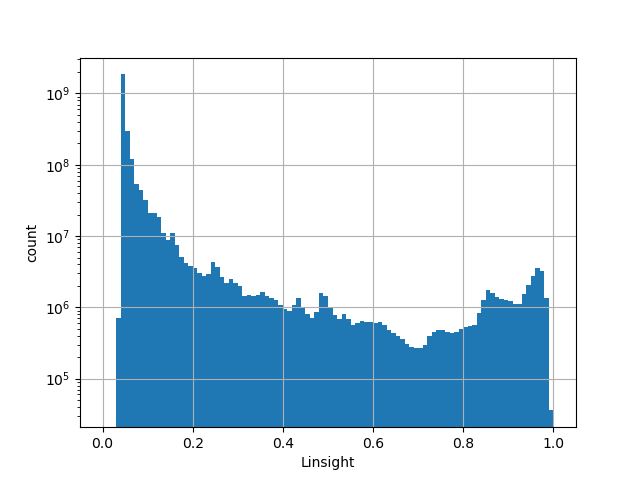
linsight
The LINSIGHT score measures the probability of negative selection on noncoding
sites.
Small values desc: negative selection is less likely
Large values desc: negative selection is more likely
×

