| id |
type |
default annotation |
description |
histogram |
range |
| ID |
str |
None
|
variant ID |
NO HISTOGRAM |
NO DOMAIN |
| AC |
int |
genome_gnomad_ac
|
Alternative allele count in the whole gnomAD genome samples v2.1.1 |
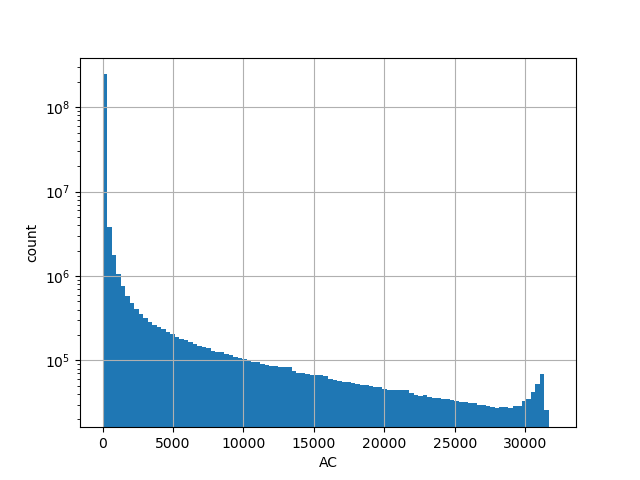 |
[0.000, 31416.000] |
| AN |
int |
genome_gnomad_an
|
Total allele count in the whole gnomAD genome samples v2.1.1 |
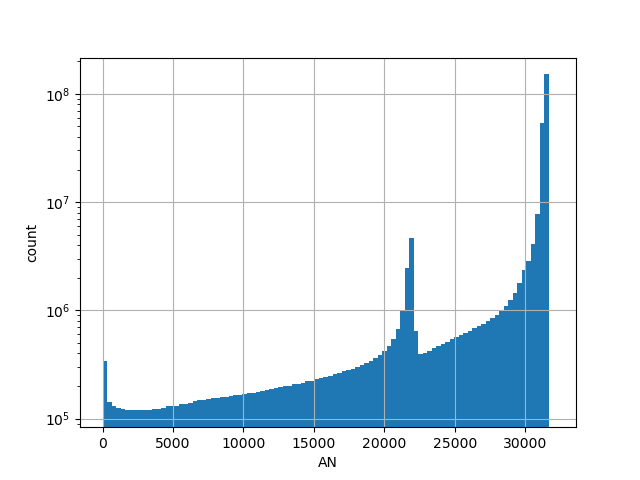 |
[0.000, 31416.000] |
| AF |
float |
genome_gnomad_af
|
Alternative allele frequency in the whole gnomAD genome samples v2.1.1 |
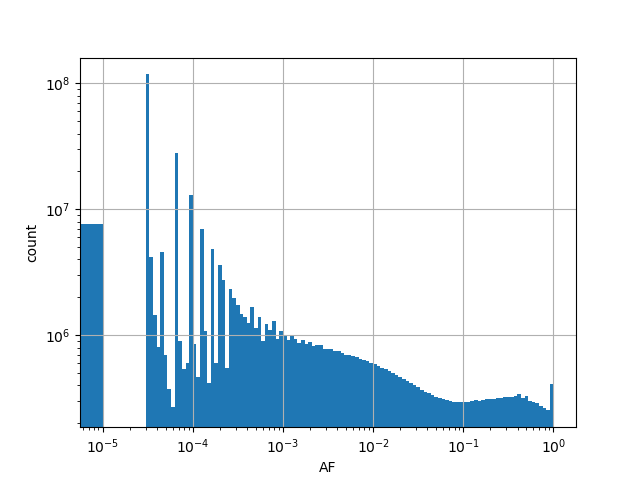 |
[0.000, 1.000] |
| AF_percent |
float |
genome_gnomad_af_percent
|
Alternative allele frequency in the whole gnomAD genome samples v2.1.1 as % |
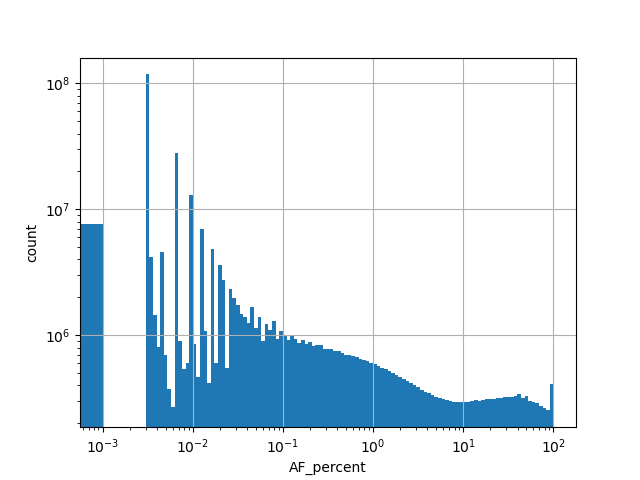 |
[0.000, 100.000] |
| controls_AC |
int |
genome_gnomad_controls_ac
|
Alternative allele count in the controls subset of whole gnomAD genome samples v2.1.1 |
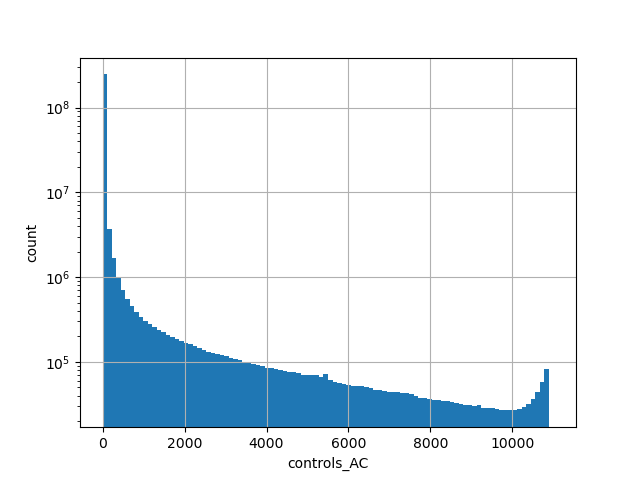 |
[0.000, 10884.000] |
| controls_AN |
int |
genome_gnomad_controls_an
|
gnomAD v2.1.1 liftover genome count of genotyped individuals in control group |
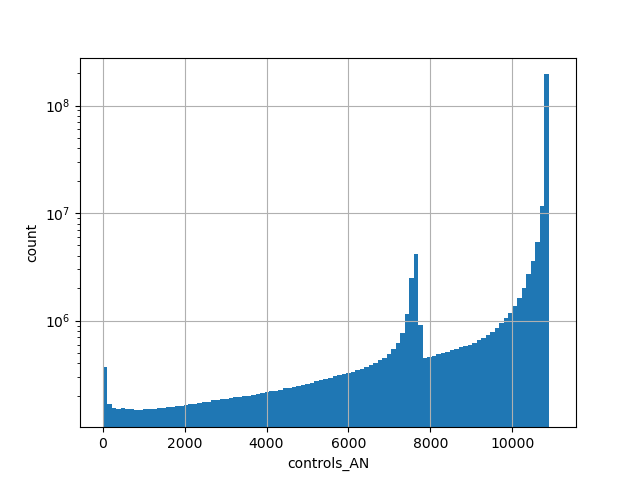 |
[0.000, 10884.000] |
| controls_AF |
float |
genome_gnomad_controls_af
|
Alternative allele frequency in the controls subset of whole gnomAD genome samples v2.1.1 |
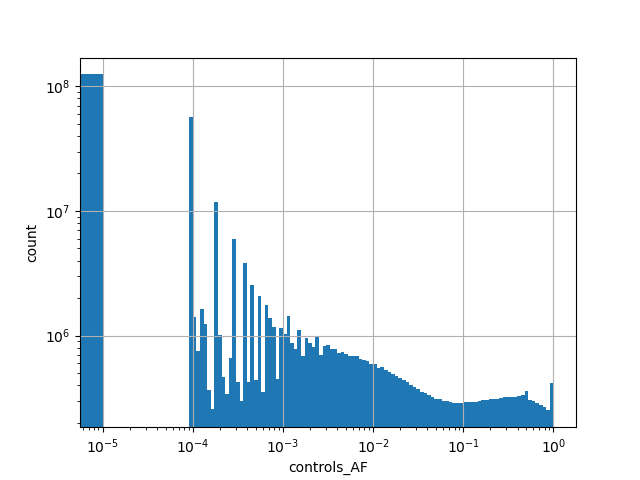 |
[0.000, 1.000] |
| controls_AF_percent |
float |
genome_gnomad_controls_af_percent
|
Alternative allele frequency in the controls subset of whole gnomAD genome samples v2.1.1 as % |
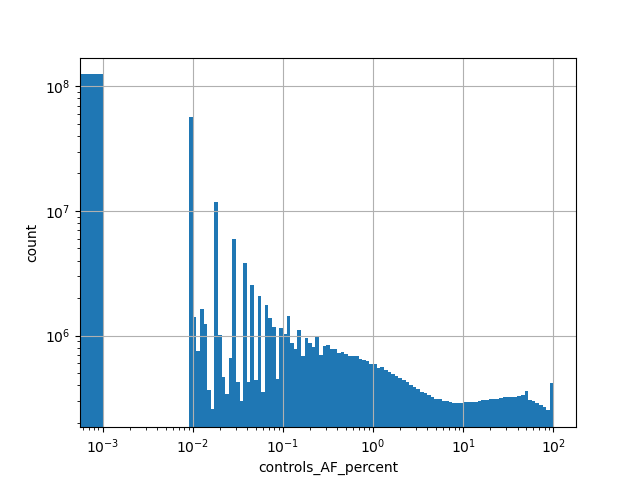 |
[0.000, 100.000] |
| non_neuro_AC |
int |
genome_gnomad_non_neuro_ac
|
Alternative allele count in the non-neuro subset of whole gnomAD genome samples v2.1.1 |
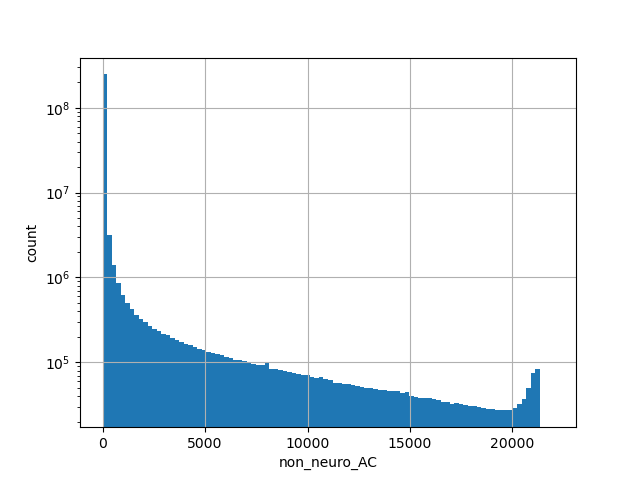 |
[0.000, 21272.000] |
| non_neuro_AN |
int |
genome_gnomad_non_neuro_an
|
Total allele count in the non-neuro subset of whole gnomAD genome samples v2.1.1 |
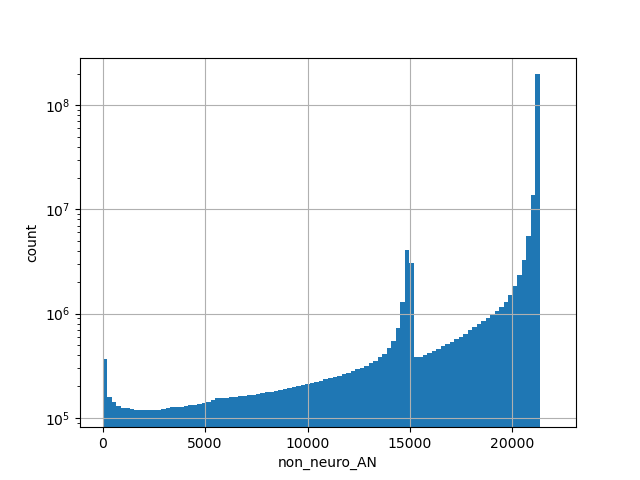 |
[0.000, 21272.000] |
| non_neuro_AF |
float |
genome_gnomad_non_neuro_af
|
Alternative allele frequency in the non-neuro subset of whole gnomAD genome samples v2.1.1 |
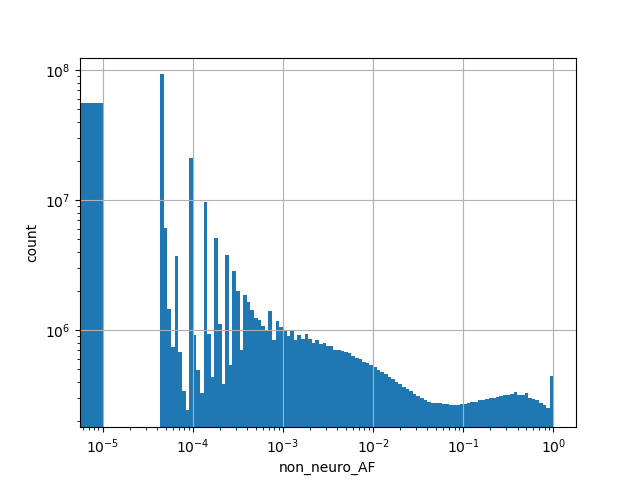 |
[0.000, 1.000] |
| non_neuro_AF_percent |
float |
genome_gnomad_non_neuro_af_percent
|
Alternative allele frequency in the non-neuro subset of whole gnomAD genome samples v2.1.1 as % |
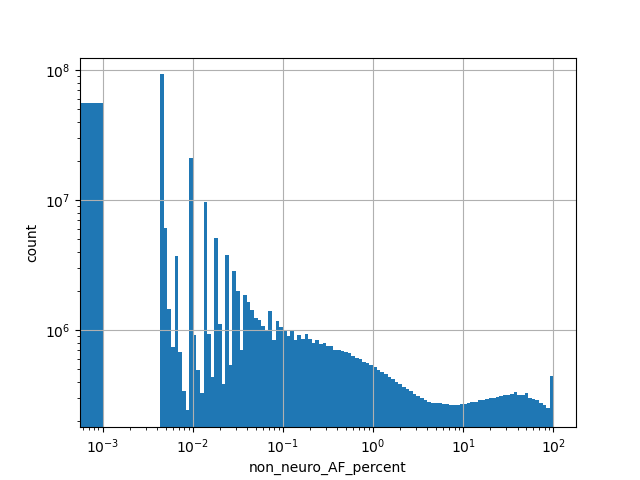 |
[0.000, 100.000] |











