-
variant ID
No histogram: Histogram for ID nullified for a region
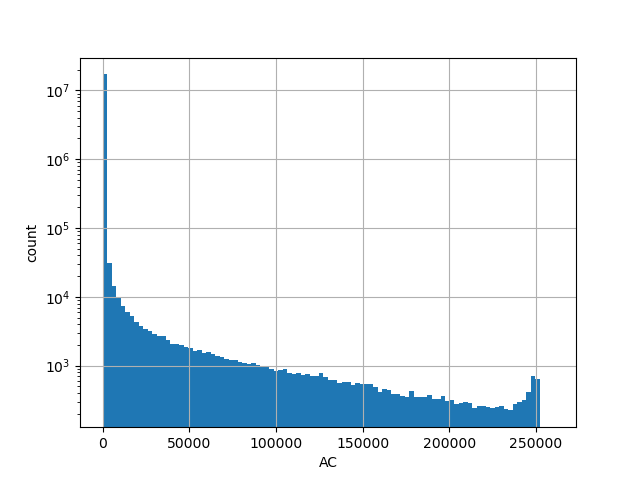
exome_gnomad_ac
Alternative allele count in the whole gnomAD exome samples v2.1.1
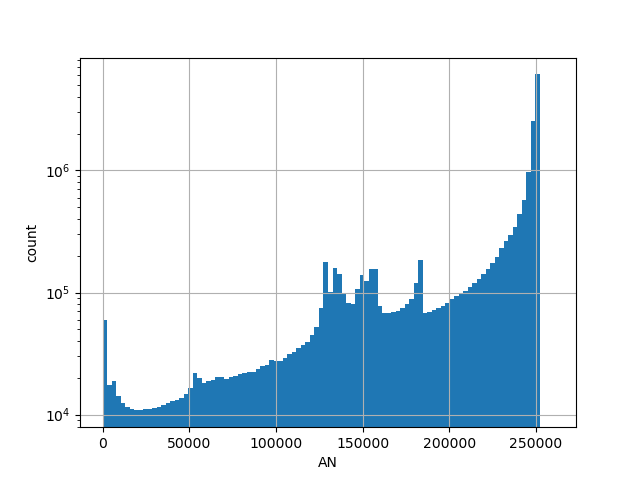
exome_gnomad_an
Total allele count in the whole gnomAD exome samples v2.1.1
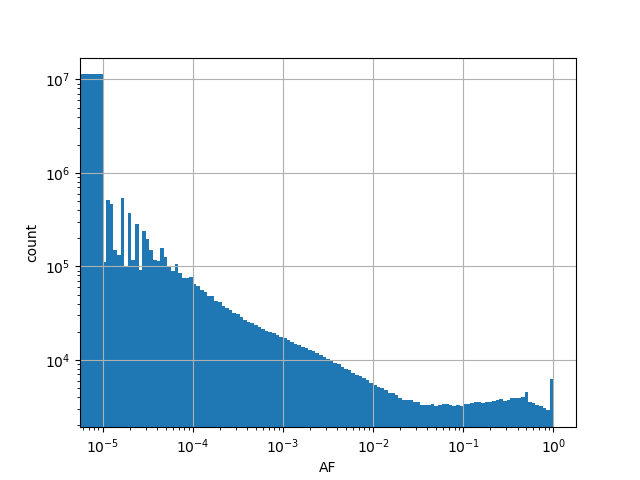
exome_gnomad_af
Alternative allele frequency in the whole gnomAD exome samples v2.1.1
exome_gnomad_af_percent
Alternative allele frequency in the whole gnomAD exome samples v2.1.1 as percent
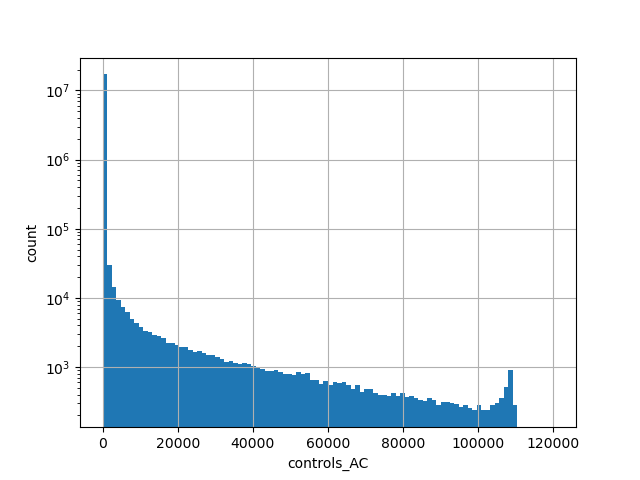
exome_gnomad_controls_ac
Alternative allele count in the controls subset of whole gnomAD exome samples v2.1.1
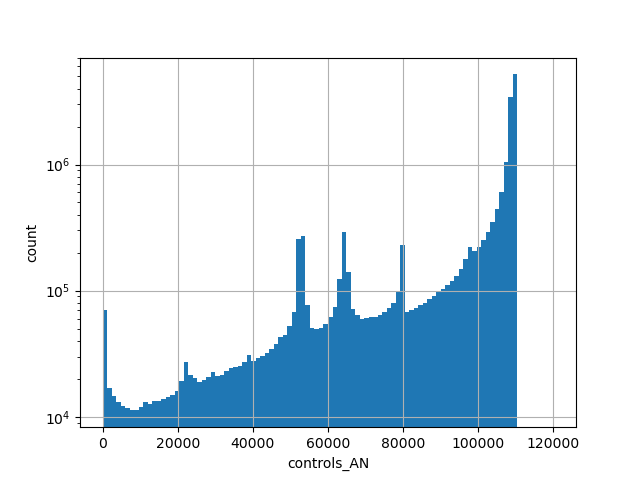
exome_gnomad_controls_an
gnomAD v2.1.1 liftover exomes count of genotyped individuals in control group
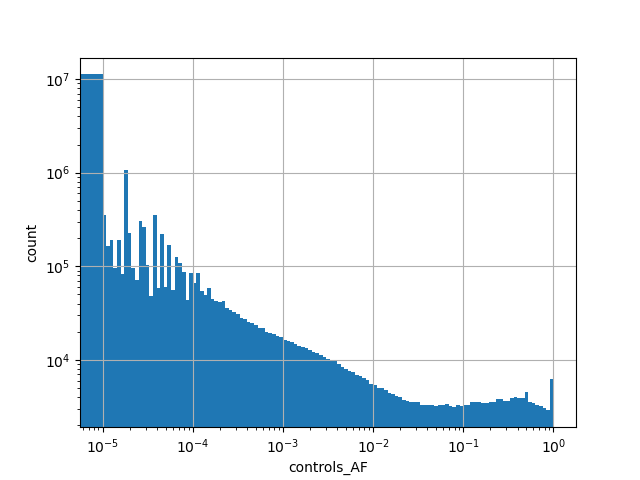
exome_gnomad_controls_af
Alternative allele frequency in the controls subset of whole gnomAD exome samples v2.1.1
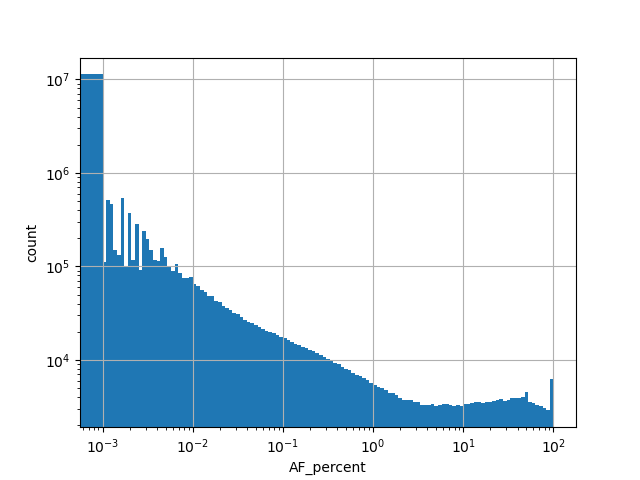
exome_gnomad_controls_af_percent
Alternative allele frequency in the controls subset of whole gnomAD exome samples v2.1.1 as %
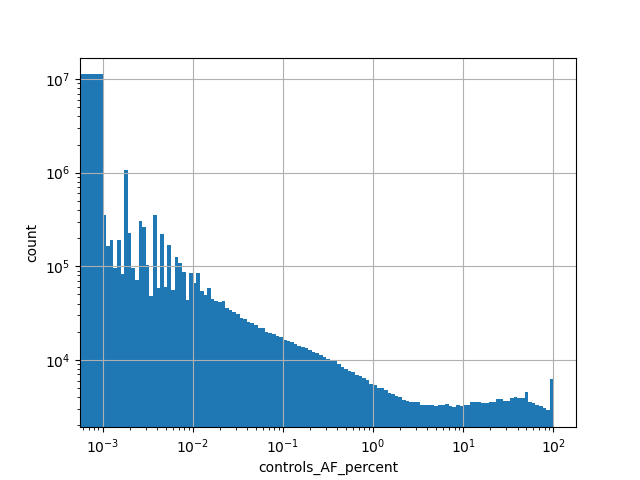
exome_gnomad_non_neuro_ac
Alternative allele count in the non-neuro subset of whole gnomAD exome samples v2.1.1
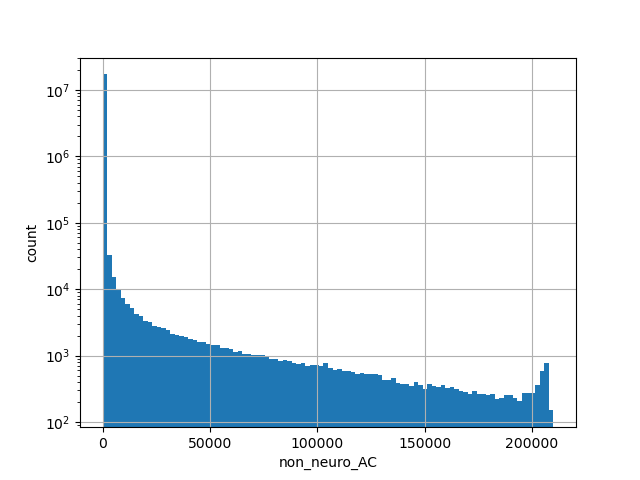
exome_gnomad_non_neuro_an
Total allele count in the non-neuro subset of whole gnomAD exome samples v2.1.1
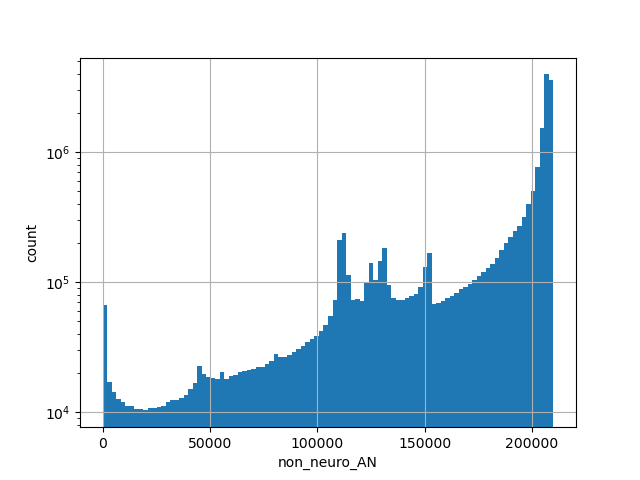
exome_gnomad_non_neuro_af
Alternative allele frequency in the non-neuro subset of whole gnomAD exome samples v2.1.1
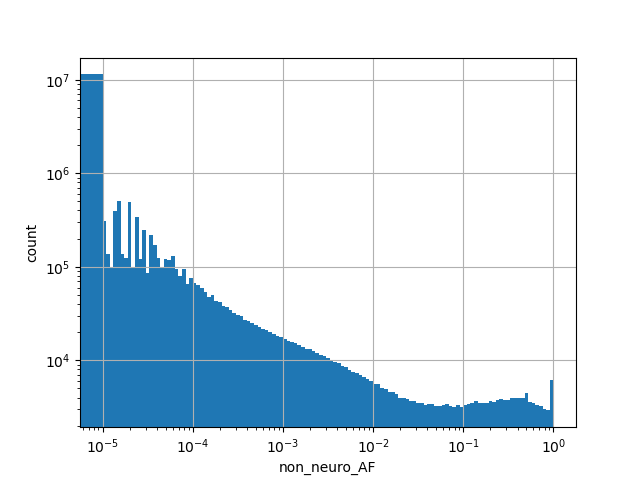
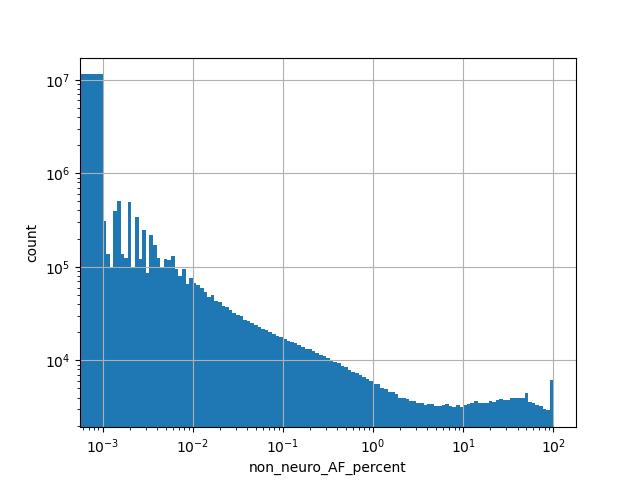
exome_gnomad_non_neuro_af_percent
Alternative allele frequency in the non-neuro subset of whole gnomAD exome samples v2.1.1 as %