-
-
-
-
CLNDN
-
-
-
-
CLNSIG
-
-
-
-
-
-
-
-
-
-
-
-
-

-
-
-
-
| Id: | hg38/scores/ClinVar_20240730 |
| Type: | allele_score |
| Version: | 0 |
| Summary: |
Measure used to assess the clinical significance of genetic variants |
| Description: |
ClinVar is a publicly available resource that collects and shares information on the relationships between human genetic variants and their associated health conditions. By providing detailed evidence and supporting data, ClinVar enables researchers, healthcare professionals, and the broader medical community to better understand how specific genetic variations impact health. Submissions to ClinVar include data from patient samples, assessments of the clinical significance of variants, and information about the organizations providing the data. All submitted variants are mapped to standardized reference sequences using HGVS nomenclature. ClinVar supports both interactive exploration and integration into clinical and research workflows, ensuring its data is accessible and useful for the advancement of precision medicine. ClinVar actively collaborates with organizations worldwide to address the evolving needs of the genetics and medical communities. Downloaded on: 08/02/24 https://ftp.ncbi.nlm.nih.gov/pub/clinvar/vcf_GRCh38/ Processing details: dataPrep.sh removes rows above header and remaps chromosome names to have |
| Labels: |
|
| ID | Type | Default annotation | Description | Histogram | Range |
|---|---|---|---|---|---|
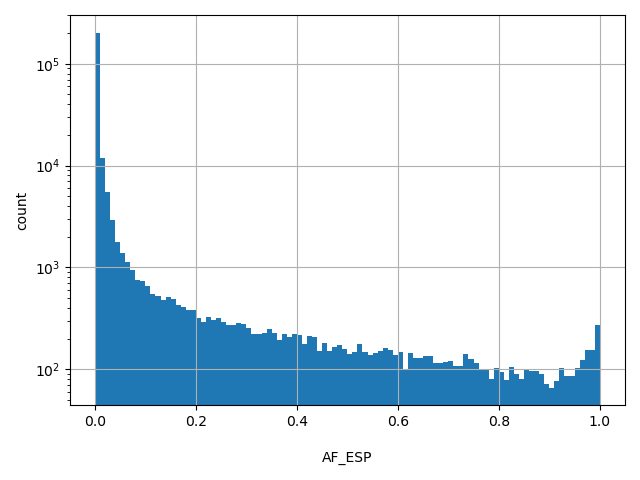
| AF_ESP | float |
- |
allele frequencies from GO-ESP
|
 |
[8e-05, 1] |
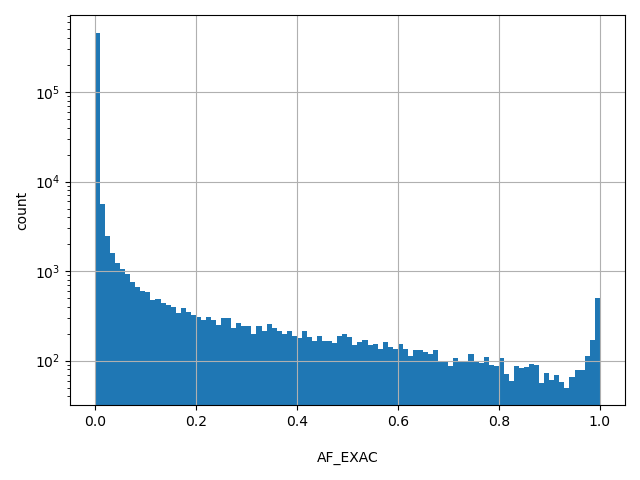
| AF_EXAC | float |
- |
allele frequencies from ExAC
|
 |
[0, 1] |
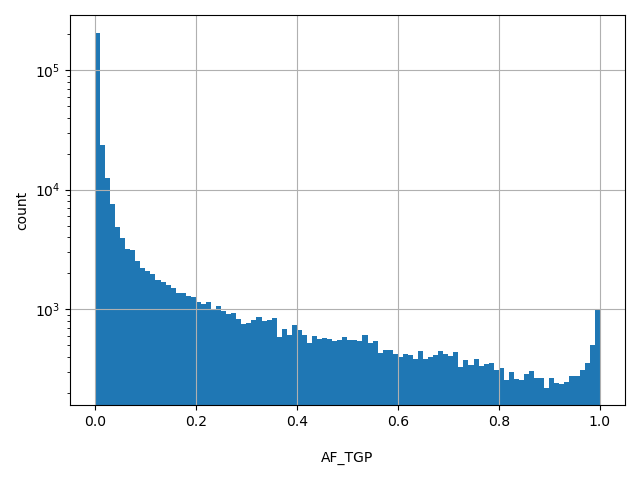
| AF_TGP | float |
- |
allele frequencies from TGP
|
 |
[0, 1] |
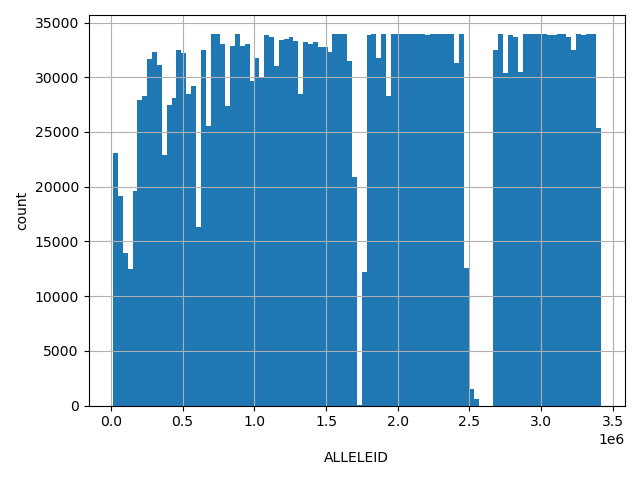
| ALLELEID | int |
- |
ClinVar Allele ID
|
 |
[1.5e+04, 3.41e+06] |
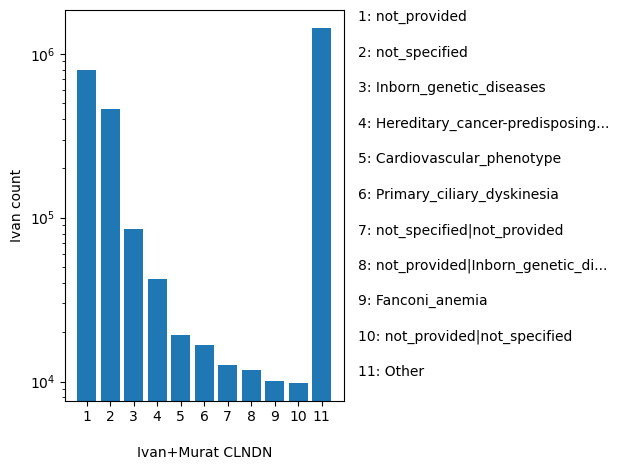
| CLNDN | str |
CLNDN |
ClinVar's preferred disease name for the concept specified by disease identifiers in CLNDISDB
|
 |
not_provided, not_specified, Inborn_genetic_diseases, Hereditary_cancer-predisposing_syndrome, Cardiovascular_phenotype, Primary_ciliary_dyskinesia, not_specified|not_provided, not_provided|Inborn_genetic_diseases, Fanconi_anemia, not_provided|not_specified, Other |
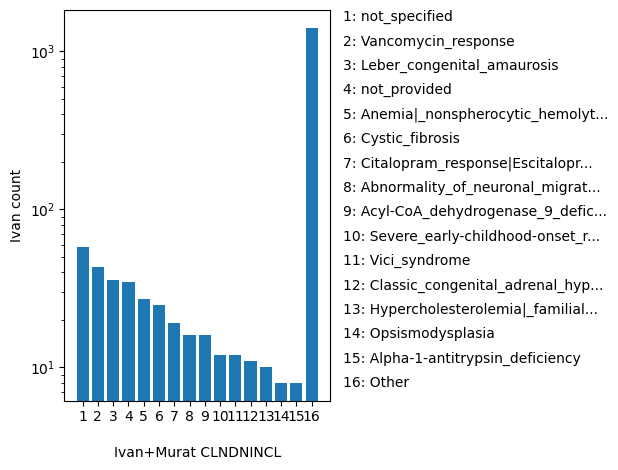
| CLNDNINCL | str |
- |
For included Variant : ClinVar's preferred disease name for the concept specified by disease identifiers in CLNDISDB
|
 |
not_specified, Vancomycin_response, Leber_congenital_amaurosis, not_provided, Anemia|_nonspherocytic_hemolytic|_due_to_G6PD_deficiency, Cystic_fibrosis, Citalopram_response|Escitalopram_response|Sertraline_response|Voriconazole_response, Abnormality_of_neuronal_migration, Acyl-CoA_dehydrogenase_9_deficiency, Severe_early-childhood-onset_retinal_dystrophy, Vici_syndrome, Classic_congenital_adrenal_hyperplasia_due_to_21-hydroxylase_deficiency, Hypercholesterolemia|_familial|_1, Opsismodysplasia, Alpha-1-antitrypsin_deficiency, Breast-ovarian_cancer|_familial|_susceptibility_to|_1, Epilepsy|_idiopathic_generalized|_susceptibility_to, Severe_combined_immunodeficiency|_autosomal_recessive|_T_cell-negative|_B_cell-negative|_NK_cell-positive, Tuberous_sclerosis_syndrome, Congenital_multicore_myopathy_with_external_ophthalmoplegia, Other |
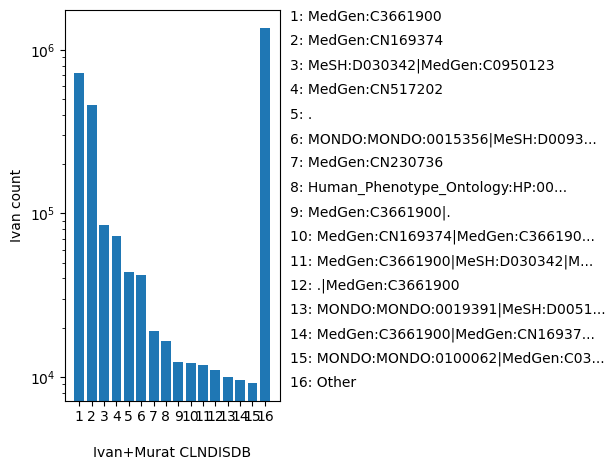
| CLNDISDB | str |
- |
Tag-value pairs of disease database name and identifier submitted for germline classifications, e.g. OMIM:NNNNNN"
|
 |
MedGen:C3661900, MedGen:CN169374, MeSH:D030342|MedGen:C0950123, MedGen:CN517202, ., MONDO:MONDO:0015356|MeSH:D009386|MedGen:C0027672|Orphanet:140162, MedGen:CN230736, Human_Phenotype_Ontology:HP:0012265|MONDO:MONDO:0016575|MedGen:C0008780|OMIM:PS244400|Orphanet:244, MedGen:C3661900|., MedGen:CN169374|MedGen:C3661900, MedGen:C3661900|MeSH:D030342|MedGen:C0950123, .|MedGen:C3661900, MONDO:MONDO:0019391|MeSH:D005199|MedGen:C0015625|OMIM:PS227650|Orphanet:84, MedGen:C3661900|MedGen:CN169374, MONDO:MONDO:0100062|MedGen:C0393706|OMIM:PS308350|Orphanet:1934, MeSH:D030342|MedGen:C0950123|MedGen:C3661900, Human_Phenotype_Ontology:HP:0001258|Human_Phenotype_Ontology:HP:0007062|Human_Phenotype_Ontology:HP:0007124|Human_Phenotype_Ontology:HP:0007216|MedGen:C0037772, MONDO:MONDO:0009725|MedGen:C1850569|OMIM:256030, MONDO:MONDO:0011400|MedGen:C1858763|OMIM:604145|Orphanet:154|MONDO:MONDO:0012127|MedGen:C1837342|OMIM:608807|Orphanet:140922, MONDO:MONDO:0018975|MedGen:C0027831|OMIM:162200|Orphanet:636, Other |
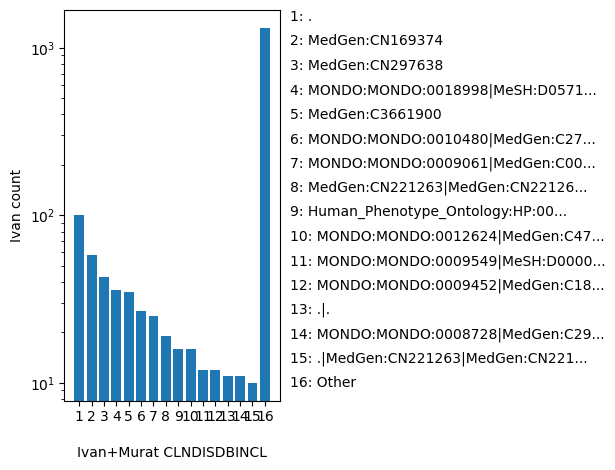
| CLNDISDBINCL | str |
- |
For included Variant: Tag-value pairs of disease database name and identifier for germline classifications, e.g. OMIM:NNNNNN
|
 |
., MedGen:CN169374, MedGen:CN297638, MONDO:MONDO:0018998|MeSH:D057130|MedGen:C0339527|OMIM:PS204000|Orphanet:65, MedGen:C3661900, MONDO:MONDO:0010480|MedGen:C2720289|OMIM:300908|Orphanet:466026, MONDO:MONDO:0009061|MedGen:C0010674|OMIM:219700|Orphanet:586, MedGen:CN221263|MedGen:CN221264|MedGen:CN221265|MedGen:CN077957, Human_Phenotype_Ontology:HP:0002269|Human_Phenotype_Ontology:HP:0007317|MedGen:C1837249, MONDO:MONDO:0012624|MedGen:C4747517|OMIM:611126|Orphanet:99901, MONDO:MONDO:0009549|MeSH:D000080362|MedGen:C1855465|OMIM:248200|Orphanet:364055|Orphanet:827, MONDO:MONDO:0009452|MedGen:C1855772|OMIM:242840|Orphanet:1493, .|., MONDO:MONDO:0008728|MedGen:C2936858|OMIM:201910|Orphanet:90794, .|MedGen:CN221263|MedGen:CN221264|MedGen:CN221265|MedGen:CN077957, MONDO:MONDO:0007750|MedGen:C0745103|OMIM:143890|Orphanet:391665, MONDO:MONDO:0009785|MedGen:C0432219|OMIM:258480|Orphanet:2746, MONDO:MONDO:0013282|MedGen:C0221757|OMIM:613490|Orphanet:60, MONDO:MONDO:0011450|MedGen:C2676676|OMIM:604370|Orphanet:145, MONDO:MONDO:0011086|MedGen:C1832322|OMIM:601457|Orphanet:331206, Other |
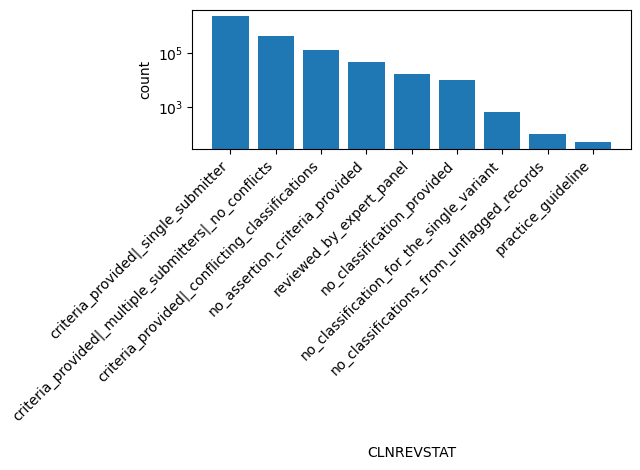
| CLNREVSTAT | str |
- |
ClinVar review status of germline classification for the Variation ID
|
 |
criteria_provided|_single_submitter, criteria_provided|_multiple_submitters|_no_conflicts, criteria_provided|_conflicting_classifications, no_assertion_criteria_provided, reviewed_by_expert_panel, no_classification_provided, no_classification_for_the_single_variant, no_classifications_from_unflagged_records, practice_guideline |
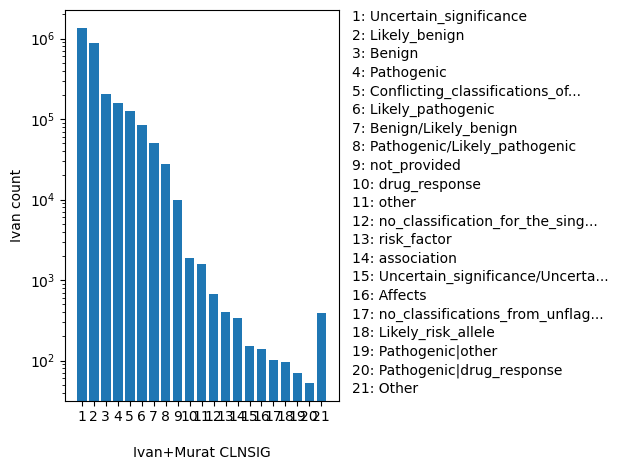
| CLNSIG | str |
CLNSIG |
Aggregate germline classification for this single variant; multiple values are separated by a vertical bar
|
 |
Uncertain_significance, Likely_benign, Benign, Pathogenic, Conflicting_classifications_of_pathogenicity, Likely_pathogenic, Benign/Likely_benign, Pathogenic/Likely_pathogenic, not_provided, drug_response, other, no_classification_for_the_single_variant, risk_factor, association, Uncertain_significance/Uncertain_risk_allele, Affects, no_classifications_from_unflagged_records, Likely_risk_allele, Pathogenic|other, Pathogenic|drug_response, Other |
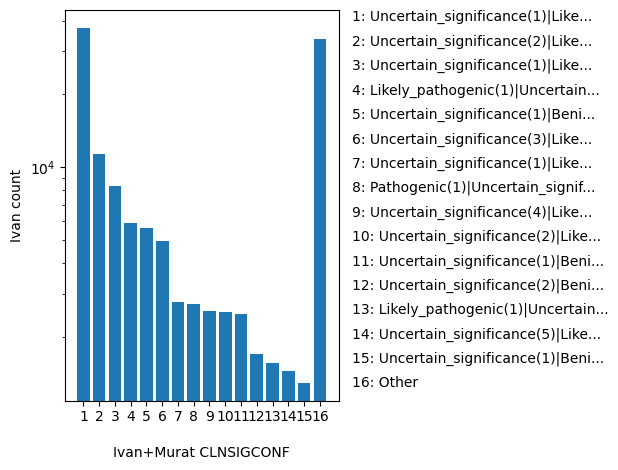
| CLNSIGCONF | str |
- |
Conflicting germline classification for this single variant; multiple values are separated by a vertical bar
|
 |
Uncertain_significance(1)|Likely_benign(1), Uncertain_significance(2)|Likely_benign(1), Uncertain_significance(1)|Likely_benign(2), Likely_pathogenic(1)|Uncertain_significance(1), Uncertain_significance(1)|Benign(1), Uncertain_significance(3)|Likely_benign(1), Uncertain_significance(1)|Likely_benign(3), Pathogenic(1)|Uncertain_significance(1), Uncertain_significance(4)|Likely_benign(1), Uncertain_significance(2)|Likely_benign(2), Uncertain_significance(1)|Benign(1)|Likely_benign(1), Uncertain_significance(2)|Benign(1), Likely_pathogenic(1)|Uncertain_significance(2), Uncertain_significance(5)|Likely_benign(1), Uncertain_significance(1)|Benign(1)|Likely_benign(2), Uncertain_significance(3)|Likely_benign(2), Uncertain_significance(1)|Likely_benign(4), Uncertain_significance(2)|Likely_benign(3), Likely_pathogenic(2)|Uncertain_significance(1), Pathogenic(1)|Likely_pathogenic(1)|Uncertain_significance(1), Other |
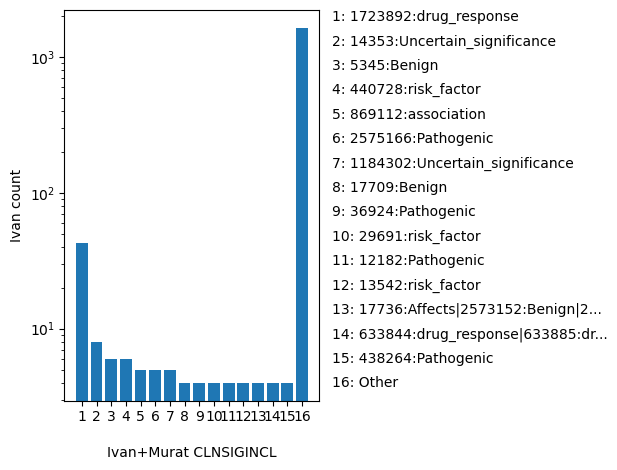
| CLNSIGINCL | str |
- |
Germline classification for a haplotype or genotype that includes this variant. Reported as pairs of VariationID:classification; multiple values are separated by a vertical bar
|
 |
1723892:drug_response, 14353:Uncertain_significance, 5345:Benign, 440728:risk_factor, 869112:association, 2575166:Pathogenic, 1184302:Uncertain_significance, 17709:Benign, 36924:Pathogenic, 29691:risk_factor, 12182:Pathogenic, 13542:risk_factor, 17736:Affects|2573152:Benign|2573153:Benign|2573154:Benign, 633844:drug_response|633885:drug_response|633887:drug_response|633888:drug_response|633904:drug_response|633905:drug_response|633910:drug_response|633913:drug_response|633916:drug_response|633925:drug_response|633927:drug_response|633930:drug_response|633941:drug_response|633952:drug_response|633958:drug_response|633965:drug_response|634060:drug_response|634114:drug_response|634132:drug_response|634136:drug_response|634137:drug_response|634138:drug_response|634205:drug_response|634212:drug_response|634213:drug_response|634232:drug_response|634272:drug_response|634298:drug_response|634355:drug_response|634365:drug_response|634375:drug_response|634413:drug_response|634414:drug_response|634427:drug_response|638789:drug_response, 438264:Pathogenic, 1722723:Likely_pathogenic, 202166:not_provided, 12304:Pathogenic, 14182:Pathogenic, 14180:Pathogenic, Other |
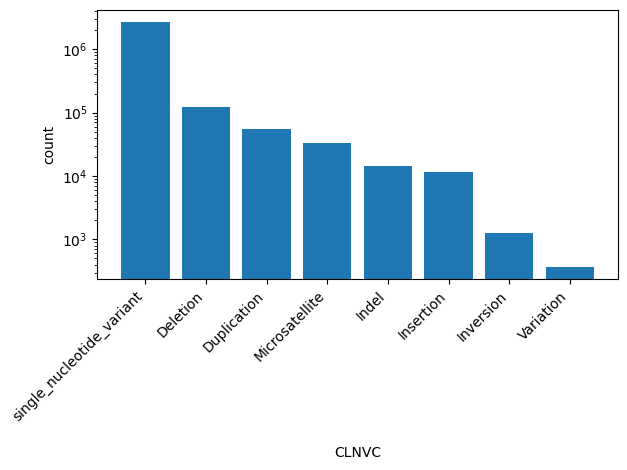
| CLNVC | str |
- |
Variant type
|
 |
single_nucleotide_variant, Deletion, Duplication, Microsatellite, Indel, Insertion, Inversion, Variation |
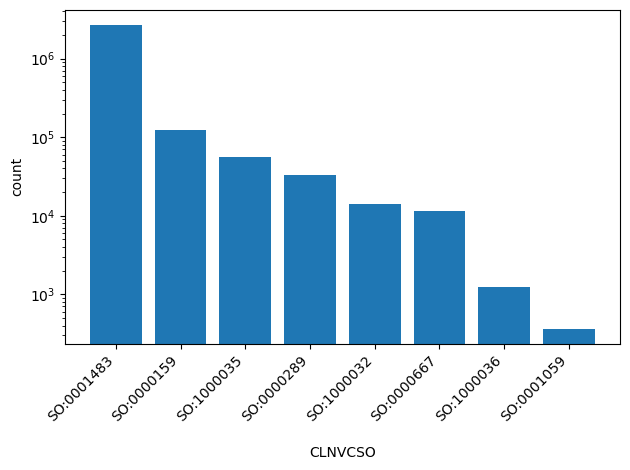
| CLNVCSO | str |
- |
Sequence Ontology id for variant type
|
 |
SO:0001483, SO:0000159, SO:1000035, SO:0000289, SO:1000032, SO:0000667, SO:1000036, SO:0001059 |
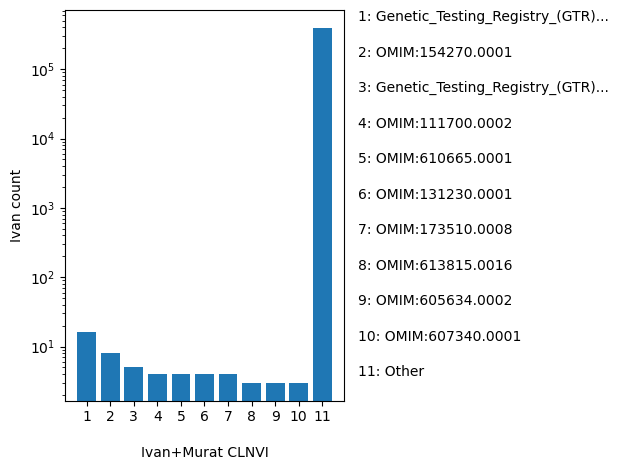
| CLNVI | str |
- |
the variant's clinical sources reported as tag-value pairs of database and variant identifier
|
 |
Genetic_Testing_Registry_(GTR):GTR000613302, OMIM:154270.0001, Genetic_Testing_Registry_(GTR):GTR000562560|Genetic_Testing_Registry_(GTR):GTR000613302, OMIM:111700.0002, OMIM:610665.0001, OMIM:131230.0001, OMIM:173510.0008, OMIM:613815.0016, OMIM:605634.0002, OMIM:607340.0001, OMIM:604824.0001, OMIM:612386.0014, OMIM:601146.0012, OMIM:147795.0004, OMIM:613890.0003, OMIM:191315.0006, OMIM:608515.0002, OMIM:600031.0002, OMIM:600713.0001, OMIM:102610.0015, Other |
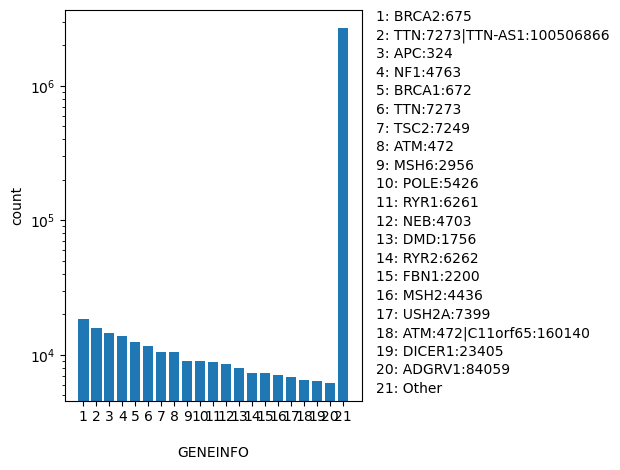
| GENEINFO | str |
- |
Gene(s) for the variant reported as gene symbol:gene id. The gene symbol and id are delimited by a colon (:) and each pair is delimited by a vertical bar (|)
|
 |
BRCA2:675, TTN:7273|TTN-AS1:100506866, APC:324, NF1:4763, BRCA1:672, TTN:7273, TSC2:7249, ATM:472, MSH6:2956, POLE:5426, RYR1:6261, NEB:4703, DMD:1756, RYR2:6262, FBN1:2200, MSH2:4436, USH2A:7399, ATM:472|C11orf65:160140, DICER1:23405, ADGRV1:84059, Other |
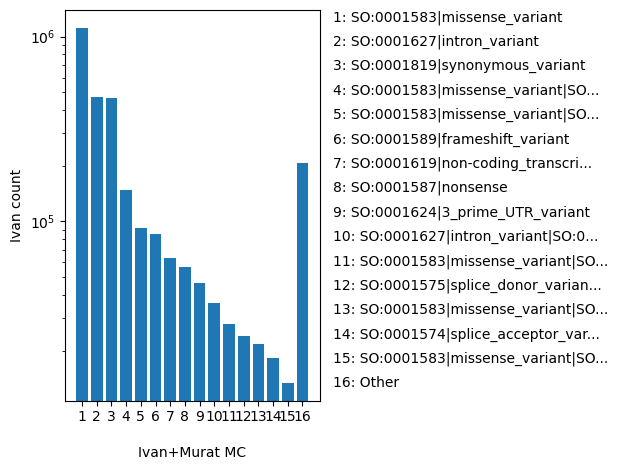
| MC | str |
- |
comma separated list of molecular consequence in the form of Sequence Ontology ID|molecular_consequence
|
 |
SO:0001583|missense_variant, SO:0001627|intron_variant, SO:0001819|synonymous_variant, SO:0001583|missense_variant|SO:0001619|non-coding_transcript_variant, SO:0001583|missense_variant|SO:0001627|intron_variant, SO:0001589|frameshift_variant, SO:0001619|non-coding_transcript_variant|SO:0001819|synonymous_variant, SO:0001587|nonsense, SO:0001624|3_prime_UTR_variant, SO:0001627|intron_variant|SO:0001819|synonymous_variant, SO:0001583|missense_variant|SO:0001623|5_prime_UTR_variant, SO:0001575|splice_donor_variant, SO:0001583|missense_variant|SO:0001619|non-coding_transcript_variant|SO:0001627|intron_variant, SO:0001574|splice_acceptor_variant, SO:0001583|missense_variant|SO:0001624|3_prime_UTR_variant, SO:0001822|inframe_deletion, SO:0001583|missense_variant|SO:0001619|non-coding_transcript_variant|SO:0001623|5_prime_UTR_variant, SO:0001623|5_prime_UTR_variant|SO:0001819|synonymous_variant, SO:0001623|5_prime_UTR_variant, SO:0001589|frameshift_variant|SO:0001619|non-coding_transcript_variant, Other |
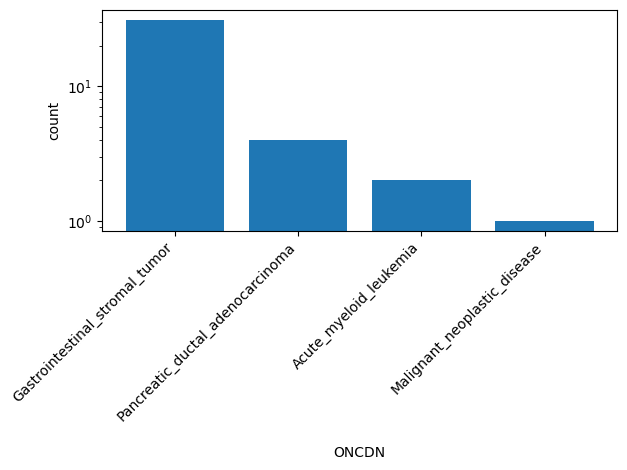
| ONCDN | str |
- |
ClinVar's preferred disease name for the concept specified by disease identifiers in ONCDISDB
|
 |
Gastrointestinal_stromal_tumor, Pancreatic_ductal_adenocarcinoma, Acute_myeloid_leukemia, Malignant_neoplastic_disease |
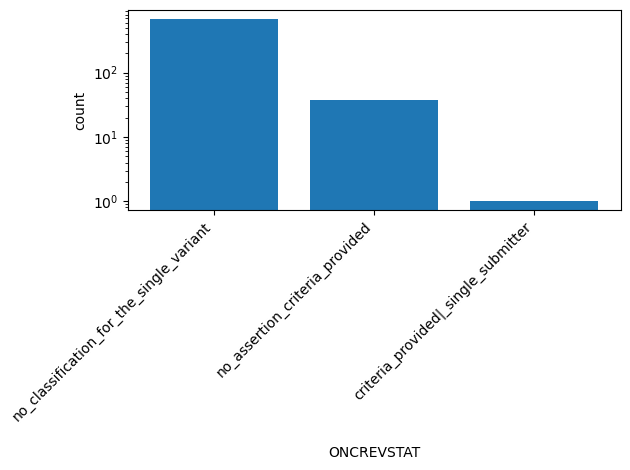
| ONCREVSTAT | str |
- |
ClinVar review status of oncogenicity classification for the Variation ID
|
 |
no_classification_for_the_single_variant, no_assertion_criteria_provided, criteria_provided|_single_submitter |
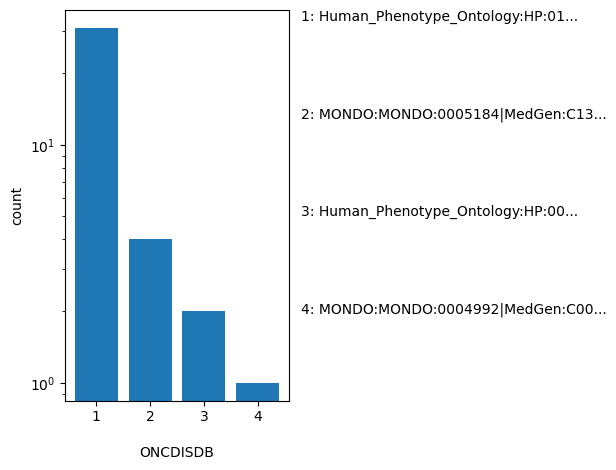
| ONCDISDB | str |
- |
Tag-value pairs of disease database name and identifier submitted for oncogenicity classifications, e.g. MedGen:NNNNNN
|
 |
Human_Phenotype_Ontology:HP:0100723|MONDO:MONDO:0011719|MeSH:D046152|MedGen:C0238198|OMIM:606764|Orphanet:44890, MONDO:MONDO:0005184|MedGen:C1335302, Human_Phenotype_Ontology:HP:0001914|Human_Phenotype_Ontology:HP:0004808|Human_Phenotype_Ontology:HP:0004843|Human_Phenotype_Ontology:HP:0005516|Human_Phenotype_Ontology:HP:0006724|Human_Phenotype_Ontology:HP:0006728|MONDO:MONDO:0018874|MeSH:D015470|MedGen:C0023467|OMIM:601626|Orphanet:519, MONDO:MONDO:0004992|MedGen:C0006826 |
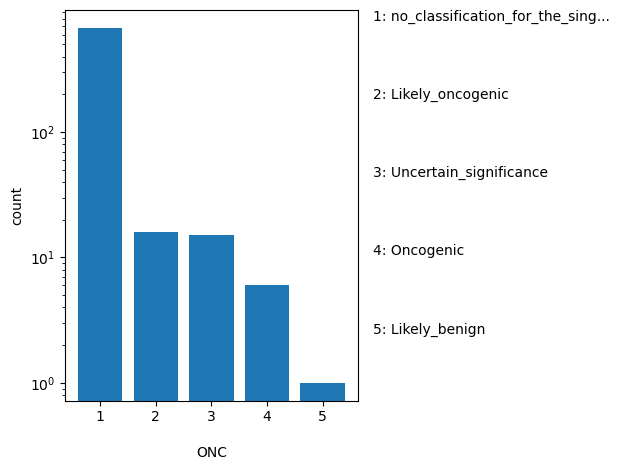
| ONC | str |
- |
Aggregate oncogenicity classification for this single variant; multiple values are separated by a vertical bar
|
 |
no_classification_for_the_single_variant, Likely_oncogenic, Uncertain_significance, Oncogenic, Likely_benign |
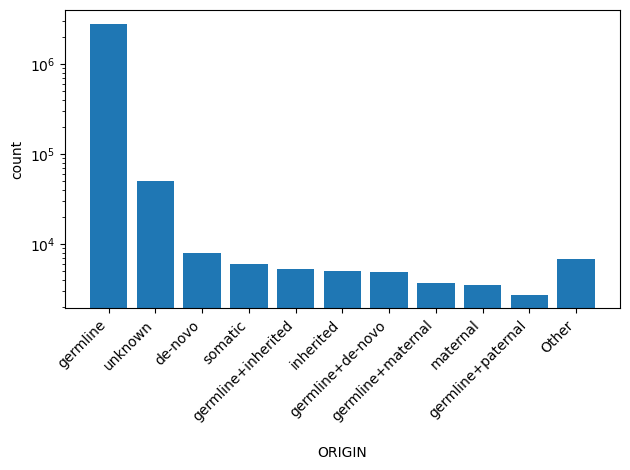
| ORIGIN | str |
- |
Allele origin
|
 |
1, 0, 32, 2, 5, 4, 33, 17, 16, 9, 3, 8, 64, 65, 21, 25, 13, 49, 35, 37, Other |
| RS | str |
- |
Gene(s) for the variant reported as gene symbol:gene id. The gene symbol and id are delimited by a colon (:) and each pair is delimited by a vertical bar (|)
|
 |
193922936, 11309117, 56986587, 535965616, 746054139, 3032358, 35896485, 200900896, 55712755, 59431308, 367885911, 3078066, 41550117, 3831942, 55889738, 371983878, 10524346, 35115360, 766545285, 746853821, Other |
| SCIDN | str |
- |
ClinVar's preferred disease name for the concept specified by disease identifiers in SCIDISDB
|
 |
Chronic_myelogenous_leukemia|_BCR-ABL1_positive, Acute_myeloid_leukemia, Diffuse_midline_glioma|_H3_K27M-mutant, Non-small_cell_lung_carcinoma, Colorectal_cancer|Melanoma, Essential_thrombocythemia|Acute_myeloid_leukemia |
| SCIDISDB | str |
- |
Tag-value pairs of disease database name and identifier submitted for somatic clinial impact classifications, e.g. MedGen:NNNNNN
|
 |
Human_Phenotype_Ontology:HP:0005506|Human_Phenotype_Ontology:HP:0005544|MONDO:MONDO:0011996|MeSH:D015464|MedGen:C0279543|OMIM:608232|Orphanet:521, Human_Phenotype_Ontology:HP:0001914|Human_Phenotype_Ontology:HP:0004808|Human_Phenotype_Ontology:HP:0004843|Human_Phenotype_Ontology:HP:0005516|Human_Phenotype_Ontology:HP:0006724|Human_Phenotype_Ontology:HP:0006728|MONDO:MONDO:0018874|MeSH:D015470|MedGen:C0023467|OMIM:601626|Orphanet:519, MONDO:MONDO:0957196|MedGen:C4289688, Human_Phenotype_Ontology:HP:0030358|MONDO:MONDO:0005233|MeSH:D002289|MedGen:C0007131, MONDO:MONDO:0005575|MedGen:C0346629|OMIM:114500|Human_Phenotype_Ontology:HP:0002861|Human_Phenotype_Ontology:HP:0002887|Human_Phenotype_Ontology:HP:0006777|Human_Phenotype_Ontology:HP:0007474|MONDO:MONDO:0005105|MeSH:D008545|MedGen:C0025202, MONDO:MONDO:0005029|MeSH:D013920|MedGen:C0040028|Orphanet:3318|Human_Phenotype_Ontology:HP:0001914|Human_Phenotype_Ontology:HP:0004808|Human_Phenotype_Ontology:HP:0004843|Human_Phenotype_Ontology:HP:0005516|Human_Phenotype_Ontology:HP:0006724|Human_Phenotype_Ontology:HP:0006728|MONDO:MONDO:0018874|MeSH:D015470|MedGen:C0023467|OMIM:601626|Orphanet:519 |
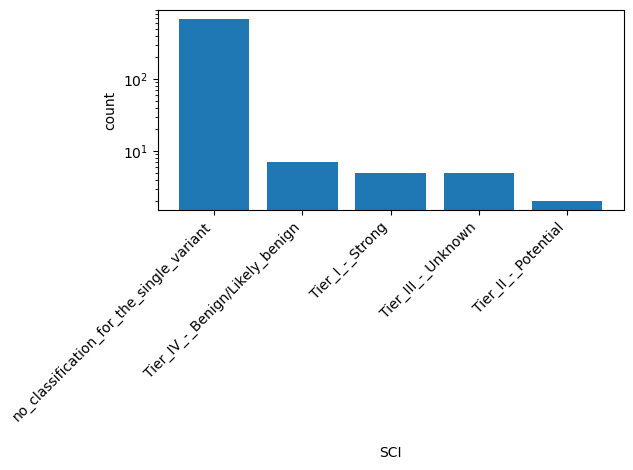
| SCI | str |
- |
Aggregate somatic clinical impact for this single variant; multiple values are separated by a vertical bar
|
 |
no_classification_for_the_single_variant, Tier_IV_-_Benign/Likely_benign, Tier_I_-_Strong, Tier_III_-_Unknown, Tier_II_-_Potential |
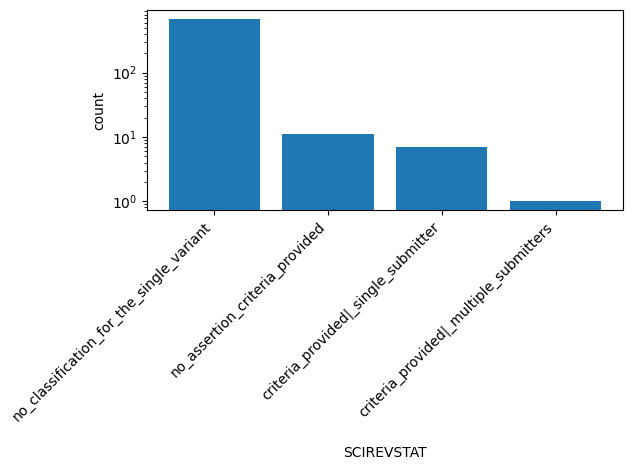
| SCIREVSTAT | str |
- |
ClinVar review status of somatic clinical impact for the Variation ID
|
 |
no_classification_for_the_single_variant, no_assertion_criteria_provided, criteria_provided|_single_submitter, criteria_provided|_multiple_submitters |
| Filename | Size | md5 |
|---|---|---|
| clinvar_20240730.vcf | 1.11 GB | 7cf71a319a358a12af1d846c9a2f7a0c |
| clinvar_20240730.vcf.dvc | 106.0 B | edda6efc4ce3674a47a6a64d148fc8d7 |
| clinvar_20240730_chr.header.vcf.gz | 1.35 KB | dd54c9ab1fb6afeea484b5e25c9c8ce0 |
| clinvar_20240730_chr.header.vcf.gz.tbi | 72.0 B | 4cb176febbc8c26d717a6c6e67b9c905 |
| clinvar_20240730_chr.vcf.gz | 96.99 MB | f08b390ca6f3e120e867c51fead6f310 |
| clinvar_20240730_chr.vcf.gz.dvc | 112.0 B | 5a22525321aca7e83801f7ab111c402e |
| clinvar_20240730_chr.vcf.gz.tbi | 523.22 KB | 1bf7469c46bcec7fbb478e30dc4e3358 |
| clinvar_20240730_chr.vcf.gz.tbi.dvc | 113.0 B | cd3f4285f0540f43b580350c77918616 |
| clinvar_20240730_chr_with_origin_name.vcf | 34.0 B | 4d757233ee10cec0a3c7a0cd3b41b41d |
| customplot1.py | 8.09 KB | 87a55cc9f852228b00b8ea19f96d9e59 |
| dataPrep.sh | 722.0 B | 787c887302b2fd4305896e50971df9a6 |
| genomic_resource.yaml | 9.78 KB | 69a2c543b960b389149636195925adba |
| statistics/ |