| ID |
Type |
Default annotation |
Description |
Histogram |
Range |
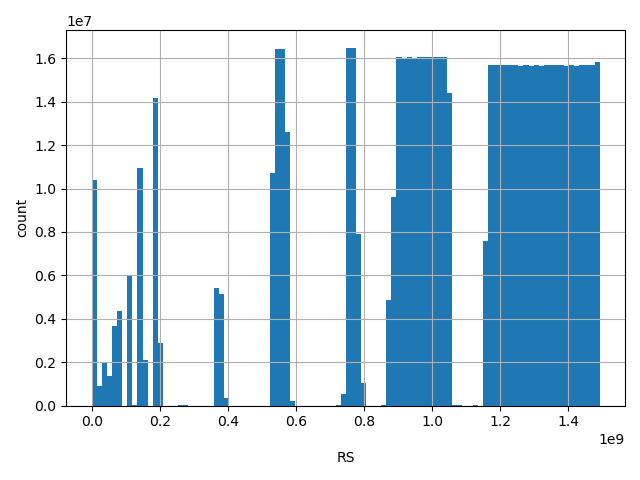
| RS |
int |
dbSNP_RS
|
dbSNP ID (i.e. rs number)
|
No histogram: Histogram is not available for this score. |
NO DOMAIN |
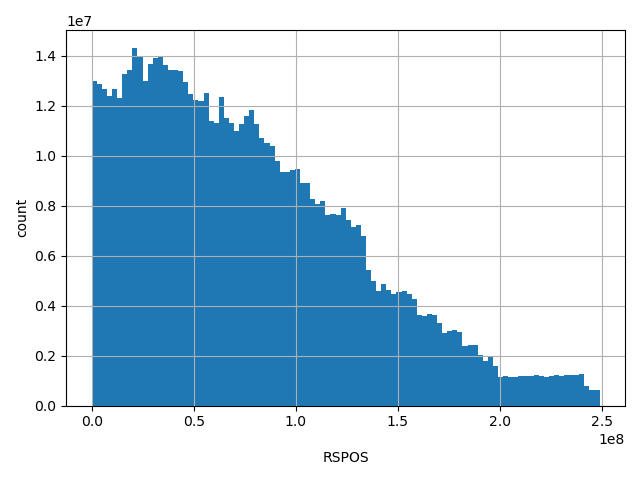
| RSPOS |
int |
-
|
Chr position reported in dbSNP
|
No histogram: Histogram is not available for this score. |
NO DOMAIN |
| RV |
bool |
-
|
RS orientation is reversed
|
 |
false, true |
| VP |
str |
-
|
Variation Property. Documentation is at ftp://ftp.ncbi.nlm.nih.gov/snp/specs/dbSNP_BitField_latest.pdf
|
 |
0x050000080005000002000100, 0x050000000005000002000100, 0x050000080005000002000200, 0x050000000005000002000200, 0x050000080005000026000100, 0x0500000a0005000002000100, 0x050000000005000026000100, 0x050000020005000002000100, 0x050000080005040026000100, 0x050000080005040036000100, Other |
| GENEINFO |
str |
-
|
Pairs each of gene symbol:gene id. The gene symbol and id are delimited by a colon (:) and each pair is delimited by a vertical bar (|)
|
 |
CSMD1:64478, RBFOX1:54715, PTPRD:5789, CNTNAP2:26047, DLG2:1740, WWOX:51741, EYS:346007, DMD:1756, LRP1B:53353, MACROD2:140733, Other |
| dbSNPBuildID |
int |
-
|
First dbSNP Build for RS
|
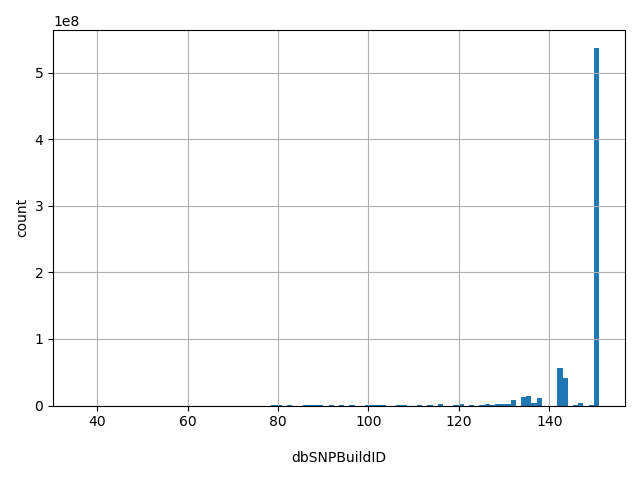 |
151, 150, 142, 144, 135, 134, 138, 132, 147, 137, Other |
| SAO |
int |
-
|
Variant Allele Origin: 0 - unspecified, 1 - Germline, 2 - Somatic, 3 - Both
|
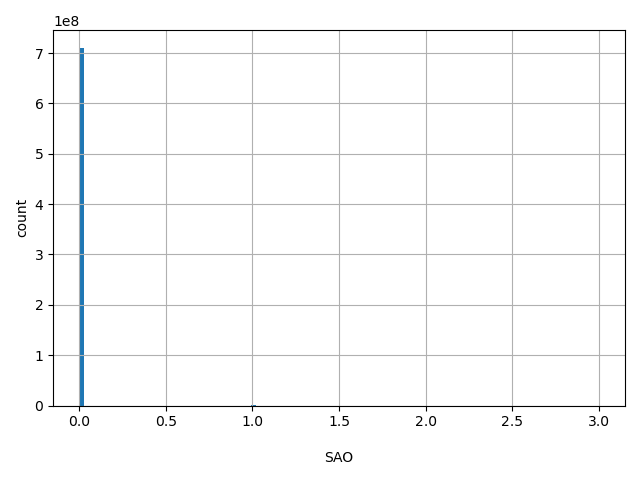 |
0, 1, 3 |
| SSR |
int |
-
|
Variant Suspect Reason Codes (may be more than one value added together) 0 - unspecified, 1 - Paralog, 2 - byEST, 4 - oldAlign, 8 - Para_EST, 16 - 1kg_failed, 1024 - other
|
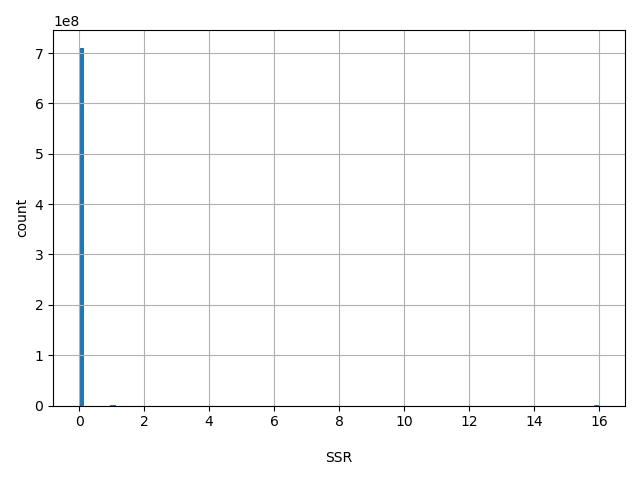 |
0, 16, 1 |
| WGT |
int |
-
|
Weight, 00 - unmapped, 1 - weight 1, 2 - weight 2, 3 - weight 3 or more
|
 |
1 |
| VC |
str |
-
|
Variation Class
|
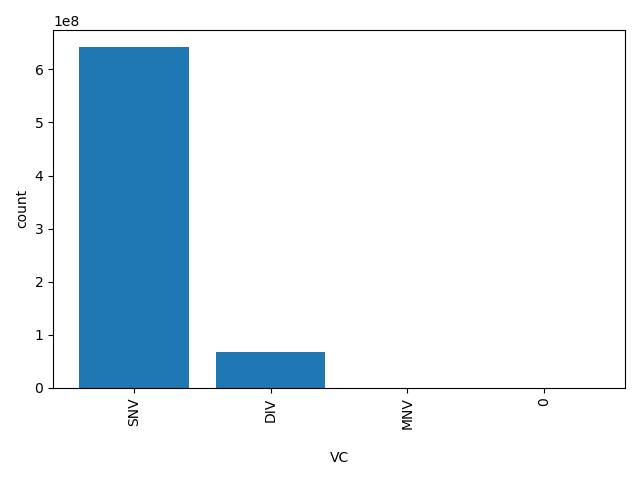 |
SNV, DIV, MNV, 0 |
| PM |
bool |
-
|
Variant is Precious(Clinical,Pubmed Cited)
|
 |
false, true |
| TPA |
bool |
-
|
Provisional Third Party Annotation(TPA) (currently rs from PHARMGKB who will give phenotype data)
|
 |
false, true |
| PMC |
bool |
-
|
Links exist to PubMed Central article
|
 |
false, true |
| S3D |
bool |
-
|
Has 3D structure - SNP3D table
|
 |
false, true |
| SLO |
bool |
-
|
Has SubmitterLinkOut - From SNP->SubSNP->Batch.link_out
|
 |
false, true |
| NSF |
bool |
-
|
Has non-synonymous frameshift A coding region variation where one allele in the set changes all downstream amino acids. FxnClass = 44
|
 |
false, true |
| NSM |
bool |
-
|
Has non-synonymous missense A coding region variation where one allele in the set changes protein peptide. FxnClass = 42
|
 |
false, true |
| NSN |
bool |
-
|
Has non-synonymous nonsense A coding region variation where one allele in the set changes to STOP codon (TER). FxnClass = 41
|
 |
false, true |
| REF |
bool |
-
|
Has reference A coding region variation where one allele in the set is identical to the reference sequence. FxnCode = 8
|
 |
false, true |
| SYN |
bool |
-
|
Has synonymous A coding region variation where one allele in the set does not change the encoded amino acid. FxnCode = 3
|
 |
false, true |
| U3 |
bool |
-
|
In 3' UTR Location is in an untranslated region (UTR). FxnCode = 53
|
 |
false, true |
| U5 |
bool |
-
|
In 5' UTR Location is in an untranslated region (UTR). FxnCode = 55
|
 |
false, true |
| ASS |
bool |
-
|
In acceptor splice site FxnCode = 73
|
 |
false, true |
| DSS |
bool |
-
|
In donor splice-site FxnCode = 75
|
 |
false, true |
| INT |
bool |
-
|
In Intron FxnCode = 6
|
 |
true, false |
| R3 |
bool |
-
|
In 3' gene region FxnCode = 13
|
 |
false, true |
| R5 |
bool |
-
|
In 5' gene region FxnCode = 15
|
 |
false, true |
| OTH |
bool |
-
|
Has other variant with exactly the same set of mapped positions on NCBI refernce assembly.
|
 |
false, true |
| CFL |
bool |
-
|
Has Assembly conflict. This is for weight 1 and 2 variant that maps to different chromosomes on different assemblies.
|
 |
false, true |
| ASP |
bool |
-
|
Is Assembly specific. This is set if the variant only maps to one assembly
|
 |
true |
| MUT |
bool |
-
|
Is mutation (journal citation, explicit fact): a low frequency variation that is cited in journal and other reputable sources
|
 |
false |
| VLD |
bool |
-
|
Is Validated. This bit is set if the variant has 2+ minor allele count based on frequency or genotype data.
|
 |
false, true |
| G5A |
bool |
-
|
>5% minor allele frequency in each and all populations
|
 |
false, true |
| G5 |
bool |
-
|
>5% minor allele frequency in 1+ populations
|
 |
false, true |
| HD |
bool |
-
|
Marker is on high density genotyping kit (50K density or greater). The variant may have phenotype associations present in dbGaP.
|
 |
false, true |
| GNO |
int |
-
|
Genotypes available. The variant has individual genotype (in SubInd table).
|
 |
0, 1 |
| KGPhase1 |
bool |
-
|
1000 Genome phase 1 (incl. June Interim phase 1)
|
 |
false, true |
| KGPhase3 |
bool |
-
|
1000 Genome phase 3
|
 |
false, true |
| CDA |
bool |
-
|
Variation is interrogated in a clinical diagnostic assay
|
 |
false |
| LSD |
bool |
-
|
Submitted from a locus-specific database
|
 |
false, true |
| MTP |
bool |
-
|
Microattribution/third-party annotation(TPA:GWAS,PAGE)
|
 |
false, true |
| OM |
bool |
-
|
Has OMIM/OMIA
|
 |
false, true |
| NOC |
bool |
-
|
Contig allele not present in variant allele list. The reference sequence allele at the mapped position is not present in the variant allele list, adjusted for orientation.
|
 |
false, true |
| WTD |
bool |
-
|
Is Withdrawn by submitter If one member ss is withdrawn by submitter, then this bit is set. If all member ss' are withdrawn, then the rs is deleted to SNPHistory
|
 |
false |
| NOV |
bool |
-
|
Rs cluster has non-overlapping allele sets. True when rs set has more than 2 alleles from different submissions and these sets share no alleles in common.
|
 |
false, true |
| CAF |
str |
-
|
An ordered, comma delimited list of allele frequencies based on 1000Genomes, starting with the reference allele followed by alternate alleles as ordered in the ALT column. Where a 1000Genomes alternate allele is not in the dbSNPs alternate allele set, the allele is added to the ALT column. The minor allele is the second largest value in the list, and was previuosly reported in VCF as the GMAF. This is the GMAF reported on the RefSNP and EntrezSNP pages and VariationReporter
|
No histogram: Histogram is not available for this score. |
NO DOMAIN |
| COMMON |
int |
-
|
RS is a common SNP. A common SNP is one that has at least one 1000Genomes population with a minor allele of frequency >= 1% and for which 2 or more founders contribute to that minor allele frequency.
|
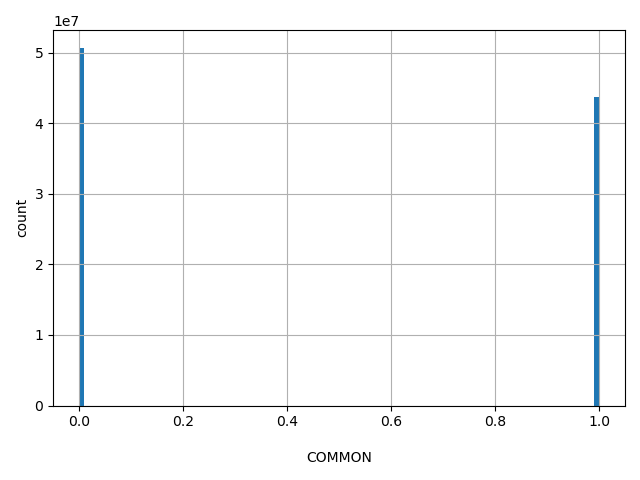 |
0, 1 |
| TOPMED |
str |
-
|
An ordered, comma delimited list of allele frequencies based on TOPMed, starting with the reference allele followed by alternate alleles as ordered in the ALT column. The TOPMed minor allele is the second largest value in the list.
|
No histogram: Histogram is not available for this score. |
NO DOMAIN |
×
×
×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×

×
















































