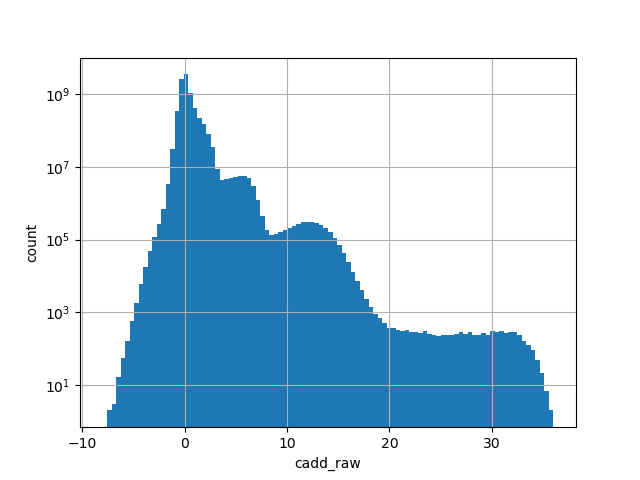
cadd_raw
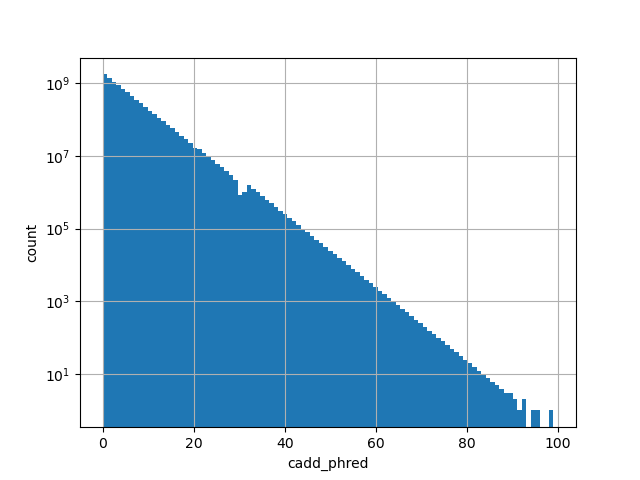
cadd_phred
| Id: | hg19/scores/CADD |
| Type: | allele_score |
| Version: | 0 |
| Summary: |
CADD score for functional prediction of a SNP |
| Description: |
CADDPlease refer to Kircher et al. (2014) Nature Genetics 46(3):310-5 for details. The larger the score the more likely the SNP has damaging effect. Downloaded from https://cadd.gs.washington.edu/download ; Higher values of raw scores have relative meaning that a variant is more likely to be simulated (or "not observed") and therefore more likely to have deleterious effects. Scaled scores are PHRED-like (-10*log10(rank/total)) scaled C-score ranking a variant relative to all possible substitutions of the human genome (8.6x10^9). Please note the following copyright statement for CADD: "CADD scores (http://cadd.gs.washington.edu/) are Copyright 2013 University of Washington and Hudson-Alpha Institute for Biotechnology (all rights reserved) but are freely available for all academic, non-commercial applications. For commercial licensing information contact Jennifer McCullar (mccullaj@uw.edu). |
| Labels: |
|
| ID | Type | Default annotation | Description | Histogram | Range |
|---|---|---|---|---|---|
| cadd_raw | float |
cadd_raw |
CADD raw score for functional prediction of a SNP. The larger the score
the more likely the SNP has damaging effect
Small values desc: less damaging
Large values desc: more damaging
|
 |
[-7.54, 35.8] |
| cadd_phred | float |
cadd_phred |
CADD phred-like score. This is phred-like rank score based on whole
genome CADD raw scores. The larger the score the more likely the SNP
has damaging effect.
Small values desc: less damaging
Large values desc: more damaging
|
 |
[0, 99] |
| Filename | Size | md5 |
|---|---|---|
| CADD.bedgraph.gz | 79.37 GB | 3a766a8e17649754371fdcafd4d1438d |
| CADD.bedgraph.gz.dvc | 91.0 B | 61b8c446d51719165655dd42b1ee5536 |
| CADD.bedgraph.gz.tbi | 2.6 MB | 757558e9cc3731ea7ce629ff35ebb7b7 |
| CADD.bedgraph.gz.tbi.dvc | 91.0 B | 89e4400ddd3349702b7654ff5ab5935c |
| genomic_resource.yaml | 2.38 KB | 09e5c171f68fccfa0d7f833cbefd7ce9 |
| statistics/ |