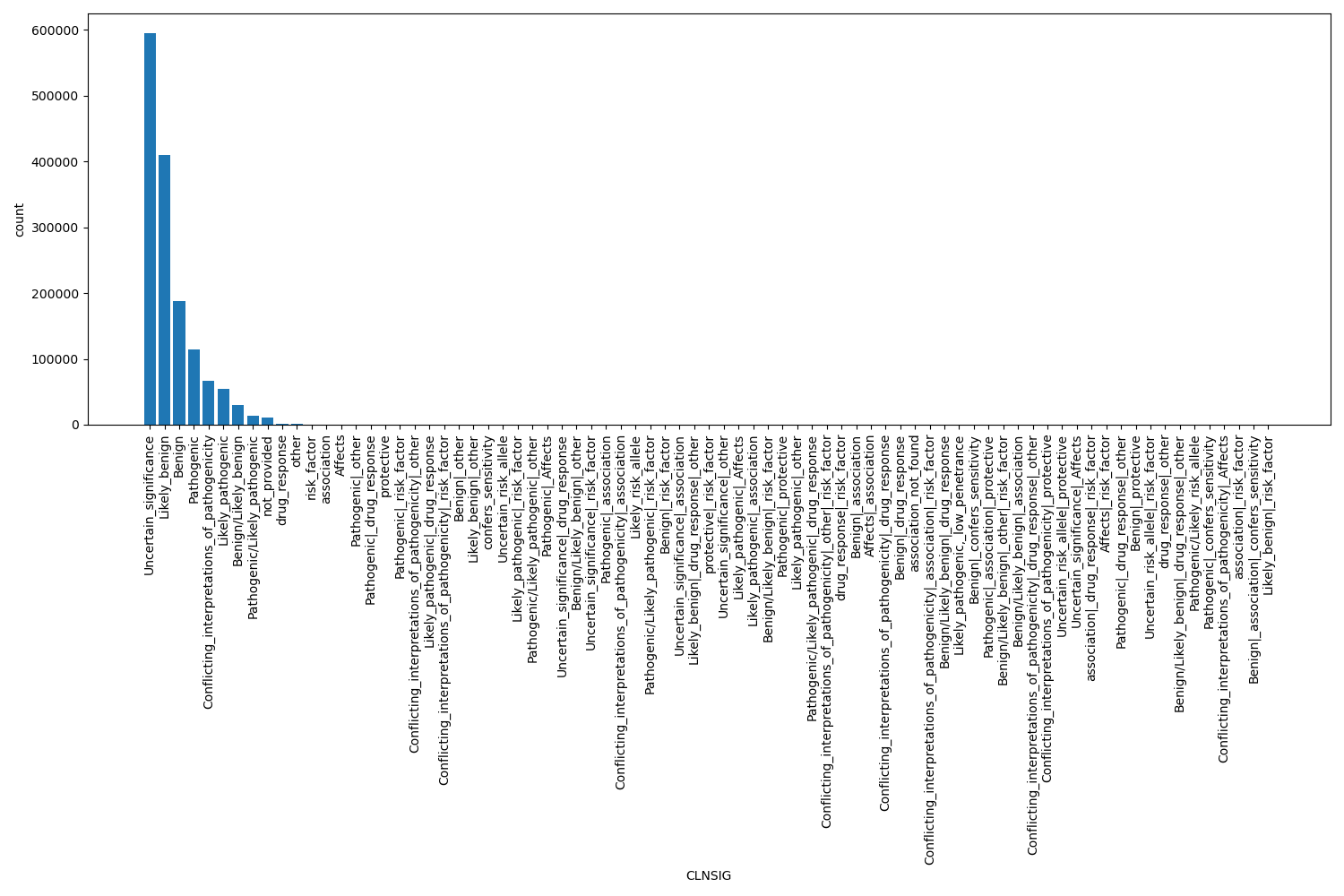
CLNSIG
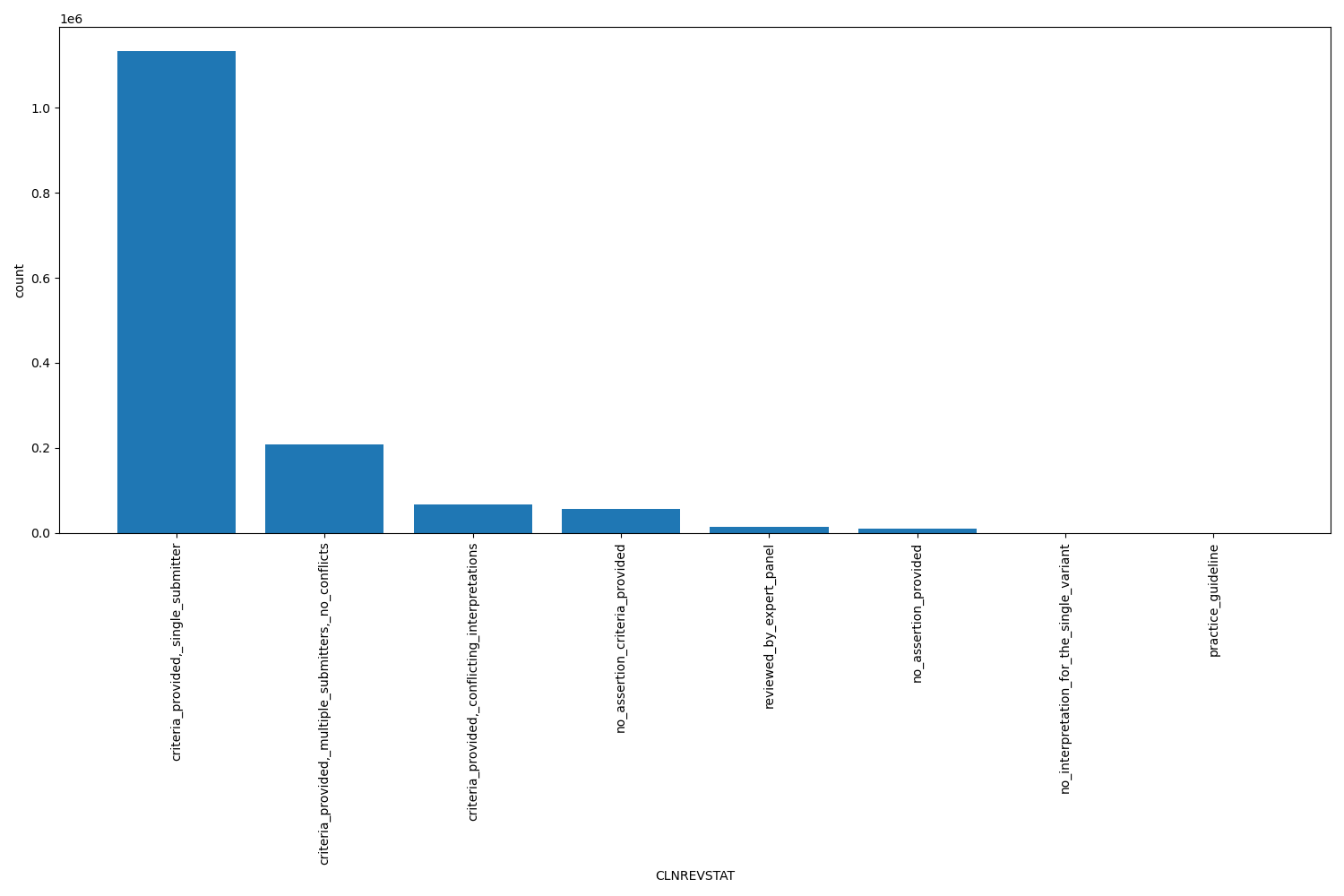
CLNREVSTAT
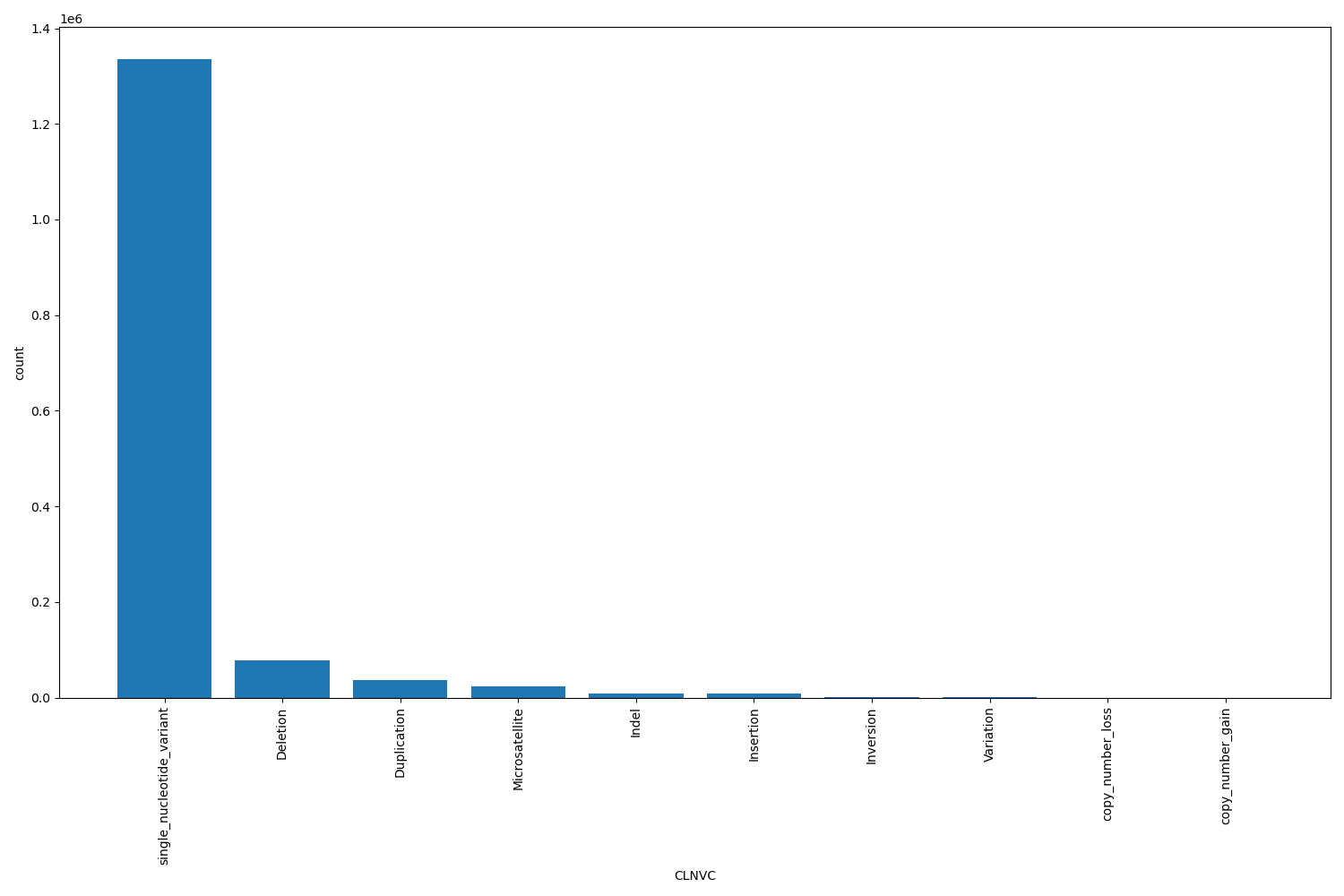
CLNVC
| Id: | hg38/scores/clinvar_20221105 |
| Type: | allele_score |
| Version: | 0 |
| Summary: |
Measure used to assess the clinical significance of genetic variants |
| Description: |
ClinVar resource downloaded on 2022-11-05. Chromosome names are
remapped to have ClinVar is a freely accessible, public archive of reports of the relationships among human variations and phenotypes, with supporting evidence. ClinVar thus facilitates access to and communication about the relationships asserted between human variation and observed health status, and the history of that interpretation. ClinVar processes submissions reporting variants found in patient samples, assertions made regarding their clinical significance, information about the submitter, and other supporting data. The alleles described in submissions are mapped to reference sequences, and reported according to the HGVS standard. ClinVar then presents the data for interactive users as well as those wishing to use ClinVar in daily workflows and other local applications. ClinVar works in collaboration with interested organizations to meet the needs of the medical genetics community as efficiently and effectively as possible |
| Labels: |
|
| ID | Type | Default annotation | Description | Histogram | Range |
|---|---|---|---|---|---|
| CLNSIG | str |
CLNSIG |
Clinical significance for this single variant; multiple values are separated by a vertical bar
|
 |
Uncertain_significance, Likely_benign, Benign, Pathogenic, Conflicting_interpretations_of_pathogenicity, Likely_pathogenic, Benign/Likely_benign, Pathogenic/Likely_pathogenic, not_provided, drug_response, other, risk_factor, association, Affects, Pathogenic|_other, Pathogenic|_drug_response, protective, Pathogenic|_risk_factor, Conflicting_interpretations_of_pathogenicity|_other, Likely_pathogenic|_drug_response, Other |
| CLNREVSTAT | str |
CLNREVSTAT |
ClinVar review status for the Variation ID
|
 |
criteria_provided|_single_submitter, criteria_provided|_multiple_submitters|_no_conflicts, criteria_provided|_conflicting_interpretations, no_assertion_criteria_provided, reviewed_by_expert_panel, no_assertion_provided, no_interpretation_for_the_single_variant, practice_guideline |
| CLNVC | str |
CLNVC |
Variant type
|
 |
single_nucleotide_variant, Deletion, Duplication, Microsatellite, Indel, Insertion, Inversion, Variation, copy_number_loss, copy_number_gain |
| Filename | Size | md5 |
|---|---|---|
| chr_rename.txt | 203.0 B | 6b381222aba146516fd282587ed8598d |
| clinvar_20221105.vcf.gz | 56.57 MB | 48ef344c7120566088450abbd9b03b3a |
| clinvar_20221105.vcf.gz.dvc | 95.0 B | d08b2c9d458728873976154de56e977e |
| clinvar_20221105.vcf.gz.tbi | 323.08 KB | 1a0236efda3814b788a72d8a519bcce7 |
| clinvar_20221105.vcf.gz.tbi.dvc | 97.0 B | b13efe1916ba28c569680903ef7374b9 |
| clinvar_20221105_chr.header.vcf.gz | 1.33 KB | 0004a6cee60b4e8d76185a9f8b9a550a |
| clinvar_20221105_chr.header.vcf.gz.tbi | 80.0 B | e7180bb953d2bd657c420a5f76a7164d |
| clinvar_20221105_chr.vcf.gz | 57.9 MB | d2f167e0c1155e416baf545205ed17b1 |
| clinvar_20221105_chr.vcf.gz.dvc | 99.0 B | 4bf30688e4fb884644fb5806d2679268 |
| clinvar_20221105_chr.vcf.gz.tbi | 320.76 KB | d1e6cc2b74018dfc994d3da648b591f6 |
| clinvar_20221105_chr.vcf.gz.tbi.dvc | 101.0 B | e067c10134471aa904d5aa815311b27a |
| genomic_resource.yaml | 1.93 KB | 1611201af472a1a39b18e38f15d9feab |
| statistics/ |