am_pathogenicity
-
| Id: | hg38/scores/AlphaMissense |
| Type: | allele_score |
| Version: | 0 |
| Summary: |
Functional impact of mutations on protein function |
| Description: |
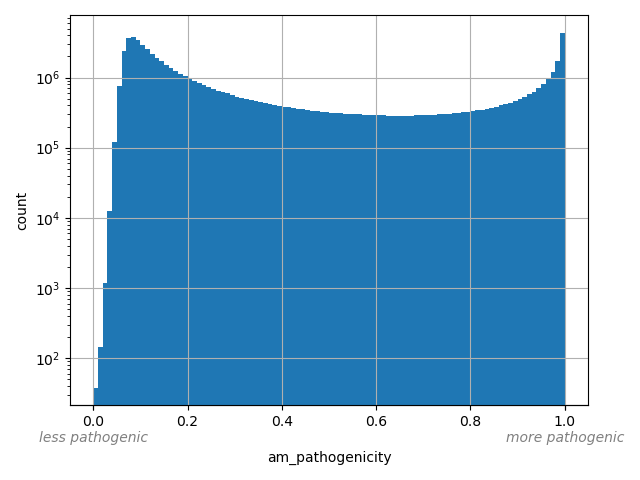
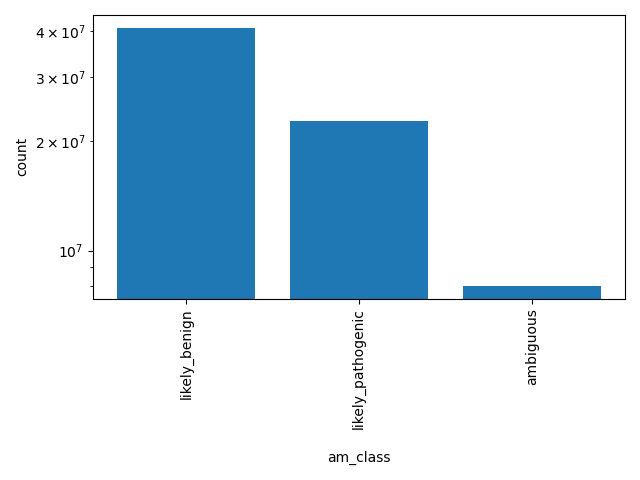
AlphaMissense is a computational tool designed to predict the functional impact of missense mutations on protein structure and function. Leveraging advanced machine learning algorithms, AlphaMissense analyzes genetic variations to determine their potential effects on protein stability, interaction, and activity. By integrating structural bioinformatics with functional annotations, it provides insights into how specific missense mutations might contribute to disease mechanisms or impact drug efficacy. This tool is particularly valuable for researchers in genomics and precision medicine, offering a detailed assessment of mutation-related risks and aiding in the identification of therapeutic targets. Scores smaller than 0.34 or larger than 0.564 are categorized as likely_benign or likely_pathogenic, respectively. Downloaded on: 07/29/24 https://zenodo.org/records/8208688 Processing details: dataPrep.sh prepares the data and index files. |
| Labels: |
|
| ID | Type | Default annotation | Description | Histogram | Range |
|---|---|---|---|---|---|
| am_pathogenicity | float |
am_pathogenicity |
AlphaMissense Pathogenicity score is a deleteriousness score for missense variants
Small values desc: less pathogenic
Large values desc: more pathogenic
|
 |
[0.0001, 1] |
| am_class | str |
- |
AlphaMissense Class is a deleteriousness category for missense variants
|
 |
likely_benign, likely_pathogenic, ambiguous |
| Filename | Size | md5 |
|---|---|---|
| AlphaMissense_hg38.tsv.gz | 599.3 MB | 46d0028375cf95088bd014ff6855cffd |
| AlphaMissense_hg38.tsv.gz.dvc | 110.0 B | b544e7e3ac8d41f519a2c4611fa6567c |
| AlphaMissense_hg38_modified.tsv.gz | 599.3 MB | 4392511387da48c919dbf9c5bcb34c62 |
| AlphaMissense_hg38_modified.tsv.gz.dvc | 119.0 B | 54e71db25713fb555d0d4eb900d06e92 |
| AlphaMissense_hg38_modified.tsv.gz.tbi | 668.87 KB | 0ab830224dba1887ac79d1a884021023 |
| AlphaMissense_hg38_modified.tsv.gz.tbi.dvc | 120.0 B | 4d74729414489de8e0620e226645f828 |
| dataPrep.sh | 390.0 B | 5c7ae6bac885219a611c1ebbb4205c75 |
| genomic_resource.yaml | 2.28 KB | 2a1221f81afbbeeec81eb9bc5ee452cd |
| statistics/ |