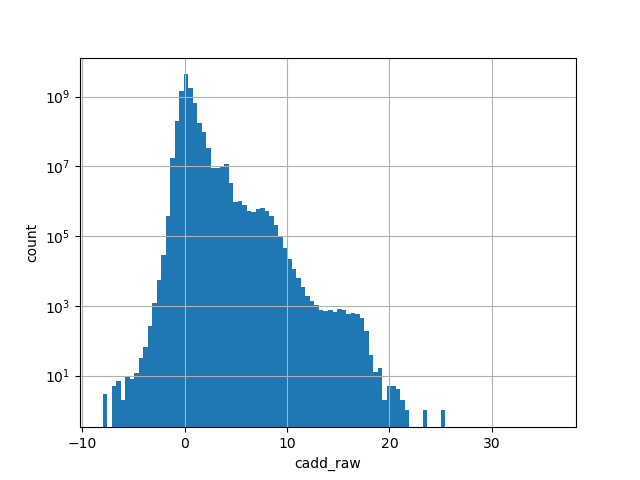
cadd_raw
CADD raw score for functional prediction of a SNP. The larger the score
the more likely the SNP has damaging effect
Small values desc: less damaging
Large values desc: more damaging
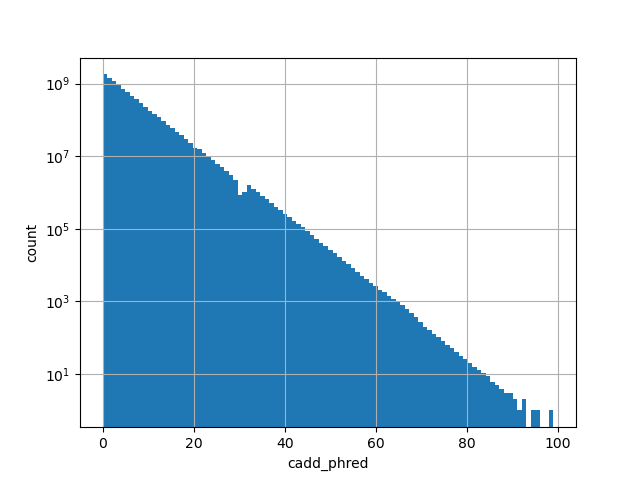
cadd_phred
CADD phred-like score. This is phred-like rank score based on whole
genome CADD raw scores. The larger the score the more likely the SNP
has damaging effect.
Small values desc: less damaging
Large values desc: more damaging