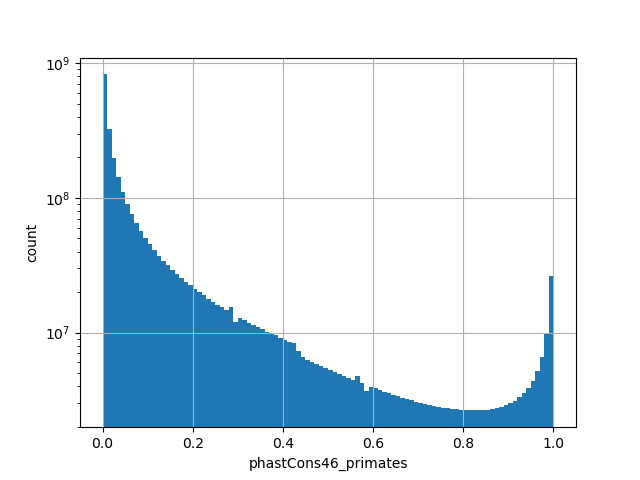
phastcons46_primates
The scores is a number that reflects the conservation at a position.
Small values desc: less conserved
Large values desc: more conserved