LINSIGHT score measures the probability of negative selection on noncoding sites.
position_aggregator: mean [default]
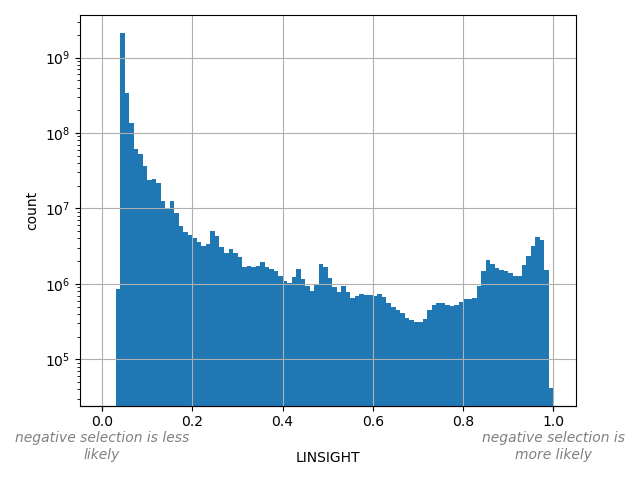
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
hg19_annotatableThe scores is the probability for a mutation to have a fitenss consequence.
position_aggregator: mean [default]
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
hg19_annotatableThe scores is the probability for a mutation to have a fitenss consequence.
position_aggregator: mean [default]
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
hg19_annotatableThe scores is the probability for a mutation to have a fitenss consequence.
position_aggregator: mean [default]
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
hg19_annotatableThe scores is the probability for a mutation to have a fitenss consequence.
position_aggregator: mean [default]
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
hg19_annotatableThe scores is the probability for a mutation to have a fitenss consequence.
position_aggregator: mean [default]
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
hg19_annotatableThe scores is the probability for a mutation to have a fitenss consequence.
position_aggregator: mean [default]
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
hg19_annotatableThe scores is the probability for a mutation to have a fitenss consequence.
position_aggregator: mean [default]
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
hg19_annotatableThe scores is the probability for a mutation to have a fitenss consequence.
position_aggregator: mean [default]
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
hg19_annotatableThe scores is the probability for a mutation to have a fitness consequence.
position_aggregator: mean [default]
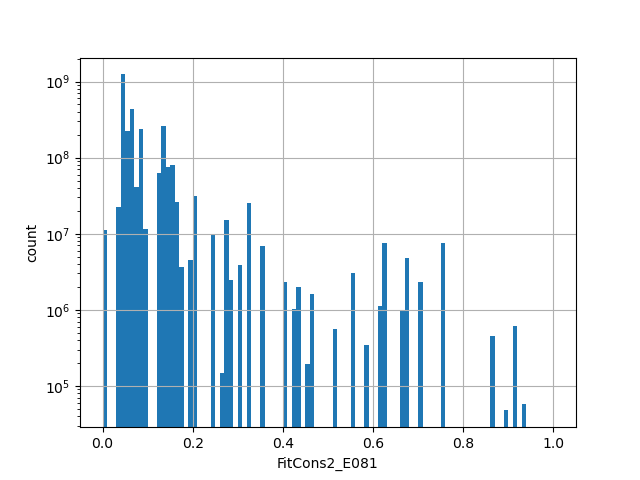
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
hg19_annotatableThe scores is the probability for a mutation to have a fitenss consequence.
position_aggregator: mean [default]
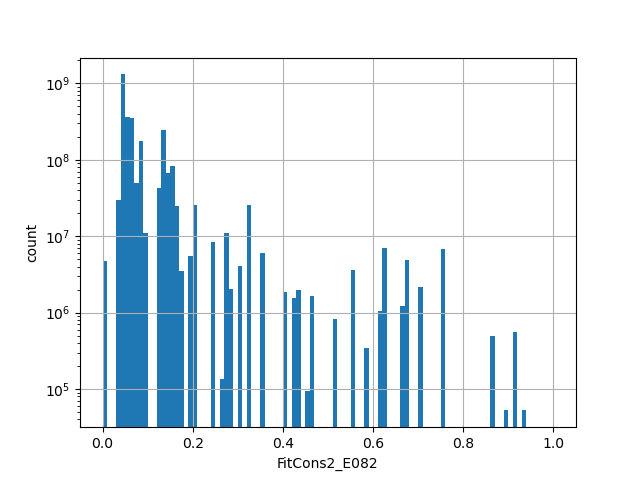
Annotator to use with genomic scores depending on genomic position like phastCons, phyloP, FitCons2, etc.
hg19_annotatableMissense badness, PolyPhen-2, and Constraint. A deleteriousness prediction score for missense variants"
position_aggregator: mean [default]
allele_aggregator: max [default]
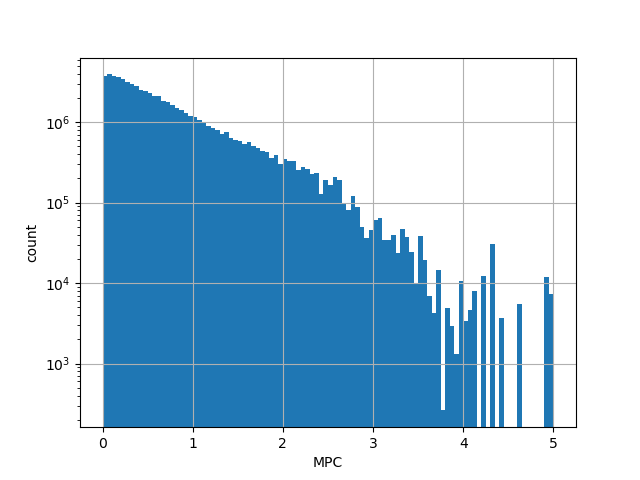
Annotator to use with scores that depend on allele like variant frequencies, etc.
hg19_annotatableNormalized allele.
SFARI SSC WGS CSHL allele frequency in %
position_aggregator: mean [default]
allele_aggregator: max [default]
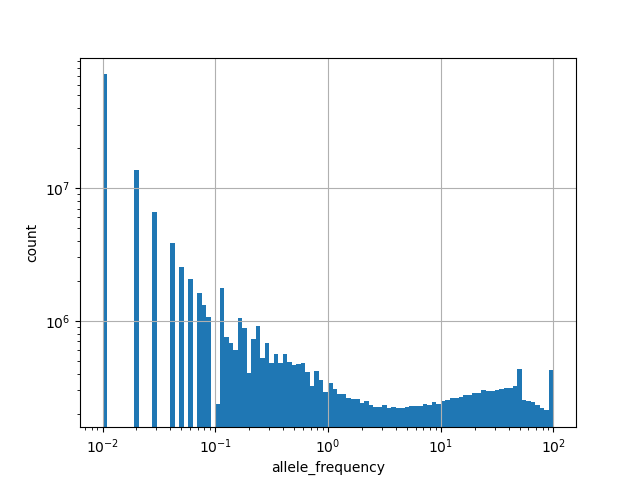
Annotator to use with scores that depend on allele like variant frequencies, etc.
Alternative allele frequency in the whole gnomAD exome samples v2.1.1 as %
position_aggregator: mean [default]
allele_aggregator: max [default]
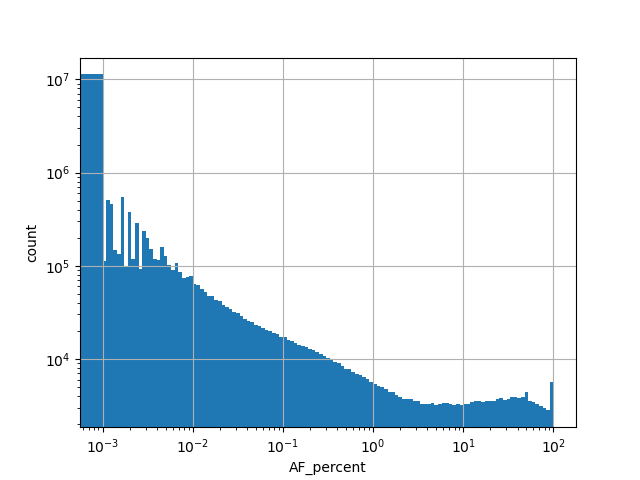
Alternative allele count in the whole gnomAD exome samples v2.1.1
position_aggregator: mean [default]
allele_aggregator: max [default]
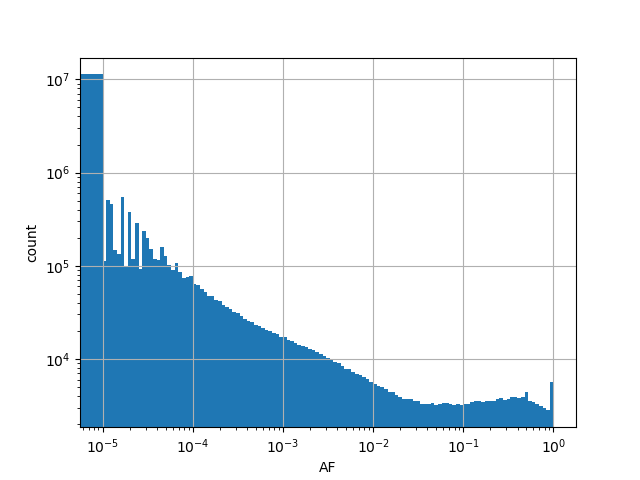
Alternative allele frequency in the whole gnomAD exome samples v2.1.1
position_aggregator: mean [default]
allele_aggregator: max [default]
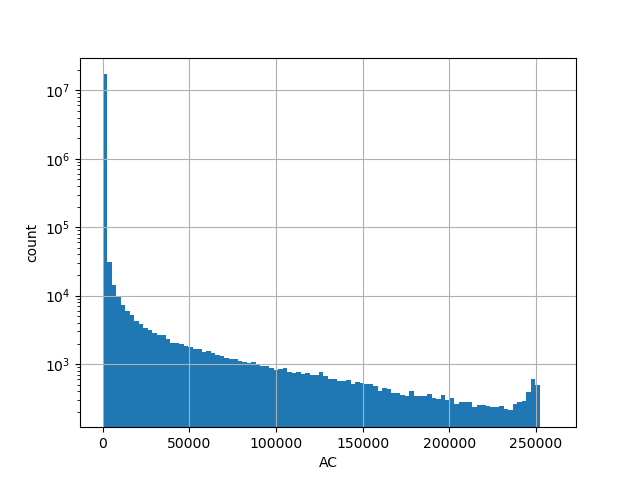
Total allele count in the whole gnomAD exome samples v2.1.1
position_aggregator: mean [default]
allele_aggregator: max [default]
Alternative allele count in the controls subset of whole gnomAD exome samples v2.1.1
position_aggregator: mean [default]
allele_aggregator: max [default]
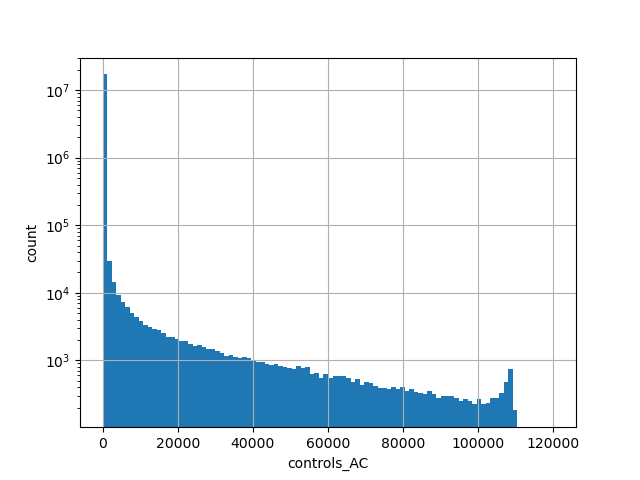
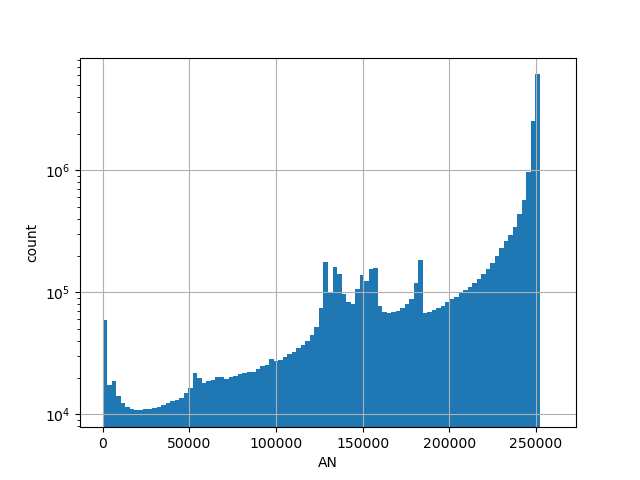
gnomAD v2.1.1 liftover exomes count of genotyped individuals in control group
position_aggregator: mean [default]
allele_aggregator: max [default]
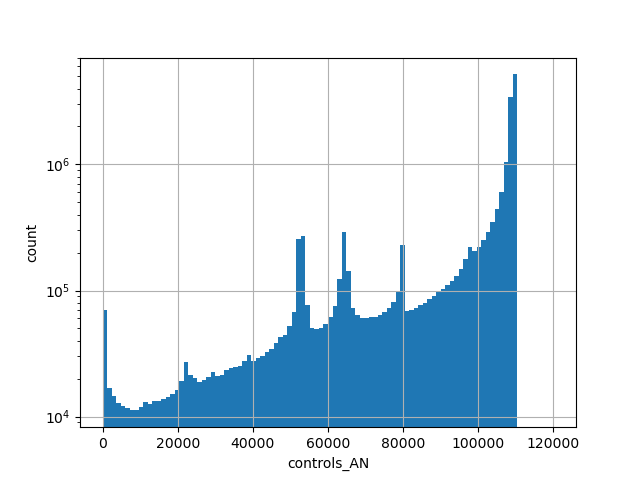
Alternative allele count in the non-neuro subset of whole gnomAD exome samples v2.1.1
position_aggregator: mean [default]
allele_aggregator: max [default]
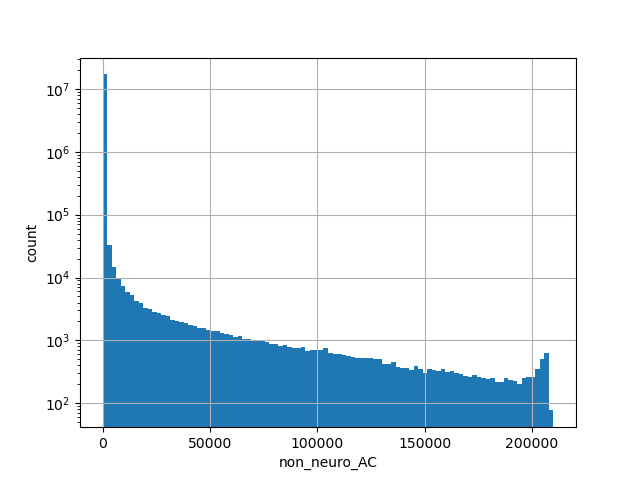
Total allele count in the non-neuro subset of whole gnomAD exome samples v2.1.1
position_aggregator: mean [default]
allele_aggregator: max [default]
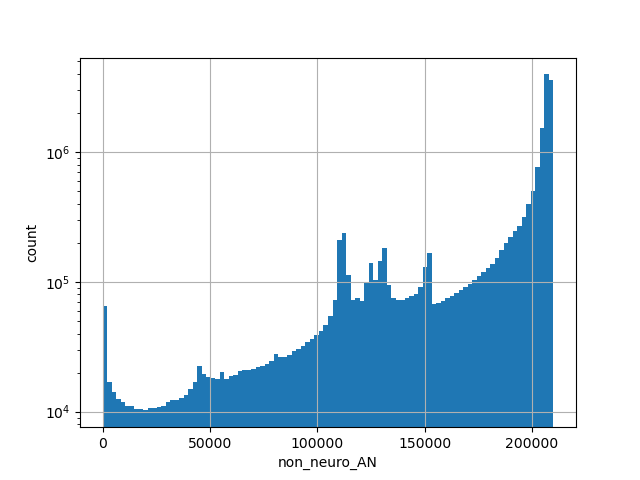
Alternative allele frequency in the controls subset of whole gnomAD exome samples v2.1.1 as %
position_aggregator: mean [default]
allele_aggregator: max [default]
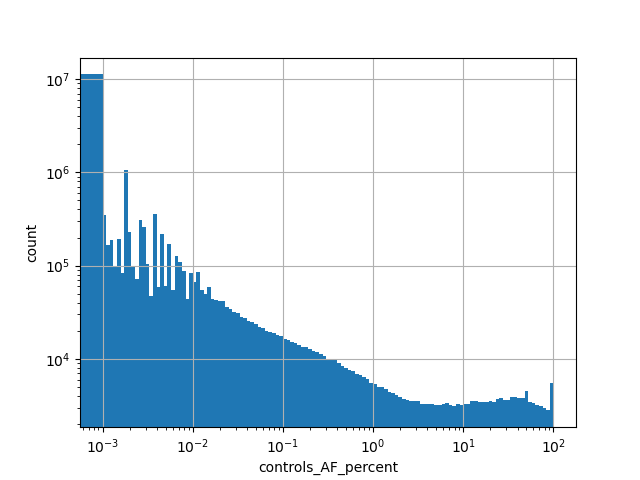
Alternative allele frequency in the non-neuro subset of whole gnomAD exome samples v2.1.1 as %
position_aggregator: mean [default]
allele_aggregator: max [default]
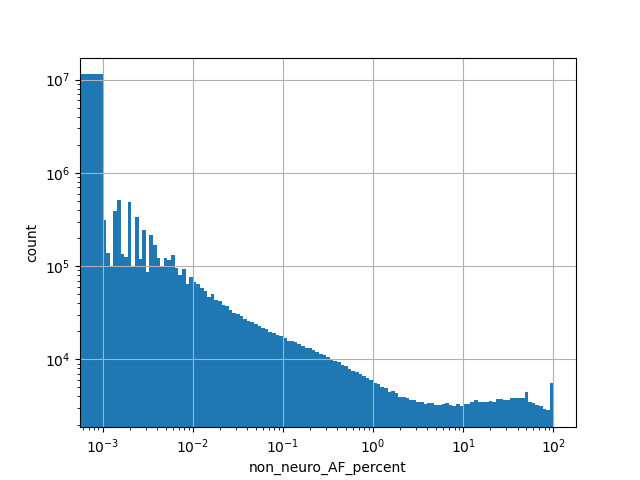
Annotator to use with scores that depend on allele like variant frequencies, etc.
normalized_alleleAlternative allele frequency in the whole gnomAD genome samples v2.1.1 as %
position_aggregator: mean [default]
allele_aggregator: max [default]
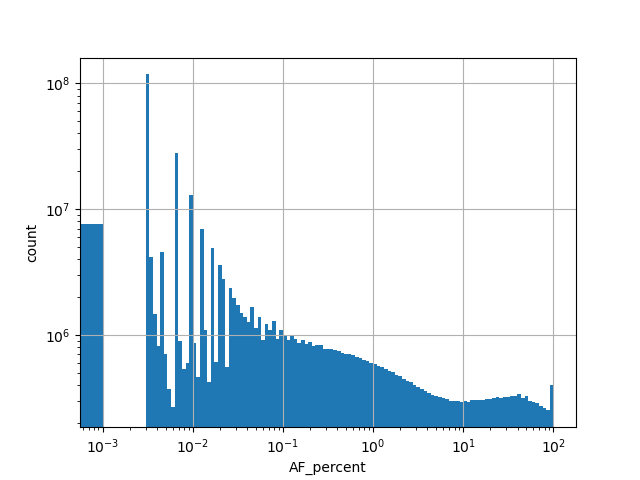
Alternative allele count in the whole gnomAD genome samples v2.1.1
position_aggregator: mean [default]
allele_aggregator: max [default]
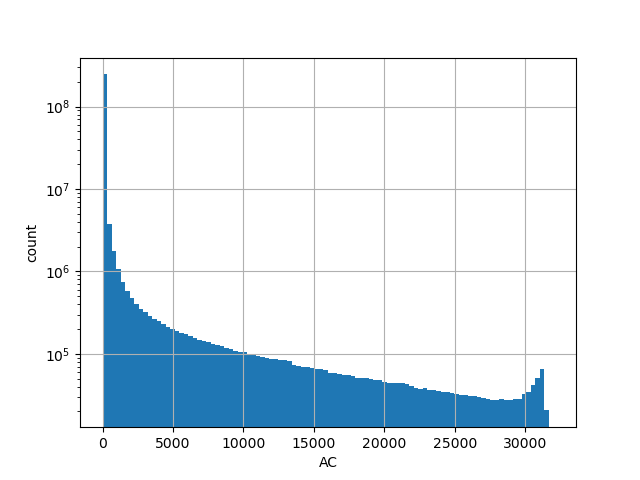
Alternative allele frequency in the whole gnomAD genome samples v2.1.1
position_aggregator: mean [default]
allele_aggregator: max [default]
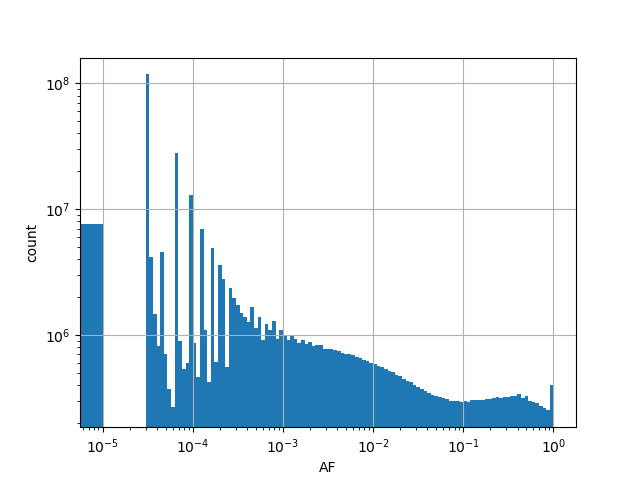
Total allele count in the whole gnomAD genome samples v2.1.1
position_aggregator: mean [default]
allele_aggregator: max [default]
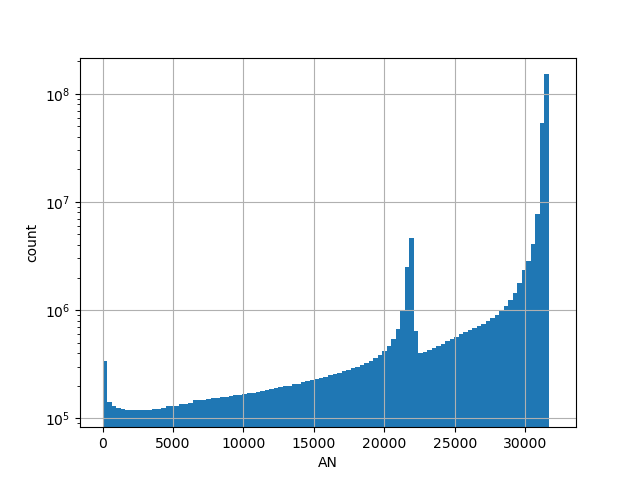
Alternative allele count in the controls subset of whole gnomAD genome samples v2.1.1
position_aggregator: mean [default]
allele_aggregator: max [default]
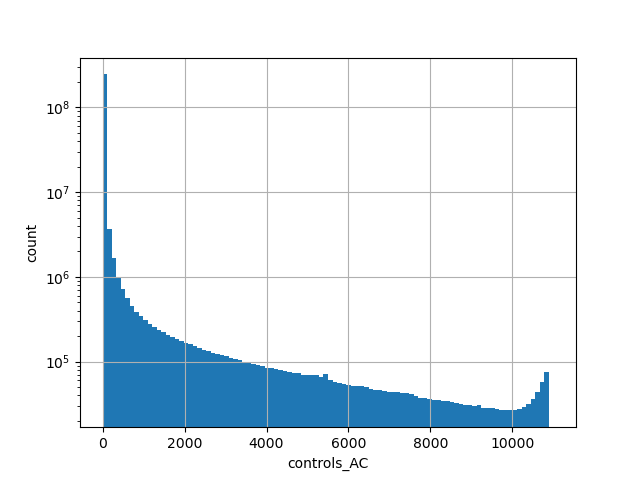
gnomAD v2.1.1 liftover genome count of genotyped individuals in control group
position_aggregator: mean [default]
allele_aggregator: max [default]
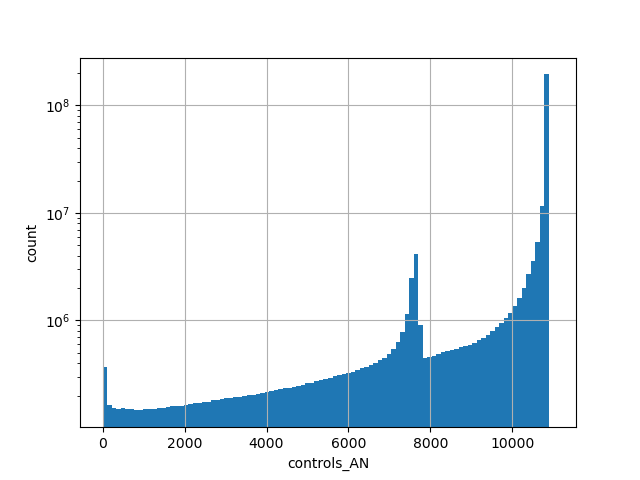
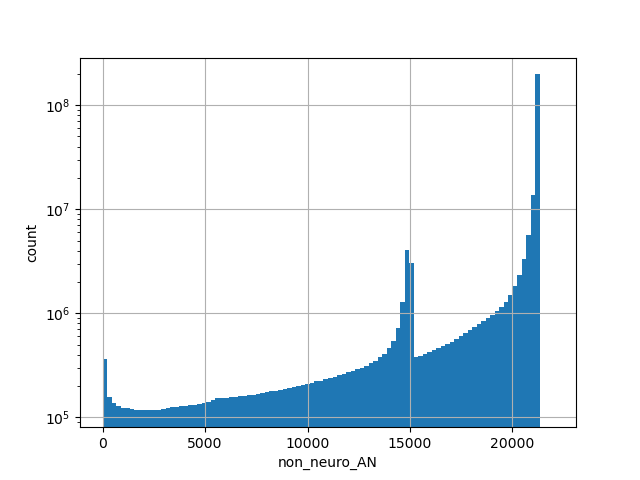
Alternative allele count in the non-neuro subset of whole gnomAD genome samples v2.1.1
position_aggregator: mean [default]
allele_aggregator: max [default]
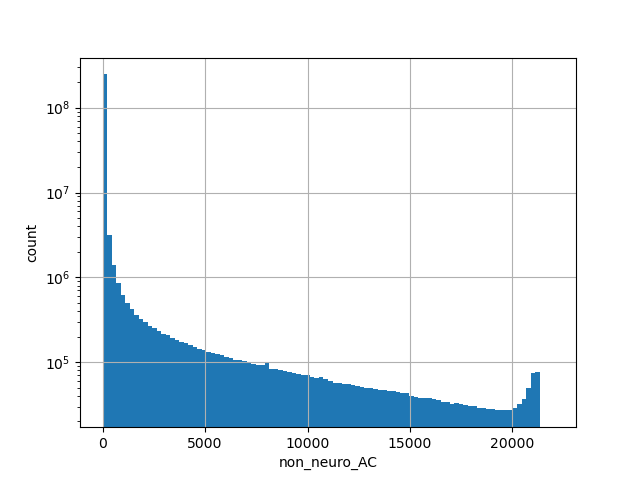
Total allele count in the non-neuro subset of whole gnomAD genome samples v2.1.1
position_aggregator: mean [default]
allele_aggregator: max [default]
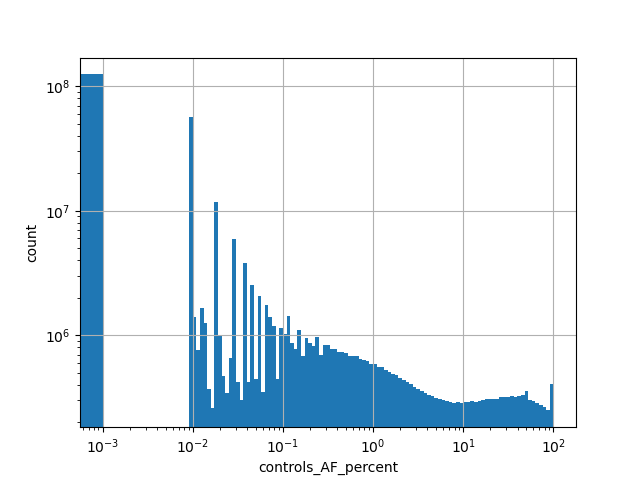
Alternative allele frequency in the controls subset of whole gnomAD genome samples v2.1.1 as %
position_aggregator: mean [default]
allele_aggregator: max [default]
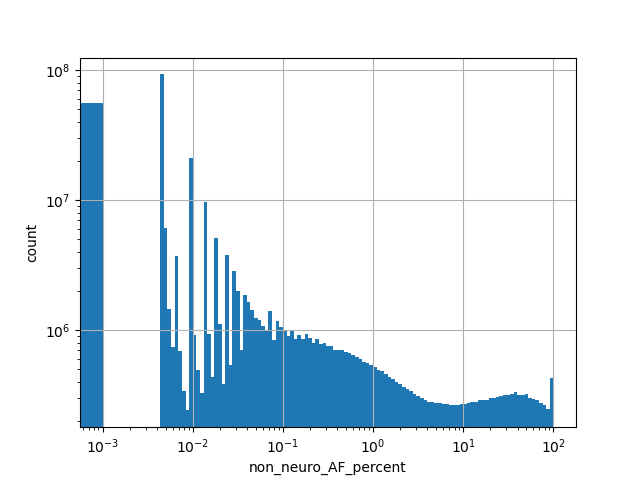
Alternative allele frequency in the non-neuro subset of whole gnomAD genome samples v2.1.1 as %
position_aggregator: mean [default]
allele_aggregator: max [default]
Annotator to use with scores that depend on allele like variant frequencies, etc.
normalized_alleleAlternative allele frequency represented as a percent in the all gnomAD v3.0 genome samples.
position_aggregator: mean [default]
allele_aggregator: max [default]
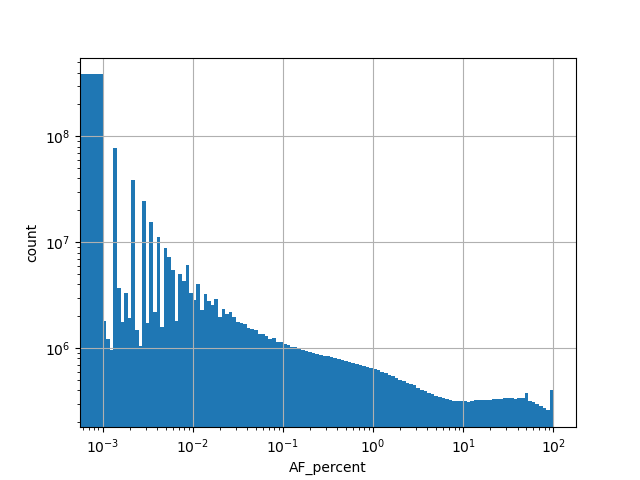
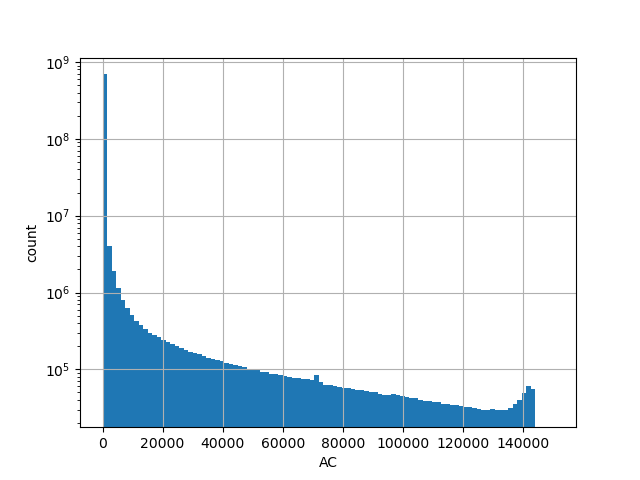
Number of alternative alleles in the all gnomAD v3.0 genome samples.
position_aggregator: mean [default]
allele_aggregator: max [default]
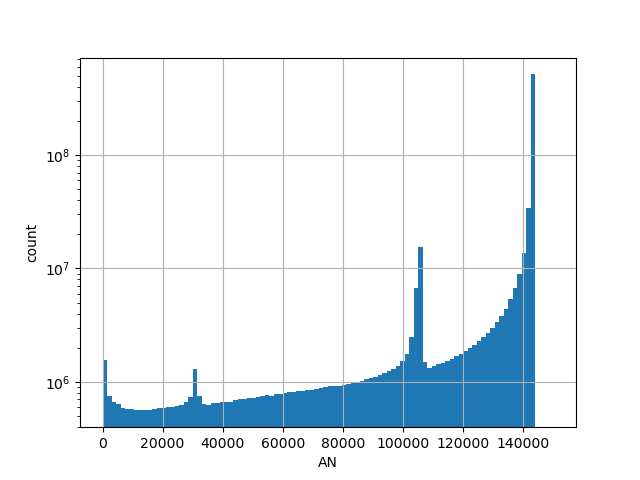
Number of genotyped individuals in the all gnomAD v3.0 genome samples.
position_aggregator: mean [default]
allele_aggregator: max [default]
Annotator to use with scores that depend on allele like variant frequencies, etc.
normalized_allele| Filename | Size | md5 |
|---|---|---|
| GPF-SFARI_annotation.yaml | 2.79 KB | 1c01dbef40729b322875b66413c93493 |
| genomic_resource.yaml | 232.0 B | ef3a0a26a02c613fe081d1c681d5ce73 |
| statistics/ |