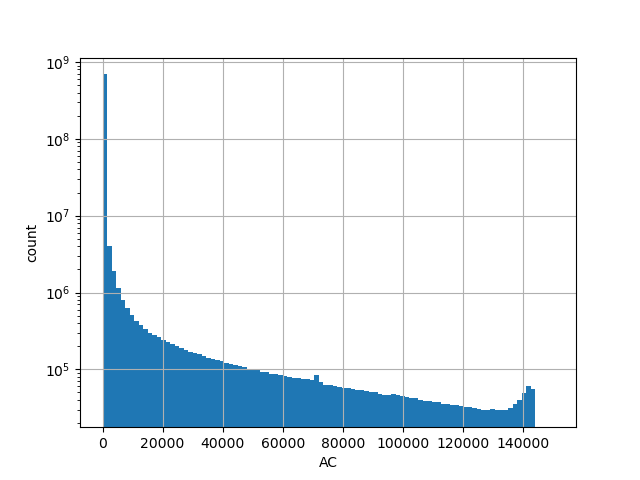
genome_gnomad_v3_ac
Number of alternative alleles in the all gnomAD v3.0 genome samples.
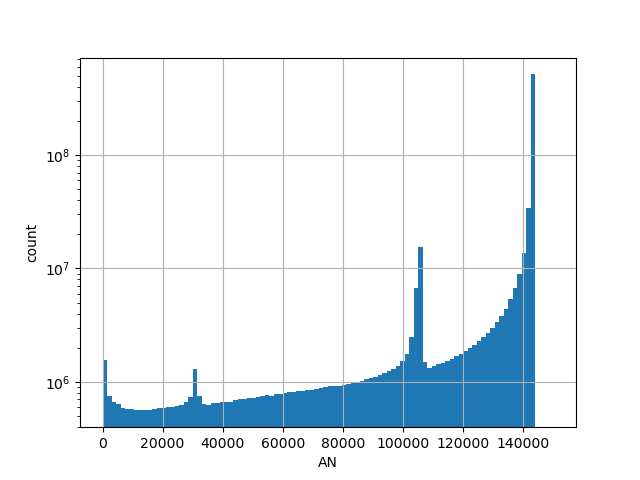
genome_gnomad_v3_an
Number of genotyped individuals in the all gnomAD v3.0 genome samples.
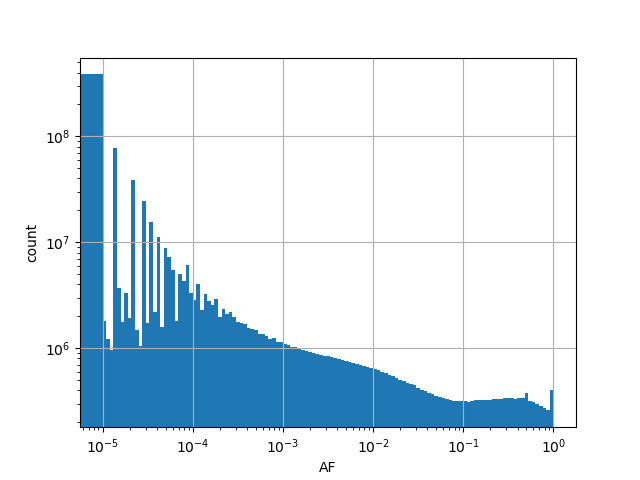
-
Alternative allele frequency in the all gnomAD v3.0 genome samples.
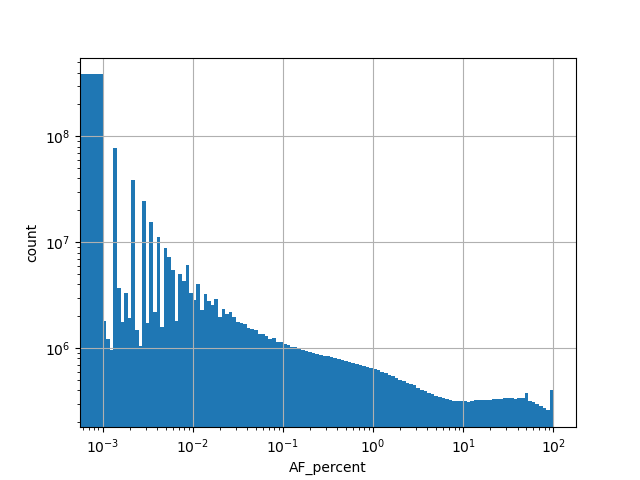
genome_gnomad_v3_af_percent
Alternative allele frequency represented as a percent in the all gnomAD v3.0 genome samples.