LINSIGHT

| Id: | hg19/scores/LINSIGHT |
| Type: | position_score |
| Version: | 0 |
| Summary: |
The likelihood of negative selection on noncoding sites |
| Description: |
LINSIGHT (Linear INSIGHT) is a computational tool designed to predict the likelihood of negative selection on noncoding nucleotide sites. It identifies noncoding sites where mutations are likely to have deleterious fitness consequences, thus playing a crucial role in phenotypic variation and disease. LINSIGHT combines a generalized linear model with a probabilistic model of molecular evolution. This integration allows it to merge functional genomic data (such as chromatin state, transcription factor binding, and other regulatory information) with evolutionary conservation data. Downloaded on: 06/07/24 |
| Labels: |
|
| ID | Type | Default annotation | Description | Histogram | Range |
|---|---|---|---|---|---|
| LINSIGHT | float |
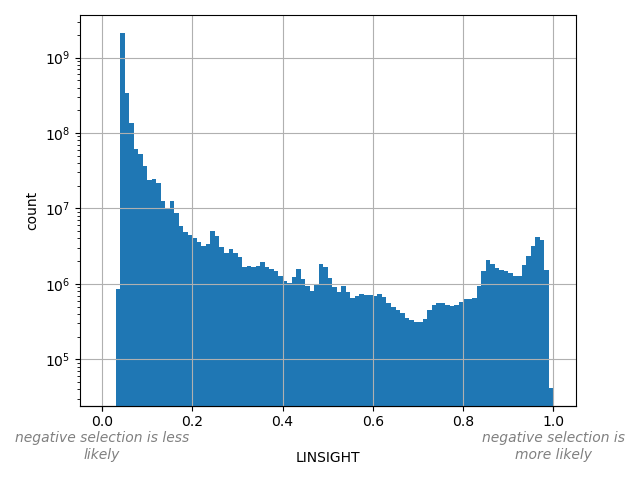
LINSIGHT |
The LINSIGHT score measures the probability of negative selection on noncoding
sites.
Small values desc: negative selection is less likely
Large values desc: negative selection is more likely
|
 |
[0.033, 0.995] |
| Filename | Size | md5 |
|---|---|---|
| LINSIGHT.bw | 1.3 GB | 210d1ac092ba5f9e3420a5b41f09aafa |
| LINSIGHT.bw.dvc | 97.0 B | e21aa95e268041fbde04101de1c25505 |
| genomic_resource.yaml | 1.84 KB | 5d340a4b53880e8df2ad26470651c4ba |
| statistics/ |