| Id: | gene_properties/gene_sets/PFAM_37.0_domains |
| Type: | gene_set_collection |
| Version: | 0 |
| Summary: |
PFAM 37.0 domains |
| Description: |
Pfam (Protein Families) is a comprehensive database of protein families that includes their annotations and multiple sequence alignments generated using hidden Markov models (HMMs). Pfam domains are conserved regions within proteins that often correlate with specific structural or functional roles. Each domain in Pfam is represented by a curated alignment of related sequences and an HMM profile, which can be used to identify and analyze these domains in other proteins. This allows researchers to predict the function of uncharacterized proteins, study protein evolution, and understand protein domain architecture across different organisms. Finn et al, Pfam: The protein families database, Nucleic Acids Research 2014 Mistry et al, Pfam: The protein families database in 2021, Nucleic Acids Research 2021 Processing DetailsDownloaded PFAM domain id - Gene name data from https://useast.ensembl.org/biomart/martview/ on 06/23/2024. Filtered to include genes with PFAM id (mart_export.xlsx). Downloaded PFAM domain id - PFAM domain name data from https://ftp.ebi.ac.uk/pub/databases/Pfam/releases/Pfam37.0/Pfam-A.clans.tsv.gz on 06/23/2024 (Pfam-A.clans.xlsx). PFAMdataprep.py merges the two files, prepares domain-map.txt listing the domains per gene and domain-mapnames.txt listing the description of each domain. |
| Labels: |
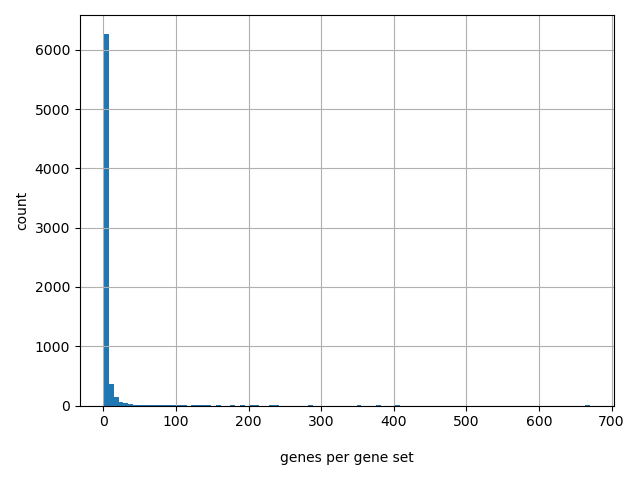
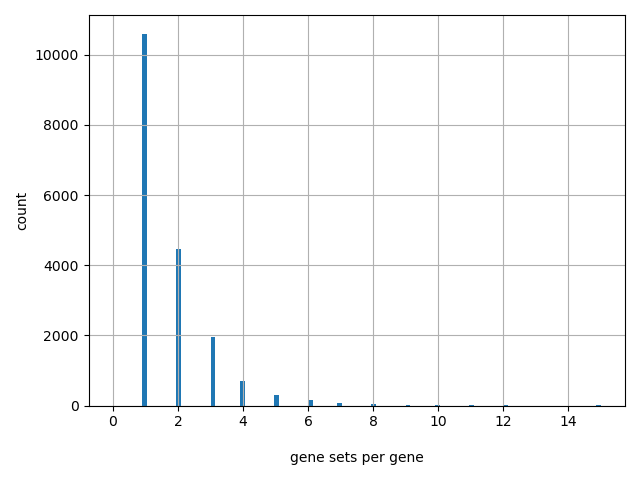
Number of gene sets: 7013
Number of unique genes: 18336


| Gene Set | Gene Count | Description |
|---|---|---|
| zf-C2H2 | 670 | Zinc finger, C2H2 type |
| 7tm_4 | 405 | Olfactory receptor |
| KRAB | 378 | KRAB box |
| Pkinase | 355 | Protein kinase domain |
| 7tm_1 | 286 | 7 transmembrane receptor (rhodopsin family) |
| WD40 | 236 | WD domain, G-beta repeat |
| Ank_2 | 235 | Ankyrin repeats (3 copies) |
| RRM_1 | 210 | RNA recognition motif |
| Homeodomain | 207 | Homeodomain |
| PH | 189 | PH domain |
| LRR_8 | 178 | Leucine rich repeat |
| V-set | 160 | Immunoglobulin V-set domain |
| Ig_3 | 147 | Immunoglobulin domain |
| PK_Tyr_Ser-Thr | 138 | Protein tyrosine and serine/threonine kinase |
| fn3 | 137 | Fibronectin type III domain |
| Ras | 136 | Ras family |
| I-set | 135 | Immunoglobulin I-set domain |
| BTB | 130 | BTB/POZ domain |
| C2 | 130 | C2 domain |
| PDZ | 127 | PDZ domain |
| Trypsin | 113 | Trypsin |
| Cadherin | 112 | Cadherin domain |
| SH3_1 | 112 | SH3 domain |
| EF-hand_7 | 109 | EF-hand domain pair |
| Ion_trans | 109 | Ion transport protein |
| Helicase_C | 106 | Helicase conserved C-terminal domain |
| HLH | 105 | Helix-loop-helix DNA-binding domain |
| Ank | 103 | Ankyrin repeat |
| SH2 | 101 | SH2 domain |
| SPRY | 94 | SPRY domain |
| zf-C2H2_6 | 93 | C2H2-type zinc finger |
| Lectin_C | 83 | Lectin C-type domain |
| Collagen | 82 | Collagen triple helix repeat (20 copies) |
| ig | 81 | Immunoglobulin domain |
| UCH | 78 | Ubiquitin carboxyl-terminal hydrolase |
| EGF | 77 | EGF-like domain |
| zf-B_box | 77 | B-box zinc finger |
| EGF_CA | 75 | Calcium-binding EGF domain |
| Filament | 73 | Intermediate filament protein |
| Ig_2 | 71 | Immunoglobulin domain |
| LIM | 71 | LIM domain |
| DEAD | 70 | DEAD/DEAH box helicase |
| RhoGEF | 70 | RhoGEF domain |
| MFS_1 | 69 | Major Facilitator Superfamily |
| Histone | 68 | Core histone H2A/H2B/H3/H4 |
| RhoGAP | 68 | RhoGAP domain |
| Ank_4 | 67 | Ankyrin repeats (many copies) |
| CH | 67 | Calponin homology (CH) domain |
| SAM_1 | 66 | SAM domain (Sterile alpha motif) |
| SH3_9 | 65 | Variant SH3 domain |
| Cadherin_2 | 64 | Cadherin-like |
| Kelch_1 | 64 | Kelch motif |
| SH3_2 | 62 | Variant SH3 domain |
| BACK | 61 | BTB And C-terminal Kelch |
| zf-H2C2_2 | 61 | Zinc-finger double domain |
| LRR_1 | 59 | Leucine Rich Repeat |
| TPR_8 | 59 | Tetratricopeptide repeat |
| p450 | 58 | Cytochrome P450 |
| C1_1 | 57 | Phorbol esters/diacylglycerol binding domain (C1 domain) |
| LRR_6 | 57 | Leucine Rich repeat |
| SCAN | 57 | SCAN domain |
| F-box-like | 56 | F-box-like |
| PHD | 54 | PHD-finger |
| Mito_carr | 53 | Mitochondrial carrier protein |
| RabGAP-TBC | 53 | Rab-GTPase-TBC domain |
| adh_short | 53 | short chain dehydrogenase |
| TSP_1 | 52 | Thrombospondin type 1 domain |
| zf-RING_2 | 52 | Ring finger domain |
| Forkhead | 51 | Forkhead domain |
| HMG_box | 51 | HMG (high mobility group) box |
| Sushi | 51 | Sushi repeat (SCR repeat) |
| BTB_2 | 50 | BTB/POZ domain |
| CUB | 50 | CUB domain |
| ABC_tran | 49 | ABC transporter |
| 7tm_2 | 48 | 7 transmembrane receptor (Secretin family) |
| AAA | 48 | ATPase family associated with various cellular activities (AAA) |
| Hormone_recep | 48 | Ligand-binding domain of nuclear hormone receptor |
| PX | 48 | PX domain |
| FERM_M | 47 | FERM central domain |
| Neur_chan_LBD | 46 | Neurotransmitter-gated ion-channel ligand binding domain |
| VWA | 46 | von Willebrand factor type A domain |
| zf-C4 | 46 | Zinc finger, C4 type (two domains) |
| DnaJ | 45 | DnaJ domain |
| IQ | 45 | IQ calmodulin-binding motif |
| Neur_chan_memb | 45 | Neurotransmitter-gated ion-channel transmembrane region |
| PRY | 45 | SPRY-associated domain |
| zf-C3HC4 | 45 | Zinc finger, C3HC4 type (RING finger) |
| IL8 | 44 | Small cytokines (intecrine/chemokine), interleukin-8 like |
| Kinesin | 44 | Kinesin motor domain |
| zf-RING_UBOX | 44 | RING-type zinc-finger |
| ubiquitin | 43 | Ubiquitin family |
| C1-set | 42 | Immunoglobulin C1-set domain |
| Ldl_recept_a | 42 | Low-density lipoprotein receptor domain class A |
| Y_phosphatase | 42 | Protein-tyrosine phosphatase |
| WW | 41 | WW domain |
| Bromodomain | 40 | Bromodomain |
| Cadherin_C_2 | 40 | Cadherin cytoplasmic C-terminal |
| DSPc | 40 | Dual specificity phosphatase, catalytic domain |
| Keratin_B2_2 | 40 | Keratin, high sulfur B2 protein |
| UQ_con | 40 | Ubiquitin-conjugating enzyme |
| zf-C3HC4_4 | 40 | zinc finger of C3HC4-type, RING |
| Myosin_head | 39 | Myosin head (motor domain) |
| zf-C3HC4_3 | 39 | Zinc finger, C3HC4 type (RING finger) |
| zf-met | 39 | Zinc-finger of C2H2 type |
| FXa_inhibition | 38 | Coagulation Factor Xa inhibitory site |
| MAGE | 38 | MAGE homology domain |
| Reprolysin | 38 | Reprolysin (M12B) family zinc metalloprotease |
| ANF_receptor | 37 | Receptor family ligand binding region |
| Cadherin_tail | 37 | Cadherin C-terminal cytoplasmic tail, catenin-binding region |
| Serpin | 37 | Serpin (serine protease inhibitor) |
| TGF_beta | 37 | Transforming growth factor beta like domain |
| Laminin_G_2 | 36 | Laminin G domain |
| PMP22_Claudin | 36 | PMP-22/EMP/MP20/Claudin family |
| SAM_2 | 36 | SAM domain (Sterile alpha motif) |
| SOCS_box | 36 | SOCS box |
| Sugar_tr | 36 | Sugar (and other) transporter |
| KH_1 | 35 | KH domain |
| Pep_M12B_propep | 35 | Reprolysin family propeptide |
| RGS | 35 | Regulator of G protein signaling domain |
| Kazal_2 | 34 | Kazal-type serine protease inhibitor domain |
| Pkinase_C | 34 | Protein kinase C terminal domain |
| RA | 34 | Ras association (RalGDS/AF-6) domain |
| SET | 34 | SET domain |
| Sulfotransfer_1 | 34 | Sulfotransferase domain |
| hEGF | 34 | Human growth factor-like EGF |
| zf-CCCH | 34 | Zinc finger C-x8-C-x5-C-x3-H type (and similar) |
| Cyclin_N | 33 | Cyclin, N-terminal domain |
| Tetraspanin | 33 | Tetraspanin family |
| Cation_ATPase | 32 | Cation transport ATPase (P-type) |
| Hydrolase | 32 | haloacid dehalogenase-like hydrolase |
| SNF2-rel_dom | 32 | SNF2-related domain |
| cNMP_binding | 32 | Cyclic nucleotide-binding domain |
| zf-C3HC4_2 | 32 | Zinc finger, C3HC4 type (RING finger) |
| Actin | 31 | Actin |
| Arf | 31 | ADP-ribosylation factor family |
| C1q | 31 | C1q domain |
| C2-set_2 | 31 | CD80-like C2-set immunoglobulin domain |
| Death | 31 | Death domain |
| E1-E2_ATPase | 31 | E1-E2 ATPase |
| FHA | 31 | FHA domain |
| GPS | 31 | GPCR proteolysis site, GPS, motif |
| Laminin_EGF | 31 | Laminin EGF domain |
| Lipocalin | 31 | Lipocalin / cytosolic fatty-acid binding protein family |
| AMP-binding | 30 | AMP-binding enzyme |
| Sema | 30 | Sema domain |
| bZIP_1 | 30 | bZIP transcription factor |
| cEGF | 30 | Complement Clr-like EGF-like |
| Defensin_beta_2 | 29 | Beta defensin |
| Ets | 29 | Ets-domain |
| FYVE | 29 | FYVE zinc finger |
| LRRNT | 29 | Leucine rich repeat N-terminal domain |
| RasGEF | 29 | RasGEF domain |
| Thioredoxin | 29 | Thioredoxin |
| FERM_N | 28 | FERM N-terminal domain |
| HECT | 28 | HECT-domain (ubiquitin-transferase) |
| Metallophos | 28 | Calcineurin-like phosphoesterase |
| PID | 28 | Phosphotyrosine interaction domain (PTB/PID) |
| PSI | 28 | Plexin repeat |
| TPR_1 | 28 | Tetratricopeptide repeat |
| TPR_16 | 28 | Tetratricopeptide repeat |
| zf-C2H2_4 | 28 | C2H2-type zinc finger |
| Abhydrolase_1 | 27 | alpha/beta hydrolase fold |
| ArfGap | 27 | Putative GTPase activating protein for Arf |
| Arm | 27 | Armadillo/beta-catenin-like repeat |
| EF-hand_5 | 27 | EF hand |
| EF-hand_8 | 27 | EF-hand domain pair |
| FERM_C | 27 | FERM C-terminal PH-like domain |
| Myb_DNA-binding | 27 | Myb-like DNA-binding domain |
| S_100 | 27 | S-100/ICaBP type calcium binding domain |
| TPR_12 | 27 | Tetratricopeptide repeat |
| VWC | 27 | von Willebrand factor type C domain |
| CARD | 26 | Caspase recruitment domain |
| Chromo | 26 | Chromo (CHRromatin Organisation MOdifier) domain |
| Glycos_transf_2 | 26 | Glycosyl transferase family 2 |
| TSP1_ADAMTS | 26 | Thrombospondin type 1 domain |
| AAA_lid_3 | 25 | AAA+ lid domain |
| ADAMTS_CR_3 | 25 | ADAMTS cysteine-rich domain |
| ADAMTS_spacer1 | 25 | ADAM-TS Spacer 1 |
| Guanylate_kin | 25 | Guanylate kinase |
| JmjC | 25 | JmjC domain, hydroxylase |
| Keratin_2_head | 25 | Keratin type II head |
| LRR_4 | 25 | Leucine Rich repeats (2 copies) |
| TAS2R | 25 | Taste receptor protein (TAS2R) |
| UBA | 25 | UBA/TS-N domain |
| DHHC | 24 | DHHC palmitoyltransferase |
| EF-hand_1 | 24 | EF hand |
| Fibrinogen_C | 24 | Fibrinogen beta and gamma chains, C-terminal globular domain |
| HRM | 24 | Hormone receptor domain |
| MARVEL | 24 | Membrane-associating domain |
| NACHT | 24 | NACHT domain |
| Pro_isomerase | 24 | Cyclophilin type peptidyl-prolyl cis-trans isomerase/CLD |
| Rhodanese | 24 | Rhodanese-like domain |
| Ricin_B_lectin | 24 | Ricin-type beta-trefoil lectin domain |
| Spectrin | 24 | Spectrin repeat |
| Tubulin | 24 | Tubulin/FtsZ family, GTPase domain |
| zf-CCHC_6 | 24 | Zinc knuckle |
| ABC_membrane | 23 | ABC transporter transmembrane region |
| F5_F8_type_C | 23 | F5/8 type C domain |
| Fz | 23 | Fz domain |
| GOLGA2L5 | 23 | Putative golgin subfamily A member 2-like protein 5 |
| Hemopexin | 23 | Hemopexin |
| Histone_H2A_C | 23 | C-terminus of histone H2A |
| Homeobox_KN | 23 | Homeobox KN domain |
| LRR_9 | 23 | Leucine-rich repeat |
| Peptidase_M10 | 23 | Matrixin |
| Peptidase_M14 | 23 | Zinc carboxypeptidase |
| 7tm_3 | 22 | 7 transmembrane sweet-taste receptor of 3 GCPR |
| ANAPC4_WD40 | 22 | Anaphase-promoting complex subunit 4 WD40 domain |
| CADH_Y-type_LIR | 22 | Cadherin, Y-type LIR-motif |
| CD20 | 22 | CD20-like family |
| DEP | 22 | Domain found in Dishevelled, Egl-10, and Pleckstrin (DEP) |
| EGF_2 | 22 | EGF-like domain |
| FCH | 22 | Fes/CIP4, and EFC/F-BAR homology domain |
| FGF | 22 | Fibroblast growth factor |
| GAGE | 22 | GAGE protein |
| Ion_trans_2 | 22 | Ion channel |
| MAGE_N | 22 | Melanoma associated antigen family N terminal |
| NUDIX | 22 | NUDIX domain |
| PLAT | 22 | PLAT/LH2 domain |
| PWWP | 22 | PWWP domain |
| PYRIN | 22 | PAAD/DAPIN/Pyrin domain |
| Tubulin_C | 22 | Tubulin C-terminal domain |
| UDPGT | 22 | UDP-glucoronosyl and UDP-glucosyl transferase |
| Connexin | 21 | Connexin |
| G-patch | 21 | G-patch domain |
| PDEase_I | 21 | 3'5'-cyclic nucleotide phosphodiesterase |
| RasGEF_N | 21 | RasGEF N-terminal motif |
| SEA | 21 | SEA domain |
| Spy1 | 21 | Cell cycle regulatory protein |
| TGFb_propeptide | 21 | TGF-beta propeptide |
| TNFR_c6 | 21 | TNFR/NGFR cysteine-rich region |
| UPAR_LY6 | 21 | u-PAR/Ly-6 domain |
| Acetyltransf_1 | 20 | Acetyltransferase (GNAT) family |
| Disintegrin | 20 | Disintegrin |
| GST_C | 20 | Glutathione S-transferase, C-terminal domain |
| GTP_EFTU | 20 | Elongation factor Tu GTP binding domain |
| Glyco_transf_29 | 20 | Glycosyltransferase family 29 (sialyltransferase) |
| PDZ_6 | 20 | PDZ domain |
| RCC1 | 20 | Regulator of chromosome condensation (RCC1) repeat |
| TIG | 20 | IPT/TIG domain |
| ADAMTS_CR_2 | 19 | ADAMTS cysteine-rich domain 2 |
| Aldedh | 19 | Aldehyde dehydrogenase family |
| CCDC144C | 19 | CCDC144C protein coiled-coil region |
| FG-GAP | 19 | FG-GAP repeat |
| G-gamma | 19 | GGL domain |
| GRAM | 19 | GRAM domain |
| HABP4_PAI-RBP1 | 19 | Hyaluronan / mRNA binding family |
| IGFBP | 19 | Insulin-like growth factor binding protein |
| NAP | 19 | Nucleosome assembly protein (NAP) |
| NPIP | 19 | Nuclear pore complex interacting protein (NPIP) |
| PA | 19 | PA domain |
| PAS | 19 | PAS fold |
| PP2C | 19 | Protein phosphatase 2C |
| Proteasome | 19 | Proteasome subunit |
| RUN | 19 | RUN domain |
| SNF | 19 | Sodium:neurotransmitter symporter family |
| SRCR | 19 | Scavenger receptor cysteine-rich domain |
| TIR | 19 | TIR domain |
| wnt | 19 | wnt family |
| zf-CCHC | 19 | Zinc knuckle |
| zf-MYND | 19 | MYND finger |
| ADAM_CR | 18 | ADAM cysteine-rich |
| Cation_ATPase_N | 18 | Cation transporter/ATPase, N-terminus |
| EGF_3 | 18 | EGF domain |
| Guanylate_cyc | 18 | Adenylate and Guanylate cyclase catalytic domain |
| HA2_C | 18 | Helicase associated domain (HA2), ratchet-like |
| Integrin_A_Ig_2 | 18 | Integrin alpha Ig-like domain 2 |
| Kunitz_BPTI | 18 | Kunitz/Bovine pancreatic trypsin inhibitor domain |
| LCE | 18 | Late cornified envelope |
| LSM | 18 | LSM domain |
| Lig_chan | 18 | Ligand-gated ion channel |
| Lig_chan-Glu_bd | 18 | Ligated ion channel L-glutamate- and glycine-binding site |
| MAM | 18 | MAM domain, meprin/A5/mu |
| MHC_I | 18 | Class I Histocompatibility antigen, domains alpha 1 and 2 |
| Myosin_tail_1 | 18 | Myosin tail |
| NOD2_WH | 18 | NOD2 winged helix domain |
| NTR | 18 | UNC-6/NTR/C345C module |
| PAP2 | 18 | PAP2 superfamily |
| PG_binding_1 | 18 | Putative peptidoglycan binding domain |
| PI3_PI4_kinase | 18 | Phosphatidylinositol 3- and 4-kinase |
| SAP | 18 | SAP domain |
| Sulfatase | 18 | Sulfatase |
| Zona_pellucida | 18 | Zona pellucida-like domain |
| zf-RanBP | 18 | Zn-finger in Ran binding protein and others |
| AMP-binding_C | 17 | AMP-binding enzyme C-terminal domain |
| Aa_trans | 17 | Transmembrane amino acid transporter protein |
| Aldo_ket_red | 17 | Aldo/keto reductase family |
| Aminotran_1_2 | 17 | Aminotransferase class I and II |
| CBS | 17 | CBS domain |
| Carb_anhydrase | 17 | Eukaryotic-type carbonic anhydrase |
| Clat_adaptor_s | 17 | Clathrin adaptor complex small chain |
| Cyclin_C | 17 | Cyclin, C-terminal domain |
| Cys_knot | 17 | Cystine-knot domain |
| Cystatin | 17 | Cystatin domain |
| DENN | 17 | DENN (AEX-3) domain |
| Exo_endo_phos | 17 | Endonuclease/Exonuclease/phosphatase family |
| GM130_C | 17 | GM130 C-terminal binding motif |
| Gal-bind_lectin | 17 | Galactoside-binding lectin |
| IBN_N | 17 | Importin-beta N-terminal domain |
| Interferon | 17 | Interferon alpha/beta domain |
| NLRC4_HD2 | 17 | NLRC4 helical domain HD2 |
| OB_NTP_bind | 17 | Oligonucleotide/oligosaccharide-binding (OB)-fold |
| PCI | 17 | PCI domain |
| PI-PLC-X | 17 | Phosphatidylinositol-specific phospholipase C, X domain |
| PRDM2_PR | 17 | PR domain zinc finger protein 2, PR domain |
| T-box | 17 | T-box |
| TNF | 17 | TNF(Tumour Necrosis Factor) family |
| Thyroglobulin_1 | 17 | Thyroglobulin type-1 repeat |
| ZZ | 17 | Zinc finger, ZZ type |
| dsrm | 17 | Double-stranded RNA binding motif |
| AAA_6 | 16 | Hydrolytic ATP binding site of dynein motor region |
| AAA_7 | 16 | P-loop containing dynein motor region |
| AAA_8 | 16 | P-loop containing dynein motor region D4 |
| ADH_zinc_N | 16 | Zinc-binding dehydrogenase |
| Acyltransferase | 16 | Acyltransferase |
| DHC_N2 | 16 | Dynein heavy chain, N-terminal region 2 |
| Dynein_C | 16 | Dynein heavy chain C-terminal domain |
| Dynein_heavy | 16 | Dynein heavy chain region D6 P-loop domain |
| ECH_1 | 16 | Enoyl-CoA hydratase/isomerase |
| FKBP_C | 16 | FKBP-type peptidyl-prolyl cis-trans isomerase |
| G-alpha | 16 | G-protein alpha subunit |
| Galactosyl_T | 16 | Galactosyltransferase |
| HA2_N | 16 | Helicase associated domain (HA2), winged-helix |
| IRK | 16 | Inward rectifier potassium channel transmembrane domain |
| IRK_C | 16 | Inward rectifier potassium channel C-terminal domain |
| Integrin_A_Ig_1 | 16 | Integrin alpha Ig-like domain 1 |
| Kringle | 16 | Kringle domain |
| Laminin_N | 16 | Laminin N-terminal (Domain VI) |
| MMR_HSR1 | 16 | 50S ribosome-binding GTPase |
| MORN | 16 | MORN repeat |
| MT | 16 | Microtubule-binding stalk of dynein motor |
| PARP | 16 | Poly(ADP-ribose) polymerase catalytic domain |
| Pou | 16 | Pou domain - N-terminal to homeobox domain |
| TRAM_LAG1_CLN8 | 16 | TLC domain |
| WAP | 16 | WAP-type (Whey Acidic Protein) 'four-disulfide core' |
| uDENN | 16 | uDENN domain |
| AAA_9 | 15 | ATP-binding dynein motor region |
| AAA_lid_11 | 15 | Dynein heavy chain AAA lid domain |
| ADH_N | 15 | Alcohol dehydrogenase GroES-like domain |
| ARID | 15 | ARID/BRIGHT DNA binding domain |
| BAR | 15 | BAR domain |
| BRCT | 15 | BRCA1 C Terminus (BRCT) domain |
| CAP | 15 | Cysteine-rich secretory protein family |
| CD225 | 15 | Interferon-induced transmembrane protein |
| CENP-T_C | 15 | Centromere kinetochore component CENP-T histone fold |
| CNH | 15 | CNH domain |
| CRAL_TRIO_2 | 15 | Divergent CRAL/TRIO domain |
| Cation_ATPase_C | 15 | Cation transporting ATPase, C-terminus |
| Cpn60_TCP1 | 15 | TCP-1/cpn60 chaperonin family |
| Crystall | 15 | Beta/Gamma crystallin |
| DAGK_cat | 15 | Diacylglycerol kinase catalytic domain |
| Dynamin_N | 15 | Dynamin family |
| Dynein_AAA_lid | 15 | Dynein heavy chain AAA lid domain |
| FH2 | 15 | Formin Homology 2 Domain |
| GST_N | 15 | Glutathione S-transferase, N-terminal domain |
| GTP_EFTU_D2 | 15 | Elongation factor Tu domain 2 |
| HTH_Tnp_Tc5 | 15 | Tc5 transposase DNA-binding domain |
| MIP | 15 | Major intrinsic protein |
| OAR | 15 | OAR motif |
| Olduvai | 15 | Olduvai domain |
| PI-PLC-Y | 15 | Phosphatidylinositol-specific phospholipase C, Y domain |
| Peptidase_C2 | 15 | Calpain family cysteine protease |
| RasGAP | 15 | GTPase-activator protein for Ras-like GTPase |
| RnaseA | 15 | Pancreatic ribonuclease |
| START | 15 | START domain |
| Sec7 | 15 | Sec7 domain |
| TAFII28 | 15 | hTAFII28-like protein conserved region |
| TTL | 15 | Tubulin-tyrosine ligase family |
| VWD | 15 | von Willebrand factor type D domain |
| Acyl-CoA_dh_M | 14 | Acyl-CoA dehydrogenase, middle domain |
| Adaptin_N | 14 | Adaptin N terminal region |
| BEX | 14 | Brain expressed X-linked like family |
| COesterase | 14 | Carboxylesterase family |
| CT47 | 14 | Cancer/testis gene family 47 |
| Cyt-b5 | 14 | Cytochrome b5-like Heme/Steroid binding domain |
| EphA2_TM | 14 | Ephrin type-A receptor 2 transmembrane domain |
| Ephrin_lbd | 14 | Ephrin receptor ligand binding domain |
| Ephrin_rec_like | 14 | Tyrosine-protein kinase ephrin type A/B receptor-like |
| GATA | 14 | GATA zinc finger |
| Gelsolin | 14 | Gelsolin repeat |
| HSP70 | 14 | Hsp70 protein |
| LRRCT | 14 | Leucine rich repeat C-terminal domain |
| Myosin_N | 14 | Myosin N-terminal SH3-like domain |
| Myotub-related | 14 | Myotubularin-like phosphatase domain |
| Na_H_Exchanger | 14 | Sodium/hydrogen exchanger family |
| PB1 | 14 | PB1 domain |
| PBD | 14 | P21-Rho-binding domain |
| Peptidase_C14 | 14 | Caspase domain |
| PhoLip_ATPase_C | 14 | Phospholipid-translocating P-type ATPase C-terminal |
| PhoLip_ATPase_N | 14 | Phospholipid-translocating ATPase N-terminal |
| TUDOR | 14 | Tudor domain |
| WH2 | 14 | WH2 motif |
| Xlink | 14 | Extracellular link domain |
| Zip | 14 | ZIP Zinc transporter |
| bZIP_2 | 14 | Basic region leucine zipper |
| fn2 | 14 | Fibronectin type II domain |
| zf-HC5HC2H_2 | 14 | PHD-zinc-finger like domain |
| zf-UBP | 14 | Zn-finger in ubiquitin-hydrolases and other protein |
| AAA_lid_1 | 13 | AAA+ lid domain |
| AA_permease_2 | 13 | Amino acid permease |
| Annexin | 13 | Annexin |
| CENP-B_N | 13 | CENP-B N-terminal DNA-binding domain |
| CRAL_TRIO | 13 | CRAL/TRIO domain |
| CS | 13 | CS domain |
| Calpain_III | 13 | Calpain large subunit, domain III |
| DUF4599 | 13 | Domain of unknown function (DUF4599) |
| EF-hand_6 | 13 | EF-hand domain |
| ELM2 | 13 | ELM2 domain |
| FA | 13 | FERM adjacent (FA) |
| FAM75 | 13 | FAM75 family |
| Globin | 13 | Globin |
| HATPase_c_3 | 13 | Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase |
| KRTAP | 13 | Keratin-associated matrix |
| LBP_BPI_CETP | 13 | LBP / BPI / CETP family, N-terminal domain |
| Ldl_recept_b | 13 | Low-density lipoprotein receptor repeat class B |
| NCD3G | 13 | Nine Cysteines Domain of family 3 GPCR |
| OLF | 13 | Olfactomedin-like domain |
| OTU | 13 | OTU-like cysteine protease |
| PHD_2 | 13 | PHD-finger |
| Peptidase_C1 | 13 | Papain family cysteine protease |
| Roc | 13 | Ras of Complex, Roc, domain of DAPkinase |
| SNARE | 13 | SNARE domain |
| Septin | 13 | Septin |
| TPR_2 | 13 | Tetratricopeptide repeat |
| Tissue_fac | 13 | Tissue factor |
| UBX | 13 | UBX domain |
| bZIP_Maf | 13 | bZIP Maf transcription factor |
| zf-CCCH_2 | 13 | RNA-binding, Nab2-type zinc finger |
| ABC2_membrane_3 | 12 | ABC-2 family transporter protein |
| Acyl-CoA_dh_1 | 12 | Acyl-CoA dehydrogenase, C-terminal domain |
| Ank_5 | 12 | Ankyrin repeats (many copies) |
| Arm_2 | 12 | Armadillo-like |
| BAR_3 | 12 | BAR domain of APPL family |
| CarboxypepD_reg | 12 | Carboxypeptidase regulatory-like domain |
| DDE_1 | 12 | DDE superfamily endonuclease |
| EF-hand_like | 12 | Phosphoinositide-specific phospholipase C, efhand-like |
| Ferric_reduct | 12 | Ferric reductase like transmembrane component |
| Gla | 12 | Vitamin K-dependent carboxylation/gamma-carboxyglutamic (GLA) domain |
| HEAT | 12 | HEAT repeat |
| His_Phos_1 | 12 | Histidine phosphatase superfamily (branch 1) |
| INT_SG_DDX_CT_C | 12 | INTS6/SAGE1/DDX26B/CT45 C-terminus |
| IRS | 12 | PTB domain (IRS-1 type) |
| JAB | 12 | JAB1/Mov34/MPN/PAD-1 ubiquitin protease |
| Kazal_1 | 12 | Kazal-type serine protease inhibitor domain |
| Kelch_3 | 12 | Galactose oxidase, central domain |
| Linker_histone | 12 | linker histone H1 and H5 family |
| MH1 | 12 | MH1 domain |
| Metallothio | 12 | Metallothionein |
| NAD_binding_1 | 12 | Oxidoreductase NAD-binding domain |
| OATP | 12 | Organic Anion Transporter Polypeptide (OATP) family |
| Oxysterol_BP | 12 | Oxysterol-binding protein |
| PLAC | 12 | PLAC (protease and lacunin) domain |
| PMG | 12 | PMG protein |
| PNMA_N | 12 | PNMA N-terminal RRM-like domain |
| Peptidase_M1 | 12 | Peptidase family M1 domain |
| Pyr_redox_2 | 12 | Pyridine nucleotide-disulphide oxidoreductase |
| RWD | 12 | RWD domain |
| Ran_BP1 | 12 | RanBP1 domain |
| SSF | 12 | Sodium:solute symporter family |
| Snf7 | 12 | Snf7 |
| Sulfotransfer_2 | 12 | Sulfotransferase family |
| THAP | 12 | THAP domain |
| TPR_19 | 12 | Tetratricopeptide repeat |
| WH1 | 12 | WH1 domain |
| WWE | 12 | WWE domain |
| dDENN | 12 | dDENN domain |
| zf-CXXC | 12 | CXXC zinc finger domain |
| 2OG-FeII_Oxy_3 | 11 | 2OG-Fe(II) oxygenase superfamily |
| AAA_11 | 11 | AAA domain |
| AAA_12 | 11 | AAA domain |
| ADK | 11 | Adenylate kinase |
| Acyl-CoA_dh_N | 11 | Acyl-CoA dehydrogenase, N-terminal domain |
| Adap_comp_sub | 11 | Adaptor complexes medium subunit family |
| BAH | 11 | BAH domain |
| Band_7 | 11 | SPFH domain / Band 7 family |
| Bcl-2 | 11 | Apoptosis regulator proteins, Bcl-2 family |
| C8 | 11 | C8 domain |
| COLFI | 11 | Fibrillar collagen C-terminal domain |
| DHR-2_Lobe_A | 11 | DHR-2, Lobe A |
| DHR-2_Lobe_B | 11 | DHR-2, Lobe B |
| DHR-2_Lobe_C | 11 | DHR-2, Lobe C |
| DOCK-C2 | 11 | C2 domain in Dock180 and Zizimin proteins |
| E2F_TDP | 11 | E2F/DP family winged-helix DNA-binding domain |
| FYVE_2 | 11 | FYVE-type zinc finger |
| Frizzled | 11 | Frizzled/Smoothened family membrane region |
| GAIN | 11 | GPCR-Autoproteolysis INducing (GAIN) domain |
| GBP | 11 | Guanylate-binding protein, N-terminal domain |
| GRIP | 11 | GRIP domain |
| Hairy_orange | 11 | Hairy Orange |
| Hist_deacetyl | 11 | Histone deacetylase domain |
| HlyIII | 11 | Haemolysin-III related |
| IBR | 11 | IBR domain, a half RING-finger domain |
| Integrin_A_Ig_3 | 11 | Integrin alpha Ig-like domain 3 |
| JmjN | 11 | jmjN domain |
| Kelch_6 | 11 | Kelch motif |
| MACPF | 11 | MAC/Perforin domain |
| MBOAT | 11 | MBOAT, membrane-bound O-acyltransferase family |
| Mab-21_C | 11 | Mab-21 protein HhH/H2TH-like domain |
| Methyltransf_16 | 11 | Lysine methyltransferase |
| PAS_11 | 11 | PAS domain |
| PKD_channel | 11 | Polycystin cation channel |
| Pentaxin | 11 | Pentaxin family |
| Peptidase_M1_N | 11 | Peptidase M1 N-terminal domain |
| Peptidase_M28 | 11 | Peptidase family M28 |
| Peptidase_S8 | 11 | Subtilase family |
| R3H | 11 | R3H domain |
| RINGv | 11 | RING-variant domain |
| Ribosomal_L7Ae | 11 | Ribosomal protein L7Ae/L30e/S12e/Gadd45 family |
| SAM_PNT | 11 | Sterile alpha motif (SAM)/Pointed domain |
| SBP_bac_3 | 11 | Bacterial extracellular solute-binding proteins, family 3 |
| TIL | 11 | Trypsin Inhibitor like cysteine rich domain |
| Transglut_core | 11 | Transglutaminase-like superfamily |
| UBA_4 | 11 | UBA-like domain |
| ZU5 | 11 | ZU5 domain |
| adh_short_C2 | 11 | Enoyl-(Acyl carrier protein) reductase |
| zf-C2H2_jaz | 11 | Zinc-finger double-stranded RNA-binding |
| zf-HC5HC2H | 11 | PHD-like zinc-binding domain |
| A2M | 10 | Alpha-2-macroglobulin family |
| A2M_BRD | 10 | Alpha-2-macroglobulin bait region domain |
| A2M_recep | 10 | A-macroglobulin receptor binding domain |
| Amidohydro_1 | 10 | Amidohydrolase family |
| Amino_oxidase | 10 | Flavin containing amine oxidoreductase |
| An_peroxidase | 10 | Animal haem peroxidase |
| Anoctamin | 10 | Calcium-activated chloride channel |
| Arrestin_C | 10 | Arrestin (or S-antigen), C-terminal domain |
| Arrestin_N | 10 | Arrestin (or S-antigen), N-terminal domain |
| Asp | 10 | Eukaryotic aspartyl protease |
| CAP_GLY | 10 | CAP-Gly domain |
| COMM_domain | 10 | COMM domain |
| Cation_efflux | 10 | Cation efflux family |
| Claudin_2 | 10 | PMP-22/EMP/MP20/Claudin tight junction |
| Cofilin_ADF | 10 | Cofilin/tropomyosin-type actin-binding protein |
| Cullin | 10 | Cullin family |
| DAGK_acc | 10 | Diacylglycerol kinase accessory domain |
| DUF4939 | 10 | Domain of unknown function (DUF4939) |
| EF-hand_4 | 10 | Cytoskeletal-regulatory complex EF hand |
| EMI | 10 | EMI domain |
| EMP24_GP25L | 10 | emp24/gp25L/p24 family/GOLD |
| Filamin | 10 | Filamin/ABP280 repeat |
| GST_N_2 | 10 | Glutathione S-transferase, N-terminal domain |
| HCO3_cotransp | 10 | HCO3- transporter integral membrane domain |
| HMG_box_2 | 10 | HMG-box domain |
| HSF_DNA-bind | 10 | HSF-type DNA-binding |
| HSP20 | 10 | Hsp20/alpha crystallin family |
| IL1 | 10 | Interleukin-1 / 18 |
| Insulin | 10 | Insulin/IGF/Relaxin family |
| KOW | 10 | KOW motif |
| Kelch_5 | 10 | Kelch motif |
| Keratin_B2 | 10 | Keratin, high sulfur B2 protein |
| L27 | 10 | L27 domain |
| LBP_BPI_CETP_C | 10 | LBP / BPI / CETP family, C-terminal domain |
| Laminin_G_1 | 10 | Laminin G domain |
| Lipase | 10 | Lipase |
| MBT | 10 | mbt repeat |
| MG2 | 10 | MG2 domain |
| MG3 | 10 | Macroglobulin domain MG3 |
| MIF4G | 10 | MIF4G domain |
| MIR | 10 | MIR domain |
| Med26 | 10 | TFIIS helical bundle-like domain |
| NDK | 10 | Nucleoside diphosphate kinase |
| Na_trans_assoc | 10 | Sodium ion transport-associated |
| PAS_3 | 10 | PAS fold |
| PAS_9 | 10 | PAS domain |
| PNMA | 10 | PNMA |
| Phosphodiest | 10 | Type I phosphodiesterase / nucleotide pyrophosphatase |
| RHD_DNA_bind | 10 | Rel homology DNA-binding domain |
| RHD_dimer | 10 | Rel homology dimerisation domain |
| RNase_T | 10 | Exonuclease |
| Rap_GAP | 10 | Rap/ran-GAP |
| SPAN-X | 10 | Sperm protein associated with nucleus, mapped to X chromosome |
| SSXRD | 10 | SSXRD motif |
| STAS | 10 | STAS domain |
| Sulfate_transp | 10 | Sulfate permease family |
| Synaptobrevin | 10 | Synaptobrevin |
| TED_complement | 10 | A-macroglobulin TED domain |
| TPR_10 | 10 | Tetratricopeptide repeat |
| ThiF | 10 | ThiF family |
| Thioredoxin_6 | 10 | Thioredoxin-like domain |
| VPS9 | 10 | Vacuolar sorting protein 9 (VPS9) domain |
| zf-RING_5 | 10 | zinc-RING finger domain |
| AAA_33 | 9 | AAA domain |
| AA_permease | 9 | Amino acid permease |
| AIG1 | 9 | AIG1 family |
| ASC | 9 | Amiloride-sensitive sodium channel |
| Activin_recp | 9 | Activin types I and II receptor domain |
| Alpha_adaptinC2 | 9 | Adaptin C-terminal domain |
| BEN | 9 | BEN domain |
| BRICHOS | 9 | BRICHOS domain |
| BTK | 9 | BTK motif |
| Band_3_cyto | 9 | Band 3 cytoplasmic domain |
| Beach | 9 | Beige/BEACH domain |
| Biotin_lipoyl | 9 | Biotin-requiring enzyme |
| CRAL_TRIO_N | 9 | CRAL/TRIO, N-terminal domain |
| Calx-beta | 9 | Calx-beta domain |
| Complex1_LYR | 9 | Complex 1 protein (LYR family) |
| Copine | 9 | Copine |
| DAO | 9 | FAD dependent oxidoreductase |
| DCX | 9 | Doublecortin |
| DHC_N1 | 9 | Dynein heavy chain, N-terminal region 1 |
| Defensin_beta | 9 | Beta defensin |
| DnaJ_C | 9 | DnaJ C terminal domain |
| Drf_FH3 | 9 | Diaphanous FH3 Domain |
| Drf_GBD | 9 | Diaphanous GTPase-binding Domain |
| Ephrin | 9 | Ephrin |
| F-box | 9 | F-box domain |
| GARIL_Rab2_bd | 9 | Golgi associated RAB2 interactor protein-like, Rab2B-binding domain |
| GBP_C | 9 | Guanylate-binding protein, C-terminal domain |
| Glyco_transf_7C | 9 | N-terminal domain of galactosyltransferase |
| Glyco_transf_8 | 9 | Glycosyl transferase family 8 |
| HGTP_anticodon | 9 | Anticodon binding domain |
| IRF | 9 | Interferon regulatory factor transcription factor |
| Integrin_alpha | 9 | Integrin alpha cytoplasmic region |
| Interfer-bind | 9 | Interferon-alpha/beta receptor, fibronectin type III |
| Lactamase_B | 9 | Metallo-beta-lactamase superfamily |
| Ldh_1_C | 9 | lactate/malate dehydrogenase, alpha/beta C-terminal domain |
| Lys | 9 | C-type lysozyme/alpha-lactalbumin family |
| MBD | 9 | Methyl-CpG binding domain |
| MCM | 9 | MCM P-loop domain |
| MCM_lid | 9 | MCM AAA-lid domain |
| MG4 | 9 | Macroglobulin domain MG4 |
| MHC_II_beta | 9 | Class II histocompatibility antigen, beta domain |
| Macro | 9 | Macro domain |
| Maestro_HEAT | 9 | Maestro, HEAT repeats |
| Methyltransf_11 | 9 | Methyltransferase domain |
| MyTH4 | 9 | MyTH4 domain |
| Myb_DNA-bind_4 | 9 | Myb/SANT-like DNA-binding domain |
| NTF2 | 9 | Nuclear transport factor 2 (NTF2) domain |
| Na_Ca_ex | 9 | Sodium/calcium exchanger protein |
| PAM2 | 9 | Ataxin-2 C-terminal region |
| PAX | 9 | 'Paired box' domain |
| PAZ | 9 | PAZ domain |
| PDGF | 9 | PDGF/VEGF domain |
| PH_12 | 9 | Pleckstrin homology domain |
| PH_9 | 9 | Pleckstrin homology domain |
| PI3Ka | 9 | Phosphoinositide 3-kinase family, accessory domain (PIK domain) |
| Patatin | 9 | Patatin-like phospholipase |
| Peptidase_M24 | 9 | Metallopeptidase family M24 |
| Peptidase_S9 | 9 | Prolyl oligopeptidase family |
| Phospholip_A2_1 | 9 | Phospholipase A2 |
| Plexin_RBD | 9 | Plexin cytoplasmic RhoGTPase-binding domain |
| Plexin_cytopl | 9 | Plexin cytoplasmic RasGAP domain |
| RAWUL | 9 | RAWUL domain RING finger- and WD40-associated ubiquitin-like |
| RBM1CTR | 9 | RBM1CTR (NUC064) family |
| RCC1_2 | 9 | Regulator of chromosome condensation (RCC1) repeat |
| RFX_DNA_binding | 9 | RFX DNA-binding domain |
| Radical_SAM | 9 | Radical SAM superfamily |
| Rhomboid | 9 | Rhomboid family |
| SAND | 9 | SAND domain |
| SHIPPO-rpt | 9 | Sperm-tail PG-rich repeat |
| Shisa_N | 9 | Shisa, N-terminal domain |
| Transglut_C | 9 | Transglutaminase family, C-terminal ig like domain |
| Transglut_N | 9 | Transglutaminase family |
| UIM | 9 | Ubiquitin interaction motif |
| Uteroglobin | 9 | Uteroglobin family |
| VHS | 9 | VHS domain |
| Voltage_CLC | 9 | Voltage gated chloride channel |
| XK-related | 9 | XK-related protein |
| AC_N | 8 | Adenylyl cyclase N-terminal extracellular and transmembrane region |
| ATP1G1_PLM_MAT8 | 8 | ATP1G1/PLM/MAT8 family |
| Alpha-amylase | 8 | Alpha amylase, catalytic domain |
| CHCH | 8 | CHCH domain |
| CHGN | 8 | Chondroitin N-acetylgalactosaminyltransferase |
| CID | 8 | CID domain |
| CSD | 8 | 'Cold-shock' DNA-binding domain |
| CUE | 8 | CUE domain |
| Chorein_N | 8 | VPS13-like, N-terminal |
| Cupin_8 | 8 | Cupin-like domain |
| DUF1899 | 8 | Domain of unknown function (DUF1899) |
| EPL1 | 8 | Enhancer of polycomb-like |
| ERAP1_C | 8 | ERAP1-like C-terminal domain |
| FAM83 | 8 | FAM83 A-H |
| Flavodoxin_1 | 8 | Flavodoxin |
| Folate_rec | 8 | Folate receptor family |
| GDA1_CD39 | 8 | GDA1/CD39 (nucleoside phosphatase) family |
| GSHPx | 8 | Glutathione peroxidase |
| GST_C_2 | 8 | Glutathione S-transferase, C-terminal domain |
| Glyco_tran_10_N | 8 | Fucosyltransferase, N-terminal |
| Glyco_transf_10 | 8 | Glycosyltransferase family 10 (fucosyltransferase) C-term |
| HATPase_c | 8 | Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase |
| HEAT_2 | 8 | HEAT repeats |
| HEAT_EZ | 8 | HEAT-like repeat |
| His_Phos_2 | 8 | Histidine phosphatase superfamily (branch 2) |
| IL6Ra-bind | 8 | Interleukin-6 receptor alpha chain, binding |
| Integrin_beta | 8 | Integrin beta chain VWA domain |
| LRAT | 8 | Lecithin retinol acyltransferase |
| LSDAT_euk | 8 | SLOG in TRPM |
| Laminin_B | 8 | Laminin B (Domain IV) |
| Laminin_G_3 | 8 | Concanavalin A-like lectin/glucanases superfamily |
| Ldh_1_N | 8 | lactate/malate dehydrogenase, NAD binding domain |
| LisH | 8 | LisH |
| MBD_C | 8 | C-terminal domain of methyl-CpG binding protein 2 and 3 |
| MBDa | 8 | p55-binding region of Methyl-CpG-binding domain proteins MBD |
| MCM_OB | 8 | MCM OB domain |
| MH2 | 8 | MH2 domain |
| MIT | 8 | MIT (microtubule interacting and transport) domain |
| Mob1_phocein | 8 | Mob1/phocein family |
| Myosin_TH1 | 8 | Unconventional myosin tail, actin- and lipid-binding |
| NAD_binding_8 | 8 | NAD(P)-binding Rossmann-like domain |
| NHL | 8 | NHL repeat |
| NIF | 8 | NLI interacting factor-like phosphatase |
| Neurexophilin | 8 | Neurexophilin |
| PI3K_C2 | 8 | Phosphoinositide 3-kinase C2 |
| PIP5K | 8 | Phosphatidylinositol-4-phosphate 5-Kinase |
| PSI_integrin | 8 | Integrin plexin domain |
| PTB | 8 | Phosphotyrosine-binding domain |
| PTEN_C2 | 8 | C2 domain of PTEN tumour-suppressor protein |
| Patched | 8 | Patched family |
| Peptidase_M13 | 8 | Peptidase family M13 |
| Peptidase_M13_N | 8 | Peptidase family M13 |
| Piwi | 8 | Piwi domain |
| Propep_M14 | 8 | Carboxypeptidase activation peptide |
| Proteasome_A_N | 8 | Proteasome subunit A N-terminal signature |
| Pyridoxal_deC | 8 | Pyridoxal-dependent decarboxylase conserved domain |
| Rab_bind | 8 | Rab binding domain |
| ResIII | 8 | Type III restriction enzyme, res subunit |
| S1 | 8 | S1 RNA binding domain |
| SLC12 | 8 | Solute carrier family 12 |
| Sec1 | 8 | Sec1 family |
| Sulfatase_C | 8 | C-terminal region of aryl-sulfatase |
| Syntaxin | 8 | Syntaxin |
| TIG_plexin | 8 | TIG domain |
| TMC | 8 | TMC domain |
| TPR_11 | 8 | TPR repeat |
| TPR_7 | 8 | Tetratricopeptide repeat |
| TPT | 8 | Triose-phosphate Transporter family |
| TRAF-mep_MATH | 8 | TRAF/meprin, MATH domain |
| Tim17 | 8 | Tim17/Tim22/Tim23/Pmp24 family |
| Transket_pyr | 8 | Transketolase, pyrimidine binding domain |
| Trefoil | 8 | Trefoil (P-type) domain |
| VHP | 8 | Villin headpiece domain |
| VIT | 8 | Vault protein inter-alpha-trypsin domain |
| Vps4_C | 8 | Vps4 C terminal oligomerisation domain |
| WD40_4 | 8 | Type of WD40 repeat |
| Yippee-Mis18 | 8 | Yippee zinc-binding/DNA-binding /Mis18, centromere assembly |
| bMERB_dom | 8 | Bivalent Mical/EHBP Rab binding domain |
| tRNA-synt_2b | 8 | tRNA synthetase class II core domain (G, H, P, S and T) |
| zf-AN1 | 8 | AN1-like Zinc finger |
| zf-CCCH_4 | 8 | CCCH-type zinc finger |
| zf-H2C2_5 | 8 | C2H2-type zinc-finger domain |
| 14-3-3 | 7 | 14-3-3 protein |
| 2OG-FeII_Oxy_2 | 7 | 2OG-Fe(II) oxygenase superfamily |
| 4HBT | 7 | Thioesterase superfamily |
| A1_Propeptide | 7 | A1 Propeptide |
| ACBP | 7 | Acyl CoA binding protein |
| ANAPC10 | 7 | Anaphase-promoting complex, subunit 10 (APC10) |
| ANATO | 7 | Anaphylotoxin-like domain |
| ATG8 | 7 | Autophagy protein Atg8 ubiquitin like |
| Abhydrolase_3 | 7 | alpha/beta hydrolase fold |
| Ank_3 | 7 | Ankyrin repeat |
| Anoct_dimer | 7 | Anoctamin, dimerisation domain |
| ApoL | 7 | Apolipoprotein L |
| Arm_3 | 7 | Atypical Arm repeat |
| BIR | 7 | Inhibitor of Apoptosis domain |
| Branch | 7 | Core-2/I-Branching enzyme |
| CBM_21 | 7 | Carbohydrate/starch-binding module (family 21) |
| CN_hydrolase | 7 | Carbon-nitrogen hydrolase |
| CPSase_L_D2 | 7 | Carbamoyl-phosphate synthase L chain, ATP binding domain |
| CTP_transf_like | 7 | Cytidylyltransferase-like |
| CUT | 7 | CUT domain |
| Ca_chan_IQ | 7 | Voltage gated calcium channel IQ domain |
| Cadherin_pro | 7 | Cadherin prodomain like |
| Carn_acyltransf | 7 | Choline/Carnitine o-acyltransferase |
| DAGAT | 7 | Diacylglycerol acyltransferase |
| DED | 7 | Death effector domain |
| DL-JAG_EGF-like | 7 | Delta-like/Jagged, EGF-like domain |
| DM | 7 | DM DNA binding domain |
| DUSP | 7 | DUSP domain |
| ELO | 7 | GNS1/SUR4 family |
| EamA | 7 | EamA-like transporter family |
| F420_oxidored | 7 | NADP oxidoreductase coenzyme F420-dependent |
| FAD_binding_8 | 7 | FAD-binding domain |
| FA_desaturase | 7 | Fatty acid desaturase |
| FGGY_N | 7 | FGGY family of carbohydrate kinases, N-terminal domain |
| Filament_head | 7 | Intermediate filament head (DNA binding) region |
| Furin-like | 7 | Furin-like cysteine rich region |
| GAF | 7 | GAF domain |
| GDPD | 7 | Glycerophosphoryl diester phosphodiesterase family |
| GPHH | 7 | Voltage-dependent L-type calcium channel, IQ-associated |
| G_glu_transpept | 7 | Gamma-glutamyltranspeptidase |
| Glutaredoxin | 7 | Glutaredoxin |
| Glyco_hydro_18 | 7 | Glycosyl hydrolases family 18 |
| Glyco_hydro_31_2nd | 7 | Glycosyl hydrolases family 31 TIM-barrel domain |
| Glyco_hydro_31_3rd | 7 | Glycosyl hydrolase family 31 C-terminal domain |
| Glyco_hydro_47 | 7 | Glycosyl hydrolase family 47 |
| Glyco_transf_7N | 7 | N-terminal region of glycosyl transferase group 7 |
| Glycos_transf_1 | 7 | Glycosyl transferases group 1 |
| GoLoco | 7 | GoLoco motif |
| Haspin_kinase | 7 | Haspin like kinase domain |
| IBB | 7 | Importin beta binding domain |
| ILEI | 7 | Interleukin-like EMT inducer |
| IPK | 7 | Inositol polyphosphate kinase |
| IRF-3 | 7 | Interferon-regulatory factor 3 |
| Inhibitor_I29 | 7 | Cathepsin propeptide inhibitor domain (I29) |
| Kelch_4 | 7 | Galactose oxidase, central domain |
| LRR_5 | 7 | BspA type Leucine rich repeat region (6 copies) |
| L_HMGIC_fpl | 7 | Lipoma HMGIC fusion partner-like protein |
| La | 7 | La domain |
| MUN | 7 | MUN domain |
| Methyltr_RsmB-F | 7 | 16S rRNA methyltransferase RsmB/F |
| Methyltransf_25 | 7 | Methyltransferase domain |
| Myb_DNA-bind_5 | 7 | Myb/SANT-like DNA-binding domain |
| NAD_binding_6 | 7 | Ferric reductase NAD binding domain |
| NTP_transf_2 | 7 | Nucleotidyltransferase domain |
| NUT | 7 | NUT protein |
| P2X_receptor | 7 | ATP P2X receptor |
| PAN_1 | 7 | PAN domain |
| PAP_assoc | 7 | Cid1 family poly A polymerase |
| PH_16 | 7 | PH domain |
| PI3K_rbd | 7 | PI3-kinase family, ras-binding domain |
| PLA2_B | 7 | Lysophospholipase catalytic domain |
| P_proprotein | 7 | Proprotein convertase P-domain |
| Peptidase_C48 | 7 | Ulp1 protease family, C-terminal catalytic domain |
| Peptidase_M16 | 7 | Insulinase (Peptidase family M16) |
| Peptidase_M16_C | 7 | Peptidase M16 inactive domain |
| Polycystin_dom | 7 | Polycystin domain |
| Pre-SET | 7 | Pre-SET motif |
| PrmA | 7 | Ribosomal protein L11 methyltransferase (PrmA) |
| Pro-rich | 7 | Proline-rich protein |
| RIIa | 7 | Regulatory subunit of type II PKA R-subunit |
| RNase_PH | 7 | 3' exoribonuclease family, domain 1 |
| RNase_Zc3h12a | 7 | Zc3h12a-like Ribonuclease NYN domain |
| RPEL | 7 | RPEL repeat |
| Rad51 | 7 | Rad51 |
| Rap-GAP_dimer | 7 | Rap/Ran-GAP protein dimerization domain |
| Recep_L_domain | 7 | Receptor L domain |
| S8_pro-domain | 7 | Peptidase S8 pro-domain |
| SAM_3 | 7 | SAM domain (Sterile alpha motif) |
| SDF | 7 | Sodium:dicarboxylate symporter family |
| SIR2 | 7 | Sir2 family |
| SIX1_SD | 7 | Transcriptional regulator, SIX1, N-terminal SD domain |
| SLFN_AlbA_2 | 7 | Schlafen, AlbA_2 |
| SPARC_Ca_bdg | 7 | Secreted protein acidic and rich in cysteine Ca binding region |
| SRCR_2 | 7 | Scavenger receptor cysteine-rich domain |
| STAT_alpha | 7 | STAT protein, all-alpha domain |
| STAT_bind | 7 | STAT protein, DNA binding domain |
| STAT_int | 7 | STAT protein, protein interaction domain |
| STAT_linker | 7 | Signal transducer and activator of transcription, linker domain |
| Sad1_UNC | 7 | Sad1 / UNC-like C-terminal |
| Ski_Sno | 7 | SKI/SNO/DAC family |
| Sprouty | 7 | Sprouty protein (Spry) |
| Ssu72 | 7 | Ssu72-like protein |
| TB | 7 | TB domain |
| TFIIS_M | 7 | Transcription factor S-II (TFIIS), central domain |
| TGF_beta_GS | 7 | Transforming growth factor beta type I GS-motif |
| Tektin | 7 | Tektin family |
| Tropomodulin | 7 | Tropomodulin |
| Trypsin_2 | 7 | Trypsin-like peptidase domain |
| Tudor_2 | 7 | Jumonji domain-containing protein 2A Tudor domain |
| U-box | 7 | U-box domain |
| VWA_3 | 7 | von Willebrand factor type A domain |
| W2 | 7 | eIF4-gamma/eIF5/eIF2-epsilon |
| tRNA_SAD | 7 | Threonyl and Alanyl tRNA synthetase second additional domain |
| tRNA_anti-codon | 7 | OB-fold nucleic acid binding domain |
| zf-3CxxC | 7 | Zinc-binding domain |
| zf-A20 | 7 | A20-like zinc finger |
| zf-CW | 7 | CW-type Zinc Finger |
| zf-Di19 | 7 | Drought induced 19 protein (Di19), zinc-binding |
| zf-GRF | 7 | GRF zinc finger |
| 3Beta_HSD | 6 | 3-beta hydroxysteroid dehydrogenase/isomerase family |
| ANAPC3 | 6 | Anaphase-promoting complex, cyclosome, subunit 3 |
| A_deamin | 6 | Adenosine-deaminase (editase) domain |
| Adcy_cons_dom | 6 | Adenylate cyclase, conserved domain |
| Alpha-amylase_C | 6 | Alpha amylase, C-terminal all-beta domain |
| Alpha_kinase | 6 | Alpha-kinase family |
| Aminotran_5 | 6 | Aminotransferase class-V |
| Anticodon_1 | 6 | Anticodon-binding domain of tRNA ligase |
| ArgoL1 | 6 | Argonaute linker 1 domain |
| Astacin | 6 | Astacin (Peptidase family M12A) |
| B3R | 6 | Poxviridae B3 protein |
| BRCT_2 | 6 | BRCT domain, a BRCA1 C-terminus domain |
| BTG | 6 | BTG family |
| Bax1-I | 6 | Inhibitor of apoptosis-promoting Bax1 |
| Brix | 6 | Brix domain |
| C4 | 6 | C-terminal tandem repeated domain in type 4 procollagen |
| CAMSAP_CH | 6 | CAMSAP CH domain |
| CBFD_NFYB_HMF | 6 | Histone-like transcription factor (CBF/NF-Y) and archaeal histone |
| CP2 | 6 | CP2 transcription factor |
| CTLH | 6 | CTLH/CRA C-terminal to LisH motif domain |
| Ca_hom_mod | 6 | Calcium homeostasis modulator |
| Calponin | 6 | Calponin family repeat |
| Cullin_Nedd8 | 6 | Cullin protein neddylation domain |
| DDHD | 6 | DDHD domain |
| DIL | 6 | DIL domain |
| DIX | 6 | DIX domain |
| DOCK_C-D_N | 6 | Dedicator of cytokinesis C/D, N terminal |
| DPPIV_N | 6 | Dipeptidyl peptidase IV (DPP IV) N-terminal region |
| DUF3456 | 6 | TLR4 regulator and MIR-interacting MSAP |
| DUF3504 | 6 | Domain of unknown function (DUF3504) |
| DUF3694 | 6 | Kinesin protein |
| Defensin_1 | 6 | Mammalian defensin |
| Defensin_propep | 6 | Defensin propeptide |
| Dimer_Tnp_hAT | 6 | hAT family C-terminal dimerisation region |
| Dynamin_M | 6 | Dynamin central region |
| E1_dh | 6 | Dehydrogenase E1 component |
| E2F_CC-MB | 6 | E2F transcription factor CC-MB domain |
| EF-hand_2 | 6 | EF hand |
| EF-hand_3 | 6 | EF-hand |
| EFG_C | 6 | Elongation factor G C-terminus |
| EFhand_Ca_insen | 6 | Ca2+ insensitive EF hand |
| ELMO_CED12 | 6 | ELMO/CED-12 family |
| ENTH | 6 | ENTH domain |
| EPTP | 6 | EPTP domain |
| Epimerase | 6 | NAD dependent epimerase/dehydratase family |
| FAD_binding_1 | 6 | FAD binding domain |
| FAD_binding_3 | 6 | FAD binding domain |
| FAST_1 | 6 | FAST kinase-like protein, subdomain 1 |
| FAST_2 | 6 | FAST kinase-like protein, subdomain 2 |
| FATC | 6 | FATC domain |
| FA_hydroxylase | 6 | Fatty acid hydroxylase |
| FBA | 6 | F-box associated region |
| FGGY_C | 6 | FGGY family of carbohydrate kinases, C-terminal domain |
| Frag1 | 6 | Frag1/DRAM/Sfk1 family |
| Fringe | 6 | Fringe-like |
| GAS2 | 6 | Growth-Arrest-Specific Protein 2 Domain |
| GAT | 6 | GAT domain |
| GDNF | 6 | GDNF/GAS1 domain |
| GED | 6 | Dynamin GTPase effector domain |
| GF_recep_IV | 6 | Growth factor receptor domain IV |
| GST_N_3 | 6 | Glutathione S-transferase, N-terminal domain |
| GTP_EFTU_D3 | 6 | Elongation factor Tu C-terminal domain |
| Gal_mutarotas_2 | 6 | Glycosyl hydrolase 31 N-terminal galactose mutarotase-like domain |
| Gasdermin | 6 | Gasdermin pore forming domain |
| Glypican | 6 | Glypican |
| HELP | 6 | HELP motif |
| HOOK_N | 6 | HOOK domain |
| Hormone_1 | 6 | Somatotropin hormone family |
| Hormone_2 | 6 | Peptide hormone |
| Hydrolase_4 | 6 | Serine aminopeptidase, S33 |
| IL17 | 6 | Interleukin-17 |
| ITAM | 6 | Immunoreceptor tyrosine-based activation motif |
| ITI_HC_C | 6 | Inter-alpha-trypsin inhibitor heavy chain C-terminus |
| Ig_6 | 6 | Immunoglobulin domain |
| Ins145_P3_rec | 6 | Inositol 1,4,5-trisphosphate/ryanodine receptor |
| Integrin_B_tail | 6 | Integrin beta tail domain |
| Integrin_b_cyt | 6 | Integrin beta cytoplasmic domain |
| Kinesin_assoc | 6 | Kinesin-associated |
| L6_membrane | 6 | L6 membrane protein |
| LCCL | 6 | LCCL domain |
| LEM | 6 | LEM domain |
| LON_substr_bdg | 6 | ATP-dependent protease La (LON) substrate-binding domain |
| Lipoxygenase | 6 | Lipoxygenase |
| M20_dimer | 6 | Peptidase dimerisation domain |
| MA3 | 6 | MA3 domain |
| MAPEG | 6 | MAPEG family |
| MCM_N | 6 | MCM N-terminal domain |
| MFS_2 | 6 | MFS/sugar transport protein |
| MHC_II_alpha | 6 | Class II histocompatibility antigen, alpha domain |
| MHC_I_3 | 6 | MHC-I family domain |
| Methyltransf_23 | 6 | Methyltransferase domain |
| Mg_trans_NIPA | 6 | Magnesium transporter NIPA |
| Mucin2_WxxW | 6 | Mucin-2 protein WxxW repeating region |
| NAD2 | 6 | Novel AID APOBEC clade 2 |
| NIDO | 6 | Nidogen-like |
| Na_trans_cytopl | 6 | Cytoplasmic domain of voltage-gated Na+ ion channel |
| Nore1-SARAH | 6 | Novel Ras effector 1 C-terminal SARAH (Sav/Rassf/Hpo) domain |
| Notch | 6 | LNR domain |
| OSR1_C | 6 | Oxidative-stress-responsive kinase 1 C-terminal domain |
| Occludin_ELL | 6 | Occludin homology domain |
| PET | 6 | PET Domain |
| PH_BEACH | 6 | PH domain associated with Beige/BEACH |
| PIP49_C | 6 | Protein-kinase domain of FAM69 |
| PRELI | 6 | PRELI-like family |
| PRR20 | 6 | Proline-rich protein family 20 |
| Pepdidase_M14_N | 6 | Cytosolic carboxypeptidase N-terminal domain |
| Peptidase_M20 | 6 | Peptidase family M20/M25/M40 |
| Plectin | 6 | Plectin repeat |
| Pribosyltran | 6 | Phosphoribosyl transferase domain |
| RAP | 6 | RAP domain |
| RBD | 6 | Raf-like Ras-binding domain |
| RBD-FIP | 6 | FIP domain |
| RIH_assoc | 6 | RyR and IP3R Homology associated |
| RNase_PH_C | 6 | 3' exoribonuclease family, domain 2 |
| RRM_5 | 6 | RNA recognition motif. (a.k.a. RRM, RBD, or RNP domain) |
| RYDR_ITPR | 6 | RIH domain |
| SBF | 6 | Sodium Bile acid symporter family |
| SEFIR | 6 | SEFIR domain |
| SH3_10 | 6 | SH3 domain |
| SLFN_GTPase-like | 6 | Schlafen, GTPase-like domain |
| SMC_N | 6 | RecF/RecN/SMC N terminal domain |
| SMC_hinge | 6 | SMC proteins Flexible Hinge Domain |
| SPOC | 6 | SPOC domain |
| SPRR2 | 6 | Small proline-rich 2 |
| SRF-TF | 6 | SRF-type transcription factor (DNA-binding and dimerisation domain) |
| SWIRM | 6 | SWIRM domain |
| Sec23_BS | 6 | Sec23/Sec24 beta-sandwich domain |
| Sec23_helical | 6 | Sec23/Sec24 helical domain |
| Sec23_trunk | 6 | Sec23/Sec24 trunk domain |
| Sel1 | 6 | Sel1 repeat |
| Somatomedin_B | 6 | Somatomedin B domain |
| Sterol-sensing | 6 | Sterol-sensing domain of SREBP cleavage-activation |
| Surp | 6 | Surp module |
| Syndecan | 6 | Syndecan domain |
| TB2_DP1_HVA22 | 6 | TB2/DP1, HVA22 family |
| TFIIS_C | 6 | Transcription factor S-II (TFIIS) |
| THB | 6 | Tri-helix bundle domain |
| TIG_2 | 6 | TIG domain found in plexin |
| TRP_2 | 6 | Transient receptor ion channel II |
| TSP1_CCN | 6 | CCN3 Nov like TSP1 domain |
| TSP1_spondin | 6 | Spondin-like TSP1 domain |
| Tctex-1 | 6 | Tctex-1 family |
| Thiolase_C | 6 | Thiolase, C-terminal domain |
| Thiolase_N | 6 | Thiolase, N-terminal domain |
| Thymosin | 6 | Thymosin beta-4 family |
| Troponin | 6 | Troponin |
| Tudor-knot | 6 | RNA binding activity-knot of a chromodomain |
| VCX_VCY | 6 | Variable charge X/Y family |
| XPG_N | 6 | XPG N-terminal domain |
| Xpo1 | 6 | Exportin 1-like protein |
| Yip1 | 6 | Yip1 domain |
| tRNA-synt_1 | 6 | tRNA synthetases class I (I, L, M and V) |
| zf-BED | 6 | BED zinc finger |
| zf-C2HC | 6 | Zinc finger, C2HC type |
| zf-CCCH_3 | 6 | Zinc-finger containing family |
| zf-FCS | 6 | MYM-type Zinc finger with FCS sequence motif |
| zf-HIT | 6 | HIT zinc finger |
| zf-MIZ | 6 | MIZ/SP-RING zinc finger |
| zf-Sec23_Sec24 | 6 | Sec23/Sec24 zinc finger |
| zf-TRAF | 6 | TRAF-type zinc finger |
| zf-Tim10_DDP | 6 | Tim10/DDP family zinc finger |
| zf-UBR | 6 | Putative zinc finger in N-recognin (UBR box) |
| 1-cysPrx_C | 5 | C-terminal domain of 1-Cys peroxiredoxin |
| 5_nucleotid | 5 | 5' nucleotidase family |
| AA_permease_C | 5 | C-terminus of AA_permease |
| ABC1 | 5 | ABC1 atypical kinase-like domain |
| ABC2_membrane | 5 | ABC-2 type transporter |
| ABC2_membrane_7 | 5 | ABC-2 type transporter |
| ATP-gua_Ptrans | 5 | ATP:guanido phosphotransferase, C-terminal catalytic domain |
| ATP-gua_PtransN | 5 | ATP:guanido phosphotransferase, N-terminal domain |
| ATP-synt_C | 5 | ATP synthase subunit C |
| ATP-synt_ab | 5 | ATP synthase alpha/beta family, nucleotide-binding domain |
| ATP-synt_ab_N | 5 | ATP synthase alpha/beta family, beta-barrel domain |
| AWS | 5 | AWS domain |
| Abhydrolase_6 | 5 | Alpha/beta hydrolase family |
| Acyltransf_C | 5 | Acyltransferase C-terminus |
| AhpC-TSA | 5 | AhpC/TSA family |
| Alpha-mann_mid | 5 | Alpha mannosidase middle domain |
| Aminotran_3 | 5 | Aminotransferase class-III |
| Ammonium_transp | 5 | Ammonium Transporter Family |
| Arfaptin | 5 | Arfaptin-like domain |
| B56 | 5 | Protein phosphatase 2A regulatory B subunit (B56 family) |
| BAAT_C | 5 | BAAT / Acyl-CoA thioester hydrolase C terminal |
| BCDHK_Adom3 | 5 | Mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase |
| BORG_CEP | 5 | Cdc42 effector |
| BRK | 5 | BRK domain |
| BRO1 | 5 | BRO1-like domain |
| Beta_helix | 5 | Right handed beta helix region |
| Bile_Hydr_Trans | 5 | Acyl-CoA thioester hydrolase/BAAT N-terminal region |
| Biotin_carb_C | 5 | Biotin carboxylase C-terminal domain |
| Biotin_carb_N | 5 | Biotin carboxylase, N-terminal domain |
| BolA | 5 | BolA-like protein |
| CAF1 | 5 | CAF1 family ribonuclease |
| CBX7_C | 5 | CBX family C-terminal motif |
| CDP-OH_P_transf | 5 | CDP-alcohol phosphatidyltransferase |
| CEP104-like_TOG | 5 | Centrosomal protein CEP104-like, TOG domain |
| CIDE-N | 5 | CIDE-N domain |
| Cadherin_3 | 5 | Cadherin-like |
| Choline_transpo | 5 | Plasma-membrane choline transporter |
| Clathrin | 5 | Region in Clathrin and VPS |
| CobW_C | 5 | Cobalamin synthesis protein cobW C-terminal domain |
| Cortexin | 5 | Cortexin of kidney |
| Cu-oxidase_3 | 5 | Multicopper oxidase |
| Cullin_binding | 5 | Cullin binding |
| Cytochrom_B561 | 5 | Eukaryotic cytochrome b561 |
| DAN | 5 | DAN domain |
| DDE_Tnp_1_7 | 5 | Transposase IS4 |
| DMAP_binding | 5 | DMAP1-binding Domain |
| DNA_pol_A_exo1 | 5 | 3'-5' exonuclease |
| DOCK_N | 5 | DOCK N-terminus |
| DTC | 5 | Deltex C-terminal domain |
| DUF2476 | 5 | Protein of unknown function (DUF2476) |
| DUF3496 | 5 | Domain of unknown function (DUF3496) |
| DUF4560 | 5 | Domain of unknown function (DUF4560) |
| DUF4749 | 5 | Domain of unknown function (DUF4749) |
| DZF_C | 5 | DZF C-terminal domain |
| DZF_N | 5 | DZF N-terminal domain |
| Daz | 5 | Daz repeat |
| Dpy-30 | 5 | Dpy-30 motif |
| Dus | 5 | Dihydrouridine synthase (Dus) |
| EBP | 5 | EXPERA (EXPanded EBP superfamily) |
| EFG_III | 5 | Elongation Factor G, domain III |
| EGF_Tenascin | 5 | Tenascin EGF domain |
| EHD_N | 5 | N-terminal EH-domain containing protein |
| ERCC4 | 5 | ERCC4 domain |
| Endonuclease_NS | 5 | DNA/RNA non-specific endonuclease |
| EpoR_lig-bind | 5 | Erythropoietin receptor, ligand binding |
| FAD_binding_6 | 5 | Oxidoreductase FAD-binding domain |
| FAM47 | 5 | FAM47 family |
| FAT | 5 | FAT domain |
| FERM_C_FAK1 | 5 | FAK1/PYK2, FERM domain C-lobe |
| FF | 5 | FF domain |
| FMO-like | 5 | Flavin-binding monooxygenase-like |
| FOLN | 5 | Follistatin/Osteonectin-like EGF domain |
| FYRC | 5 | F/Y rich C-terminus |
| FYRN | 5 | F/Y-rich N-terminus |
| FerB | 5 | FerB (NUC096) domain |
| Ferlin_C | 5 | Ferlin C-terminus |
| FtsJ | 5 | FtsJ-like methyltransferase |
| GATase | 5 | Glutamine amidotransferase class-I |
| GCP_C_terminal | 5 | Gamma tubulin complex component C-terminal |
| GCP_N_terminal | 5 | Gamma tubulin complex component N-terminal |
| GHMP_kinases_N | 5 | GHMP kinases N terminal domain |
| GKAP | 5 | Guanylate-kinase-associated protein (GKAP) protein |
| GST_C_3 | 5 | Glutathione S-transferase, C-terminal domain |
| Gasdermin_C | 5 | Gasdermin PUB domain |
| Gly_acyl_tr_C | 5 | Aralkyl acyl-CoA:amino acid N-acyltransferase, C-terminal region |
| Gly_acyl_tr_N | 5 | Aralkyl acyl-CoA:amino acid N-acyltransferase |
| Glyco_hydro_1 | 5 | Glycosyl hydrolase family 1 |
| Glyco_hydro_38C | 5 | Glycosyl hydrolases family 38 C-terminal domain |
| Glyco_hydro_38N | 5 | Glycosyl hydrolases family 38 N-terminal domain |
| Glyco_hydro_56 | 5 | Hyaluronidase |
| Glyco_transf_64 | 5 | Glycosyl transferase family 64 domain |
| Glyoxalase | 5 | Glyoxalase/Bleomycin resistance protein/Dioxygenase superfamily |
| Got1 | 5 | Got1/Sft2-like family |
| HIG_1_N | 5 | Hypoxia induced protein conserved region |
| HMG14_17 | 5 | HMG14 and HMG17 |
| HNOBA | 5 | Heme NO binding associated |
| HR1 | 5 | Hr1 repeat |
| HSR | 5 | HSR domain |
| Hexokinase_1 | 5 | Hexokinase |
| Hexokinase_2 | 5 | Hexokinase |
| HnRNPA1_LC | 5 | Heterogeneous nuclear ribonucleoprotein A1, LC domain |
| Hydrolase_6 | 5 | Haloacid dehalogenase-like hydrolase |
| I-EGF_1 | 5 | Integrin beta epidermal growth factor like domain 1 |
| ICAM_N | 5 | Intercellular adhesion molecule (ICAM), N-terminal domain |
| IMD | 5 | IRSp53/MIM homology domain |
| ING | 5 | Inhibitor of growth proteins N-terminal histone-binding |
| IP_trans | 5 | Phosphatidylinositol transfer protein |
| Inositol_P | 5 | Inositol monophosphatase family |
| Iso_dh | 5 | Isocitrate/isopropylmalate dehydrogenase |
| JHD | 5 | Jumonji helical domain |
| KA1 | 5 | Kinase associated domain 1 |
| KCNQ_channel | 5 | KCNQ voltage-gated potassium channel |
| LRRC37AB_C | 5 | LRRC37A/B like protein 1 C-terminal domain |
| LTD | 5 | Lamin Tail Domain |
| Laminin_I | 5 | Laminin Domain I |
| Laminin_II | 5 | Laminin Domain II |
| Lamp2-like_luminal | 5 | Lysosome-associated membrane glycoprotein 2-like, luminal domains |
| Longin | 5 | Regulated-SNARE-like domain |
| LysM | 5 | LysM domain |
| Lysyl_oxidase | 5 | Lysyl oxidase |
| MNNL | 5 | N terminus of Notch ligand C2-like domain |
| MOEP19 | 5 | KH-like RNA-binding domain |
| MOZ_SAS | 5 | MOZ/SAS family |
| Mab-21 | 5 | Mab-21 protein nucleotidyltransferase domain |
| Meis_PKNOX_N | 5 | N-terminal of Homeobox Meis and PKNOX1 |
| Methyltransf_12 | 5 | Methyltransferase domain |
| Methyltransf_31 | 5 | Methyltransferase domain |
| Motile_Sperm | 5 | MSP (Major sperm protein) domain |
| MutS_III | 5 | MutS domain III |
| MutS_V | 5 | MutS domain V |
| Myb_DNA-bind_6 | 5 | Myb-like DNA-binding domain |
| NAC | 5 | NAC domain |
| NPB13-l_MII_rpt | 5 | Nuclear pore complex-interacting protein B13-like, MII repeat |
| Na_K-ATPase | 5 | Sodium / potassium ATPase beta chain |
| Na_sulph_symp | 5 | Sodium:sulfate symporter transmembrane region |
| Neuralized | 5 | Neuralized |
| Nuc_sug_transp | 5 | Nucleotide-sugar transporter |
| PABP | 5 | Poly-adenylate binding protein, unique domain |
| PAD | 5 | Protein-arginine deiminase (PAD) |
| PAD_M | 5 | Protein-arginine deiminase (PAD) middle domain |
| PAD_N | 5 | Protein-arginine deiminase (PAD) N-terminal domain |
| PALP | 5 | Pyridoxal-phosphate dependent enzyme |
| PGM_PMM_I | 5 | Phosphoglucomutase/phosphomannomutase, alpha/beta/alpha domain I |
| PHD_5 | 5 | PhD finger domain |
| PHR | 5 | PHR domain |
| PH_19 | 5 | PH domain |
| PKD | 5 | PKD domain |
| PLDc_2 | 5 | PLD-like domain |
| POM121 | 5 | POM121 family |
| PP-binding | 5 | Phosphopantetheine attachment site |
| PP1_inhibitor | 5 | PKC-activated protein phosphatase-1 inhibitor |
| PQ-loop | 5 | PQ loop repeat |
| PTR2 | 5 | POT family |
| PWI | 5 | PWI domain |
| Pannexin_like | 5 | Pannexin-like TM region of LRRC8 |
| Peptidase_A22B | 5 | Signal peptide peptidase |
| Perilipin | 5 | Perilipin family |
| Porin_3 | 5 | Eukaryotic porin |
| Prefoldin_2 | 5 | Prefoldin subunit |
| Pribosyl_synth | 5 | Phosphoribosyl synthetase-associated domain |
| Pribosyltran_N | 5 | N-terminal domain of ribose phosphate pyrophosphokinase |
| Prot_ATP_ID_OB_C | 5 | Proteasomal ATPase OB C-terminal domain |
| Protocadherin | 5 | Protocadherin |
| Pyr_redox_dim | 5 | Pyridine nucleotide-disulphide oxidoreductase, dimerisation domain |
| RDM | 5 | RFPL defining motif (RDM) |
| RH_dom | 5 | Rubicon Homology Domain |
| RII_binding_1 | 5 | RII binding domain |
| RRM_8 | 5 | RRM-like domain |
| Rad60-SLD | 5 | Ubiquitin-2 like Rad60 SUMO-like |
| Rdx | 5 | Rdx family |
| SCAMP | 5 | SCAMP family |
| SCP2 | 5 | SCP-2 sterol transfer family |
| SERTA | 5 | SERTA motif |
| SFXNs | 5 | Sideroflexins |
| SH3BGR | 5 | SH3-binding, glutamic acid-rich protein |
| SKICH | 5 | SKICH domain |
| SLED | 5 | SLED domain |
| SOXp | 5 | SOX transcription factor |
| SWIB | 5 | SWIB/MDM2 domain |
| SWIM | 5 | SWIM zinc finger |
| Scramblase | 5 | Scramblase |
| Sec6 | 5 | Exocyst complex component Sec6 |
| Serinc | 5 | Serine incorporator (Serinc) |
| Sortilin-Vps10 | 5 | Sortilin, neurotensin receptor 3, |
| Sortilin_C | 5 | Sortilin, neurotensin receptor 3, C-terminal |
| Spin-Ssty | 5 | Spin/Ssty Family |
| Stathmin | 5 | Stathmin family |
| Steroid_dh | 5 | 3-oxo-5-alpha-steroid 4-dehydrogenase |
| Suppressor_APC | 5 | Adenomatous polyposis coli tumour suppressor protein |
| Syja_N | 5 | SacI homology domain |
| TAFA | 5 | TAFA family |
| TAFH | 5 | NHR1 homology to TAF |
| TF_AP-2 | 5 | Transcription factor AP-2 |
| TGS | 5 | TGS domain |
| TIR_2 | 5 | TIR domain |
| TLD | 5 | TLD |
| TLE_N | 5 | Groucho/TLE N-terminal Q-rich domain |
| TMEM132 | 5 | Transmembrane protein family 132 |
| TMEM132D_C | 5 | Mature oligodendrocyte transmembrane protein, TMEM132D, C-term |
| TMEM132D_N | 5 | Mature oligodendrocyte transmembrane protein, TMEM132D, N-term |
| TPH | 5 | Trichohyalin-plectin-homology domain |
| TRAPP | 5 | Transport protein particle (TRAPP) component |
| TSP_3 | 5 | Thrombospondin type 3 repeat |
| TSP_C | 5 | Thrombospondin C-terminal region |
| Tcp11 | 5 | T-complex protein 11 |
| Torsin | 5 | Torsin |
| Transketolase_C | 5 | Transketolase, C-terminal domain |
| Tub | 5 | Tub family |
| UPA | 5 | UPA domain |
| VWA_2 | 5 | von Willebrand factor type A domain |
| Vinculin | 5 | Vinculin family |
| Vps5 | 5 | Vps5 C terminal like |
| WD40_2 | 5 | WD40 repeated domain |
| WHEP-TRS | 5 | WHEP-TRS domain |
| WSC | 5 | WSC domain |
| WSD | 5 | Williams-Beuren syndrome DDT (WSD), D-TOX E motif |
| XPG_I | 5 | XPG I-region |
| YTH | 5 | YT521-B-like domain |
| cPLA2_C2 | 5 | Cytosolic phospholipases A2 C2-domain |
| cobW | 5 | CobW/HypB/UreG, nucleotide-binding domain |
| tRNA-synt_2 | 5 | tRNA synthetases class II (D, K and N) |
| zf-C2H2_12 | 5 | Spin-doc zinc-finger |
| zf-C2HC_2 | 5 | zinc-finger of a C2HC-type |
| zf-C5HC2 | 5 | C5HC2 zinc finger |
| zf-FCS_1 | 5 | Zinc finger, FCS-type |
| zf-MYST | 5 | MYST family zinc finger domain |
| zf_ZIC | 5 | Zic proteins zinc finger domain |
| 2-Hacid_dh | 4 | D-isomer specific 2-hydroxyacid dehydrogenase, catalytic domain |
| 2-Hacid_dh_C | 4 | D-isomer specific 2-hydroxyacid dehydrogenase, NAD binding domain |
| 2-oxoacid_dh | 4 | 2-oxoacid dehydrogenases acyltransferase (catalytic domain) |
| 3HCDH | 4 | 3-hydroxyacyl-CoA dehydrogenase, C-terminal domain |
| 3HCDH_N | 4 | 3-hydroxyacyl-CoA dehydrogenase, NAD binding domain |
| 4_1_CTD | 4 | 4.1 protein C-terminal domain (CTD) |
| 6PF2K | 4 | 6-phosphofructo-2-kinase |
| AAA_18 | 4 | AAA domain |
| ABC_membrane_2 | 4 | ABC transporter transmembrane region 2 |
| ACAS_N | 4 | Acetyl-coenzyme A synthetase N-terminus |
| ACOX | 4 | Acyl-CoA oxidase |
| ADH_zinc_N_2 | 4 | Zinc-binding dehydrogenase |
| ADK_lid | 4 | Adenylate kinase, active site lid |
| AF-4 | 4 | AF-4 proto-oncoprotein N-terminal region |
| AF4_int | 4 | AF4 interaction motif |
| AFF4_CHD | 4 | AFF4, C-terminal homology domain |
| AIRS_C | 4 | AIR synthase related protein, C-terminal domain |
| ANTH | 4 | ANTH domain |
| ART | 4 | NAD:arginine ADP-ribosyltransferase |
| ASD2 | 4 | Apx/Shroom domain ASD2 |
| ATG2-VPS13_C | 4 | ATG2/VPS13, C terminal domain |
| ATP_Ca_trans_C | 4 | Plasma membrane calcium transporter ATPase C terminal |
| AbLIM_anchor | 4 | Putative adherens-junction anchoring region of AbLIM |
| Aldolase_II | 4 | Class II Aldolase and Adducin N-terminal domain |
| Alk_phosphatase | 4 | Alkaline phosphatase |
| Amidase_2 | 4 | N-acetylmuramoyl-L-alanine amidase |
| Apolipoprotein | 4 | Apolipoprotein A1/A4/E domain |
| ArgoL2 | 4 | Argonaute linker 2 domain |
| ArgoMid | 4 | Mid domain of argonaute |
| ArgoN | 4 | N-terminal domain of argonaute |
| Asp_protease | 4 | Aspartyl protease |
| BAG | 4 | BAG domain |
| BET | 4 | Bromodomain extra-terminal - transcription regulation |
| BH4 | 4 | Bcl-2 homology region 4 |
| BK_channel_a | 4 | Calcium-activated BK potassium channel alpha subunit |
| BNIP2 | 4 | Bcl2-/adenovirus E1B nineteen kDa-interacting protein 2 |
| BNR_2 | 4 | BNR repeat-like domain |
| Basic | 4 | Myogenic Basic domain |
| Bestrophin | 4 | Bestrophin, RFP-TM, chloride channel |
| Beta-Casp | 4 | Beta-Casp domain |
| BetaGal_ABD_1 | 4 | Beta-galactosidase, first all-beta domain |
| BetaGal_gal-bd | 4 | Beta-galactosidase, galactose-binding domain |
| Biopterin_H | 4 | Biopterin-dependent aromatic amino acid hydroxylase |
| Bravo_FIGEY | 4 | Bravo-like intracellular region |
| CABIT | 4 | Cell-cycle sustaining, positive selection, |
| CAS_C | 4 | Crk-Associated Substrate C-terminal domain |
| CCDC92 | 4 | Coiled-coil domain of unknown function |
| CEBP_ZZ | 4 | Cytoplasmic polyadenylation element-binding protein ZZ domain |
| CFC | 4 | Cripto_Frl-1_Cryptic (CFC) |
| CLASP_N | 4 | CLASP N terminal |
| CLZ | 4 | C-terminal leucine zipper domain of cyclic nucleotide-gated channels |
| COE1_DBD | 4 | Transcription factor COE1 DNA-binding domain |
| COE1_HLH | 4 | Transcription factor COE1 helix-loop-helix domain |
| COMM_HN | 4 | COMMD2-7/10, HN domain |
| COS | 4 | TRIM C-terminal subgroup One Signature domain |
| COX6B | 4 | Cytochrome oxidase c subunit VIb |
| CRF | 4 | Corticotropin-releasing factor family |
| CTF_NFI | 4 | CTF/NF-I family transcription modulation region |
| CTNNB1_binding | 4 | N-terminal CTNNB1 binding |
| CUPID | 4 | Cytohesin Ubiquitin Protein Inducing Domain |
| CYRIA-B_Rac1-bd | 4 | CYRIA/CYRIB Rac1 binding domain |
| CaKB | 4 | Calcium-activated potassium channel, beta subunit |
| CaMBD | 4 | Calmodulin binding domain |
| CaMKII_AD | 4 | Calcium/calmodulin dependent protein kinase II association domain |
| Calc_CGRP_IAPP | 4 | Calcitonin / CGRP / IAPP family |
| Calreticulin | 4 | Calreticulin family |
| Carboxyl_trans | 4 | Carboxyl transferase domain |
| Choline_kinase | 4 | Choline/ethanolamine kinase |
| Cornichon | 4 | Cornichon protein |
| CortBP2 | 4 | Cortactin-binding protein-2 |
| Crystallin | 4 | Alpha crystallin A chain, N terminal |
| Cu-oxidase_2 | 4 | Multicopper oxidase |
| DCB | 4 | Mon2/Sec7/BIG1-like, dimerisation and cyclophilin-binding domain |
| DC_STAMP | 4 | DC-STAMP-like protein |
| DDT | 4 | DDT domain |
| DEAD_2 | 4 | DEAD_2 |
| DEK_C | 4 | DEK C terminal domain |
| DMPK_coil | 4 | DMPK coiled coil domain like |
| DNA_methylase | 4 | C-5 cytosine-specific DNA methylase |
| DNA_mis_repair | 4 | DNA mismatch repair protein, C-terminal domain |
| DNA_pol_B | 4 | DNA polymerase family B |
| DNA_pol_B_exo1 | 4 | DNA polymerase family B, exonuclease domain |
| DNA_pol_B_thumb | 4 | DNA polymerase beta thumb |
| DNA_pol_lambd_f | 4 | Fingers domain of DNA polymerase lambda |
| DND1_DSRM | 4 | double strand RNA binding domain from DEAD END PROTEIN 1 |
| DOMON | 4 | DOMON domain |
| DSL | 4 | Delta serrate ligand |
| DUF1908 | 4 | Domain of unknown function (DUF1908) |
| DUF3371 | 4 | Domain of unknown function (DUF3371) |
| DUF3377 | 4 | Domain of unknown function (DUF3377) |
| DUF4195 | 4 | Domain of unknown function (DUF4195) |
| DUF4200 | 4 | Domain of unknown function (DUF4200) |
| DUF4217 | 4 | Domain of unknown function (DUF4217) |
| DUF4704 | 4 | Neurobeachin/BDCP, DUF4704 alpha solenoid region |
| DUF5600 | 4 | Domain of unknown function (DUF5600) |
| DUF5917 | 4 | Family of unknown function (DUF5917) |
| DUF758 | 4 | Domain of unknown function (DUF758) |
| DUF846 | 4 | Eukaryotic protein of unknown function (DUF846) |
| Dickkopf_N | 4 | Dickkopf N-terminal cysteine-rich region |
| DnaJ_CXXCXGXG | 4 | DnaJ central domain |
| Dpy19 | 4 | Q-cell neuroblast polarisation |
| E1_DerP2_DerF2 | 4 | ML domain |
| EFG_IV | 4 | Elongation factor G, domain IV |
| EMP70 | 4 | Endomembrane protein 70 |
| ERM_C | 4 | Ezrin/radixin/moesin family C terminal |
| ERM_helical | 4 | Ezrin/radixin/moesin, alpha-helical domain |
| Enolase_C | 4 | Enolase, C-terminal TIM barrel domain |
| Erf4 | 4 | Golgin subfamily A member 7/ERF4 family |
| Exostosin | 4 | Exostosin family |
| FAA_hydrolase | 4 | Fumarylacetoacetate (FAA) hydrolase family |
| FAD_binding_4 | 4 | FAD binding domain |
| FAM110_C | 4 | Centrosome-associated C terminus |
| FAM110_N | 4 | Centrosome-associated N terminus |
| FERM_F1 | 4 | FERM F1 ubiquitin-like domain |
| FERM_F2 | 4 | FERM F2 acyl-CoA binding protein-like domain |
| FOXO-TAD | 4 | Transactivation domain of FOXO protein family |
| FOXP-CC | 4 | FOXP coiled-coil domain |
| FRG2 | 4 | Facioscapulohumeral muscular dystrophy candidate 2 |
| Fasciclin | 4 | Fasciclin domain |
| Fer2 | 4 | 2Fe-2S iron-sulfur cluster binding domain |
| FerI | 4 | FerI (NUC094) domain |
| Ferritin | 4 | Ferritin-like domain |
| Fez1 | 4 | Fez1 |
| Formyl_trans_N | 4 | Formyl transferase |
| Fumble | 4 | Fumble |
| Furin-like_2 | 4 | Furin-like repeat, cysteine-rich |
| Fuz_longin_1 | 4 | First Longin domain of FUZ, MON1 and HPS1 |
| Fuz_longin_2 | 4 | Second Longin domain of FUZ, MON1 and HPS1 |
| Fuz_longin_3 | 4 | Third Longin domain of FUZ, MON1 and HPS1 |
| G2F | 4 | G2F domain |
| G8 | 4 | G8 domain |
| GCV_T | 4 | Aminomethyltransferase folate-binding domain |
| GCV_T_C | 4 | Glycine cleavage T-protein C-terminal barrel domain |
| GDI | 4 | GDP dissociation inhibitor |
| GDP_Man_Dehyd | 4 | GDP-mannose 4,6 dehydratase |
| GFA | 4 | Glutathione-dependent formaldehyde-activating enzyme |
| GFO_IDH_MocA | 4 | Oxidoreductase family, NAD-binding Rossmann fold |
| GHMP_kinases_C | 4 | GHMP kinases C terminal |
| GLTP | 4 | Glycolipid transfer protein (GLTP) |
| GOST_TM | 4 | GOST, seven transmembrane domain |
| GPCR_chapero_1 | 4 | GPCR-chaperone |
| GST_C_6 | 4 | Glutathione S-transferase, C-terminal domain |
| GTF2I | 4 | GTF2I-like repeat |
| Gal-3-0_sulfotr | 4 | Galactose-3-O-sulfotransferase |
| Gal_Lectin | 4 | Galactose binding lectin domain |
| Glucosamine_iso | 4 | Glucosamine-6-phosphate isomerases/6-phosphogluconolactonase |
| Glyco_hydro_35 | 4 | Glycosyl hydrolases family 35 |
| Glyco_transf_22 | 4 | Alg9-like mannosyltransferase family |
| Glyco_transf_54 | 4 | N-Acetylglucosaminyltransferase-IV (GnT-IV) conserved region |
| Glyco_transf_6 | 4 | Glycosyltransferase family 6 |
| Gtr1_RagA | 4 | Gtr1/RagA G protein conserved region |
| HHH_3 | 4 | Helix-hairpin-helix motif |
| HHH_5 | 4 | Helix-hairpin-helix domain |
| HHH_8 | 4 | Helix-hairpin-helix domain |
| HIN | 4 | HIN-200/IF120x domain |
| HJURP_C | 4 | Holliday junction regulator protein family C-terminal repeat |
| HMA | 4 | Heavy-metal-associated domain |
| HORMA | 4 | HORMA domain |
| HSA | 4 | HSA domain |
| HSNSD | 4 | heparan sulfate-N-deacetylase |
| HSP90 | 4 | Hsp90 protein |
| HYPK_UBA | 4 | HYPK UBA domain |
| Helicase_C_2 | 4 | Helicase C-terminal domain |
| Hox9_act | 4 | Hox9 activation region |
| HoxA13_N | 4 | Hox protein A13 N terminal |
| Hydrolase_like | 4 | HAD-hyrolase-like |
| IF-2B | 4 | Initiation factor 2 subunit family |
| IF4E | 4 | Eukaryotic initiation factor 4E |
| IGFL | 4 | Insulin growth factor-like family |
| IL10 | 4 | Interleukin 10 |
| IL28A | 4 | Interleukin-28A |
| IMPDH | 4 | IMP dehydrogenase / GMP reductase domain |
| IMS | 4 | impB/mucB/samB family |
| IMS_C | 4 | impB/mucB/samB family C-terminal domain |
| ISK_Channel | 4 | Slow voltage-gated potassium channel |
| IZUMO | 4 | Izumo sperm-egg fusion, Ig domain-associated |
| I_LWEQ | 4 | I/LWEQ domain |
| Ifi-6-16 | 4 | Interferon-induced 6-16 family |
| Intu_longin_1 | 4 | First Longin domain of INTU, CCZ1 and HPS4 |
| Intu_longin_3 | 4 | Intu longin-like domain 3 |
| Ion_trans_N | 4 | Ion transport protein N-terminal |
| Jak1_Phl | 4 | Jak1 pleckstrin homology-like domain |
| Josephin | 4 | Josephin |
| KASH | 4 | Nuclear envelope localisation domain |
| KDM5_C-hel | 4 | Lysine-specific demethylase 5, C-terminal helical domain |
| KELAA | 4 | KELAA motif |
| KIF1B | 4 | Kinesin protein 1B |
| KIND | 4 | Kinase non-catalytic C-lobe domain |
| KN_motif | 4 | KN motif |
| LLGL | 4 | LLGL2 |
| LMBR1 | 4 | LMBR1-like membrane protein |
| LRRC37 | 4 | Leucine-rich repeat-containing protein 37 family |
| LUC7 | 4 | LUC7 N_terminus |
| Lactamase_B_6 | 4 | Metallo-beta-lactamase superfamily domain |
| Lectin_leg-like | 4 | Legume-like lectin family |
| Lep_receptor_Ig | 4 | Ig-like C2-type domain |
| Leuk-A4-hydro_C | 4 | Leukotriene A4 hydrolase, C-terminal |
| Ligase_CoA | 4 | CoA-ligase |
| MAGUK_N_PEST | 4 | Polyubiquitination (PEST) N-terminal domain of MAGUK |
| MANEC | 4 | MANEC domain |
| MAP7 | 4 | MAP7 (E-MAP-115) family |
| MATH | 4 | MATH domain |
| MHC_I_C | 4 | MHC_I C-terminus |
| MIB_HERC2 | 4 | Mib_herc2 |
| MTS | 4 | Methyltransferase small domain |
| Maf_N | 4 | Maf N-terminal region |
| Matrilin_ccoil | 4 | Trimeric coiled-coil oligomerisation domain of matrilin |
| Methyltransf_3 | 4 | O-methyltransferase |
| Mod_r | 4 | Modifier of rudimentary (Mod(r)) protein |
| Morc6_S5 | 4 | Morc6 ribosomal protein S5 domain 2-like |
| Mpv17_PMP22 | 4 | Mpv17 / PMP22 family |
| MutS_II | 4 | MutS domain II |
| MutS_IV | 4 | MutS family domain IV |
| NBCH_WD40 | 4 | Neurobeachin beta propeller domain |
| NGF | 4 | Nerve growth factor family |
| NHS | 4 | NHS-like protein |
| NIPSNAP | 4 | NIPSNAP |
| NKAIN | 4 | Na,K-Atpase Interacting protein |
| NOD | 4 | NOTCH protein |
| NODP | 4 | NOTCH protein |
| NT-C2 | 4 | N-terminal C2 in EEIG1 and EHBP1 proteins |
| NTP_transf_7 | 4 | Nucleotidyltransferase |
| Ndr | 4 | Ndr family |
| Nebulin | 4 | Nebulin repeat |
| Neuro_bHLH | 4 | Neuronal helix-loop-helix transcription factor |
| Neurobeachin | 4 | Neurobeachin alpha solenoid region |
| Nexin_C | 4 | Sorting nexin C terminal |
| NfI_DNAbd_pre-N | 4 | Nuclear factor I protein pre-N-terminus |
| Nucleoside_tran | 4 | Nucleoside transporter |
| OAS1_C | 4 | 2'-5'-oligoadenylate synthetase 1, domain 2, C-terminus |
| OCRE | 4 | OCRE domain |
| P5-ATPase | 4 | P5-type ATPase cation transporter |
| PBC | 4 | PBC domain |
| PDCD10_N | 4 | Programmed cell death protein 10, dimerisation domain |
| PDE4_UCR | 4 | Phosphodiesterase 4 upstream conserved regions (UCR) |
| PDZ_assoc | 4 | PDZ-associated domain of NMDA receptors |
| PGM_PMM_II | 4 | Phosphoglucomutase/phosphomannomutase, alpha/beta/alpha domain II |
| PGM_PMM_III | 4 | Phosphoglucomutase/phosphomannomutase, alpha/beta/alpha domain III |
| PH-GRAM_MTMR6-like | 4 | GRAM domain-like |
| PH_14 | 4 | PH domain |
| PH_8 | 4 | Pleckstrin homology domain |
| PH_RBD | 4 | Rab-binding domain (RBD) |
| PI3K_P85_iSH2 | 4 | Phosphatidylinositol 3-kinase regulatory subunit P85 inter-SH2 domain |
| PIH1_CS | 4 | PIH1 CS-like domain |
| PINIT | 4 | PINIT domain |
| PIN_4 | 4 | PIN domain |
| PKD_4 | 4 | PKD domain |
| PLU-1 | 4 | PLU-1-like protein |
| PNP_UDP_1 | 4 | Phosphorylase superfamily |
| PPR_3 | 4 | Pentatricopeptide repeat domain |
| PROPPIN | 4 | PROPPIN |
| PTCB-BRCT | 4 | twin BRCT domain |
| PTPLA | 4 | Protein tyrosine phosphatase-like protein, PTPLA |
| PTRF_SDPR | 4 | PTRF/SDPR family |
| PXA | 4 | PXA domain |
| Paralemmin | 4 | Paralemmin |
| Pcc1 | 4 | Transcription factor Pcc1 |
| Pecanex_C | 4 | Pecanex protein (C-terminus) |
| Peptidase_C12 | 4 | Ubiquitin carboxyl-terminal hydrolase, family 1 |
| Peptidase_C54 | 4 | Peptidase family C54 |
| Phosducin | 4 | Phosducin |
| Prefoldin | 4 | Prefoldin subunit |
| Prenyltrans | 4 | Prenyltransferase and squalene oxidase repeat |
| Profilin | 4 | Profilin |
| PseudoU_synth_2 | 4 | RNA pseudouridylate synthase |
| RAI16-like | 4 | Retinoic acid induced 16-like protein |
| REJ | 4 | REJ domain |
| RF-1 | 4 | RF-1 domain |
| RGS_DHEX | 4 | Regulator of G-protein signalling DHEX domain |
| RH1 | 4 | RILP homology 1 domain |
| RNA_pol_L_2 | 4 | RNA polymerase Rpb3/Rpb11 dimerisation domain |
| RNB | 4 | RNB domain |
| RRM_7 | 4 | RNA recognition motif |
| RTT107_BRCT_5 | 4 | Regulator of Ty1 transposition protein 107 BRCT domain |
| RecQ_Zn_bind | 4 | RecQ zinc-binding |
| Reticulon | 4 | Reticulon |
| SAA | 4 | Serum amyloid A protein |
| SAB | 4 | SAB domain |
| SAMD1_WH | 4 | SAM domain-containing protein 1, WH domain |
| SCA7 | 4 | SCA7, zinc-binding domain |
| SEP | 4 | SEP domain |
| SERF-like_N | 4 | Small EDRK-rich factor 1/2-like, N-terminal |
| SK_channel | 4 | Calcium-activated SK potassium channel |
| SUI1 | 4 | Translation initiation factor SUI1 |
| SUZ | 4 | SUZ domain |
| SapB_2 | 4 | Saposin-like type B, region 2 |
| Sarcoglycan_1 | 4 | Sarcoglycan complex subunit protein |
| Sec63 | 4 | Sec63 Brl domain |
| Sec7-like_HDS | 4 | Mon2/Sec7/BIG1-like, HDS |
| Sec7-like_HUS | 4 | Mon2/Sec7/BIG1-like, HUS domain |
| Serum_albumin | 4 | Serum albumin family |
| Sld5 | 4 | GINS complex protein helical bundle domain |
| Solute_trans_a | 4 | Organic solute transporter Ostalpha |
| Sox17_18_mid | 4 | Sox 17/18 central domain |
| Spectrin_2 | 4 | Spectrin like domain |
| Spectrin_3 | 4 | Spectrin-like repeat |
| Spectrin_4 | 4 | Spectrin-like repeat |
| Synaphin | 4 | Synaphin protein |
| TEA | 4 | TEA/ATTS domain |
| TEX13 | 4 | Testis-expressed sequence 13 protein family |
| TFR_dimer | 4 | Transferrin receptor-like dimerisation domain |
| TF_Zn_Ribbon | 4 | TFIIB zinc-binding |
| THF_DHG_CYH | 4 | Tetrahydrofolate dehydrogenase/cyclohydrolase, catalytic domain |
| THF_DHG_CYH_C | 4 | Tetrahydrofolate dehydrogenase/cyclohydrolase, NAD(P)-binding domain |
| TIMP | 4 | Tissue inhibitor of metalloproteinase |
| TLV_coat | 4 | ENV polyprotein (coat polyprotein) |
| TM2 | 4 | TM2 domain |
| TMTC_DUF1736 | 4 | Protein O-mannosyl-transferase TMTC, DUF1736 |
| TOR1A_C | 4 | Torsin-1A, C-terminal domain |
| TPD52 | 4 | Tumour protein D52 family |
| TPR_6 | 4 | Tetratricopeptide repeat |
| TRPM_tetra | 4 | Tetramerisation domain of TRPM |
| TSC22 | 4 | TSC-22/dip/bun family |
| Tap-RNA_bind | 4 | Tap, RNA-binding |
| Ten_N | 4 | Teneurin Intracellular Region |
| Thioredoxin_7 | 4 | Thioredoxin-like |
| Thioredoxin_8 | 4 | Thioredoxin-like |
| Tmemb_cc2 | 4 | Predicted transmembrane and coiled-coil 2 protein |
| Toprim | 4 | Toprim domain |
| Tox-GHH | 4 | GHH signature containing HNH/Endo VII superfamily nuclease toxin |
| Tropomyosin | 4 | Tropomyosin |
| UAA | 4 | UAA transporter family |
| UBA_6 | 4 | UBA-like domain |
| Ubiquitin_3 | 4 | Ubiquitin-like domain |
| V-SNARE_C | 4 | Snare region anchored in the vesicle membrane C-terminus |
| V1R | 4 | Vomeronasal organ pheromone receptor family, V1R |
| VEGFR-1-like_Ig-like | 4 | Vascular endothelial growth factor receptor 1-like, Ig-like domain |
| VGCC_alpha2 | 4 | Neuronal voltage-dependent calcium channel alpha 2acd |
| VGCC_beta4Aa_N | 4 | Voltage gated calcium channel subunit beta domain 4Aa N terminal |
| VPS13_DH-like | 4 | Vacuolar-sorting-associated 13 protein, DH-like domain |
| VPS13_VAB | 4 | Vacuolar-sorting associated protein 13, adaptor binding domain |
| VTT_dom | 4 | VTT domain |
| VWA_N | 4 | VWA N-terminal |
| V_ATPase_I | 4 | V-type ATPase 116kDa subunit family |
| WHIM1 | 4 | WSTF, HB1, Itc1p, MBD9 motif 1 |
| WRNPLPNID | 4 | Putative WW-binding domain and destruction box |
| YBD | 4 | YAP binding domain |
| YEATS | 4 | YEATS family |
| YPEH2ZP | 4 | Yippee homolog 2 zinc finger protein |
| ZSWIM4-8_C | 4 | ZSWIM4-8, C-terminal |
| c-SKI_SMAD_bind | 4 | c-SKI Smad4 binding domain |
| dCMP_cyt_deam_1 | 4 | Cytidine and deoxycytidylate deaminase zinc-binding region |
| dNK | 4 | Deoxynucleoside kinase |
| eIF2A | 4 | Eukaryotic translation initiation factor eIF2A |
| mTERF | 4 | mTERF |
| polyprenyl_synt | 4 | Polyprenyl synthetase |
| tRNA-synt_1b | 4 | tRNA synthetases class I (W and Y) |
| zf-C2H2_11 | 4 | zinc-finger C2H2-type |
| zf-C2H2_5 | 4 | C2H2-type zinc finger |
| zf-RING_11 | 4 | RING-like zinc finger |
| zf-U11-48K | 4 | U11-48K-like CHHC zinc finger |
| zf_C2HC_14 | 4 | C2HC Zing finger domain |
| 2OG-FeII_Oxy | 3 | 2OG-Fe(II) oxygenase superfamily |
| 3-PAP | 3 | Myotubularin-associated protein |
| 40S_S4_C | 3 | 40S ribosomal protein S4 C-terminus |
| 5-FTHF_cyc-lig | 3 | 5-formyltetrahydrofolate cyclo-ligase family |
| 5-nucleotidase | 3 | 5'-nucleotidase |
| AA_permease_N | 3 | Amino acid permease N-terminal |
| ABC_tran_Xtn | 3 | ABC transporter |
| ABM | 3 | Antibiotic biosynthesis monooxygenase |
| ADD_DNMT3 | 3 | DNMT3, cysteine rich ADD domain, GATA1-like zinc finger |
| ADP_ribosyl_GH | 3 | ADP-ribosylglycohydrolase |
| AGRB_N | 3 | Adhesion GPCR B N-terminal region |
| AIF-1 | 3 | Allograft inflammatory factor 1 |
| AIRS | 3 | AIR synthase related protein, N-terminal domain |
| AKAP2_C | 3 | A-kinase anchor protein 2 C-terminus |
| AKAP7_NLS | 3 | AKAP7 2'5' RNA ligase-like domain |
| AKAP95 | 3 | A-kinase anchoring protein 95 (AKAP95) |
| AKAP_110 | 3 | A-kinase anchor protein 110 kDa (AKAP 110) |
| ALMS_motif | 3 | ALMS motif |
| AMER | 3 | APC membrane recruitment protein |
| AMOP | 3 | AMOP domain |
| AMP_deaminase | 3 | AMP deaminase |
| ANP | 3 | Atrial natriuretic peptide |
| APH | 3 | Phosphotransferase enzyme family |
| APP_Cu_bd | 3 | Copper-binding of amyloid precursor, CuBD |
| APP_E2 | 3 | E2 domain of amyloid precursor protein |
| APP_N | 3 | Amyloid A4 N-terminal heparin-binding |
| APP_amyloid | 3 | Beta-amyloid precursor protein C-terminus |
| ASD1 | 3 | Apx/Shroom domain ASD1 |
| ASH | 3 | Abnormal spindle-like microcephaly-assoc'd, ASPM-SPD-2-Hydin |
| ASK_PH | 3 | ASK kinase PH domain |
| ASMase_C | 3 | Acid sphingomyelin phosphodiesterase C-terminal region |
| ASXH | 3 | Asx homology domain |
| ATAD3_N | 3 | ATPase family AAA domain-containing protein 3, N-terminal |
| ATG4_LIR | 3 | ATG4, F-type LIR motif |
| ATP_bind_1 | 3 | Conserved hypothetical ATP binding protein |
| AT_hook | 3 | AT hook motif |
| AXH | 3 | Ataxin-1 and HBP1 module (AXH) |
| A_deaminase | 3 | Adenosine deaminase |
| Abhydrolase_2 | 3 | Phospholipase/Carboxylesterase |
| Abi_HHR | 3 | Abl-interactor HHR |
| Aconitase | 3 | Aconitase family (aconitate hydratase) |
| Aconitase_C | 3 | Aconitase C-terminal domain |
| AdoHcyase | 3 | S-adenosyl-L-homocysteine hydrolase |
| AdoHcyase_NAD | 3 | S-adenosyl-L-homocysteine hydrolase, NAD binding domain |
| Agenet | 3 | Agenet domain |
| Ago_hook | 3 | Argonaute hook |
| AhpC-TSA_2 | 3 | AhpC/TSA antioxidant enzyme |
| Amidase | 3 | Amidase |
| Angiomotin_C | 3 | Angiomotin C terminal |
| Anillin | 3 | Cell division protein anillin |
| Anth_Ig | 3 | Anthrax receptor extracellular domain |
| Arginase | 3 | Arginase family |
| Ariadne | 3 | Ariadne domain |
| Arylesterase | 3 | Arylesterase |
| Asp_Arg_Hydrox | 3 | Aspartyl/Asparaginyl beta-hydroxylase |
| Asparaginase_2 | 3 | Asparaginase |
| Auts2 | 3 | Autism susceptibility gene 2 protein |
| B12D | 3 | NADH-ubiquinone reductase complex 1 MLRQ subunit |
| B2-adapt-app_C | 3 | Beta2-adaptin appendage, C-terminal sub-domain |
| B9-C2 | 3 | Ciliary basal body-associated, B9 protein |
| BAR_3_WASP_bdg | 3 | WASP-binding domain of Sorting nexin protein |
| BAT2_N | 3 | BAT2 N-terminus |
| BCL_N | 3 | BCL7, N-terminal conserver region |
| BIG2_C | 3 | BIG2 C-terminal domain |
| BIRC2-3-like_UBA | 3 | BIRC2/3-like, UBA domain |
| BPS | 3 | BPS (Between PH and SH2) |
| BRINP | 3 | BMP/retinoic acid-inducible neural-specific protein |
| BSD | 3 | BSD domain |
| Bap31 | 3 | Bap31/Bap29 transmembrane region |
| Bap31_Bap29_C | 3 | Bap31/Bap29 cytoplasmic coiled-coil domain |
| Bromo_TP | 3 | Bromodomain associated |
| C1_4 | 3 | TFIIH C1-like domain |
| C2-set | 3 | Immunoglobulin C2-set domain |
| C4_MG1 | 3 | Complement C4, MG1 domain |
| C5HCH | 3 | NSD Cys-His rich domain |
| C8A_B_C6_EGF-like | 3 | Complement components C8A/B/C6, EGF-like domain |
| CAC1F_C | 3 | Voltage-gated calcium channel subunit alpha, C-term |
| CAF1C_H4-bd | 3 | Histone-binding protein RBBP4 or subunit C of CAF1 complex |
| CAMSAP_CC1 | 3 | Spectrin-binding region of Ca2+-Calmodulin |
| CAMSAP_CKK | 3 | Microtubule-binding calmodulin-regulated spectrin-associated |
| CAP-ZIP_m | 3 | WASH complex subunit CAP-Z interacting, central region |
| CARD_2 | 3 | Caspase recruitment domain |
| CARMIL_C | 3 | CARMIL C-terminus |
| CBF | 3 | CBF/Mak21 family |
| CBM_20 | 3 | Starch binding domain |
| CC2-LZ | 3 | Leucine zipper of domain CC2 of NEMO, NF-kappa-B essential modulator |
| CCDC85 | 3 | CCDC85 family |
| CD34_antigen | 3 | CD34/Podocalyxin family |
| CD36 | 3 | CD36 family |
| CD99L2 | 3 | CD99 antigen like protein 2 |
| CDI | 3 | Cyclin-dependent kinase inhibitor |
| CFA20_dom | 3 | CFA20 domain |
| CHD1-like_C | 3 | Chromodomain-helicase-DNA-binding protein 1-like, C-terminal |
| CHDCT2 | 3 | CHDCT2 (NUC038) domain |
| CHDII_SANT-like | 3 | CHD subfamily II, SANT-like domain |
| CHDNT | 3 | CHDNT (NUC034) domain |
| CH_2 | 3 | CH-like domain in sperm protein |
| CITED | 3 | CITED |
| CK1gamma_C | 3 | Casein kinase 1 gamma C terminal |
| CLAMP | 3 | Flagellar C1a complex subunit C1a-32 |
| CLCA | 3 | Calcium-activated chloride channel N terminal |
| CLSTN_C | 3 | Calsyntenin C-terminal |
| CMI2B-like | 3 | Ciliary microtubule inner protein 2B-like |
| CNNM | 3 | Cyclin M transmembrane N-terminal domain |
| COMP | 3 | Cartilage oligomeric matrix protein |
| COMPASS-Shg1 | 3 | COMPASS (Complex proteins associated with Set1p) component shg1 |
| COPIIcoated_ERV | 3 | Endoplasmic reticulum vesicle transporter |
| COX7a | 3 | Cytochrome c oxidase subunit VII |
| CPSF_A | 3 | CPSF A subunit region |
| CPT_N | 3 | Carnitine O-palmitoyltransferase N-terminus |
| CRIC_ras_sig | 3 | Connector enhancer of kinase suppressor of ras |
| CSN8_PSD8_EIF3K | 3 | CSN8/PSMD8/EIF3K family |
| CSRNP_N | 3 | Cysteine/serine-rich nuclear protein N-terminus |
| Calcipressin | 3 | Calcipressin |
| Calcyon | 3 | D1 dopamine receptor-interacting protein (calcyon) |
| Calsarcin | 3 | Calcineurin-binding protein (Calsarcin) |
| Carm_PH | 3 | Carmil pleckstrin homology domain |
| Caudal_act | 3 | Caudal like protein activation region |
| Caveolin | 3 | Caveolin |
| Cbl_N | 3 | CBL proto-oncogene N-terminal domain 1 |
| Cbl_N2 | 3 | CBL proto-oncogene N-terminus, EF hand-like domain |
| Cbl_N3 | 3 | CBL proto-oncogene N-terminus, SH2-like domain |
| Ceramidase | 3 | Ceramidase |
| Chibby | 3 | Chibby family |
| Chromo_shadow | 3 | Chromo shadow domain |
| Cir_N | 3 | N-terminal domain of CBF1 interacting co-repressor CIR |
| Clathrin_H_link | 3 | Clathrin-H-link |
| Cmyb_C | 3 | C-myb, C-terminal |
| Cobalamin_bind | 3 | Eukaryotic cobalamin-binding protein |
| Complex1_LYR_2 | 3 | Complex1_LYR-like |
| Cor1 | 3 | Cor1/Xlr/Xmr conserved region |
| Cornifin | 3 | Cornifin (SPRR) family |
| Crisp | 3 | Crisp |
| Cse1 | 3 | Cse1 |
| Cu2_monoox_C | 3 | Copper type II ascorbate-dependent monooxygenase, C-terminal domain |
| Cu2_monooxygen | 3 | Copper type II ascorbate-dependent monooxygenase, N-terminal domain |
| Cu_amine_oxid | 3 | Copper amine oxidase, enzyme domain |
| Cu_amine_oxidN2 | 3 | Copper amine oxidase, N2 domain |
| Cu_amine_oxidN3 | 3 | Copper amine oxidase, N3 domain |
| Cul7 | 3 | Mouse development and cellular proliferation protein Cullin-7 |
| CycT2-like_C | 3 | Cyclin-T2-like, C-terminal domain |
| DAB2P_C | 3 | Disabled homolog 2-interacting protein, C-terminal domain |
| DAG_kinase_N | 3 | Diacylglycerol kinase N-terminus |
| DALR_1 | 3 | DALR anticodon binding domain |
| DARPP-32 | 3 | Protein phosphatase inhibitor 1/DARPP-32 |
| DER1 | 3 | Der1-like family |
| DIKK1-2-4_C-subdom1 | 3 | Dickkopf-related protein 1/2/4, C-terminal subdomain 1 |
| DIKK1-2-4_C-subdom2 | 3 | Dickkopf-related protein 1/2/4, C-terminal subdomain 2 |
| DLIC | 3 | Dynein light intermediate chain (DLIC) |
| DLL_N | 3 | Homeobox protein distal-less-like N terminal |
| DMA | 3 | DMRTA motif |
| DMRT-like | 3 | Doublesex-and mab-3-related transcription factor C1 and C2 |
| DNA_ligase_A_C | 3 | ATP dependent DNA ligase C terminal region |
| DNA_ligase_A_M | 3 | ATP dependent DNA ligase domain |
| DNA_ligase_A_N | 3 | DNA ligase N terminus |
| DNA_pol3_delta2 | 3 | DNA polymerase III, delta subunit |
| DNA_pol_B_palm | 3 | DNA polymerase beta palm |
| DNA_pol_E_B | 3 | DNA polymerase alpha/epsilon subunit B |
| DNMT3_ADD_GATA1-like | 3 | DNMT3, ADD PHD zinc finger |
| DP | 3 | Transcription factor DP |
| DPF1-3_N | 3 | DPF1-3, N-terminal |
| DRHyd-ASK | 3 | Deoxyribohydrolase (DRHyd) domain of the ASK signalosome |
| DRMBL | 3 | DNA repair metallo-beta-lactamase |
| DSPn | 3 | Dual specificity protein phosphatase, N-terminal half |
| DTW | 3 | DTW domain |
| DUF1087 | 3 | CHD subfamily II, DUF1087 |
| DUF1180 | 3 | Membrane protein FAM174-like |
| DUF1518 | 3 | Nuclear receptor coactivator, DUF1518 |
| DUF1977 | 3 | Domain of unknown function (DUF1977) |
| DUF2371 | 3 | Uncharacterised conserved protein (DUF2371) |
| DUF2464 | 3 | Multivesicular body subunit 12 |
| DUF2465 | 3 | Protein of unknown function (DUF2465) |
| DUF3399 | 3 | Domain of unknown function (DUF3399) |
| DUF3446 | 3 | Early growth response N-terminal domain |
| DUF3452 | 3 | Domain of unknown function (DUF3452) |
| DUF3517 | 3 | Domain of unknown function (DUF3517) |
| DUF3522 | 3 | Protein of unknown function (DUF3522) |
| DUF3528 | 3 | Protein of unknown function (DUF3528) |
| DUF3715 | 3 | Protein of unknown function (DUF3715) |
| DUF4074 | 3 | Domain of unknown function (DUF4074) |
| DUF4174 | 3 | Domain of unknown function (DUF4174) |
| DUF4211 | 3 | Domain of unknown function (DUF4211) |
| DUF4515 | 3 | Domain of unknown function (DUF4515) |
| DUF4585 | 3 | Domain of unknown function (DUF4585) |
| DUF4592 | 3 | Domain of unknown function (DUF4592) |
| DUF4683 | 3 | Domain of unknown function (DUF4683) |
| DUF716 | 3 | Family of unknown function (DUF716) |
| Dapper | 3 | Dapper |
| DcpS_C | 3 | Scavenger mRNA decapping enzyme C-term binding |
| Dishevelled | 3 | Dishevelled specific domain |
| Dsh_C | 3 | Segment polarity protein dishevelled (Dsh) C terminal |
| Dynein_light | 3 | Dynein light chain type 1 |
| Dysbindin | 3 | Dysbindin (Dystrobrevin binding protein 1) |
| E1_4HB | 3 | Ubiquitin-activating enzyme E1 four-helix bundle |
| E1_FCCH | 3 | Ubiquitin-activating enzyme E1 FCCH domain |
| E1_UFD | 3 | Ubiquitin fold domain |
| E3_binding | 3 | e3 binding domain |
| EB1 | 3 | EB1-like C-terminal motif |
| EF-hand_11 | 3 | EF-hand domain |
| EF-hand_13 | 3 | EF-hand domain |
| ELL | 3 | RNA polymerase II elongation factor ELL |
| ELMO_ARM | 3 | ELMO, armadillo-like helical domain |
| EMC6 | 3 | EMC6 |
| EME1-MUS81_C | 3 | EME1/MUS81, C-terminal |
| ERG4_ERG24 | 3 | Ergosterol biosynthesis ERG4/ERG24 family |
| ERGIC_N | 3 | Endoplasmic Reticulum-Golgi Intermediate Compartment (ERGIC) |
| ER_lumen_recept | 3 | ER lumen protein retaining receptor |
| EST1 | 3 | Telomerase activating protein Est1 |
| EST1_DNA_bind | 3 | Est1 DNA/RNA binding domain |
| ETS_PEA3_N | 3 | PEA3 subfamily ETS-domain transcription factor N terminal domain |
| Elf-1_N | 3 | Transcription factor protein N terminal |
| Endothelin | 3 | Endothelin family |
| Enolase_N | 3 | Enolase, N-terminal domain |
| Evr1_Alr | 3 | Erv1 / Alr family |
| F-actin_cap_A | 3 | F-actin capping protein alpha subunit |
| FAD-oxidase_C | 3 | FAD linked oxidases, C-terminal domain |
| FAM117 | 3 | Protein Family FAM117 |
| FAM131 | 3 | Putative cell signalling |
| FAM171A1-2-B_C | 3 | FAM171A1/A2/B C-terminal |
| FAM171A1-2-B_N | 3 | FAM171A1/A2/B, N-terminal |
| FAM176 | 3 | FAM176 family |
| FAM194 | 3 | FAM194 protein |
| FAM25 | 3 | FAM25 family |
| FAM53 | 3 | Family of FAM53 |
| FAM86 | 3 | Family of unknown function |
| FAO_M | 3 | FAD dependent oxidoreductase central domain |
| FBN_EGF_st1 | 3 | Fibrillin, first EGF domain |
| FDF | 3 | FDF domain |
| FERM_f0 | 3 | N-terminal or F0 domain of Talin-head FERM |
| FGF-BP1 | 3 | FGF binding protein 1 (FGF-BP1) |
| FHA_2 | 3 | FHA domain |
| FOXO_KIX_bdg | 3 | KIX-binding domain of forkhead box O, CR2 |
| FXMRP1_C_core | 3 | Fragile X-related 1 protein core C terminal |
| Fam20C | 3 | Golgi casein kinase, C-terminal, Fam20 |
| Fascin | 3 | Fascin domain |
| FerA | 3 | FerA (NUC095) domain |
| Fib_alpha | 3 | Fibrinogen alpha/beta chain family |
| Fibrillin_U_N | 3 | Fibrillin 1 unique N-terminal domain |
| Folate_carrier | 3 | Reduced folate carrier |
| Forkhead_N | 3 | Forkhead N-terminal region |
| Formyl_trans_C | 3 | Formyl transferase, C-terminal domain |
| Fox-1_C | 3 | Calcitonin gene-related peptide regulator C terminal |
| Frem_N | 3 | Frem protein N-terminal domain |
| GATA-N | 3 | GATA-type transcription activator, N-terminal |
| GATase_6 | 3 | Glutamine amidotransferase domain |
| GCFC | 3 | GC-rich sequence DNA-binding factor-like protein |
| GGA_N-GAT | 3 | N-terminal extension of GAT domain |
| GMP_PDE_delta | 3 | GMP-PDE, delta subunit |
| GPAT_C | 3 | Glycerol-3-phosphate acyltransferase C-terminal region |
| GRIN_C | 3 | G protein-regulated inducer of neurite outgrowth C-terminus |
| GSDH | 3 | Glucose / Sorbosone dehydrogenase |
| GSG-1 | 3 | GSG1-like protein |
| GYF | 3 | GYF domain |
| Gate | 3 | Nucleoside recognition |
| Glyco_hydro_15 | 3 | Glycosyl hydrolases family 15 |
| Glyco_hydro_20 | 3 | Glycosyl hydrolase family 20, catalytic domain |
| Glyco_transf_25 | 3 | Glycosyltransferase family 25 (LPS biosynthesis protein) |
| Glyco_transf_43 | 3 | Glycosyltransferase family 43 |
| Glyco_transf_49 | 3 | Glycosyl-transferase for dystroglycan |
| Glyco_transf_90 | 3 | Glycosyl transferase family 90 |
| Glycolytic | 3 | Fructose-bisphosphate aldolase class-I |
| Granin | 3 | Granin (chromogranin or secretogranin) |
| HAGH_C | 3 | Hydroxyacylglutathione hydrolase C-terminus |
| HAP1_N | 3 | HAP1 N-terminal conserved region |
| HARE-HTH | 3 | HB1, ASXL, restriction endonuclease HTH domain |
| HDAC4_Gln | 3 | Glutamine rich N terminal domain of histone deacetylase 4 |
| HHH | 3 | Helix-hairpin-helix motif |
| HH_signal | 3 | Hedgehog amino-terminal signalling domain |
| HIF-1 | 3 | Hypoxia-inducible factor-1 |
| HIT | 3 | HIT domain |
| HMGL-like | 3 | HMGL-like |
| HNF-1_N | 3 | Hepatocyte nuclear factor 1 (HNF-1), N terminus |
| HOOK | 3 | HOOK protein coiled-coil region |
| HRDC | 3 | HRDC domain |
| HTH_48 | 3 | HTH domain in Mos1 transposase |
| HYR | 3 | HYR domain |
| HbrB | 3 | HbrB-like |
| HhH-GPD | 3 | HhH-GPD superfamily base excision DNA repair protein |
| Hint | 3 | Hint module |
| HisK-N-like | 3 | HisK-N-like globin domain of the ASK signalosome |
| Hmw_CFAP97 | 3 | Hemingway/CFA97 |
| Homez | 3 | Homeodomain leucine-zipper encoding, Homez |
| Hormone_3 | 3 | Pancreatic hormone peptide |
| ICAM1_3_5_D2 | 3 | ICAM-1/3/5, D2 domain |
| IER | 3 | Immediate early response protein (IER) |
| IF5A-like_N | 3 | Translation initiation factor 5A-like, N-terminal |
| IL17_R_N | 3 | Interleukin-17 receptor extracellular region |
| IPP-2 | 3 | Protein phosphatase inhibitor 2 (IPP-2) |
| IQ_SEC7_PH | 3 | PH domain |
| IRF-2BP1_2 | 3 | Interferon regulatory factor 2-binding protein zinc finger |
| IRK_N | 3 | Inward rectifier potassium channel N-terminal |
| ITGAX-like_Ig_3 | 3 | Integrin alpha-X-like, Ig-like domain 3 |
| Ig_C17orf99 | 3 | C17orf99 Ig domain |
| Iml2-TPR_39 | 3 | Iml2/Tetratricopeptide repeat protein 39 |
| Innexin | 3 | Innexin |
| Integrase_H2C2 | 3 | Integrase zinc binding domain |
| Intu_longin_2 | 3 | Intu longin-like domain 2 |
| JAKMIP_CC3 | 3 | JAKMIP CC3 domain |
| Jun | 3 | Jun-like transcription factor |
| KDM6_C-hel | 3 | Lysine-specific demethylase 6/UTY, C-terminal helical domain |
| KDM6_GATAL | 3 | KDM6, GATA-like |
| KH_9 | 3 | FMRP KH0 domain |
| KPBB_C | 3 | Phosphorylase b kinase C-terminal domain |
| KRAP_IP3R_bind | 3 | Ki-ras-induced actin-interacting protein-IP3R-interacting domain |
| Kelch_2 | 3 | Kelch motif |
| Keratin_matx | 3 | Keratin, high-sulphur matrix protein |
| Kindlin_2_N | 3 | Kindlin-2 N-terminal domain |
| L51_S25_CI-B8 | 3 | Mitochondrial ribosomal protein L51 / S25 / CI-B8 domain |
| LAMININ_IV_B | 3 | Laminin IV type B domain |
| LANC_like | 3 | Lanthionine synthetase C-like protein |
| LETM1_RBD | 3 | LETM1-like, RBD |
| LNS2 | 3 | LNS2 (Lipin/Ned1/Smp2) |
| LRRCT_2 | 3 | Leucine rich repeat C-terminal motif |
| LSM14 | 3 | Scd6-like Sm domain |
| LSR | 3 | Lipolysis stimulated receptor (LSR) |
| LURAP | 3 | Leucine rich adaptor protein |
| Latrophilin | 3 | Latrophilin Cytoplasmic C-terminal region |
| Lbh | 3 | Cardiac transcription factor regulator, Developmental protein |
| Lectin_N | 3 | Hepatic lectin, N-terminal domain |
| Lipase_GDSL | 3 | GDSL-like Lipase/Acylhydrolase |
| Lipin_N | 3 | lipin, N-terminal conserved region |
| Lipin_mid | 3 | Lipin/Ned1/Smp2 multi-domain protein middle domain |
| LisH_TPL | 3 | LisH-like dimerisation domain |
| Lyase_1 | 3 | Lyase |
| M-inducer_phosp | 3 | M-phase inducer phosphatase |
| M60-like_N | 3 | N-terminal domain of M60-like peptidases |
| MAP3K_TRAF_bd | 3 | MAP3K TRAFs-binding domain |
| MCLN_ECD | 3 | Mucolipin, extracytosolic domain |
| MDFI | 3 | MyoD family inhibitor |
| MGC-24 | 3 | Multi-glycosylated core protein 24 (MGC-24), sialomucin |
| MGS | 3 | MGS-like domain |
| MIF | 3 | Macrophage migration inhibitory factor (MIF) |
| MINDY-3_4_CD | 3 | Deubiquitinating enzyme MINDY-3/4, conserved domain |
| MITF_TFEB_C_3_N | 3 | MITF/TFEB/TFEC/TFE3 N-terminus |
| MMS1_N | 3 | Mono-functional DNA-alkylating methyl methanesulfonate N-term |
| MOSC | 3 | MOSC domain |
| MOSC_N | 3 | MOSC N-terminal beta barrel domain |
| MPC | 3 | Mitochondrial pyruvate carriers |
| MRFAP1 | 3 | MORF4 family-associated protein1 |
| MRG | 3 | MRG |
| MT-A70 | 3 | MT-A70 |
| MTA_R1 | 3 | MTA R1 domain |
| MUM1-like_PWWP | 3 | MUM1-like, PWWP domain |
| MYT1 | 3 | Myelin transcription factor 1 |
| Malic_M | 3 | Malic enzyme, NAD binding domain |
| MamL-1 | 3 | MamL-1 domain |
| Methyltransf_32 | 3 | Methyltransferase domain |
| MgtE | 3 | Divalent cation transporter |
| MitMem_reg | 3 | Maintenance of mitochondrial structure and function |
| Mito_fiss_reg | 3 | Mitochondrial fission regulator |
| Mtf2_C | 3 | Polycomb-like MTF2 factor 2 |
| Mtp | 3 | Golgi 4-transmembrane spanning transporter |
| MutS_I | 3 | MutS domain I |
| Myc_N | 3 | Myc amino-terminal region |
| Myelin_PLP | 3 | Myelin proteolipid protein (PLP or lipophilin) |
| NAD_binding_2 | 3 | NAD binding domain of 6-phosphogluconate dehydrogenase |
| NCOA_u2 | 3 | Unstructured region on nuclear receptor coactivator protein |
| NEXCaM_BD | 3 | Regulatory region of Na+/H+ exchanger NHE binds to calmodulin |
| NHR2 | 3 | NHR2 domain like |
| NKAP | 3 | NF-kappa-B-activating protein |
| NMDAR2_C | 3 | N-methyl D-aspartate receptor 2B3 C-terminus |
| NNMT_PNMT_TEMT | 3 | NNMT/PNMT/TEMT family |
| NOPS | 3 | NOPS (NUC059) domain |
| NO_synthase | 3 | Nitric oxide synthase, oxygenase domain |
| NTP_transferase | 3 | Nucleotidyl transferase |
| NYAP_C | 3 | Neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter |
| NYAP_N | 3 | Neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter |
| Na_Ca_ex_C | 3 | C-terminal extension of sodium/calcium exchanger domain |
| Na_Pi_cotrans | 3 | Na+/Pi-cotransporter |
| Neurensin | 3 | Neurensin/Transmembrane protein 74 |
| Noelin-1 | 3 | Neurogenesis glycoprotein |
| Nop | 3 | snoRNA binding domain, fibrillarin |
| Nuc_rec_co-act | 3 | Nuclear receptor coactivator |
| Nucleoplasmin | 3 | Nucleoplasmin/nucleophosmin domain |
| Nucleos_tra2_C | 3 | Na+ dependent nucleoside transporter C-terminus |
| Nucleos_tra2_N | 3 | Na+ dependent nucleoside transporter N-terminus |
| OB_Dis3 | 3 | Dis3-like cold-shock domain 2 (CSD2) |
| ODC_AZ | 3 | Ornithine decarboxylase antizyme |
| ORMDL | 3 | ORMDL family |
| OST-HTH | 3 | OST-HTH/LOTUS domain |
| OST3_OST6 | 3 | OST3 / OST6 family, transporter family |
| Opiods_neuropep | 3 | Vertebrate endogenous opioids neuropeptide |
| Orai-1 | 3 | Mediator of CRAC channel activity |
| Orn_Arg_deC_N | 3 | Pyridoxal-dependent decarboxylase, pyridoxal binding domain |
| Orn_DAP_Arg_deC | 3 | Pyridoxal-dependent decarboxylase, C-terminal sheet domain |
| Otopetrin | 3 | Otopetrin |
| OxoGdeHyase_C | 3 | 2-oxoglutarate dehydrogenase C-terminal |
| P4Ha_N | 3 | Prolyl 4-Hydroxylase alpha-subunit, N-terminal region |
| P53 | 3 | P53 DNA-binding domain |
| P53_tetramer | 3 | P53 tetramerisation motif |
| P5CR_dimer | 3 | Pyrroline-5-carboxylate reductase dimerisation |
| PA26 | 3 | PA26 p53-induced protein (sestrin) |
| PA28_C | 3 | Proteasome activator PA28, C-terminal |
| PA28_N | 3 | Proteasome activator PA28, N-terminal |
| PAH | 3 | Paired amphipathic helix repeat |
| PAP2_C | 3 | PAP2 superfamily C-terminal |
| PAP_NTPase | 3 | Poly(A) polymerase nucleotidyltransferase domain |
| PAP_RNA-bind | 3 | Poly(A) polymerase predicted RNA binding domain |
| PAP_central | 3 | Poly(A) polymerase central domain |
| PARP_reg | 3 | Poly(ADP-ribose) polymerase, regulatory domain |
| PBP | 3 | Phosphatidylethanolamine-binding protein |
| PCMT | 3 | Protein-L-isoaspartate(D-aspartate) O-methyltransferase (PCMT) |
| PDEase_I_N | 3 | 3'5'-cyclic nucleotide phosphodiesterase N-terminal |
| PDS5 | 3 | Sister chromatid cohesion protein PDS5 protein |
| PEHE | 3 | PEHE domain |
| PEX11 | 3 | Peroxisomal biogenesis factor 11 (PEX11) |
| PFK | 3 | Phosphofructokinase |
| PGC7_Stella | 3 | PGC7/Stella/Dppa3 domain |
| PHD_3 | 3 | PHD domain of transcriptional enhancer, Asx |
| PHD_4 | 3 | PHD-finger |
| PHM7_cyt | 3 | Cytosolic domain of 10TM putative phosphate transporter |
| PH_13 | 3 | Pleckstrin homology domain |
| PH_17 | 3 | PH domain |
| PI3K_p85B | 3 | PI3-kinase family, p85-binding domain |
| PID_2 | 3 | Phosphotyrosine interaction domain (PTB/PID) |
| PIH1 | 3 | PIH1 N-terminal domain |
| PIP49_N | 3 | N-term cysteine-rich ER, FAM69 |
| PKAT_KLD | 3 | PKAT, KLD domain |
| PKI | 3 | cAMP-dependent protein kinase inhibitor |
| PL48 | 3 | Filopodia upregulated, FAM65 |
| PLAC8 | 3 | PLAC8 family |
| PLC-beta_C | 3 | PLC-beta C terminal |
| PLDc | 3 | Phospholipase D Active site motif |
| PLDc_3 | 3 | PLD-like domain |
| POLO_box | 3 | POLO box duplicated region |
| PP2C_C | 3 | Protein serine/threonine phosphatase 2C, C-terminal domain |
| PP4R3 | 3 | Phosphatase 4 regulatory subunit 3 |
| PPP5 | 3 | PPP5 TPR repeat region |
| PPR_long | 3 | Pentacotripeptide-repeat region of PRORP |
| PPTA | 3 | Protein prenyltransferase alpha subunit repeat |
| PRA1 | 3 | PRA1 family protein |
| PRK | 3 | Phosphoribulokinase / Uridine kinase family |
| PRKCSH | 3 | Glucosidase II beta subunit-like protein |
| PRKG1_interact | 3 | cGMP-dependent protein kinase interacting domain |
| PROL5-SMR | 3 | Proline-rich submaxillary gland androgen-regulated family |
| PRT6_C | 3 | Proteolysis_6 C-terminal |
| PUA | 3 | PUA domain |
| PUB | 3 | PUB domain |
| PWP3A-B_C | 3 | PWWP domain-containing DNA repair factor 3A/B, C-terminal |
| PWP3A-B_N | 3 | PWWP domain-containing DNA repair factor 3A/B, N-terminal |
| ParA | 3 | NUBPL iron-transfer P-loop NTPase |
| Pax2_C | 3 | Paired-box protein 2 C terminal |
| Pellino_FHA | 3 | Pellino, FHA domain |
| Pellino_RING | 3 | Pellino, RING domain |
| Peptidase_C69 | 3 | Peptidase family C69 |
| Peptidase_C78 | 3 | Peptidase family C78 |
| Peptidase_M19 | 3 | Membrane dipeptidase (Peptidase family M19) |
| Peptidase_M3 | 3 | Peptidase family M3 |
| Peptidase_M41 | 3 | Peptidase family M41 |
| Peptidase_M60 | 3 | Peptidase M60, enhancin and enhancin-like |
| Peptidase_S10 | 3 | Serine carboxypeptidase |
| Peptidase_S28 | 3 | Serine carboxypeptidase S28 |
| Per3-like_PAS-A | 3 | Period circadian protein homolog 3-like, PAS-A domain |
| Period_C | 3 | Period protein 2/3C-terminal region |
| Pex2_Pex12 | 3 | Pex2 / Pex12 amino terminal region |
| PfkB | 3 | pfkB family carbohydrate kinase |
| Phe_ZIP | 3 | Phenylalanine zipper |
| Phosphorylase | 3 | Carbohydrate phosphorylase |
| Popeye | 3 | Popeye protein conserved region |
| Pre-PUA | 3 | Pre-PUA-like domain |
| Proton_antipo_M | 3 | Proton-conducting membrane transporter |
| Prp3_C | 3 | Small nuclear ribonucleoprotein Prp3, C-terminal domain |
| PseudoU_synth_1 | 3 | tRNA pseudouridine synthase |
| PurA | 3 | PurA ssDNA and RNA-binding protein |
| Qua1 | 3 | Qua1 domain |
| RALGAPB_N | 3 | RALGAPB N-terminal domain |
| RAMP | 3 | Receptor activity modifying family |
| RBR | 3 | RNA binding Region |
| RB_A | 3 | Retinoblastoma-associated protein A domain |
| RB_B | 3 | Retinoblastoma-associated protein B domain |
| RELT | 3 | Tumour necrosis factor receptor superfamily member 19 |
| RESP18 | 3 | RESP18 domain |
| REST_helical | 3 | Helical region in REST corepressor |
| RFX1_trans_act | 3 | RFX1 transcription activation region |
| RGM_C | 3 | Repulsive guidance molecule (RGM) C-terminus |
| RGM_N | 3 | Repulsive guidance molecule (RGM) N-terminus |
| RGS-like | 3 | Regulator of G protein signalling-like domain |
| RHIM | 3 | RIP homotypic interaction motif |
| RIG-I_C | 3 | RIG-I receptor C-terminal domain |
| RIG-I_C-RD | 3 | C-terminal domain of RIG-I |
| RILP | 3 | Rab interacting lysosomal protein |
| RIO1 | 3 | RIO1 family |
| RMD1-3 | 3 | Regulator of microtubule dynamics protein 1-3 |
| RMMBL | 3 | Zn-dependent metallo-hydrolase RNA specificity domain |
| RNA_pol_Rpb1_1 | 3 | RNA polymerase Rpb1, domain 1 |
| RNA_pol_Rpb1_2 | 3 | RNA polymerase Rpb1, domain 2 |
| RNA_pol_Rpb1_3 | 3 | RNA polymerase Rpb1, domain 3 |
| RNA_pol_Rpb1_4 | 3 | RNA polymerase Rpb1, domain 4 |
| RNA_pol_Rpb1_5 | 3 | RNA polymerase Rpb1, domain 5 |
| RNA_pol_Rpb2_1 | 3 | RNA polymerase beta subunit |
| RNA_pol_Rpb2_2 | 3 | RNA polymerase Rpb2, domain 2 |
| RNA_pol_Rpb2_3 | 3 | RNA polymerase Rpb2, domain 3 |
| RNA_pol_Rpb2_6 | 3 | RNA polymerase Rpb2, domain 6 |
| RNA_pol_Rpb2_7 | 3 | RNA polymerase Rpb2, domain 7 |
| RPAP3_C | 3 | Potential Monad-binding region of RPAP3 |
| RPE65 | 3 | Retinal pigment epithelial membrane protein |
| RR_TM4-6 | 3 | Ryanodine Receptor TM 4-6 |
| RS4NT | 3 | RS4NT (NUC023) domain |
| RSN1_7TM | 3 | Calcium-dependent channel, 7TM region, putative phosphate |
| RSN1_TM | 3 | Late exocytosis, associated with Golgi transport |
| RYDR_Jsol | 3 | Ryanodine receptor junctional solenoid repeat |
| Rad17 | 3 | Rad17 P-loop domain |
| Rad21_Rec8 | 3 | Conserved region of Rad21 / Rec8 like protein |
| Rad21_Rec8_N | 3 | N terminus of Rad21 / Rec8 like protein |
| RasGAP_C | 3 | RasGAP C-terminus |
| Reeler | 3 | Reeler domain |
| Regnase_1_C | 3 | Endoribonuclease Regnase 1/ ZC3H12 C-terminal domain |
| Rep_fac_C | 3 | Replication factor C C-terminal domain |
| Rho_GDI | 3 | RHO protein GDP dissociation inhibitor |
| Ribonuc_2-5A | 3 | Ribonuclease 2-5A |
| Ribosomal_60s | 3 | 60s Acidic ribosomal protein |
| Ribosomal_L1 | 3 | Ribosomal protein L1p/L10e family |
| Ribosomal_L10 | 3 | Ribosomal protein L10 |
| Ribosomal_L16 | 3 | Ribosomal protein L16p/L10e |
| Ribosomal_L22 | 3 | Ribosomal protein L22p/L17e |
| Ribosomal_L3 | 3 | Ribosomal protein L3 |
| Ribosomal_L30 | 3 | Ribosomal protein L30p/L7e |
| Ribosomal_L44 | 3 | Ribosomal protein L44 |
| Ribosomal_S10 | 3 | Ribosomal protein S10p/S20e |
| Ribosomal_S18 | 3 | Ribosomal protein S18 |
| Ribosomal_S2 | 3 | Ribosomal protein S2 |
| Ribosomal_S4e | 3 | Ribosomal family S4e |
| Rieske | 3 | Rieske [2Fe-2S] domain |
| Ripply | 3 | Transcription Regulator |
| Robl_LC7 | 3 | Roadblock/LC7 domain |
| Rootletin | 3 | Ciliary rootlet component, centrosome cohesion |
| RrnaAD | 3 | Ribosomal RNA adenine dimethylase |
| Rrp44_CSD1 | 3 | Rrp44-like cold shock domain |
| Rubis-subs-bind | 3 | Rubisco LSMT substrate-binding |
| Runt | 3 | Runt domain |
| RunxI | 3 | Runx inhibition domain |
| RyR | 3 | RyR domain |
| S-methyl_trans | 3 | Homocysteine S-methyltransferase |
| S1-like | 3 | S1-like |
| S10_plectin | 3 | Plectin/S10 domain |
| SAC3_GANP | 3 | SAC3/GANP family |
| SAPS | 3 | SIT4 phosphatase-associated protein |
| SCD | 3 | Stromalin conservative domain |
| SHS2_Rpb7-N | 3 | SHS2 domain found in N terminus of Rpb7p/Rpc25p/MJ0397 |
| SIT | 3 | SHP2-interacting transmembrane adaptor protein, SIT |
| SLC35F | 3 | Solute carrier family 35 |
| SLC52_ribofla_tr | 3 | Solute carrier family 52, riboflavin transporter |
| SLFN-g3_helicase | 3 | Schlafen group 3, DNA/RNA helicase domain |
| SMAP | 3 | Small acidic protein family |
| SMIM5_18_22 | 3 | Small integral membrane protein 5/18/22 |
| SMN_Tudor | 3 | Survival motor neuron protein (SMN), Tudor domain |
| SMP_LBD | 3 | Synaptotagmin-like mitochondrial-lipid-binding domain |
| SNAP | 3 | Soluble NSF attachment protein, SNAP |
| SOGA | 3 | Protein SOGA |
| SOGA1-2-like_CC | 3 | SOGA 1/2-like, coiled-coil |
| SPAR_C | 3 | C-terminal domain of SPAR protein |
| SPIDER | 3 | SLy Proteins Associated Disordered Region |
| SRC-1 | 3 | Steroid receptor coactivator |
| SR_plectin_7 | 3 | Spectrin repeat |
| SSDP | 3 | Single-stranded DNA binding protein, SSDP |
| SSXT | 3 | SSXT protein (N-terminal region) |
| STAC2_u1 | 3 | Unstructured on SH3 and cysteine-rich domain-containing protein 2 |
| STAG | 3 | STAG domain |
| STPPase_N | 3 | Serine-threonine protein phosphatase N-terminal domain |
| SUZ-C | 3 | SUZ-C motif |
| Sam68-YY | 3 | Tyrosine-rich domain of Sam68 |
| SapA | 3 | Saposin A-type domain |
| SapB_1 | 3 | Saposin-like type B, region 1 |
| Sas10_Utp3 | 3 | Sas10/Utp3/C1D family |
| Sds3 | 3 | Sds3-like |
| Sec16_C | 3 | Sec23-binding domain of Sec16 |
| Sec3-PIP2_bind | 3 | Exocyst complex component SEC3 N-terminal PIP2 binding PH |
| Sedlin_N | 3 | Sedlin, N-terminal conserved region |
| SelR | 3 | SelR domain |
| Serine_rich | 3 | Serine rich protein interaction domain |
| Shal-type | 3 | Shal-type voltage-gated potassium channels, N-terminal |
| Sina_TRAF | 3 | Sina, TRAF-like domain |
| Sod_Cu | 3 | Copper/zinc superoxide dismutase (SODC) |
| Somatostatin | 3 | Somatostatin/Cortistatin family |
| Sorb | 3 | Sorbin homologous domain |
| Sox_N | 3 | Sox developmental protein N terminal |
| SpaA | 3 | Prealbumin-like fold domain |
| Spem1 | 3 | Spermatid maturation protein 1 |
| Speriolin_N | 3 | Speriolin N terminus |
| Spermine_synth | 3 | Spermine/spermidine synthase domain |
| SpoU_methylase | 3 | SpoU rRNA Methylase family |
| Spt20_SEP | 3 | Spt20, SEP domain |
| Spt46 | 3 | Spermatogenesis-associated protein 46 |
| Ssl1 | 3 | Ssl1-like |
| Striatin | 3 | Striatin family |
| Sulfotransfer_3 | 3 | Sulfotransferase family |
| Synapsin | 3 | Synapsin, N-terminal domain |
| Synapsin_C | 3 | Synapsin, ATP binding domain |
| Synapsin_N | 3 | Synapsin N-terminal |
| Syntaxin_2 | 3 | Syntaxin-like protein |
| Synuclein | 3 | Synuclein |
| T4_deiodinase | 3 | Iodothyronine deiodinase |
| TACC_C | 3 | Transforming acidic coiled-coil-containing protein (TACC), C-terminal |
| TAP_C | 3 | TAP C-terminal domain |
| TBCC | 3 | Tubulin binding cofactor C |
| TBD | 3 | TBD domain |
| TBP | 3 | Transcription factor TFIID (or TATA-binding protein, TBP) |
| TCL1_MTCP1 | 3 | TCL1/MTCP1 family |
| TCTN_DUF1619 | 3 | Tectonic domain DUF1619 |
| TEX13_ZnF | 3 | Putative testis-expressed protein 13C, zinc finger domain |
| TFIIA | 3 | Transcription factor IIA, alpha/beta subunit |
| TFIIB | 3 | Transcription factor TFIIB repeat |
| TF_Otx | 3 | Otx1 transcription factor |
| THRAP3_BCLAF1 | 3 | THRAP3/BCLAF1 family |
| THUMP | 3 | THUMP domain |
| TILa | 3 | TILa domain |
| TMEM106 | 3 | TM106 protein C-terminal domain |
| TMEM106_N | 3 | Transmembrane protein 106 N-terminal region |
| TMEM70 | 3 | TMEM70/TMEM186/TMEM223 protein family |
| TM_ErbB1 | 3 | Epidermal growth factor receptor transmembrane-juxtamembrane segment |
| TNRC6-PABC_bdg | 3 | TNRC6-PABC binding domain |
| TORC_C | 3 | Transducer of regulated CREB activity, C terminus |
| TORC_M | 3 | Transducer of regulated CREB activity middle domain |
| TORC_N | 3 | Transducer of regulated CREB activity, N terminus |
| TSNAXIP1_N | 3 | Translin-associated factor X-interacting N-terminus |
| TatD_DNase | 3 | TatD related DNase |
| Taxilin | 3 | Myosin-like coiled-coil protein |
| Tet_JBP | 3 | Oxygenase domain of the 2OGFeDO superfamily |
| Tmemb_14 | 3 | Transmembrane proteins 14C |
| Tmemb_185A | 3 | Transmembrane Fragile-X-F protein |
| Tom37 | 3 | Outer mitochondrial membrane transport complex protein |
| Toxin_TOLIP | 3 | Snake toxin and toxin-like protein |
| TraB_PrgY_gumN | 3 | TraB/PrgY/gumN family |
| Transferrin | 3 | Transferrin |
| Transketolase_N | 3 | Transketolase, thiamine diphosphate binding domain |
| Transmemb_17 | 3 | Predicted membrane protein |
| TruB_N | 3 | TruB family pseudouridylate synthase (N terminal domain) |
| Tub_N | 3 | Tubby N-terminal |
| Tubulin-binding | 3 | Tau and MAP protein, tubulin-binding repeat |
| Tudor_FRX1 | 3 | Fragile X messenger ribonucleoprotein 1, Tudor domain |
| Tweety | 3 | Tweety |
| Tyrosinase | 3 | Common central domain of tyrosinase |
| UBA_E1_SCCH | 3 | Ubiquitin-activating enzyme, SCCH domain |
| UCH_N | 3 | N-terminal of ubiquitin carboxyl-terminal hydrolase 37 |
| UDG | 3 | Uracil DNA glycosylase superfamily |
| UDPGP | 3 | UTP--glucose-1-phosphate uridylyltransferase |
| UNC-93 | 3 | Ion channel regulatory protein UNC-93 |
| UPA_2 | 3 | UPA domain |
| UPF0020 | 3 | RMKL-like, methyltransferase domain |
| USP7_C2 | 3 | Ubiquitin-specific protease C-terminal |
| USP8_dimer | 3 | USP8 dimerisation domain |
| UbiA | 3 | UbiA prenyltransferase family |
| Ubiquitin_2 | 3 | Ubiquitin-like domain |
| Utp12 | 3 | Dip2/Utp12 Family |
| V-ATPase_G | 3 | Vacuolar (H+)-ATPase G subunit |
| VASP_tetra | 3 | VASP tetramerisation domain |
| VASt | 3 | VAD1 Analog of StAR-related lipid transfer domain |
| VEGFR-2_TMD | 3 | VEGFR-2 Transmembrane domain |
| VPS13_C | 3 | Intermembrane lipid transfer protein VPS13, C-terminal |
| VPS13_ext_chorein | 3 | Vacuolar sorting-associated protein 13, extended-chorein |
| VPS13_mid_rpt | 3 | VPS13, central RBG modules |
| VPS51_Exo84_N | 3 | Vps51/EXO84/COG1 N-terminal |
| Vanin_C | 3 | Vanin C-terminal domain |
| Vg_Tdu | 3 | Vestigial/Tondu family |
| Vps26 | 3 | Vacuolar protein sorting-associated protein 26 |
| WBP-1 | 3 | WW domain-binding protein 1 |
| WGR | 3 | WGR domain |
| XMAP215_CLASP_TOG | 3 | XMAP215/Dis1/CLASP, TOG domain |
| XTBD | 3 | XRN-Two Binding Domain, XTBD |
| Xan_ur_permease | 3 | Permease family |
| YjeF_N | 3 | YjeF-related protein N-terminus |
| Zfx_Zfy_act | 3 | Zfx / Zfy transcription activation region |
| cwf21 | 3 | cwf21 domain |
| eIF-1a | 3 | Translation initiation factor 1A / IF-1 |
| eIF-5a | 3 | Eukaryotic elongation factor 5A hypusine, DNA-binding OB fold |
| eIF_4EBP | 3 | Eukaryotic translation initiation factor 4E binding protein (EIF4EBP) |
| fn1 | 3 | Fibronectin type I domain |
| hSH3 | 3 | Helically-extended SH3 domain |
| hSac2 | 3 | Inositol phosphatase |
| malic | 3 | Malic enzyme, N-terminal domain |
| muHD | 3 | Muniscin C-terminal mu homology domain |
| p25-alpha | 3 | p25-alpha |
| pKID | 3 | pKID domain |
| tRNA-synt_1c | 3 | tRNA synthetases class I (E and Q), catalytic domain |
| tRNA-synt_His | 3 | Histidyl-tRNA synthetase |
| tRNA_m1G_MT | 3 | tRNA (Guanine-1)-methyltransferase |
| zf-CDGSH | 3 | Iron-binding zinc finger CDGSH type |
| zf-DBF | 3 | DBF zinc finger |
| zf-LITAF-like | 3 | LITAF-like zinc ribbon domain |
| zf-RNPHF | 3 | RNPHF zinc finger |
| zf-U1 | 3 | U1 zinc finger |
| zf-nanos | 3 | Nanos RNA binding domain |
| zf_C2H2_ZHX | 3 | Zinc-fingers and homeoboxes C2H2 finger domain |
| zf_CCCH_4 | 3 | Zinc finger domain |
| 2-oxogl_dehyd_N | 2 | 2-oxoglutarate dehydrogenase N-terminus |
| 40S_SA_C | 2 | 40S ribosomal protein SA C-terminus |
| 60KD_IMP | 2 | 60Kd inner membrane protein |
| AAA_16 | 2 | AAA ATPase domain |
| AAA_2 | 2 | AAA domain (Cdc48 subfamily) |
| AAA_23 | 2 | AAA domain |
| AAA_34 | 2 | P-loop containing NTP hydrolase pore-1 |
| AAA_5 | 2 | AAA domain (dynein-related subfamily) |
| AAA_lid_10 | 2 | AAA lid domain |
| AARP2CN | 2 | AARP2CN (NUC121) domain |
| AA_kinase | 2 | Amino acid kinase family |
| ABA_GPCR | 2 | Abscisic acid G-protein coupled receptor |
| ACAD9-ACADV_C | 2 | ACAD9/ACADV, C-terminal domain |
| ACCA_BT | 2 | Acetyl-CoA carboxylase, BT domain |
| ACC_central | 2 | Acetyl-CoA carboxylase, central region |
| ACT_7 | 2 | ACT domain |
| AChE_tetra | 2 | Acetylcholinesterase tetramerisation domain |
| ADH_N_2 | 2 | N-terminal domain of oxidoreductase |
| ADNP_N | 2 | Activity-dependent neuroprotector homeobox protein N-terminal |
| AHD | 2 | ANC1 homology domain (AHD) |
| AIF-MLS | 2 | Mitochondria Localisation Sequence |
| AIP3 | 2 | Actin interacting protein 3 |
| AJAP1_PANP_C | 2 | AJAP1/PANP C-terminus |
| AKAP7_RIRII_bdg | 2 | PKA-RI-RII subunit binding domain of A-kinase anchor protein |
| AKNA | 2 | AT-hook-containing transcription factor |
| ALIX_LYPXL_bnd | 2 | ALIX V-shaped domain binding to HIV |
| ALKL1_2 | 2 | ALK and LTK ligand 1/2 |
| AMMECR1 | 2 | AMMECR1 |
| AMPK1_CBM | 2 | Glycogen recognition site of AMP-activated protein kinase |
| AMPKBI | 2 | 5'-AMP-activated protein kinase beta subunit, interaction domain |
| AMPK_alpha_AID | 2 | AMP-activated protein kinase, alpha subunit, autoinhibitory domain |
| AMP_N | 2 | Aminopeptidase P, N-terminal domain |
| AP3B1_C | 2 | Clathrin-adaptor complex-3 beta-1 subunit C-terminal |
| APCDDC | 2 | Adenomatosis polyposis coli down-regulated 1 |
| APC_N_CC | 2 | Coiled-coil N-terminus of APC, dimerisation domain |
| APC_basic | 2 | APC basic domain |
| APC_r | 2 | APC repeat |
| APC_rep | 2 | Adenomatous polyposis coli (APC) repeat |
| APG6 | 2 | Apg6 BARA domain |
| APG6_N | 2 | Apg6 coiled-coil region |
| APOC4 | 2 | Apolipoprotein C4 |
| APS_kinase | 2 | Adenylylsulphate kinase |
| ARF7EP_C | 2 | ARF7 effector protein C-terminus |
| ARHGEF5_35 | 2 | RhoGEF 5/35 N-terminal disordered region |
| ARL17 | 2 | ADP-ribosylation factor-like protein 17 |
| ARL2_Bind_BART | 2 | The ARF-like 2 binding protein BART |
| ARMET_C | 2 | ARMET, C-terminal |
| ARMET_N | 2 | ARMET, N-terminal |
| ARMT1-like_dom | 2 | Damage-control phosphatase ARMT1-like domain |
| ARPC4 | 2 | ARP2/3 complex 20 kDa subunit (ARPC4) |
| ASF1_hist_chap | 2 | ASF1 like histone chaperone |
| ASPP2-like_RA | 2 | Apoptosis-stimulating of p53 protein 2-like, N-terminal RA domain |
| ASTN1_2_fn3 | 2 | ASTN1/2 Fn3 domain |
| ASTN_1_2_N | 2 | Astrotactin 1/2 N-terminal |
| ATAD4 | 2 | ATPase family AAA domain containing 4 |
| ATF7IP_BD | 2 | ATF-interacting protein binding domain |
| ATG1-like_MIT1 | 2 | Atg1-like, MIT domain 1 |
| ATG1-like_MIT2 | 2 | ATG1-like, MIT domain 2 |
| ATG14 | 2 | Vacuolar sorting 38 and autophagy-related subunit 14 |
| ATG16 | 2 | Autophagy protein 16 (ATG16) |
| ATG9 | 2 | Autophagy protein ATG9 |
| ATP-grasp_2 | 2 | ATP-grasp domain |
| ATP-sulfurylase | 2 | ATP-sulfurylase |
| ATP-synt_G | 2 | Mitochondrial ATP synthase g subunit |
| ATP_synt_H | 2 | ATP synthase subunit H |
| ATXN-1_C | 2 | Ataxin-1 like family |
| AXIN1_TNKS_BD | 2 | Axin-1 tankyrase binding domain |
| AbfB | 2 | Alpha-L-arabinofuranosidase B (ABFB) domain |
| Abhydro_lipase | 2 | Partial alpha/beta-hydrolase lipase region |
| Abhydrolase_11 | 2 | Alpha/beta hydrolase domain |
| Acetyltransf_13 | 2 | ESCO1/2 acetyl-transferase |
| Acetyltransf_2 | 2 | N-acetyltransferase |
| Acid_PPase | 2 | Acid Phosphatase |
| Acyl-CoA_ox_N | 2 | Acyl-coenzyme A oxidase N-terminal |
| Acyl_transf_1 | 2 | Acyl transferase domain |
| Acylphosphatase | 2 | Acylphosphatase |
| AdenylateSensor | 2 | Adenylate sensor of SNF1-like protein kinase |
| Adenylsucc_synt | 2 | Adenylosuccinate synthetase |
| Agouti | 2 | Agouti protein |
| AlaDh_PNT_N | 2 | Alanine dehydrogenase/PNT, N-terminal domain |
| Alba | 2 | Alba |
| Ald_Xan_dh_C | 2 | Aldehyde oxidase and xanthine dehydrogenase, a/b hammerhead domain |
| Alg6_Alg8 | 2 | ALG6, ALG8 glycosyltransferase family |
| Alpha_L_fucos | 2 | Alpha-L-fucosidase |
| Alpha_adaptin_C | 2 | Alpha adaptin AP2, C-terminal domain |
| Amelogenin | 2 | Amelogenin |
| Aminotran_4 | 2 | Amino-transferase class IV |
| Annexin_2 | 2 | Annexin-like domain |
| Ant_C | 2 | Anthrax receptor C-terminus region |
| Anticodon_3 | 2 | Anticodon binding domain of methionyl tRNA ligase |
| Aph-1 | 2 | Aph-1 protein |
| Apo-CII | 2 | Apolipoprotein C-II |
| ApoO | 2 | Apolipoprotein O |
| Arm_APC_u3 | 2 | Armadillo-associated region on APC |
| Asn_synthase | 2 | Asparagine synthase |
| Asp-B-Hydro_N | 2 | Aspartyl beta-hydroxylase N-terminal region |
| AstE_AspA | 2 | Succinylglutamate desuccinylase / Aspartoacylase family |
| Atrophin-1 | 2 | Atrophin-1 family |
| Autophagy_act_C | 2 | Autophagocytosis associated protein, active-site domain |
| Avl9 | 2 | Transport protein Avl9 |
| Ax_dynein_light | 2 | Axonemal dynein light chain |
| Axin_b-cat_bind | 2 | Axin beta-catenin binding motif |
| B12-binding | 2 | B12 binding domain |
| BAF | 2 | Barrier to autointegration factor |
| BAF250_C | 2 | SWI/SNF-like complex subunit BAF250/Osa |
| BAHCC1-like_Tudor | 2 | BAHCC1-like, Tudor domain |
| BCL9 | 2 | B-cell lymphoma 9 protein |
| BCLP | 2 | Beta-casein like protein |
| BDV_P40 | 2 | Borna disease virus P40 protein |
| BMP2K_C | 2 | BMP-2-inducible protein kinase C-terminus |
| BNIP3 | 2 | BNIP3 |
| BRD4_CDT | 2 | C-terminal domain of bromodomain protein 4 |
| BRI3BP | 2 | Negative regulator of p53/TP53 |
| BSMAP | 2 | Brain specific membrane anchored protein |
| BTBD8_C | 2 | BTB/POZ domain-containing protein 8, C-terminal |
| BTB_3 | 2 | BTB/POZ domain |
| BTD | 2 | Beta-trefoil DNA-binding domain |
| BTHB | 2 | Basic tilted helix bundle domain |
| BURAN | 2 | BURAN domain |
| Bcr-Abl_Oligo | 2 | Bcr-Abl oncoprotein oligomerisation domain |
| Beta-TrCP_D | 2 | D domain of beta-TrCP |
| Beta-lactamase | 2 | Beta-lactamase |
| BicD | 2 | Microtubule-associated protein Bicaudal-D |
| Big_2 | 2 | Bacterial Ig-like domain (group 2) |
| Bin3 | 2 | Bicoid-interacting protein 3 (Bin3) |
| Bombesin | 2 | Bombesin-like peptide |
| Btz | 2 | CASC3/Barentsz eIF4AIII binding |
| C2-C2_1 | 2 | First C2 domain of RPGR-interacting protein 1 |
| CALCOCO1 | 2 | Calcium binding and coiled-coil domain (CALCOCO1) like |
| CANIN | 2 | Coiled-coil and Nse Interacting (CANIN) domain |
| CAP_C | 2 | Adenylate cyclase associated (CAP) C terminal |
| CAP_N-CM | 2 | CAP N-terminal conserved motif |
| CASTOR1_ACT-like | 2 | Cytosolic arginine sensor for mTORC1 subunit 1/2, ACT-like |
| CATSPERD_beta-prop | 2 | CATSPERD, beta-propeller domain |
| CBAH | 2 | Linear amide C-N hydrolases, choloylglycine hydrolase family |
| CBFNT | 2 | CBFNT (NUC161) domain |
| CBM_14 | 2 | Chitin binding Peritrophin-A domain |
| CC2D1A-B_DM14 | 2 | Coiled-coil and C2 domain-containing protein 1, DM14 domain |
| CC2D2AN-C2 | 2 | CC2D2A N-terminal C2 domain |
| CCDC28 | 2 | Coiled-coil domain-containing protein 28 |
| CCDC71L | 2 | Coiled-coil domain-containing protein 71L |
| CCDC74_C | 2 | Coiled coil protein 74, C terminal |
| CCDC90-like | 2 | Coiled-coil domain-containing protein 90-like |
| CCM2_C | 2 | Cerebral cavernous malformation protein, harmonin-homology |
| CDC37_M | 2 | Cdc37 Hsp90 binding domain |
| CDC48_2 | 2 | Cell division protein 48 (CDC48), domain 2 |
| CDC48_N | 2 | Cell division protein 48 (CDC48), N-terminal domain |
| CDC50 | 2 | LEM3 (ligand-effect modulator 3) family / CDC50 family |
| CDH1_2_SANT_HL1 | 2 | CDH1/2 SANT-Helical linker 1 |
| CDK2AP | 2 | Cyclin-dependent kinase 2-associated protein |
| CDK5_activator | 2 | Cyclin-dependent kinase 5 activator protein |
| CECR6_TMEM121 | 2 | CECR6/TMEM121 family |
| CENP-S | 2 | CENP-S protein |
| CEP170_C | 2 | CEP170 C-terminus |
| CEP63 | 2 | Centrosomal protein of 63 kDa |
| CERK_C | 2 | Ceramide kinase C-terminal domain |
| CFAP298 | 2 | Cilia- and flagella-associated protein 298 |
| CG-1 | 2 | CG-1 domain |
| CHD5 | 2 | CHD5-like protein |
| CHORD | 2 | CHORD |
| CHRDL_1_2_C | 2 | Chordin-like protein 1/2 C-terminal |
| CKAP2_C | 2 | Cytoskeleton-associated protein 2 C-terminus |
| CKS | 2 | Cyclin-dependent kinase regulatory subunit |
| CLP1_P | 2 | mRNA cleavage and polyadenylation factor CLP1 P-loop |
| CLPTM1 | 2 | Cleft lip and palate transmembrane protein 1 (CLPTM1) |
| CNK2_3_dom | 2 | Connector enhancer of kinase suppressor of ras 2/3 domain |
| COP-gamma_platf | 2 | Coatomer gamma subunit appendage platform subdomain |
| COR | 2 | C-terminal of Roc, COR, domain |
| COX16 | 2 | Cytochrome c oxidase assembly protein COX16 |
| COX4 | 2 | Cytochrome c oxidase subunit IV |
| COX6A | 2 | Cytochrome c oxidase subunit VIa |
| COX7B | 2 | Cytochrome C oxidase chain VIIB |
| COX8 | 2 | Cytochrome oxidase c subunit VIII |
| CO_deh_flav_C | 2 | CO dehydrogenase flavoprotein C-terminal domain |
| CPSase_L_D3 | 2 | Carbamoyl-phosphate synthetase large chain, oligomerisation domain |
| CPSase_sm_chain | 2 | Carbamoyl-phosphate synthase small chain, CPSase domain |
| CREPT | 2 | Cell-cycle alteration and expression-elevated protein in tumour |
| CRLF2-like_D2 | 2 | Cytokine receptor-like factor 2-like, D2 domain |
| CRLF2_D1 | 2 | Cytokine receptor-like factor 2-like, D1 domain |
| CRM1_C | 2 | CRM1 C terminal |
| CSN7a_helixI | 2 | COP9 signalosome complex subunit 7a helix I domain |
| CSTF2_hinge | 2 | Hinge domain of cleavage stimulation factor subunit 2 |
| CSTF_C | 2 | Transcription termination and cleavage factor C-terminal |
| CTP_synth_N | 2 | CTP synthase N-terminus |
| CTP_transf_1 | 2 | Cytidylyltransferase family |
| CUTL | 2 | CUT1-like DNA-binding domain of SATB |
| CXCXC | 2 | CXCXC repeat |
| CaM-KIIN | 2 | Calcium/calmodulin-dependent protein kinase II inhibitor |
| Caldesmon | 2 | Caldesmon |
| Calsequestrin | 2 | Calsequestrin |
| Caprin-1_C | 2 | Cytoplasmic activation/proliferation-associated protein-1 C term |
| Caprin-1_dimer | 2 | Caprin-1 dimerization domain |
| Caskin-Pro-rich | 2 | Proline rich region of Caskin proteins |
| Caskin-tail | 2 | C-terminal region of Caskin |
| Caskin1-CID | 2 | Caskin1 CASK-interaction domain |
| Cast | 2 | RIM-binding protein of the cytomatrix active zone |
| Castor1_N | 2 | Cytosolic arginine sensor for mTORC1 subunit 1 N-terminal domain |
| Cathelicidins | 2 | Cathelicidin |
| Cauli_VI | 2 | Caulimovirus viroplasmin |
| Cdc6_C | 2 | CDC6, C terminal winged helix domain |
| Cep57_CLD | 2 | Centrosome localisation domain of Cep57 |
| Cep57_MT_bd | 2 | Centrosome microtubule-binding domain of Cep57 |
| Ceramidse_alk_C | 2 | Neutral/alkaline non-lysosomal ceramidase, C-terminal |
| ChaC | 2 | ChaC-like protein |
| Chitin_synth_2 | 2 | Chitin synthase |
| Chromosome_seg | 2 | Chromosome segregation during meiosis |
| Churchill | 2 | Churchill protein |
| Citrate_synt | 2 | Citrate synthase, C-terminal domain |
| Clathrin-link | 2 | Clathrin, heavy-chain linker |
| Clathrin_bdg | 2 | Clathrin-binding box of Aftiphilin, vesicle trafficking |
| Clathrin_lg_ch | 2 | Clathrin light chain |
| Clathrin_propel | 2 | Clathrin propeller repeat |
| ClpB_D2-small | 2 | C-terminal, D2-small domain, of ClpB protein |
| ClpS | 2 | ATP-dependent Clp protease adaptor protein ClpS |
| Clusterin | 2 | Clusterin |
| Cmc1 | 2 | Cytochrome c oxidase biogenesis protein Cmc1 like |
| Cnd1 | 2 | non-SMC mitotic condensation complex subunit 1 |
| Cnn_1N | 2 | Centrosomin N-terminal motif 1 |
| CoA_binding | 2 | CoA binding domain |
| CoA_trans | 2 | Coenzyme A transferase |
| CoA_transf_3 | 2 | CoA-transferase family III |
| CoaE | 2 | Dephospho-CoA kinase |
| Coatomer_WDAD | 2 | Coatomer WD associated region |
| Coatomer_g_Cpla | 2 | Coatomer subunit gamma-1 C-terminal appendage platform |
| Cobl | 2 | Cordon-bleu ubiquitin-like domain |
| Collagen_trimer | 2 | Collagen trimerization domain |
| Collectrin | 2 | Renal amino acid transporter |
| Costars | 2 | Costars |
| Cpn10 | 2 | Chaperonin 10 Kd subunit |
| Creatinase_N | 2 | Creatinase/Prolidase N-terminal domain |
| Creatinase_N_2 | 2 | Creatinase/Prolidase N-terminal domain |
| Creb_binding | 2 | Creb binding |
| CtIP_N | 2 | Tumour-suppressor protein CtIP N-terminal domain |
| Ctr | 2 | Ctr copper transporter family |
| Cu-oxidase | 2 | Multicopper oxidase |
| CwfJ_C_1 | 2 | Protein similar to CwfJ C-terminus 1 |
| CwfJ_C_2 | 2 | Protein similar to CwfJ C-terminus 2 |
| Cyclin_C_2 | 2 | Cyclin C-terminal domain |
| Cyclin_N2 | 2 | Cyclin-A N-terminal APC/C binding region |
| Cylicin_N | 2 | Cylicin N-terminus |
| DAB2_SBM | 2 | Disabled homolog 2-like, sulfatide-binding motifs |
| DAP | 2 | Death-associated protein |
| DBB | 2 | Dof, BCAP, and BANK (DBB) motif, |
| DBC1 | 2 | DBC1 |
| DCP1 | 2 | Dcp1-like decapping family |
| DCR | 2 | Dppa2/4 conserved region |
| DDR1-2_DS-like | 2 | Discoidin domain-containing receptor 1/2, DS-like domain |
| DFRP_C | 2 | DRG Family Regulatory Proteins, Tma46 |
| DGCR6 | 2 | DiGeorge syndrome critical region 6 (DGCR6) protein |
| DHDPS | 2 | Dihydrodipicolinate synthetase family |
| DHFR_1 | 2 | Dihydrofolate reductase |
| DHHA2 | 2 | DHHA2 domain |
| DHO_dh | 2 | Dihydroorotate dehydrogenase |
| DIE2_ALG10 | 2 | DIE2/ALG10 family |
| DIM1 | 2 | Mitosis protein DIM1 |
| DM10_dom | 2 | DM10 domain |
| DMRT5_DMB | 2 | DMRT5, DMB domain |
| DNA_gyraseB | 2 | DNA gyrase B |
| DNA_photolyase | 2 | DNA photolyase |
| DNA_pol_A | 2 | DNA polymerase family A |
| DNA_primase_S | 2 | DNA primase small subunit |
| DNA_topoisoIV | 2 | DNA gyrase/topoisomerase IV, subunit A |
| DNase_II | 2 | Deoxyribonuclease II |
| DOR | 2 | DOR family |
| DRY_EERY | 2 | Alternative splicing regulator |
| DSHCT | 2 | DSHCT (NUC185) domain |
| DTHCT | 2 | DTHCT (NUC029) region |
| DUF1075 | 2 | Protein of unknown function (DUF1075) |
| DUF1088 | 2 | Neurobeachin-like, DUF1088 |
| DUF1151 | 2 | Protein of unknown function (DUF1151) |
| DUF1242 | 2 | Protein of unknown function (DUF1242) |
| DUF1681 | 2 | Protein of unknown function (DUF1681) |
| DUF1866 | 2 | Domain of unknown function (DUF1866) |
| DUF1897 | 2 | Domain of unknown function (DUF1897) |
| DUF2370 | 2 | Protein of unknown function (DUF2370) |
| DUF3350 | 2 | Domain of unknown function (DUF3350) |
| DUF3402 | 2 | Domain of unknown function (DUF3402) |
| DUF3481 | 2 | C-terminal domain of neuropilin glycoprotein |
| DUF3512 | 2 | Domain of unknown function (DUF3512) |
| DUF3591 | 2 | Protein of unknown function (DUF3591) |
| DUF3657 | 2 | Protein FAM135 |
| DUF3719 | 2 | Protein of unknown function (DUF3719) |
| DUF3740 | 2 | Sulfatase protein |
| DUF4201 | 2 | Domain of unknown function (DUF4201) |
| DUF4203 | 2 | Domain of unknown function (DUF4203) |
| DUF4210 | 2 | Domain of unknown function (DUF4210) |
| DUF4371 | 2 | Domain of unknown function (DUF4371) |
| DUF4430 | 2 | Domain of unknown function (DUF4430) |
| DUF4525 | 2 | Domain of unknown function (DUF4525) |
| DUF4533 | 2 | Protein of unknown function (DUF4533) |
| DUF4534 | 2 | Protein of unknown function (DUF4534) |
| DUF4537 | 2 | Domain of unknown function (DUF4537) |
| DUF4553 | 2 | Domain of unknown function (DUF4553) |
| DUF4587 | 2 | Domain of unknown function (DUF4587) |
| DUF4589 | 2 | Domain of unknown function (DUF4589) |
| DUF4594 | 2 | Domain of unknown function (DUF4594) |
| DUF4600 | 2 | Domain of unknown function (DUF4600) |
| DUF4605 | 2 | Domain of unknown function (DUF4605) |
| DUF4641 | 2 | Domain of unknown function (DUF4641) |
| DUF4659 | 2 | Domain of unknown function (DUF4659) |
| DUF4682 | 2 | Domain of unknown function (DUF4682) |
| DUF4685 | 2 | Domain of unknown function (DUF4685) |
| DUF4757 | 2 | Domain of unknown function (DUF4757) |
| DUF4795 | 2 | Domain of unknown function (DUF4795) |
| DUF4800 | 2 | Domain of unknown function (DUF4800) |
| DUF4927 | 2 | Domain of unknown function (DUF4927) |
| DUF498 | 2 | Protein of unknown function (DUF498/DUF598) |
| DUF5050 | 2 | Domain of unknown function (DUF5050) |
| DUF525 | 2 | ApaG domain |
| DUF5523 | 2 | Family of unknown function (DUF5523) |
| DUF5588 | 2 | Family of unknown function (DUF5588) |
| DUF5601 | 2 | Domain of unknown function (DUF5601) |
| DUF676 | 2 | Putative serine esterase (DUF676) |
| DUF719 | 2 | Protein of unknown function (DUF719) |
| Dimerisation2 | 2 | Dimerisation domain |
| Diphthamide_syn | 2 | Putative diphthamide synthesis protein |
| Dopey_N | 2 | Dopey, N-terminal |
| Dpp_8_9_N | 2 | Dipeptidyl peptidase 8 and 9 N-terminal |
| Dppa2_A | 2 | Dppa2/4 conserved region in higher vertebrates |
| DuoxA | 2 | Dual oxidase maturation factor |
| Dynein_IC2 | 2 | Cytoplasmic dynein 1 intermediate chain 2 |
| Dzip-like_N | 2 | Iguana/Dzip1-like DAZ-interacting protein N-terminal |
| EABR | 2 | TSG101 and ALIX binding domain of CEP55 |
| EAF | 2 | RNA polymerase II transcription elongation factor |
| EAP30 | 2 | EAP30/Vps36 family |
| EBP50_C | 2 | EBP50, C-terminal |
| ECR1_N | 2 | Exosome complex exonuclease RRP4 N-terminal region |
| EF-1_beta_acid | 2 | Eukaryotic elongation factor 1 beta central acidic region |
| EF-hand_14 | 2 | EF-hand domain |
| EF-hand_9 | 2 | EF-hand domain |
| EF-hand_Zip | 2 | Zip protein EF-hand |
| EF1_GNE | 2 | EF-1 guanine nucleotide exchange domain |
| EFR3_ARM | 2 | EFR3, armadillo repeat |
| EF_assoc_1 | 2 | EF hand associated |
| EF_assoc_2 | 2 | EF hand associated |
| EHMT1-2_CRR | 2 | Histone-lysine N-methyltransferase EHMT1/EHMT2, Cys-rich region |
| ELFV_dehydrog | 2 | Glutamate/Leucine/Phenylalanine/Valine dehydrogenase |
| ELFV_dehydrog_N | 2 | Glu/Leu/Phe/Val dehydrogenase, dimerisation domain |
| EMC3_TMCO1 | 2 | Integral membrane protein EMC3/TMCO1-like |
| EPCAM-Trop-2_C | 2 | EPCAM/Trop-2, C-terminal |
| EPOP | 2 | Elongin BC and Polycomb repressive complex 2-associated protein |
| EPO_TPO | 2 | Erythropoietin/thrombopoietin |
| ERO1 | 2 | Endoplasmic Reticulum Oxidoreductin 1 (ERO1) |
| ETF | 2 | Electron transfer flavoprotein domain |
| EXOC6_Sec15_N | 2 | Exocyst complex component EXOC6/Sec15, N-terminal |
| EZH2_WD-Binding | 2 | WD repeat binding protein EZH2 |
| E_Pc_C | 2 | Enhancer of Polycomb C-terminus |
| Elongin_A | 2 | RNA polymerase II transcription factor SIII (Elongin) subunit A |
| Endostatin | 2 | Collagenase NC10 and Endostatin |
| Endosulfine | 2 | cAMP-regulated phosphoprotein/endosulfine conserved region |
| Engrail_1_C_sig | 2 | Engrailed homeobox C-terminal signature domain |
| Enkurin | 2 | Calmodulin-binding |
| EpCAM_N | 2 | Epithelial cell adhesion molecule, N-terminal domain |
| Epiglycanin_C | 2 | Mucin, cleavage site, TM and cytoplasmic tail region |
| Ets1_N_flank | 2 | Ets1 N-terminal flanking region of Ets domain |
| Ezh2_MCSS | 2 | Ezh2, MCSS domain |
| F-box_4 | 2 | F-box |
| FAD_SOX | 2 | Flavin adenine dinucleotide (FAD)-dependent sulfhydryl oxidase |
| FAD_binding_5 | 2 | FAD binding domain in molybdopterin dehydrogenase |
| FAD_binding_7 | 2 | FAD binding domain of DNA photolyase |
| FAF1 | 2 | Fas-associated factor 1 |
| FAM101 | 2 | FAM101 family |
| FAM104 | 2 | Family 104 |
| FAM124 | 2 | FAM124 domain |
| FAM153 | 2 | FAM153 family |
| FAM161A_B | 2 | Protein FAM161A/B |
| FAM163 | 2 | FAM163 family |
| FAM167 | 2 | FAM167 |
| FAM177 | 2 | FAM177 family |
| FAM180 | 2 | FAM180 family |
| FAM184 | 2 | Family with sequence similarity 184, A and B |
| FAM186A-B_C | 2 | FAM186A/B, C-terminal |
| FAM186A-B_N | 2 | FAM186A/B, N-terminal |
| FAM193_C | 2 | FAM193 family C-terminal |
| FAM195 | 2 | FAM195 family |
| FAM196 | 2 | FAM196 family |
| FAM198 | 2 | FAM198 protein |
| FAM209 | 2 | FAM209 family |
| FAM210A-B_dom | 2 | FAM210A/B-like domain |
| FAM212 | 2 | FAM212 family |
| FAM216B | 2 | FAM216B protein family |
| FAM217 | 2 | FAM217 family |
| FAM219A | 2 | Protein family FAM219A |
| FAM221 | 2 | Protein FAM221A/B |
| FAM222A | 2 | Protein family of FAM222A |
| FAM24 | 2 | FAM24 family |
| FAM70 | 2 | FAM70 protein |
| FAM76 | 2 | FAM76 protein |
| FAM92 | 2 | FAM92 protein |
| FBPase | 2 | Fructose-1-6-bisphosphatase, N-terminal domain |
| FBPase_C | 2 | Fructose-1-6-bisphosphatase, C-terminal domain |
| FDX-ACB | 2 | Ferredoxin-fold anticodon binding domain |
| FERM_N_2 | 2 | FERM N-terminal domain |
| FEZ | 2 | FEZ-like protein |
| FG-GAP_3 | 2 | FG-GAP-like repeat |
| FGE-sulfatase | 2 | Sulfatase-modifying factor enzyme 1 |
| FIIND | 2 | Function to find |
| FISNA | 2 | Fish-specific NACHT associated domain |
| FIT | 2 | Fat storage-inducing transmembrane protein |
| FLYWCH_N | 2 | FLYWCH-type zinc finger-containing protein |
| FMN_dh | 2 | FMN-dependent dehydrogenase |
| FNIP_C | 2 | Folliculin-interacting protein C-terminus |
| FNIP_M | 2 | Folliculin-interacting protein middle domain |
| FNIP_N | 2 | Folliculin-interacting protein N-terminus |
| FOG1-like_PR | 2 | Zinc finger protein ZFPM1-like, PR domain |
| FOP_dimer | 2 | FOP N terminal dimerisation domain |
| FST_N | 2 | Follistatin, N-terminal |
| FTCD_N | 2 | Formiminotransferase domain, N-terminal subdomain |
| FTHFS | 2 | Formate--tetrahydrofolate ligase |
| FUN14 | 2 | FUN14 family |
| FWWh | 2 | Protein of unknown function |
| FXR_C1 | 2 | Fragile X-related 1 protein C-terminal region 2 |
| FYVE_CARP1-2 | 2 | E3 ubiquitin-protein ligase CARP1/2, FYVE/PHD zinc finger |
| F_actin_bind | 2 | F-actin binding |
| Far-17a_AIG1 | 2 | FAR-17a/AIG1-like protein |
| Fcf1 | 2 | Fcf1 |
| Fe-S_biosyn | 2 | Iron-sulphur cluster biosynthesis |
| FeS_assembly_P | 2 | Iron-sulfur cluster assembly protein |
| Fe_hyd_SSU | 2 | Iron hydrogenase small subunit |
| Fe_hyd_lg_C | 2 | Iron only hydrogenase large subunit, C-terminal domain |
| Fer2_2 | 2 | [2Fe-2S] binding domain |
| Fer4 | 2 | 4Fe-4S binding domain |
| Fer4_12 | 2 | 4Fe-4S single cluster domain |
| Fer4_24 | 2 | Ferredoxin I 4Fe-4S cluster domain |
| Fes1 | 2 | Nucleotide exchange factor Fes1 |
| Fibrillarin | 2 | Fibrillarin |
| Flavodoxin_2 | 2 | Flavodoxin-like fold |
| Flot | 2 | Flotillin |
| FoP_duplication | 2 | C-terminal duplication domain of Friend of PRMT1 |
| Focal_AT | 2 | Focal adhesion targeting region |
| Folliculin | 2 | Vesicle coat protein involved in Golgi to plasma membrane transport |
| Formin_GBD_N | 2 | Formin N-terminal GTPase-binding domain |
| FragX_IP | 2 | Cytoplasmic Fragile-X interacting family |
| Fructosamin_kin | 2 | Fructosamine kinase |
| Fry_C | 2 | Furry protein C-terminal |
| FtsH_ext | 2 | FtsH Extracellular |
| Fucokinase | 2 | L-fucokinase |
| Fucosidase_C | 2 | Alpha-L-fucosidase C-terminal domain |
| Fzo_mitofusin | 2 | fzo-like conserved region |
| G6PD_C | 2 | Glucose-6-phosphate dehydrogenase, C-terminal domain |
| G6PD_N | 2 | Glucose-6-phosphate dehydrogenase, NAD binding domain |
| GATase_7 | 2 | Glutamine amidotransferase domain |
| GBBH-like_N | 2 | Gamma-butyrobetaine hydroxylase-like, N-terminal |
| GCD14 | 2 | tRNA methyltransferase complex GCD14 subunit |
| GCM | 2 | GCM motif protein |
| GCOM2 | 2 | Putative GRINL1B complex locus protein 2 |
| GCV_H | 2 | Glycine cleavage H-protein |
| GDPD_2 | 2 | Glycerophosphoryl diester phosphodiesterase family |
| GFO_IDH_MocA_C | 2 | Oxidoreductase family, C-terminal alpha/beta domain |
| GIT1_C | 2 | G protein-coupled receptor kinase-interacting protein 1 C term |
| GIT_CC | 2 | GIT coiled-coil Rho guanine nucleotide exchange factor |
| GIT_SHD | 2 | Spa2 homology domain (SHD) of GIT |
| GIY-YIG | 2 | GIY-YIG catalytic domain |
| GLTSCR1 | 2 | Conserved region of unknown function on GLTSCR protein |
| GNT-I | 2 | GNT-I family |
| GOLD_2 | 2 | Golgi-dynamics membrane-trafficking |
| GON | 2 | GON domain |
| GPHR_N | 2 | The Golgi pH Regulator (GPHR) Family N-terminal |
| GPP34 | 2 | Golgi phosphoprotein 3 (GPP34) |
| GPR180-TMEM145_TM | 2 | GPR180/TMEM145, transmembrane domain |
| GPS2_interact | 2 | G-protein pathway suppressor 2-interacting domain |
| GRASP55_65 | 2 | GRASP55/65 PDZ-like domain |
| GREB1_2nd | 2 | GREB1, second domain |
| GREB1_C | 2 | GREB1 C-terminal region |
| GREB1_N | 2 | GREB1 N-terminal domain |
| GSK-3_bind | 2 | Glycogen synthase kinase-3 binding |
| GST_N_4 | 2 | Glutathione S-transferase N-terminal domain |
| GTP1_OBG | 2 | GTP1/OBG |
| GTPase_binding | 2 | GTPase binding |
| GTSE1_N | 2 | G-2 and S-phase expressed 1 |
| GUCT | 2 | GUCT (NUC152) domain |
| GalKase_gal_bdg | 2 | Galactokinase galactose-binding signature |
| Galanin | 2 | Galanin |
| Gar1 | 2 | Gar1/Naf1 RNA binding region |
| Gastrin | 2 | Gastrin/cholecystokinin family |
| Gb3_synth | 2 | Alpha 1,4-glycosyltransferase conserved region |
| Geminin | 2 | Geminin |
| Gln-synt_C | 2 | Glutamine synthetase, catalytic domain |
| GluR_Homer-bdg | 2 | Homer-binding domain of metabotropic glutamate receptor |
| Glutaminase | 2 | Glutaminase |
| Gly_rich | 2 | Glycine rich protein |
| Gly_transf_sug | 2 | Glycosyltransferase sugar-binding region containing DXD motif |
| Glyco_hydro_2_C | 2 | Glycosyl hydrolases family 2, TIM barrel domain |
| Glyco_hydro_79n | 2 | Glycosyl hydrolase family 79, N-terminal domain |
| Glyco_hydro_99 | 2 | Glycosyl hydrolase family 99 |
| Glyco_tranf_2_3 | 2 | Glycosyltransferase like family 2 |
| Glyco_tranf_2_4 | 2 | Glycosyl transferase family 2 |
| Glyco_transf_11 | 2 | Glycosyl transferase family 11 |
| Glyco_transf_18 | 2 | Glycosyltransferase family 18 |
| Glyco_transf_24 | 2 | Glucosyltransferase 24 |
| Glyco_transf_61 | 2 | Glycosyltransferase 61 |
| Glycogen_syn | 2 | Glycogen synthase |
| Glycohydro_20b2 | 2 | beta-acetyl hexosaminidase like |
| Glycophorin_A | 2 | Glycophorin A |
| GnRH | 2 | Gonadotropin-releasing hormone |
| Gp_dh_C | 2 | Glyceraldehyde 3-phosphate dehydrogenase, C-terminal domain |
| Gp_dh_N | 2 | Glyceraldehyde 3-phosphate dehydrogenase, NAD binding domain |
| GrpE | 2 | GrpE |
| Guanylin | 2 | Guanylin precursor |
| H2TH | 2 | Formamidopyrimidine-DNA glycosylase H2TH domain |
| HAND | 2 | HAND |
| HAT_KAT11 | 2 | Histone acetylation protein |
| HDNR | 2 | Domain of unknown function with conserved HDNR motif |
| HECW1_helix | 2 | Helical box domain of E3 ubiquitin-protein ligase HECW1 |
| HECW_N | 2 | N-terminal domain of E3 ubiquitin-protein ligase HECW1 and 2 |
| HEXIM | 2 | Hexamethylene bis-acetamide-inducible protein |
| HGAL | 2 | Germinal center-associated lymphoma |
| HHH_9 | 2 | HHH domain |
| HIF-1a_CTAD | 2 | HIF-1 alpha C terminal transactivation domain |
| HIP1_clath_bdg | 2 | Clathrin-binding domain of Huntingtin-interacting protein 1 |
| HMG_CoA_synt_C | 2 | Hydroxymethylglutaryl-coenzyme A synthase C terminal |
| HMG_CoA_synt_N | 2 | Hydroxymethylglutaryl-coenzyme A synthase N terminal |
| HMMR_C | 2 | Hyaluronan mediated motility receptor C-terminal |
| HNF-1B_C | 2 | Hepatocyte nuclear factor 1 (HNF-1), beta isoform C terminus |
| HNF_C | 2 | HNF3 C-terminal domain |
| HNOB | 2 | Haem-NO-binding |
| HPD | 2 | Homeo-prospero domain |
| HS1_rep | 2 | Repeat in HS1/Cortactin |
| HSBP1 | 2 | Heat shock factor binding protein 1 |
| HTH_53 | 2 | Zap helix turn helix N-terminal domain |
| HTH_61 | 2 | Helix-turn-helix domain |
| HTH_SUN2 | 2 | SUN2 helix-turn-helix domain |
| HTH_psq | 2 | helix-turn-helix, Psq domain |
| HUN | 2 | HPC2 and ubinuclein domain |
| Helicase_C_4 | 2 | C-terminal domain on Strawberry notch homologue |
| Heme_oxygenase | 2 | Heme oxygenase |
| Hexapep | 2 | Bacterial transferase hexapeptide (six repeats) |
| Hormone_4 | 2 | Neurohypophysial hormones, N-terminal Domain |
| Hormone_5 | 2 | Neurohypophysial hormones, C-terminal Domain |
| Hus1 | 2 | Hus1-like protein |
| Hyccin | 2 | Hyccin |
| Hyd_WA | 2 | Propeller |
| ICA69 | 2 | Islet cell autoantigen ICA69, C-terminal domain |
| ICAT | 2 | Beta-catenin-interacting protein ICAT |
| IDO | 2 | Indoleamine 2,3-dioxygenase |
| IF-2 | 2 | Translation-initiation factor 2 |
| IFP_35_N | 2 | Interferon-induced 35 kDa protein (IFP 35) N-terminus |
| IFRD | 2 | Interferon-related developmental regulator (IFRD) |
| IFRD_C | 2 | Interferon-related protein conserved region |
| IHABP4_N | 2 | Intracellular hyaluronan-binding protein 4 N-terminal |
| IIGP | 2 | Interferon-inducible GTPase (IIGP) |
| IKBKB_SDD | 2 | IQBAL scaffold dimerization domain |
| IKKbetaNEMObind | 2 | I-kappa-kinase-beta NEMO binding domain |
| IL15 | 2 | Interleukin 15 |
| IL17R_fnIII_D1 | 2 | Interleukin-17 receptor, fibronectin-III-like domain 1 |
| IL17R_fnIII_D2 | 2 | Interleukin 17 receptor D |
| IL1_propep | 2 | Interleukin-1 propeptide |
| IL3Ra_N | 2 | IL-3 receptor alpha chain N-terminal domain |
| IMS_HHH | 2 | IMS family HHH motif |
| INSIG | 2 | Insulin-induced protein (INSIG) |
| IQCJ-SCHIP1 | 2 | Fusion protein IQCJ-SCHIP1 with IQ-like motif |
| Ig_4 | 2 | T-cell surface glycoprotein CD3 delta chain |
| Il2rg | 2 | Putative Interleukin 2 receptor, gamma chain |
| Importin_rep_4 | 2 | Importin repeat |
| Importin_rep_6 | 2 | Importin repeat 6 |
| InaF-motif | 2 | TRP-interacting helix |
| Inhibitor_Mig-6 | 2 | EGFR receptor inhibitor Mig-6 |
| Isochorismatase | 2 | Isochorismatase family |
| Isy1 | 2 | Isy1-like splicing family |
| JIP_LZII | 2 | JNK-interacting protein leucine zipper II |
| JMY | 2 | Junction-mediating and -regulatory protein |
| JUPITER | 2 | Microtubule-Associated protein Jupiter |
| Jacalin | 2 | Jacalin-like lectin domain |
| JmjC_2 | 2 | JmjC domain |
| KA1_BRSK | 2 | BRSK1/2 KA1 domain |
| KAP_NTPase | 2 | KAP family P-loop domain |
| KASH_CCD | 2 | Coiled-coil region of CCDC155 or KASH |
| KATNA1_MIT | 2 | Katanin p60 subunit A1, MIT domain |
| KBTB_W-LIR | 2 | KBTB, W-type LIR motif |
| KCNQC3-Ank-G_bd | 2 | Ankyrin-G binding motif of KCNQ2-3 |
| KCT2 | 2 | Keratinocyte-associated gene product |
| KCTD1-15_CTD | 2 | BTB/POZ domain-containing protein KCTD1/15, C-terminal domain |
| KCTD11_21_C | 2 | BTB/POZ domain-containing protein KCTD11/21 C-terminus |
| KELK | 2 | KELK-motif containing domain of MRCK Ser/Thr protein kinase |
| KH_6 | 2 | KH domain |
| KIAA1549 | 2 | UPF0606 protein KIAA1549 |
| KIX | 2 | KIX domain |
| KLHL33-like_BTB_POZ | 2 | Kelch-like protein 33-like, BTB/POZ domain |
| KRTDAP | 2 | Keratinocyte differentiation-associated |
| KTI12 | 2 | Chromatin associated protein KTI12 |
| Katanin_con80 | 2 | con80 domain of Katanin |
| Ketoacyl-synt_C | 2 | Beta-ketoacyl synthase, C-terminal domain |
| Ku | 2 | Ku70/Ku80 beta-barrel domain |
| Ku_C | 2 | Ku70/Ku80 C-terminal arm |
| Ku_N | 2 | Ku70/Ku80 N-terminal alpha/beta domain |
| Kv2channel | 2 | Kv2 voltage-gated K+ channel |
| L27_1 | 2 | L27_1 |
| L27_2 | 2 | L27_2 |
| LAG1-DNAbind | 2 | LAG1, DNA binding |
| LAIKA | 2 | LAIKA domain |
| LAP1_C | 2 | Lamina-associated polypeptide 1, AAA+ activator domain |
| LAP1_N | 2 | Lamina-associated polypeptide 1, N-terminal |
| LAP2alpha | 2 | Lamina-associated polypeptide 2 alpha |
| LARP1_HEAT | 2 | LARP1 HEAT repeat region |
| LCAT | 2 | Lecithin:cholesterol acyltransferase |
| LCM | 2 | Leucine carboxyl methyltransferase |
| LEDGF | 2 | Lens epithelium-derived growth factor (LEDGF) |
| LID | 2 | LIM interaction domain (LID) |
| LIF-R_Ig-like | 2 | Leukemia inhibitory factor receptor, Ig-like domain |
| LIFR_D2 | 2 | Leukemia inhibitory factor receptor D2 domain |
| LIF_OSM | 2 | LIF / OSM family |
| LIM_bind | 2 | LIM-domain binding protein |
| LIX1 | 2 | Limb expression 1 |
| LMF1 | 2 | Lipase maturation factor |
| LMSTEN | 2 | LMSTEN motif |
| LRRFIP | 2 | LRRFIP family |
| LZ3wCH | 2 | Leucine zipper with capping helix domain |
| Laa1_Sip1_HTR5 | 2 | Laa1/Sip1/HEATR5 HEAT repeat region |
| Lactamase_B_2 | 2 | Beta-lactamase superfamily domain |
| Laman-like_dom | 2 | Lysosomal alpha-mannosidase-like, central domain |
| Lamp2_2nd | 2 | Lysosome-associated membrane glycoprotein 2, transmembrane segment |
| Latexin_C | 2 | Latexin, C-terminal |
| Latexin_N | 2 | Latexin N-terminal |
| Lebercilin | 2 | Ciliary protein causing Leber congenital amaurosis disease |
| Lgl_C | 2 | Lethal giant larvae(Lgl) like, C-terminal |
| LicD | 2 | LicD family |
| Lipase_3 | 2 | Lipase (class 3) |
| Lipase_GDSL_2 | 2 | GDSL-like Lipase/Acylhydrolase family |
| Lipid_DES | 2 | Sphingolipid Delta4-desaturase (DES) |
| Lipid_desat | 2 | Lipid desaturase domain |
| Lipocalin_7 | 2 | Lipocalin / cytosolic fatty-acid binding protein family |
| Lon_C | 2 | Lon protease (S16) C-terminal proteolytic domain |
| LsmAD | 2 | LsmAD domain |
| Ly49 | 2 | Ly49-like protein, N-terminal region |
| MAGI_u1 | 2 | Unstructured region on MAGI |
| MAGP | 2 | Microfibril-associated glycoprotein (MAGP) |
| MAML1_3_TAD1 | 2 | Mastermind-like 1/3, transactivation domain 1 |
| MAML1_3_TAD2 | 2 | Mastermind-like 1/3, transactivation domain 2 |
| MAP17 | 2 | Membrane-associated protein 117 kDa, PDZK1-interacting protein 1 |
| MAP6 | 2 | Microtubule-associated protein 6 |
| MAP65_ASE1 | 2 | Microtubule associated protein (MAP65/ASE1 family) |
| MARCKS | 2 | MARCKS family |
| MAS20 | 2 | MAS20 protein import receptor |
| MATCAP | 2 | Microtubule-associated tyrosine carboxypeptidase |
| MCU | 2 | Mitochondrial calcium uniporter |
| MENTAL | 2 | Cholesterol-capturing domain |
| MFS_1_like | 2 | MFS_1 like family |
| MG1 | 2 | Macroglobulin domain MG1 |
| MIC19_MIC25 | 2 | MICOS complex subunit MIC19/MIC25 |
| MID51-like_C | 2 | Mitochondrial dynamics protein MID51-like, C-terminal domain |
| MID_MedPIWI | 2 | MID domain of medPIWI |
| MIER1_3_C | 2 | Mesoderm induction early response protein 1/3 C-terminal |
| MINAR1_C | 2 | MINAR1 C-terminal domain |
| MINDY_DUB | 2 | MINDY deubiquitinase |
| MIOS_WD40 | 2 | MIOS, WD40 repeat |
| MKRN1_C | 2 | E3 ubiquitin-protein ligase makorin, C-terminal |
| MMR_HSR1_Xtn | 2 | C-terminal region of MMR_HSR1 domain |
| MOR2-PAG1_C | 2 | Cell morphogenesis C-terminal |
| MOR2-PAG1_N | 2 | Cell morphogenesis N-terminal |
| MOR2-PAG1_mid | 2 | Cell morphogenesis central region |
| MOV-10_beta-barrel | 2 | Helicase MOV-10, beta-barrel domain |
| MOZART2 | 2 | Mitotic-spindle organizing gamma-tubulin ring associated |
| MPN_2A_DUB_like | 2 | BRCA1-A complex subunit Abraxas 1 MPN domain |
| MRAP | 2 | Melanocortin-2 receptor accessory protein family |
| MRF_C2 | 2 | Myelin gene regulatory factor C-terminal domain 2 |
| MRP-S27 | 2 | Mitochondrial 28S ribosomal protein S27 |
| MRVI1 | 2 | MRVI1 protein |
| MSC | 2 | Man1-Src1p-C-terminal domain |
| MSS51_C | 2 | MSS51 C-terminal domain |
| MTR4-like_stalk | 2 | Exosome RNA helicase MTR4-like, stalk |
| MTR4_beta-barrel | 2 | Mtr4-like, beta-barrel domain |
| MYRF_ICA | 2 | Myelin regulatory factor ICA domain |
| Mad3_BUB1_I | 2 | Mad3/BUB1 homology region 1 |
| Mago_nashi | 2 | Mago nashi protein |
| MaoC_dehydratas | 2 | MaoC like domain |
| MatE | 2 | MatE |
| Med12 | 2 | Transcription mediator complex subunit Med12 |
| Med12-LCEWAV | 2 | Eukaryotic Mediator 12 subunit domain |
| Med12-PQL | 2 | Eukaryotic Mediator 12 catenin-binding domain |
| Med13_C | 2 | Mediator complex subunit 13 C-terminal domain |
| Med25 | 2 | Mediator complex subunit 25 PTOV activation and synapsin 2 |
| Melibiase_2 | 2 | Alpha galactosidase A |
| Melibiase_2_C | 2 | Alpha galactosidase A C-terminal beta sandwich domain |
| Menorin | 2 | Menorin |
| Mesothelin | 2 | Mesothelin |
| Met_10 | 2 | Met-10+ like-protein |
| Methyltransf_2 | 2 | O-methyltransferase domain |
| Methyltransf_PK | 2 | AdoMet dependent proline di-methyltransferase |
| Miff | 2 | Mitochondrial and peroxisomal fission factor Mff |
| Miga | 2 | Mitoguardin |
| Milton | 2 | Kinesin associated protein |
| MitoNEET_N | 2 | Iron-containing outer mitochondrial membrane protein N-terminus |
| Mlf1IP | 2 | Myelodysplasia-myeloid leukemia factor 1-interacting protein |
| Mo25 | 2 | Mo25-like |
| MoCF_biosynth | 2 | Probable molybdopterin binding domain |
| MoCoBD_1 | 2 | Molybdopterin cofactor-binding domain |
| MoCoBD_2 | 2 | Molybdopterin cofactor-binding domain |
| Motilin_assoc | 2 | Motilin/ghrelin-associated peptide |
| Motilin_ghrelin | 2 | Motilin/ghrelin |
| Mst1_SARAH | 2 | C terminal SARAH domain of Mst1 |
| Mustang | 2 | Musculoskeletal, temporally activated-embryonic nuclear protein 1 |
| MutL_C | 2 | MutL C terminal dimerisation domain |
| Myb_DNA-binding_7 | 2 | Myb-like DNA-binding domain |
| Myf5 | 2 | Myogenic determination factor 5 |
| N-SET | 2 | COMPASS (Complex proteins associated with Set1p) component N |
| N1221 | 2 | N1221-like protein |
| N6-adenineMlase | 2 | Probable N6-adenine methyltransferase |
| NAAA-beta | 2 | beta subunit of N-acylethanolamine-hydrolyzing acid amidase |
| NAD1 | 2 | Novel AID APOBEC clade 1 |
| NAD_Gly3P_dh_C | 2 | NAD-dependent glycerol-3-phosphate dehydrogenase C-terminus |
| NAD_Gly3P_dh_N | 2 | NAD-dependent glycerol-3-phosphate dehydrogenase N-terminus |
| NAD_binding_10 | 2 | NAD(P)H-binding |
| NAD_binding_11 | 2 | NAD-binding of NADP-dependent 3-hydroxyisobutyrate dehydrogenase |
| NAD_binding_4 | 2 | Male sterility protein |
| NAD_kinase | 2 | ATP-NAD kinase N-terminal domain |
| NAD_synthase | 2 | NAD synthase |
| NAPRTase | 2 | Nicotinate phosphoribosyltransferase (NAPRTase) family |
| NCD1 | 2 | NAB conserved region 1 (NCD1) |
| NCD2 | 2 | NAB conserved region 2 (NCD2) |
| NCKAP5 | 2 | Nck-associated protein 5, Peripheral clock protein |
| NDT80_PhoG | 2 | NDT80 / PhoG like DNA-binding family |
| NDUFA12 | 2 | NADH ubiquinone oxidoreductase subunit NDUFA12 |
| NDUF_C2 | 2 | NADH-ubiquinone oxidoreductase subunit b14.5b (NDUFC2) |
| NEMO | 2 | NF-kappa-B essential modulator NEMO |
| NEMP | 2 | NEMP family |
| NFACT-R_1 | 2 | NFACT protein RNA binding domain |
| NID | 2 | Nmi/IFP 35 domain (NID) |
| NLE | 2 | NLE (NUC135) domain |
| NLRC4_HD | 2 | NLRC4 helical domain |
| NMT | 2 | Myristoyl-CoA:protein N-myristoyltransferase, N-terminal domain |
| NMT_C | 2 | Myristoyl-CoA:protein N-myristoyltransferase, C-terminal domain |
| NOP5NT | 2 | NOP5NT (NUC127) domain |
| NOT2_3_5_C | 2 | NOT2/NOT3/NOT5 C-terminal |
| NPBW | 2 | Neuropeptides B and W |
| NPC1_N | 2 | Niemann-Pick C1 N terminus |
| NRN1 | 2 | Neuritin protein family |
| NSUN5_fdxn-like | 2 | NOL1/NOP2/Sun domain family member 5, ferredoxin-like domain |
| NT5C | 2 | 5' nucleotidase, deoxy (Pyrimidine), cytosolic type C protein (NT5C) |
| NUC153 | 2 | NUC153 domain |
| NUDE_C | 2 | NUDE protein, C-terminal conserved region |
| NUDIX-like | 2 | NADH pyrophosphatase-like rudimentary NUDIX domain |
| NYD-SP28 | 2 | Sperm tail |
| N_BRCA1_IG | 2 | Ig-like domain from next to BRCA1 gene |
| NatA_aux_su | 2 | N-terminal acetyltransferase A, auxiliary subunit |
| Nckap1 | 2 | Nck-associated protein 1 |
| Neogenin_C | 2 | Neogenin C-terminus |
| Neuregulin | 2 | Neuregulin intracellular region |
| Ninjurin | 2 | Ninjurin |
| Nkap_C | 2 | NF-kappa-B-activating protein C-terminal domain |
| Nnf1 | 2 | Nnf1 |
| Nop52 | 2 | Nucleolar protein,Nop52 |
| NosD | 2 | Periplasmic copper-binding protein (NosD) |
| Nramp | 2 | Natural resistance-associated macrophage protein-like |
| Nse4-Nse3_bdg | 2 | Binding domain of Nse4/EID3 to Nse3-MAGE |
| Nse4_C | 2 | Nse4 C-terminal |
| Nsp1_C | 2 | Nsp1-like C-terminal region |
| Nuc_recep-AF1 | 2 | Nuclear/hormone receptor activator site AF-1 |
| Nudc_N | 2 | N-terminal conserved domain of Nudc. |
| NumbF | 2 | NUMB domain |
| O-FucT | 2 | GDP-fucose protein O-fucosyltransferase |
| OCIA | 2 | Ovarian carcinoma immunoreactive antigen (OCIA) |
| OCRL-like_ASH | 2 | Inositol polyphosphate 5-phosphatase OCRL-like, ASH domain |
| ODAD1_CC | 2 | ODAD1 central coiled coil region |
| OGFr_N | 2 | Opioid growth factor receptor (OGFr) conserved region |
| OTCace | 2 | Aspartate/ornithine carbamoyltransferase, Asp/Orn binding domain |
| OTCace_N | 2 | Aspartate/ornithine carbamoyltransferase, carbamoyl-P binding domain |
| OTU1_UBXL | 2 | OTU1, UBXL domain |
| Oxidored-like | 2 | Oxidoreductase-like protein, N-terminal |
| P16-Arc | 2 | ARP2/3 complex 16 kDa subunit (p16-Arc) |
| P2R3B_EF-hand | 2 | P2R3B, EF-hand |
| P66_CC | 2 | Coiled-coil and interaction region of P66A and P66B with MBD2 |
| PA14 | 2 | PA14 domain |
| PACT_coil_coil | 2 | Pericentrin-AKAP-450 domain of centrosomal targeting protein |
| PAF-AH_p_II | 2 | Platelet-activating factor acetylhydrolase, isoform II |
| PAT1 | 2 | Topoisomerase II-associated protein PAT1 |
| PCAF_N | 2 | PCAF (P300/CBP-associated factor) N-terminal domain |
| PCRF | 2 | PCRF domain |
| PC_rep | 2 | Proteasome/cyclosome repeat |
| PDCD2_C | 2 | Programmed cell death protein 2, C-terminal putative domain |
| PDCD9 | 2 | Mitochondrial 28S ribosomal protein S30 (PDCD9) |
| PDE6_gamma | 2 | Retinal cGMP phosphodiesterase, gamma subunit |
| PDGF_N | 2 | Platelet-derived growth factor, N terminal region |
| PDZ_5 | 2 | PDZ domain |
| PEPCK_GTP | 2 | Phosphoenolpyruvate carboxykinase C-terminal P-loop domain |
| PEPCK_N | 2 | Phosphoenolpyruvate carboxykinase N-terminal domain |
| PGK | 2 | Phosphoglycerate kinase |
| PHC2_SAM_assoc | 2 | Unstructured region on Polyhomeotic-like protein 1 and 2 |
| PHO4 | 2 | Phosphate transporter family |
| PHTF1-2_N | 2 | Homeodomain containing protein PHTF1/2, N-terminal |
| PHYHIP_C | 2 | Phytanoyl-CoA hydroxylase-interacting protein C-terminus |
| PI31_Prot_N | 2 | PI31 proteasome regulator N-terminal |
| PI3K_1B_p101 | 2 | Phosphoinositide 3-kinase gamma adapter protein p101 subunit |
| PIEZO | 2 | Piezo |
| PIRC1_2 | 2 | Piercer of microtubule wall 1/2 |
| PITH | 2 | PITH domain |
| PK | 2 | Pyruvate kinase, barrel domain |
| PKC_C2 | 2 | Protein kinase C delta/epsilon/eta/theta, C2 domain |
| PKK | 2 | Polo kinase kinase |
| PK_C | 2 | Pyruvate kinase, alpha/beta domain |
| PLA2G12 | 2 | Group XII secretory phospholipase A2 precursor (PLA2G12) |
| PMM | 2 | Eukaryotic phosphomannomutase |
| PMT | 2 | Dolichyl-phosphate-mannose-protein mannosyltransferase |
| PMT_4TMC | 2 | C-terminal four TMM region of protein-O-mannosyltransferase |
| PNRC | 2 | Proline-rich nuclear receptor coactivator motif |
| POLH-Rev1_HhH | 2 | DNApol eta/Rev1, HhH motif |
| POLQ_helical | 2 | DNA_pol_Q helicase like region helical domain |
| POU2F1_C | 2 | POU domain, class 2, transcription factor 1 C-terminal |
| PP1_bind | 2 | Protein phosphatase 1 binding |
| PP1c_bdg | 2 | Phosphatase-1 catalytic subunit binding region |
| PPDFL | 2 | Differentiation and proliferation regulator |
| PPIP5K2_N | 2 | Diphosphoinositol pentakisphosphate kinase 2 N-terminal domain |
| PQQ_2 | 2 | PQQ-like domain |
| PRAP | 2 | Proline-rich acidic protein 1, pregnancy-specific uterine |
| PRC2_HTH_1 | 2 | Polycomb repressive complex 2 tri-helical domain |
| PRP38 | 2 | PRP38 family |
| PRP4 | 2 | pre-mRNA processing factor 4 (PRP4) like |
| PRT_C | 2 | Plant phosphoribosyltransferase C-terminal |
| PSP | 2 | PSP |
| PSP94 | 2 | Beta-microseminoprotein (PSP-94) |
| PSS | 2 | Phosphatidyl serine synthase |
| PTH2 | 2 | Peptidyl-tRNA hydrolase PTH2 |
| PTN_MK_C | 2 | PTN/MK heparin-binding protein family, C-terminal domain |
| PTN_MK_N | 2 | PTN/MK heparin-binding protein family, N-terminal domain |
| PTP_tm | 2 | TM proximal of protein tyrosine phosphatase, receptor type J |
| PUA_2 | 2 | PUA-like domain |
| PUF | 2 | Pumilio-family RNA binding repeat |
| PUFD | 2 | BCORL-PCGF1-binding domain |
| Pacs-1 | 2 | PACS-1 cytosolic sorting protein |
| Palm_thioest | 2 | Palmitoyl protein thioesterase |
| Pam16 | 2 | Pam16 |
| PapD-like | 2 | Flagellar-associated PapD-like |
| Par3_HAL_N_term | 2 | N-terminal of Par3 and HAL proteins |
| Parathyroid | 2 | Parathyroid hormone family |
| ParcG | 2 | Parkin co-regulated protein |
| Pax7 | 2 | Paired box protein 7 |
| Paxillin | 2 | Paxillin family |
| Pentapeptide_4 | 2 | Pentapeptide repeats (9 copies) |
| Peptidase_C101 | 2 | Peptidase family C101 |
| Peptidase_C13 | 2 | Peptidase C13 family |
| Peptidase_C15 | 2 | Pyroglutamyl peptidase |
| Peptidase_C50 | 2 | Peptidase family C50 |
| Peptidase_C65 | 2 | Peptidase C65 Otubain |
| Peptidase_C97 | 2 | PPPDE putative peptidase domain |
| Peptidase_M16_M | 2 | Middle or third domain of peptidase_M16 |
| Peptidase_M17 | 2 | Cytosol aminopeptidase family, catalytic domain |
| Peptidase_M2 | 2 | Angiotensin-converting enzyme |
| Peptidase_M24_C | 2 | C-terminal region of peptidase_M24 |
| Peptidase_M43 | 2 | Pregnancy-associated plasma protein-A |
| Peptidase_M48 | 2 | Peptidase family M48 |
| Peptidase_M8 | 2 | Leishmanolysin |
| Peptidase_S24 | 2 | Peptidase S24-like |
| Peptidase_S26 | 2 | Signal peptidase, peptidase S26 |
| Peptidase_S74 | 2 | Chaperone of endosialidase |
| Peptidase_S9_N | 2 | Prolyl oligopeptidase, N-terminal beta-propeller domain |
| Phospho_p8 | 2 | DNA-binding nuclear phosphoprotein p8 |
| Phospholip_A2_2 | 2 | Phospholipase A2 |
| Phospholip_B | 2 | Phospholipase B |
| Phostensin | 2 | Phostensin PP1-binding and SH3-binding region |
| Phostensin_N | 2 | PP1-regulatory protein, Phostensin N-terminal |
| PhyH | 2 | Phytanoyl-CoA dioxygenase (PhyH) |
| Piezo_RRas_bdg | 2 | Piezo non-specific cation channel, R-Ras-binding domain |
| Plug_translocon | 2 | Plug domain of Sec61p |
| Polyketide_cyc | 2 | Polyketide cyclase / dehydrase and lipid transport |
| Pr_beta_C | 2 | Proteasome beta subunits C terminal |
| Prenylcys_lyase | 2 | Prenylcysteine lyase |
| Prenyltransf | 2 | Putative undecaprenyl diphosphate synthase |
| Presenilin | 2 | Presenilin |
| Preseq_ALAS | 2 | 5-aminolevulinate synthase presequence |
| Prion | 2 | Prion/Doppel alpha-helical domain |
| Pro_dh | 2 | Proline dehydrogenase |
| Prokineticin | 2 | Prokineticin |
| Prominin | 2 | Prominin |
| Prothymosin | 2 | Prothymosin/parathymosin family |
| Pterin_4a | 2 | Pterin 4 alpha carbinolamine dehydratase |
| Put_Phosphatase | 2 | Putative Phosphatase |
| Pyrid_oxidase_2 | 2 | Pyridoxamine 5'-phosphate oxidase |
| Pyrophosphatase | 2 | Inorganic pyrophosphatase |
| QLQ | 2 | QLQ |
| QSOX_Trx1 | 2 | QSOX Trx-like domain |
| RAD51_interact | 2 | RAD51 interacting motif |
| RAM | 2 | mRNA cap methylation, RNMT-activating mini protein |
| RAMA | 2 | Restriction Enzyme Adenine Methylase Associated |
| RAMP4 | 2 | Ribosome associated membrane protein RAMP4 |
| RASSF8-10_RA | 2 | Ras association domain-containing protein 8/10, RA domain |
| RBB1NT | 2 | RBB1NT (NUC162) domain |
| RBM39linker | 2 | linker between RRM2 and RRM3 domains in RBM39 protein |
| RD3 | 2 | RD3 protein |
| RFX5_N | 2 | RFX5 N-terminal domain |
| RHS_repeat | 2 | RHS Repeat |
| RIB43A | 2 | RIB43A |
| RIBIOP_C | 2 | 40S ribosome biogenesis protein Tsr1 and BMS1 C-terminal |
| RING_CBP-p300 | 2 | CREB-binding protein/p300, atypical RING domain |
| RL10P_insert | 2 | Insertion domain in 60S ribosomal protein L10P |
| RLI | 2 | Possible Fer4-like domain in RNase L inhibitor, RLI |
| RMI1_N_C | 2 | RMI1, N-terminal OB-fold domain |
| RNA_POL_M_15KD | 2 | RNA polymerases M/15 Kd subunit |
| RNA_pol_3_Rpc31 | 2 | DNA-directed RNA polymerase III subunit Rpc31 |
| RNA_pol_A_bac | 2 | RNA polymerase Rpb3/RpoA insert domain |
| RNA_pol_L | 2 | RNA polymerase Rpb3/Rpb11 dimerisation domain |
| RNA_pol_Rpb1_R | 2 | RNA polymerase Rpb1 C-terminal repeat |
| RNA_pol_Rpb2_4 | 2 | RNA polymerase Rpb2, domain 4 |
| RNA_pol_Rpb2_5 | 2 | RNA polymerase Rpb2, domain 5 |
| RNA_pol_Rpb4 | 2 | RNA polymerase Rpb4 |
| RNF111_N | 2 | E3 ubiquitin-protein ligase Arkadia N-terminus |
| RNase_H | 2 | RNase H |
| RNase_P_Rpp14 | 2 | Rpp14/Pop5 family |
| ROQ_II | 2 | Roquin II domain |
| RPA_C | 2 | Replication protein A C terminal |
| RPGR1_C | 2 | Retinitis pigmentosa G-protein regulator interacting C-terminal |
| RPN7 | 2 | 26S proteasome subunit RPN7 |
| RQC | 2 | RQC domain |
| RRM_3 | 2 | RNA binding motif |
| RSRP | 2 | Arginine/Serine-Rich protein 1 |
| RTC | 2 | RNA 3'-terminal phosphate cyclase |
| RTC_insert | 2 | RNA 3'-terminal phosphate cyclase (RTC), insert domain |
| RTP801_C | 2 | RTP801 C-terminal region |
| RT_RNaseH_2 | 2 | RNase H-like domain found in reverse transcriptase |
| RUBC_PIKBD | 2 | Rubicon, PI3K-binding domain |
| Rab5-bind | 2 | Rabaptin-like protein |
| Rab_eff_C | 2 | Rab effector MyRIP/melanophilin C-terminus |
| Rabaptin | 2 | Rabaptin |
| Rad4 | 2 | Rad4 transglutaminase-like domain |
| Rad9 | 2 | Rad9 |
| Radial_spoke | 2 | Radial spokehead-like protein |
| Raftlin | 2 | Raftlin |
| Rav1p_C | 2 | RAVE protein 1 C terminal |
| Rb_C | 2 | Rb C-terminal domain |
| Receptor_IA-2 | 2 | Protein-tyrosine phosphatase receptor IA-2 |
| Rep_fac-A_C | 2 | Replication factor-A C terminal domain |
| Reprolysin_2 | 2 | Metallo-peptidase family M12B Reprolysin-like |
| Resistin | 2 | Resistin |
| RhoGAP-FF1 | 2 | p190-A and -B Rho GAPs FF domain |
| RhoGAP_pG1_pG2 | 2 | p190RhoGAP, pG1 and pG2 domains |
| RhoGEF67_u1 | 2 | Unstructured region one on RhoGEF 6 and 7 |
| RhoGEF67_u2 | 2 | Unstructured region two on RhoGEF 6 and 7 |
| Rho_Binding | 2 | Rho Binding |
| Rib_hydrolayse | 2 | ADP-ribosyl cyclase |
| Rib_recp_KP_reg | 2 | Ribosome receptor lysine/proline rich region |
| Ribonuc_red_sm | 2 | Ribonucleotide reductase, small chain |
| Ribonuclease_3 | 2 | Ribonuclease III domain |
| Ribosom_S12_S23 | 2 | Ribosomal protein S12/S23 |
| Ribosomal_L11 | 2 | Ribosomal protein L11, RNA binding domain |
| Ribosomal_L11_N | 2 | Ribosomal protein L11, N-terminal domain |
| Ribosomal_L13 | 2 | Ribosomal protein L13 |
| Ribosomal_L14 | 2 | Ribosomal protein L14p/L23e |
| Ribosomal_L2 | 2 | Ribosomal Proteins L2, RNA binding domain |
| Ribosomal_L22e | 2 | Ribosomal L22e protein family |
| Ribosomal_L23 | 2 | Ribosomal protein L23 |
| Ribosomal_L24e | 2 | Ribosomal protein L24e |
| Ribosomal_L26 | 2 | Ribosomal proteins L26 eukaryotic, L24P archaeal |
| Ribosomal_L27A | 2 | Ribosomal proteins 50S-L15, 50S-L18e, 60S-L27A |
| Ribosomal_L28e | 2 | Ribosomal L28e protein family |
| Ribosomal_L2_C | 2 | Ribosomal Proteins L2, C-terminal domain |
| Ribosomal_L30_N | 2 | Ribosomal L30 N-terminal domain |
| Ribosomal_L39 | 2 | Ribosomal L39 protein |
| Ribosomal_L4 | 2 | Ribosomal protein L4/L1 family |
| Ribosomal_S11 | 2 | Ribosomal protein S11 |
| Ribosomal_S14 | 2 | Ribosomal protein S14p/S29e |
| Ribosomal_S15 | 2 | Ribosomal protein S15 |
| Ribosomal_S17 | 2 | Ribosomal protein S17 |
| Ribosomal_S21e | 2 | Ribosomal protein S21e |
| Ribosomal_S27e | 2 | Ribosomal protein S27 |
| Ribosomal_S4 | 2 | Ribosomal protein S4/S9 N-terminal domain |
| Ribosomal_S5 | 2 | Ribosomal protein S5, N-terminal domain |
| Ribosomal_S5_C | 2 | Ribosomal protein S5, C-terminal domain |
| Ribosomal_S7 | 2 | Ribosomal protein S7p/S5e |
| Ribosomal_S8e | 2 | Ribosomal protein S8e |
| Ribosomal_S9 | 2 | Ribosomal protein S9/S16 |
| Ribul_P_3_epim | 2 | Ribulose-phosphate 3 epimerase family |
| Ric8 | 2 | Guanine nucleotide exchange factor synembryn |
| RimK | 2 | RimK-like ATP-grasp domain |
| Roquin_1_2-like_ROQ | 2 | Roquin 1/2-like, ROQ domain |
| Rpr2 | 2 | RNAse P Rpr2/Rpp21/SNM1 subunit domain |
| Rrp44_S1 | 2 | S1 domain |
| Rttp106-like_middle | 2 | Histone chaperone Rttp106-like, middle domain |
| S-AdoMet_synt_C | 2 | S-adenosylmethionine synthetase, C-terminal domain |
| S-AdoMet_synt_M | 2 | S-adenosylmethionine synthetase, central domain |
| S-AdoMet_synt_N | 2 | S-adenosylmethionine synthetase, N-terminal domain |
| S4 | 2 | S4 domain |
| SAD_SRA | 2 | SAD/SRA domain |
| SAMP | 2 | SAMP Motif |
| SAM_KSR1_N | 2 | Kinase suppressor RAS 1 N-terminal helical hairpin |
| SAP30_Sin3_bdg | 2 | Sin3 binding region of histone deacetylase complex subunit SAP30 |
| SARA_C | 2 | Smad anchor for receptor activation-like, C-terminal |
| SARG | 2 | Specifically androgen-regulated gene protein |
| SAXO1-2 | 2 | Stabilizer of axonemal microtubules 1/2 |
| SBF2 | 2 | Myotubularin protein |
| SCGB3A | 2 | Secretoglobin 3A |
| SCHIP-1 | 2 | Schwannomin-interacting protein 1 |
| SCO1-SenC | 2 | SCO1/SenC |
| SDE2_2C | 2 | Replication stress response SDE2 C-terminal |
| SENP3_5_N | 2 | Sentrin-specific protease 3/5 N-terminal |
| SGS | 2 | SGS domain |
| SGSH_C | 2 | N-sulphoglucosamine sulphohydrolase, C-terminal |
| SGTA_dimer | 2 | Homodimerisation domain of SGTA |
| SH3-RhoG_link | 2 | SH3-RhoGEF linking unstructured region |
| SH3BP5 | 2 | SH3 domain-binding protein 5 (SH3BP5) |
| SH3_15 | 2 | Mind bomb SH3 repeat domain |
| SHMT | 2 | Serine hydroxymethyltransferase |
| SID-1_RNA_chan | 2 | dsRNA-gated channel SID-1 |
| SIKE | 2 | SIKE family |
| SIM_C | 2 | Single-minded protein C-terminus |
| SIR2_2 | 2 | SIR2-like domain |
| SIS | 2 | SIS domain |
| SKI | 2 | Shikimate kinase |
| SLAIN | 2 | SLAIN motif-containing family |
| SLIDE | 2 | SLIDE |
| SLX1_C | 2 | Structure-specific endonuclease subunit SLX1, C-terminal |
| SM-ATX | 2 | Ataxin 2 SM domain |
| SMI1_KNR4 | 2 | SMI1 / KNR4 family (SUKH-1) |
| SMN_G2-BD | 2 | Survival Motor Neuron, Gemin2-binding domain |
| SMN_YG-box | 2 | Survival Motor Neuron, YG-box |
| SMP_C2CD2L | 2 | Synaptotagmin-like, mitochondrial and lipid-binding domain |
| SNAP-25 | 2 | SNAP-25 family |
| SNX17-27-31_F1_FERM | 2 | Sortin nexin 17/27/31, FERM domain, F1 lobe |
| SNX17-31_F2_FERM | 2 | Sortin nexin 17/31, FERM domain, F2 lobe |
| SNX17_FERM_C | 2 | Sorting Nexin 17 FERM C-terminal domain |
| SOAR | 2 | STIM1 Orai1-activating region |
| SOBP | 2 | Sine oculis-binding protein |
| SOCS | 2 | Suppressor of cytokine signalling |
| SOUL | 2 | SOUL heme-binding protein |
| SPA | 2 | Stabilization of polarity axis |
| SPATA2_PUB-like | 2 | Spermatogenesis-associated protein 2, PUB-like domain |
| SPATA6 | 2 | Spermatogenesis-assoc protein 6 |
| SPATIAL | 2 | SPATIAL |
| SPATS2-like | 2 | Spermatogenesis-associated serine-rich protein 2-like |
| SPT_ssu-like | 2 | Small subunit of serine palmitoyltransferase-like |
| SQS_PSY | 2 | Squalene/phytoene synthase |
| SRP54 | 2 | SRP54-type protein, GTPase domain |
| SRP54_N | 2 | SRP54-type protein, helical bundle domain |
| SRRM_C | 2 | Serine/arginine repetitive matrix protein C-terminus |
| SRTM1 | 2 | Serine-rich and transmembrane domain-containing protein 1 |
| SSFA2_C | 2 | Sperm-specific antigen 2 C-terminus |
| ST7 | 2 | ST7 protein |
| STI1-HOP_DP | 2 | STI1/HOP, DP domain |
| STIMATE | 2 | STIMATE family |
| STT3 | 2 | Oligosaccharyl transferase STT3, N-terminal |
| STT3-PglB_core | 2 | STT3/PglB/AglB core domain |
| SUIM_assoc | 2 | Unstructured region C-term to UIM in Ataxin3 |
| SWIRM-assoc_1 | 2 | SWIRM-associated region 1 |
| SWIRM-assoc_2 | 2 | SWIRM-associated domain at the N-terminal |
| SWIRM-assoc_3 | 2 | SWIRM-associated domain at the C-terminal |
| SYCE1 | 2 | Synaptonemal complex central element protein 1 |
| SYCP2_ARLD | 2 | Synaptonemal complex 2 armadillo-repeat-like domain |
| SYCP2_SLD | 2 | Synaptonemal complex 2 Spt16M-like domain |
| Sacchrp_dh_NADP | 2 | Saccharopine dehydrogenase NADP binding domain |
| Saf4_Yju2 | 2 | Saf4/Yju2 protein |
| Sarcoglycan_2 | 2 | Sarcoglycan alpha/epsilon N-terminal domain |
| Sarcoglycan_2_C | 2 | Sarcoglycan alpha/epsilon second domain |
| Sclerostin | 2 | Sclerostin (SOST) |
| Sec15_C | 2 | Exocyst complex subunit Sec15 C-terminal |
| Sec16 | 2 | Vesicle coat trafficking protein Sec16 mid-region |
| Sec2p | 2 | GDP/GTP exchange factor Sec2p |
| Sec5 | 2 | Exocyst complex component Sec5 |
| SecY | 2 | SecY |
| Secretogranin_V | 2 | Neuroendocrine protein 7B2 precursor (Secretogranin V) |
| Securin | 2 | Securin sister-chromatid separation inhibitor |
| Semenogelin | 2 | Semenogelin |
| Sep15_SelM | 2 | Sep15/SelM redox domain |
| Shadoo | 2 | Shadow of prion protein, neuroprotective |
| Sin3_corepress | 2 | Sin3 family co-repressor |
| Sin3a_C | 2 | C-terminal domain of Sin3a protein |
| Sina_RING | 2 | E3 ubiquitin-protein ligase sina/sinah, RING finger |
| Sina_ZnF | 2 | Sina, zinc finger |
| Skp1_POZ | 2 | Skp1 family, tetramerisation domain |
| Slowpoke_C | 2 | Ca2+-activated K+ channel Slowpoke, TrkA_C like domain |
| Smg4_UPF3 | 2 | Smg-4/UPF3 family |
| SnAC | 2 | Snf2-ATP coupling, chromatin remodelling complex |
| Snapin_Pallidin | 2 | Snapin/Pallidin |
| Sorting_nexin | 2 | Sorting nexin, N-terminal domain |
| Sp38 | 2 | Zona-pellucida-binding protein (Sp38) N-terminal Ig domain |
| Sp38_C | 2 | Zona-pellucida-binding protein (Sp38) C-terminal EGF-like domain |
| Speriolin_C | 2 | Speriolin C-terminus |
| Sperm_Ag_HE2 | 2 | Sperm antigen HE2 |
| Spermine_synt_N | 2 | Spermidine synthase tetramerisation domain |
| Spond_N | 2 | Spondin_N |
| Spot_14 | 2 | Thyroid hormone-inducible hepatic protein Spot 14 |
| SprT-like | 2 | SprT-like family |
| Ssb-like_OB | 2 | Single-stranded DNA binding protein Ssb-like, OB fold |
| Stanniocalcin | 2 | Stanniocalcin family |
| Staufen_C | 2 | Staufen C-terminal domain |
| Sterile | 2 | Male sterility protein |
| Stork_head | 2 | Winged helix Storkhead-box1 domain |
| Strabismus | 2 | Strabismus protein |
| Suf | 2 | Suppressor of forked protein (Suf) |
| Sybindin | 2 | Sybindin-like family |
| Syntaphilin | 2 | Golgi-localised syntaxin-1-binding clamp |
| Syntaxin-6_N | 2 | Syntaxin 6, N-terminal |
| T-box_assoc | 2 | T-box transcription factor-associated |
| TAF | 2 | TATA box binding protein associated factor (TAF) |
| TAF4 | 2 | Transcription initiation factor TFIID component TAF4 family |
| TAF6_C | 2 | TAF6 C-terminal HEAT repeat domain |
| TAFII55_N | 2 | TAFII55 protein conserved region |
| TAGT | 2 | TET-Associated Glycosyltransferase |
| TAP42 | 2 | TAP42-like family |
| TAPR1-like | 2 | Telomere attrition and p53 response 1 protein-like |
| TARSH_C | 2 | Target of Nesh-SH3 C-terminal domain |
| TBK1_CCD1 | 2 | TANK-binding kinase 1 coiled-coil domain 1 |
| TBK1_ULD | 2 | TANK binding kinase 1 ubiquitin-like domain |
| TBP-binding | 2 | TATA box-binding protein binding |
| TBX2-3_RD | 2 | T-box transcription factor 2/3, repressor domain |
| TBX2-3_TAD | 2 | T-box transcription factor 2/3, TAD domain |
| TC1 | 2 | Thyroid cancer protein 1 |
| TCR | 2 | Tesmin/TSO1-like CXC domain, cysteine-rich domain |
| TCRP1 | 2 | Tongue Cancer Chemotherapy Resistant Protein 1 |
| TCR_zetazeta | 2 | T-cell surface glycoprotein CD3 zeta chain |
| TEADIR3 | 2 | Omega loop, TEAD interating region 3 |
| TECR_N | 2 | TECR, ubiquitin-like domain |
| TFIID-18kDa | 2 | Transcription initiation factor IID, 18kD subunit |
| TFIID-31kDa | 2 | Transcription initiation factor IID, 31kD subunit |
| TFIID_NTD2 | 2 | WD40 associated region in TFIID subunit, NTD2 domain |
| TGT | 2 | Queuine tRNA-ribosyltransferase |
| THEG | 2 | Testicular haploid expressed repeat |
| TIG_SUH | 2 | TIG domain |
| TIP120 | 2 | TATA-binding protein interacting (TIP20) |
| TIP49 | 2 | TIP49 P-loop domain |
| TIP49_C | 2 | TIP49 AAA-lid domain |
| TIR_3 | 2 | Toll/interleukin-1 receptor domain |
| TMA7 | 2 | Translation machinery associated TMA7 |
| TMCO5 | 2 | TMCO5 family |
| TMEM126 | 2 | Transmembrane protein 126 |
| TMEM131_like | 2 | Transmembrane protein 131-like |
| TMEM131_like_N | 2 | Transmembrane protein 131-like N-terminal |
| TMEM151 | 2 | TMEM151 family |
| TMEM191C | 2 | TMEM191C family |
| TMEM219 | 2 | Transmembrane 219 |
| TMEM238 | 2 | TMEM238 protein family |
| TMEM52 | 2 | Transmembrane 52 |
| TMEM_230_134 | 2 | Transmembrane proteins 230/134 |
| TMPIT | 2 | TMPIT-like protein |
| TOPRIM_C | 2 | C-terminal associated domain of TOPRIM |
| TPP_enzyme_C | 2 | Thiamine pyrophosphate enzyme, C-terminal TPP binding domain |
| TPP_enzyme_M | 2 | Thiamine pyrophosphate enzyme, central domain |
| TPP_enzyme_N | 2 | Thiamine pyrophosphate enzyme, N-terminal TPP binding domain |
| TPR_17 | 2 | Tetratricopeptide repeat |
| TRADD-like_N | 2 | TRADD-like N-terminal domain |
| TRAF3_RING | 2 | TNF receptor-associated factor 2/3/5, RING domain |
| TRAFD1-XIAF1_ZnF | 2 | TRAFD1/XAF1, zinc finger |
| TRAF_BIRC3_bd | 2 | TNF receptor-associated factor BIRC3 binding domain |
| TRAM | 2 | TRAM domain |
| TRC8_N | 2 | TRC8 N-terminal domain |
| TRF | 2 | Telomere repeat binding factor (TRF) |
| TRIC | 2 | TRIC channel |
| TRM | 2 | N2,N2-dimethylguanosine tRNA methyltransferase |
| TROVE | 2 | TROVE domain |
| TTC3_DZIP3_dom | 2 | E3 ubiquitin-protein ligase TTC3/DZIP3 domain |
| TTC7_N | 2 | Tetratricopeptide repeat protein 7 N-terminal |
| TTD | 2 | Tandem tudor domain within UHRF1 |
| TUTase | 2 | TUTase nucleotidyltransferase domain |
| TX13_rpt | 2 | Testis-expressed protein 13, MVPLGDSxSHS repeat |
| Talin_R4 | 2 | Talin, R4 domain |
| Talin_middle | 2 | Talin, middle domain |
| Tankyrase_bdg_C | 2 | Tankyrase binding protein C terminal domain |
| Tantalus | 2 | Drosophila Tantalus-like |
| TauD | 2 | Taurine catabolism dioxygenase TauD, TfdA family |
| Tectonin | 2 | Tectonin domain |
| Thioesterase | 2 | Thioesterase domain |
| Thioredoxin_12 | 2 | Thioredoxin-like domain |
| Thioredoxin_13 | 2 | Thioredoxin-like domain |
| Thioredoxin_14 | 2 | Thioredoxin-like domain |
| Thioredoxin_15 | 2 | Thioredoxin-like domain |
| Thr_synth_N | 2 | Threonine synthase N terminus |
| Thymidylate_kin | 2 | Thymidylate kinase |
| Thyroglob_assoc | 2 | Thyroglobulin_1 repeat associated disordered domain |
| Tiam_CC_Ex | 2 | T-lymphoma invasion and metastasis CC-Ex domain |
| Tim44 | 2 | Tim44-like domain |
| Tis11B_N | 2 | Tis11B like protein, N terminus |
| Tmemb_161AB | 2 | Predicted transmembrane protein 161AB |
| Tmemb_170 | 2 | Putative transmembrane protein 170 |
| Tmemb_55A | 2 | Transmembrane protein 55A |
| Tmemb_9 | 2 | TMEM9 |
| Tmp39 | 2 | Putative transmembrane protein |
| Topo_C_assoc | 2 | C-terminal topoisomerase domain |
| Topoisom_I | 2 | Eukaryotic DNA topoisomerase I, catalytic core |
| Topoisom_I_N | 2 | Eukaryotic DNA topoisomerase I, DNA binding fragment |
| Topoisom_bac | 2 | DNA topoisomerase |
| Translin | 2 | Translin family |
| Trm112p | 2 | Trm112p-like protein |
| Trnau1ap | 2 | Selenocysteine tRNA 1 associated proteins |
| TruD | 2 | tRNA pseudouridine synthase D (TruD) |
| TsaD | 2 | tRNA N6-adenosine threonylcarbamoyltransferase |
| TspO_MBR | 2 | TspO/MBR family |
| Tugs | 2 | Tethering Ubl4a to BAGS domain |
| Tyr_Deacylase | 2 | D-Tyr-tRNA(Tyr) deacylase |
| UBAP-1_UBA2 | 2 | Ubiquitin-associated protein 1, UBA2 domain |
| UBAP2-Lig | 2 | UBAP2/protein lingerer |
| UBA_BRSK | 2 | BRSK1/2 UBA domain |
| UBD | 2 | Ubiquitin-binding domain |
| UBM | 2 | Ubiquitin binding region |
| UBN_AB | 2 | Ubinuclein conserved middle domain |
| UCH_C | 2 | Ubiquitin carboxyl-terminal hydrolases |
| UCR_hinge | 2 | Ubiquinol-cytochrome C reductase hinge protein |
| UDP-g_GGTase | 2 | UDP-glucose:Glycoprotein Glucosyltransferase |
| UEV | 2 | UEV domain |
| ULD | 2 | Ubiquitin-like oligomerisation domain of SATB |
| UMPH-1 | 2 | Pyrimidine 5'-nucleotidase (UMPH-1) |
| UNC45-central | 2 | Myosin-binding striated muscle assembly central |
| UPAR_LY6_2 | 2 | Ly6/PLAUR domain-containing protein 6, Lypd6 |
| UPF0004 | 2 | Uncharacterized protein family UPF0004 |
| UPF0172 | 2 | Uncharacterised protein family (UPF0172) |
| UPF0220 | 2 | Uncharacterised protein family (UPF0220) |
| UPF0552 | 2 | Arp2/3-interacting proteins Arpin |
| UPF0731 | 2 | UPF0731 family |
| UPRTase | 2 | Uracil phosphoribosyltransferase |
| USHBP1_PDZ-bd | 2 | Harmonin-binding protein USHBP1, PDZ-binding domain |
| UT | 2 | Urea transporter |
| Ufd2P_core | 2 | Ubiquitin elongating factor core |
| Usp38-like_N | 2 | Ubiquitin carboxyl-terminal hydrolase 38-like, N-terminal domain |
| Utp14 | 2 | Utp14 protein |
| UvrD-helicase | 2 | UvrD/REP helicase N-terminal domain |
| V-ATPase_C | 2 | V-ATPase subunit C |
| V-SNARE | 2 | Vesicle transport v-SNARE protein N-terminus |
| V-set_2 | 2 | ICOS V-set domain |
| VBS | 2 | Vinculin Binding Site |
| VHL | 2 | VHL beta domain |
| VIT_2 | 2 | Vault protein inter-alpha-trypsin domain |
| VKOR | 2 | Vitamin K epoxide reductase family |
| VMA21 | 2 | VMA21-like domain |
| Vac14_Fab1_bd | 2 | Vacuolar 14 Fab1-binding region |
| Vasculin | 2 | Vascular protein family Vasculin-like 1 |
| Vasohibin | 2 | Vasohibin |
| Vert_HS_TF | 2 | Vertebrate heat shock transcription factor |
| Vitellogenin_N | 2 | Lipoprotein amino terminal region |
| Vps16_C | 2 | Vps16, C-terminal region |
| Vps39_1 | 2 | Vacuolar sorting protein 39 domain 1 |
| Vps39_2 | 2 | Vacuolar sorting protein 39 domain 2 |
| Vps54_N | 2 | Vacuolar-sorting protein 54, of GARP complex |
| Vps55 | 2 | Vacuolar protein sorting 55 |
| WAC_Acf1_DNA_bd | 2 | ATP-utilising chromatin assembly and remodelling N-terminal |
| WASH_WAHD | 2 | WAHD domain of WASH complex |
| WHAMM-JMY_N | 2 | N-terminal of Junction-mediating and WASP homolog-associated |
| WIF | 2 | WIF domain |
| WLS-like_TM | 2 | Wntless-like, transmembrane domain |
| WSK | 2 | WSK motif |
| Wyosine_form | 2 | Wyosine base formation |
| XAP5 | 2 | XAP5, circadian clock regulator C-terminal domain |
| XPC-binding | 2 | XPC-binding domain |
| XRCC1_N | 2 | XRCC1 N terminal domain |
| XRN_M | 2 | Xrn1 helical domain |
| XRN_N | 2 | XRN 5'-3' exonuclease N-terminus |
| Xin | 2 | Xin repeat |
| Xylo_C | 2 | Xylosyltransferase C terminal |
| YAF2_RYBP | 2 | Yaf2/RYBP C-terminal binding motif |
| YIF1 | 2 | YIF1 |
| YL1_C | 2 | YL1 nuclear protein C-terminal domain |
| Yae1_N | 2 | Essential protein Yae1, N terminal |
| YccV-like | 2 | Hemimethylated DNA-binding protein YccV like |
| YhhN | 2 | YhhN family |
| ZIP4_domain | 2 | Zinc transporter ZIP4 domain |
| ZNF512_C2HC | 2 | Zinc finger protein 512, C2HC zinc finger |
| ZNRF_3_ecto | 2 | ZNRF-3 Ectodomain |
| ZSWIM1-3_C | 2 | Zinc finger SWIM domain-containing protein 1/3, C-terminal |
| ZSWIM1-3_RNaseH-like | 2 | Zinc finger SWIM domain-containing protein 1/3, RNaseH-like domain |
| ZSWIM1-3_helical | 2 | ZSWIM1/3, helical domain |
| Zn-C2H2_12 | 2 | Autophagy receptor zinc finger-C2H2 domain |
| ZnF_RZ-type | 2 | RZ type zinc finger domain |
| betaPIX_CC | 2 | betaPIX coiled coil |
| cpSF2-GREB1 | 2 | Circularly permuted SF2 helicase found in GREB1 proteins |
| eIF-3c_N | 2 | Eukaryotic translation initiation factor 3 subunit 8 N-terminus |
| eIF-5_eIF-2B | 2 | Domain found in IF2B/IF5 |
| eIF2_C | 2 | Initiation factor eIF2 gamma, C terminal |
| eRF1_1 | 2 | eRF1 domain 1 |
| eRF1_2 | 2 | eRF1 domain 2 |
| eRF1_3 | 2 | eRF1 domain 3 |
| fn3_4 | 2 | Fibronectin-III type domain |
| hnRNP_Q_AcD | 2 | Heterogeneous nuclear ribonucleoprotein Q acidic domain |
| iRhom1-2_N | 2 | Inactive rhomboid proteins 1/2, N-terminal |
| ketoacyl-synt | 2 | Beta-ketoacyl synthase, N-terminal domain |
| mRNA_decap_C | 2 | mRNA-decapping enzyme C-terminus |
| nlz1 | 2 | NocA-like zinc-finger protein 1 |
| preSET_CXC | 2 | CXC domain |
| tRNA-synt_1c_C | 2 | tRNA synthetases class I (E and Q), anti-codon binding domain |
| tRNA-synt_1c_C2 | 2 | tRNA synthetases class I (E and Q), anti-codon binding domain |
| tRNA-synt_1d | 2 | tRNA synthetases class I (R) |
| tRNA-synt_1e | 2 | tRNA synthetases class I (C) catalytic domain |
| tRNA-synt_1g | 2 | tRNA synthetases class I (M) |
| tRNA-synt_2c | 2 | tRNA synthetases class II (A) |
| tRNA-synt_2d | 2 | tRNA synthetases class II core domain (F) |
| tRNA_U5-meth_tr | 2 | tRNA (Uracil-5-)-methyltransferase |
| tRNA_bind | 2 | Putative tRNA binding domain |
| tRNA_int_endo | 2 | tRNA intron endonuclease, catalytic C-terminal domain |
| vATP-synt_AC39 | 2 | ATP synthase (C/AC39) subunit |
| vATP-synt_E | 2 | ATP synthase (E/31 kDa) subunit |
| z-alpha | 2 | Adenosine deaminase z-alpha domain |
| zf-4CXXC_R1 | 2 | Zinc-finger domain of monoamine-oxidase A repressor R1 |
| zf-AD | 2 | Zinc-finger associated domain (zf-AD) |
| zf-ANAPC11 | 2 | Anaphase-promoting complex subunit 11 RING-H2 finger |
| zf-C2H2_2 | 2 | C2H2 type zinc-finger (2 copies) |
| zf-C2H2_3 | 2 | zinc-finger of acetyl-transferase ESCO |
| zf-C2H2_7 | 2 | Zinc-finger |
| zf-C3Hc3H | 2 | Potential DNA-binding domain |
| zf-C4pol | 2 | C4-type zinc-finger of DNA polymerase delta |
| zf-CCHC_2 | 2 | Zinc knuckle |
| zf-CCHC_3 | 2 | Zinc knuckle |
| zf-CSL | 2 | CSL zinc finger |
| zf-NADH-PPase | 2 | NADH pyrophosphatase zinc ribbon domain |
| zf-NF-X1 | 2 | NF-X1 type zinc finger |
| zf-PARP | 2 | Poly(ADP-ribose) polymerase and DNA-Ligase Zn-finger region |
| zf-SAP30 | 2 | SAP30 zinc-finger |
| zf-TAZ | 2 | TAZ zinc finger |
| zf-TRAF_2 | 2 | TRAF-like zinc-finger |
| zf-UBP_var | 2 | Variant UBP zinc finger |
| zf-piccolo | 2 | Piccolo Zn-finger |
| zf-rbx1 | 2 | RING-H2 zinc finger domain |
| zf_C2H2_10 | 2 | C2H2 type zinc-finger |
| zf_CCCH_5 | 2 | Unkempt Zinc finger domain 1 (Znf1) |
| zf_Hakai | 2 | C2H2 Hakai zinc finger domain |
| 23ISL | 1 | Unstructured linker between I-set domains 2 and 3 on MYLCK |
| 2Fe-2S_thioredx | 1 | Thioredoxin-like [2Fe-2S] ferredoxin |
| 2OG-FeII_Oxy_4 | 1 | 2OG-Fe(II) oxygenase superfamily |
| 3-HAO | 1 | 3-hydroxyanthranilic acid dioxygenase |
| 4HBT_2 | 1 | Thioesterase-like superfamily |
| 4HBT_3 | 1 | Acyl-CoA thioesterase N-terminal domain |
| 53-BP1_Tudor | 1 | Tumour suppressor p53-binding protein-1 Tudor |
| 5HT_transport_N | 1 | Serotonin (5-HT) neurotransmitter transporter, N-terminus |
| 5_nucleotid_C | 1 | 5'-nucleotidase, C-terminal domain |
| 6PGD | 1 | 6-phosphogluconate dehydrogenase, C-terminal domain |
| AAA_21 | 1 | AAA domain, putative AbiEii toxin, Type IV TA system |
| AAA_22 | 1 | AAA domain |
| AAA_25 | 1 | AAA domain |
| AAA_30 | 1 | AAA domain |
| AAA_assoc_2 | 1 | AAA C-terminal domain |
| AAA_lid_5 | 1 | Midasin AAA lid domain |
| AAA_lid_7 | 1 | Midasin AAA lid domain |
| AAR2 | 1 | AAR2 C-terminal repeat region |
| AAR2_1st | 1 | AAR2 N-terminal domain |
| AAT | 1 | Acyl-coenzyme A:6-aminopenicillanic acid acyl-transferase |
| AATF-Che1 | 1 | Apoptosis antagonizing transcription factor |
| ABC_trans_CmpB | 1 | Putative ABC-transporter type IV |
| ABC_transp_aux | 1 | ABC-type uncharacterized transport system |
| ABHD18 | 1 | Alpha/beta hydrolase domain containing 18 |
| ACPS | 1 | 4'-phosphopantetheinyl transferase superfamily |
| ACT | 1 | ACT domain |
| ACTH_domain | 1 | Corticotropin ACTH domain |
| ACTL7A_N | 1 | Actin-like protein 7A N-terminus |
| ACTMAP-like_C | 1 | Actin maturation protease-like, C-terminal domain |
| AD | 1 | Anticodon-binding domain |
| ADAM10_Cys-rich | 1 | ADAM10, cysteine-rich domain |
| ADAM17_MPD | 1 | Membrane-proximal domain, switch, for ADAM17 |
| ADD_ATRX | 1 | Cysteine Rich ADD domain |
| ADIP | 1 | Afadin- and alpha -actinin-Binding |
| ADP_PFK_GK | 1 | ADP-specific Phosphofructokinase/Glucokinase conserved region |
| ADSL_C | 1 | Adenylosuccinate lyase C-terminus |
| AFG1_ATPase | 1 | AFG1-like ATPase |
| AGK_C | 1 | Acylglycerol kinase C-terminal |
| AGTRAP | 1 | Angiotensin II, type I receptor-associated protein (AGTRAP) |
| AHSA1 | 1 | Activator of Hsp90 ATPase homolog 1-like protein |
| AHSP | 1 | Alpha-haemoglobin stabilising protein |
| AI-2E_transport | 1 | AI-2E family transporter |
| AIB | 1 | Aurora kinase A and ninein interacting protein |
| AICARFT_IMPCHas | 1 | AICARFT/IMPCHase bienzyme |
| AID | 1 | Activation induced deaminase |
| AIF_C | 1 | Apoptosis-inducing factor, mitochondrion-associated, C-term |
| AIG2_2 | 1 | AIG2-like family |
| AIMP2_LysRS_bd | 1 | AIMP2 lysyl-tRNA synthetase binding domain |
| AIRC | 1 | AIR carboxylase |
| AKAP28 | 1 | 28 kDa A-kinase anchor |
| ALAD | 1 | Delta-aminolevulinic acid dehydratase |
| ALG11_N | 1 | ALG11 mannosyltransferase N-terminus |
| ALG3 | 1 | ALG3 protein |
| ALKBH8_N | 1 | Alkylated DNA repair protein alkB homolog 8, N-terminal |
| ALMS_repeat | 1 | Alstrom syndrome repeat |
| ALS2CR8 | 1 | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 8 |
| AMG1_II | 1 | Phosphoacetylglucosamine mutase AMG1, domain II |
| AMG1_III | 1 | Phosphoacetylglucosamine mutase AMG1, domain III |
| AMH_N | 1 | Anti-Mullerian hormone, N terminal region |
| ANAPC1 | 1 | Anaphase-promoting complex subunit 1 WD40 beta-propeller domain |
| ANAPC15 | 1 | Anaphase-promoting complex subunit 15 |
| ANAPC16 | 1 | Anaphase-promoting complex, subunit 16 |
| ANAPC2 | 1 | Anaphase promoting complex (APC) subunit 2 |
| ANAPC4 | 1 | Anaphase-promoting complex, cyclosome, subunit 4 |
| ANAPC5 | 1 | Anaphase-promoting complex subunit 5 |
| ANAPC8 | 1 | Anaphase promoting complex subunit 8 / Cdc23 |
| ANAPC_CDC26 | 1 | Anaphase-promoting complex APC subunit CDC26 |
| ANGEL2_N | 1 | Protein angel homolog 2 N-terminal |
| ANKH | 1 | Progressive ankylosis protein (ANKH) |
| ANM3_C2H2_Zf | 1 | Protein arginine N-methyltransferase 3, C2H2 zinc finger domain |
| ANM3_zf-C2H2 | 1 | Protein arginine N-methyltransferase 3, C2H2 zinc finger |
| ANXA2R | 1 | Annexin-2 receptor |
| AP-5_subunit_s1 | 1 | AP-5 complex subunit sigma-1 |
| AP1AR | 1 | AP-1 complex-associated regulatory protein |
| AP3D1 | 1 | AP-3 complex subunit delta-1 |
| AP4E_app_platf | 1 | Adaptin AP4 complex epsilon appendage platform |
| AP5B1_C | 1 | AP5B1, C-terminal |
| AP5B1_N | 1 | AP-5 complex subunit beta-1, N-terminal |
| AP5B1_barrel | 1 | AP-5 complex subunit beta-1, beta-barrel |
| AP5B1_middle | 1 | AP5B1, middle domain |
| APAF-1-like_WHD | 1 | Apoptotic protease-activating factor 1-like, winged-helix domain |
| APAF1_C | 1 | APAF-1 helical domain |
| APC1_3rd | 1 | APC1 beta sandwich domain |
| APC1_C | 1 | Anaphase-promoting complex sub unit 1 C-terminal domain |
| APC_15aa | 1 | APC 15 residue motif |
| APC_u13 | 1 | Unstructured region on APC between APC_crr and SAMP |
| APC_u14 | 1 | Unstructured region on APC between SAMP and APC_crr |
| APC_u15 | 1 | Unstructured region on APC between APC_crr regions 5 and 6 |
| APC_u5 | 1 | Unstructured region on APC between 1st and 2nd catenin-bdg motifs |
| APC_u9 | 1 | Unstructured region on APC between 1st two creatine-rich regions |
| APEH_N | 1 | Acylamino-acid-releasing enzyme, N-terminal domain |
| APG12 | 1 | Ubiquitin-like autophagy protein Apg12 |
| API5 | 1 | Apoptosis inhibitory protein 5 (API5) |
| APOBEC2 | 1 | APOBEC2 |
| APOBEC3 | 1 | APOBEC3 |
| APOBEC4_like | 1 | APOBEC4-like -AID/APOBEC-deaminase |
| APOBEC_N | 1 | APOBEC-like N-terminal domain |
| AP_endonuc_2 | 1 | Xylose isomerase-like TIM barrel |
| ARA70 | 1 | Nuclear coactivator |
| ARD | 1 | ARD/ARD' family |
| ARGLU | 1 | Arginine and glutamate-rich 1 |
| ARI1_UBAl | 1 | E3 ubiquitin-protein ligase ARIH1, UBA-like domain |
| ARL6IP6 | 1 | Haemopoietic lineage transmembrane helix |
| ARMC9_ARM | 1 | LisH domain-containing protein ARMC9, ARM repeats |
| ARMC9_LisH | 1 | ARMC9, LIS1 homology motif |
| ARMH2 | 1 | Armadillo-like helical domain-containing protein 2 |
| ARMH3_C | 1 | Armadillo-like helical domain-containing protein 3, C-terminal |
| ARMH4 | 1 | Armadillo-like helical domain-containing protein 4 |
| AROS | 1 | Active regulator of SIRT1, or 40S ribosomal protein S19-binding 1 |
| ARS2 | 1 | Arsenite-resistance protein 2 |
| ARTD15_N | 1 | ARTD15 N-terminal domain |
| ASCH | 1 | ASCH domain |
| ASH2L-like_WH | 1 | Set1/Ash2 histone methyltransferase complex subunit ASH2-like, WH |
| ASL_C2 | 1 | Argininosuccinate lyase C-terminal |
| ASTER | 1 | PAT complex subunit Asterix |
| ASTN_2_hairpin | 1 | Astrotactin-2 C-terminal beta-hairpin domain |
| ATE_C | 1 | Arginine-tRNA-protein transferase, C terminus |
| ATE_N | 1 | Arginine-tRNA-protein transferase, N terminus |
| ATG101 | 1 | Autophagy-related protein 101 |
| ATG11 | 1 | Autophagy-related protein 11 |
| ATG13 | 1 | Autophagy-related protein 13 |
| ATG2_CAD | 1 | Autophagy-related protein 2, middle RBG modules |
| ATG5_HBR | 1 | Autophagy protein ATG5, alpha-helical bundle region |
| ATG5_UblA | 1 | Autophagy protein ATG5, UblA domain |
| ATG5_UblB | 1 | Autophagy protein ATG5, UblB domain |
| ATG7_N | 1 | Ubiquitin-like modifier-activating enzyme ATG7 N-terminus |
| ATP-cone | 1 | ATP cone domain |
| ATP-synt | 1 | ATP synthase |
| ATP-synt_8 | 1 | ATP synthase protein 8 |
| ATP-synt_A | 1 | ATP synthase A chain |
| ATP-synt_D | 1 | ATP synthase subunit D |
| ATP-synt_DE_N | 1 | ATP synthase, Delta/Epsilon chain, beta-sandwich domain |
| ATP-synt_E | 1 | ATP synthase E chain |
| ATP-synt_Eps | 1 | Mitochondrial ATP synthase epsilon chain |
| ATP-synt_F | 1 | ATP synthase (F/14-kDa) subunit |
| ATP-synt_F6 | 1 | Mitochondrial ATP synthase coupling factor 6 |
| ATP-synt_ab_C | 1 | ATP synthase alpha/beta chain, C terminal domain |
| ATP-synt_ab_Xtn | 1 | ATPsynthase alpha/beta subunit N-term extension |
| ATP11 | 1 | ATP11 protein |
| ATP12 | 1 | ATP12 chaperone protein |
| ATPD_C_metazoa | 1 | Metazoan delta subunit of F1F0-ATP synthase, C-terminal domain |
| ATP_bind_3 | 1 | PP-loop family |
| ATP_synth_reg | 1 | ATP synthase regulation |
| ATPgrasp_N | 1 | ATP-grasp N-terminal domain |
| ATPgrasp_Ter | 1 | ATP-grasp in the biosynthetic pathway with Ter operon |
| AZUL | 1 | Amino-terminal Zinc-binding domain of ubiquitin ligase E3A |
| A_deaminase_N | 1 | Adenosine/AMP deaminase N-terminal |
| Ac45-VOA1_TM | 1 | V0 complex accessory subunit Ac45/VOA1 transmembrane domain |
| Acatn | 1 | Acetyl-coenzyme A transporter 1 |
| Acetyltransf_16 | 1 | GNAT acetyltransferase, Mec-17 |
| Acetyltransf_3 | 1 | Acetyltransferase (GNAT) domain |
| Acetyltransf_CG | 1 | GCN5-related N-acetyl-transferase |
| Acyl_CoA_thio | 1 | Acyl-CoA thioesterase |
| Ada3 | 1 | Histone acetyltransferases subunit 3 |
| Adaptin_binding | 1 | Alpha and gamma adaptin binding protein p34 |
| Adhes-Ig_like | 1 | Adhesion molecule, immunoglobulin-like |
| Adipogenin | 1 | Adipogenin |
| AdoMet_MTase | 1 | Predicted AdoMet-dependent methyltransferase |
| Afaf | 1 | Acrosome formation-associated factor |
| Aha1_N | 1 | Activator of Hsp90 ATPase, N-terminal |
| Aida_C2 | 1 | Axin interactor dorsalisation-associated protein, C-terminal |
| Aida_N | 1 | Aida N-terminus |
| AlaDh_PNT_C | 1 | Alanine dehydrogenase/PNT, C-terminal domain |
| Ala_racemase_N | 1 | Alanine racemase, N-terminal domain |
| Aldose_epim | 1 | Aldose 1-epimerase |
| Alg14 | 1 | Oligosaccharide biosynthesis protein Alg14 like |
| Allantoicase | 1 | Allantoicase repeat |
| Alpha-2-MRAP_C | 1 | Alpha-2-macroglobulin RAP, C-terminal domain |
| Alpha-2-MRAP_N | 1 | Alpha-2-macroglobulin RAP, N-terminal domain |
| Alveol-reg_P311 | 1 | Neuronal protein 3.1 (p311) |
| Amelin | 1 | Ameloblastin precursor (Amelin) |
| Amelotin | 1 | Amelotin |
| Amidohydro_2 | 1 | Amidohydrolase |
| Amnionless | 1 | Amnionless |
| Anamorsin_N | 1 | Anamorsin, N-terminal |
| Androgen_recep | 1 | Androgen receptor |
| Anillin_N | 1 | Anillin N-terminus |
| AnkUBD | 1 | Ankyrin ubiquitin-binding domain |
| Anticodon_2 | 1 | Anticodon binding domain |
| Antistasin | 1 | Antistasin family |
| Apc13p | 1 | Apc13p protein |
| Apc1_MidN | 1 | Anaphase-promoting complex subunit 1 middle domain |
| Apc5_N | 1 | Anaphase-promoting complex subunit 5, N-terminal domain |
| Apelin | 1 | APJ endogenous ligand |
| Apo-CIII | 1 | Apolipoprotein CIII (Apo-CIII) |
| ApoA-II | 1 | Apolipoprotein A-II (ApoA-II) |
| ApoB100_C | 1 | Apolipoprotein B100 C terminal |
| ApoC-I | 1 | Apolipoprotein C-I (ApoC-1) |
| ApoM | 1 | ApoM domain |
| Apolipo_F | 1 | Apolipoprotein F |
| Apyrase | 1 | Apyrase |
| Aquarius_N_1st | 1 | Intron-binding protein aquarius N-terminal |
| Aquarius_N_2nd | 1 | Intron-binding protein aquarius, beta-barrel |
| Aquarius_N_3rd | 1 | Intron-binding protein aquarius insert domain |
| Arb2-like | 1 | Arb2-like domain |
| Arc_C | 1 | Arc C-lobe |
| Arc_MA | 1 | Arc MA domain |
| Arc_capsid_dom | 1 | Activity-regulated cytoskeleton-associated protein, capsid domain |
| Archease | 1 | Archease protein family (MTH1598/TM1083) |
| Arg_tRNA_synt_N | 1 | Arginyl tRNA synthetase N terminal domain |
| Arginosuc_syn_C | 1 | Arginosuccinate synthase C-terminal domain |
| Arginosuc_synth | 1 | Arginosuccinate synthase N-terminal HUP domain |
| Arm_vescicular | 1 | Armadillo tether-repeat of vescicular transport factor |
| ArsA_ATPase | 1 | Anion-transporting ATPase |
| Arv1 | 1 | Arv1-like family |
| Ashwin | 1 | Developmental protein |
| AsnRS_N | 1 | Asparaginal-tRNA synthetase, N-terminal domain |
| Asp_DH_C | 1 | Aspartate dehydrogenase, C-terminal |
| Asp_Glu_race_2 | 1 | Putative aspartate racemase |
| Asparaginase | 1 | Asparaginase, N-terminal |
| Asparaginase_C | 1 | Glutaminase/Asparaginase C-terminal domain |
| Atthog | 1 | Attenuator of Hedgehog |
| Atx10homo_assoc | 1 | Spinocerebellar ataxia type 10 protein domain |
| Atypical_Card | 1 | Atypical caspase recruitment domain |
| Augurin | 1 | Oesophageal cancer-related gene 4 |
| Aurora-A_bind | 1 | Aurora-A binding |
| Auto_anti-p27 | 1 | Sjogren's syndrome/scleroderma autoantigen 1 (Autoantigen p27) |
| B-block_TFIIIC | 1 | B-block binding subunit of TFIIIC |
| B12-binding_2 | 1 | B12 binding domain |
| B3GALT2_N | 1 | Beta-1,3-galactosyltransferase 2 N-terminus |
| B3_4 | 1 | B3/4 domain |
| B5 | 1 | tRNA synthetase B5 domain |
| BAALC_N | 1 | BAALC N-terminus |
| BAG6 | 1 | BCL2-associated athanogene 6 |
| BAMBI | 1 | BMP and activin membrane-bound inhibitor (BAMBI) N-terminal domain |
| BAMBI_C | 1 | BMP and activin membrane-bound inhibitor homolog C-terminus |
| BASP1 | 1 | Brain acid soluble protein 1 (BASP1 protein) |
| BBIP10 | 1 | Cilia BBSome complex subunit 10 |
| BBL5 | 1 | Bardet-Biedl syndrome 5 protein |
| BBS1 | 1 | Ciliary BBSome complex subunit 1 |
| BBS2_C | 1 | Ciliary BBSome complex subunit 2, C-terminal |
| BBS2_Mid | 1 | Ciliary BBSome complex subunit 2, middle region |
| BBS2_N | 1 | Ciliary BBSome complex subunit 2, N-terminal |
| BC10 | 1 | Bladder cancer-related protein BC10 |
| BCAS2 | 1 | Breast carcinoma amplified sequence 2 (BCAS2) |
| BCAS3 | 1 | BCAS3 microtubule associated cell migration factor, C-terminal |
| BCAS3_WD40 | 1 | BCAS3, WD40 repeat |
| BCCIP | 1 | BCCIP |
| BCMA-Tall_bind | 1 | BCMA, TALL-1 binding |
| BCNT | 1 | Bucentaur or craniofacial development |
| BCOR | 1 | BCL-6 co-repressor, non-ankyrin-repeat region |
| BCS1_N | 1 | BCS1 N terminal |
| BD-FAE | 1 | BD-FAE |
| BDHCT | 1 | BDHCT (NUC031) domain |
| BDHCT_assoc | 1 | BDHCT-box associated domain on Bloom syndrome protein |
| BH3 | 1 | Beclin-1 BH3 domain, Bcl-2-interacting |
| BHD_1 | 1 | Rad4 beta-hairpin domain 1 |
| BHD_2 | 1 | Rad4 beta-hairpin domain 2 |
| BHD_3 | 1 | Rad4 beta-hairpin domain 3 |
| BID | 1 | BH3 interacting domain (BID) |
| BING4CT | 1 | BING4CT (NUC141) domain |
| BIRC6 | 1 | Baculoviral IAP repeat-containing protein 6 |
| BLACT_WH | 1 | Beta-lactamase associated winged helix domain |
| BLM10_mid | 1 | Proteasome-substrate-size regulator, mid region |
| BLM_N | 1 | N-terminal region of Bloom syndrome protein |
| BLOC1S3 | 1 | Biogenesis of lysosome-related organelles complex 1 subunit 3 |
| BLOC1_2 | 1 | Biogenesis of lysosome-related organelles complex-1 subunit 2 |
| BLTP1_C | 1 | Bridge-like lipid transfer protein family member 1, C-terminal |
| BLTP1_N | 1 | BLTP1 N-terminal region |
| BLTP1_mid | 1 | BLTP1, middle RBG modules |
| BLTP2_RBG-6 | 1 | BLTP2, sixth RBG module |
| BMF | 1 | Bcl-2-modifying factor, apoptosis |
| BMT5-like | 1 | rRNA (uridine-N3-)-methyltransferase BTM5-like |
| BNR_6 | 1 | BNR-Asp box repeat |
| BOP1NT | 1 | BOP1NT (NUC169) domain |
| BORA_N | 1 | Protein aurora borealis N-terminus |
| BORCS6 | 1 | BLOC-1-related complex sub-unit 6 C-terminal helix |
| BORCS7 | 1 | BLOC-1-related complex sub-unit 7 |
| BORCS8 | 1 | BLOC-1-related complex sub-unit 8 |
| BP28CT | 1 | BP28CT (NUC211) domain |
| BPL_C | 1 | Biotin protein ligase C terminal domain |
| BPL_LplA_LipB | 1 | Biotin/lipoate A/B protein ligase family |
| BRAP2 | 1 | BRCA1-associated protein 2 |
| BRCA-2_OB1 | 1 | BRCA2, oligonucleotide/oligosaccharide-binding, domain 1 |
| BRCA-2_OB3 | 1 | BRCA2, oligonucleotide/oligosaccharide-binding, domain 3 |
| BRCA-2_helical | 1 | BRCA2, helical |
| BRCA2 | 1 | BRCA2 repeat |
| BRCA2DBD_OB2 | 1 | BRCA2, OB2 |
| BRCC36_C | 1 | BRCC36 C-terminal helical domain |
| BRCT_3 | 1 | BRCA1 C Terminus (BRCT) domain |
| BRCT_assoc | 1 | Serine-rich domain associated with BRCT |
| BRE | 1 | Brain and reproductive organ-expressed protein (BRE) |
| BRE1 | 1 | BRE1 E3 ubiquitin ligase |
| BRF1 | 1 | Brf1-like TBP-binding domain |
| BRI3 | 1 | Brain protein I3 |
| BROMI | 1 | Broad-minded protein |
| BRR2_plug | 1 | Pre-mRNA-splicing helicase BRR2 plug domain |
| BSP_II | 1 | Bone sialoprotein II (BSP-II) |
| BST2 | 1 | Bone marrow stromal antigen 2 |
| BTBD16_C | 1 | BTB/POZ domain-containing protein 16, C-terminal domain |
| BUD22 | 1 | BUD22 |
| BUD31 | 1 | Pre-mRNA-splicing factor BUD31 |
| BaffR-Tall_bind | 1 | BAFF-R, TALL-1 binding |
| Bag6_BAGS | 1 | Bag6, BAG-similar domain |
| Band_7_C | 1 | C-terminal region of band_7 |
| Barttin | 1 | Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| Bcl-2_BAD | 1 | Pro-apoptotic Bcl-2 protein, BAD |
| Bclt | 1 | Putative Bcl-2 like protein of testis |
| Bclx_interact | 1 | Bcl-x interacting, BH3 domain |
| BcrAD_BadFG | 1 | BadF/BadG/BcrA/BcrD ATPase family |
| Beta-APP | 1 | Beta-amyloid peptide (beta-APP) |
| Beta_elim_lyase | 1 | Beta-eliminating lyase |
| Biliv-reduc_cat | 1 | Biliverdin reductase, catalytic |
| Bim_N | 1 | Bim protein N-terminus |
| Bin2_C | 1 | Bridging integrator 2, C-terminal tail |
| BioT2 | 1 | Spermatogenesis family BioT2 |
| Bles03 | 1 | Basophilic leukemia-expressed protein Bles03 |
| Bmt2 | 1 | 25S rRNA (adenine(2142)-N(1))-methyltransferase, Bmt2 |
| Borealin | 1 | Cell division cycle-associated protein 8 |
| BrkDBD | 1 | Brinker DNA-binding domain |
| Brme1 | 1 | Break repair meiotic recombinase recruitment factor 1 |
| Bud13 | 1 | Pre-mRNA-splicing factor of RES complex |
| Bystin | 1 | Bystin |
| C12orf66_like | 1 | KICSTOR complex C12orf66 like |
| C19orf12 | 1 | C19orf12-like protein |
| C1ORF64 | 1 | Steroid receptor-associated and regulated protein |
| C2orf69 | 1 | C2orf69 |
| C3_CUB1 | 1 | Complement component 3, CUB domain, first segment |
| C3_CUB2 | 1 | Complement component 3, CUB domain, second segment |
| C4bp_oligo | 1 | Oligomerization domain of C4b-binding protein alpha |
| C5-epim_C | 1 | D-glucuronyl C5-epimerase C-terminus |
| C5_CUB | 1 | Complement component 5, CUB domain |
| C6_DPF | 1 | Cysteine-rich domain |
| C6_KAZAL | 1 | Complement component C6, KAZAL domain |
| C7_FIM2_N | 1 | Complement component C7, FIM2 N-terminal |
| C7_KAZAL | 1 | Complement component C7, Kazal domain |
| C9orf72-like | 1 | C9orf72-like protein family |
| CA109-like | 1 | Ribosome biogenesis protein C1orf109-like |
| CAAP1 | 1 | Caspase activity and apoptosis inhibitor 1 |
| CABS1 | 1 | Calcium-binding and spermatid-specific protein 1 |
| CAF-1_p150 | 1 | Chromatin assembly factor 1 complex p150 subunit, N-terminal |
| CAF-1_p60_C | 1 | Chromatin assembly factor complex 1 subunit p60, C-terminal |
| CAF1-p150_C2 | 1 | CAF1 complex subunit p150, region binding to CAF1-p60 at C-term |
| CAF1-p150_N | 1 | CAF1 complex subunit p150, region binding to PCNA |
| CAF1A | 1 | Chromatin assembly factor 1 subunit A |
| CAGE1 | 1 | Cancer-associated gene protein 1 family |
| CAP18_C | 1 | LPS binding domain of CAP18 (C terminal) |
| CARM1 | 1 | Coactivator-associated arginine methyltransferase 1 N terminal |
| CARME | 1 | Carnosine N-methyltransferase |
| CART | 1 | Cocaine and amphetamine regulated transcript protein (CART) |
| CASP_C | 1 | CASP C terminal |
| CAS_CSE1 | 1 | CAS/CSE protein, C-terminus |
| CATIP_N | 1 | Ciliogenesis-associated TTC17-interacting protein, N-terminal domain |
| CATSPERB_1st | 1 | CATSPER protein subunit beta protein 1st domain |
| CATSPERB_2nd | 1 | CATSPER subunit beta protein 2nd domain |
| CATSPERB_C | 1 | CATSPERB, C-terminal domain |
| CATSPERG_beta-prop | 1 | CATSPERG beta-propeller domain |
| CBFB_NFYA | 1 | CCAAT-binding transcription factor (CBF-B/NF-YA) subunit B |
| CBF_beta | 1 | Core binding factor beta subunit |
| CBM_48 | 1 | Carbohydrate-binding module 48 (Isoamylase N-terminal domain) |
| CC149 | 1 | Coiled-coil domain-containing protein 149-A |
| CC190 | 1 | Coiled-coil domain-containing protein 190 |
| CCCAP | 1 | Centrosomal colon cancer autoantigen protein family |
| CCD48 | 1 | Coiled-coil domain-containing protein 48 |
| CCD97-like_C | 1 | Coiled-coil domain containing protein 97-like, C-terminal |
| CCDC-167 | 1 | Coiled-coil domain-containing protein 167 |
| CCDC106 | 1 | Coiled-coil domain-containing protein 106 |
| CCDC117 | 1 | Coiled-coil domain-containing protein 117 |
| CCDC138_C | 1 | Coiled-coil domain-containing protein 138, C-terminal domain |
| CCDC138_cc | 1 | Coiled-coil-domain-containing protein 138, coiled-coil |
| CCDC14 | 1 | Coiled-coil domain-containing protein 14 |
| CCDC142 | 1 | Coiled-coil protein 142 |
| CCDC154 | 1 | Coiled-coil domain-containing protein 154 |
| CCDC158 | 1 | Coiled-coil domain-containing protein 158 |
| CCDC168_N | 1 | Coiled-coil domain-containing protein 168 |
| CCDC22_CC | 1 | CCDC22 protein coiled-coil region |
| CCDC22_N | 1 | CCDC22 protein N-terminal domain |
| CCDC23 | 1 | Coiled-coil domain-containing protein 23 |
| CCDC24 | 1 | Coiled-coil domain-containing protein 24 family |
| CCDC32 | 1 | Coiled-coil domain containing 32 |
| CCDC34 | 1 | Coiled-coil domain-containing protein 34 |
| CCDC47 | 1 | PAT complex subunit CCDC47 |
| CCDC50_N | 1 | Coiled-coil domain-containing protein 50 N-terminus |
| CCDC53 | 1 | Subunit CCDC53 of WASH complex |
| CCDC66 | 1 | Coiled-coil domain-containing protein 66 |
| CCDC73 | 1 | Coiled-coil domain-containing protein 73 family |
| CCDC84 | 1 | Coiled coil protein 84 |
| CCDC93_CC | 1 | CCDC93 coiled-coil domain |
| CCDC93_N | 1 | CCDC93 protein N-terminal domain |
| CCER1 | 1 | Coiled-coil domain-containing glutamate-rich protein family 1 |
| CCSAP | 1 | Centriole, cilia and spindle-associated |
| CCSMST1 | 1 | CCSMST1 family |
| CD24 | 1 | CD24 protein |
| CD4-extracel | 1 | CD4, extracellular |
| CD45 | 1 | Leukocyte receptor CD45 |
| CD47 | 1 | CD47 transmembrane region |
| CD52 | 1 | CAMPATH-1 antigen |
| CDC27 | 1 | DNA polymerase subunit Cdc27 |
| CDC37_C | 1 | Cdc37 C terminal domain |
| CDC37_N | 1 | Cdc37 N terminal kinase binding |
| CDC45 | 1 | CDC45-like protein |
| CDC73_C | 1 | RNA pol II accessory factor, Cdc73 family, C-terminal |
| CDC73_N | 1 | Paf1 complex subunit CDC73 N-terminal |
| CDK5RAP3 | 1 | CDK5 regulatory subunit-associated protein 3 |
| CDKN3 | 1 | Cyclin-dependent kinase inhibitor 3 (CDKN3) |
| CDO_I | 1 | Cysteine dioxygenase type I |
| CDRT4 | 1 | CMT1A duplicated region transcript 4 protein |
| CDT1 | 1 | DNA replication factor CDT1 like |
| CDT1_C | 1 | DNA replication factor Cdt1 C-terminal domain |
| CDV3 | 1 | Carnitine deficiency-associated protein 3 |
| CEBP1_N | 1 | Cytoplasmic polyadenylation element-binding protein 1 N-terminus |
| CEND1 | 1 | Cell cycle exit and neuronal differentiation protein 1 |
| CENP-B_dimeris | 1 | Centromere protein B dimerisation domain |
| CENP-C_C | 1 | Mif2/CENP-C like |
| CENP-C_mid | 1 | Centromere assembly component CENP-C middle DNMT3B-binding region |
| CENP-F_C_Rb_bdg | 1 | Rb-binding domain of kinetochore protein Cenp-F/LEK1 |
| CENP-F_N | 1 | Cenp-F N-terminal domain |
| CENP-F_leu_zip | 1 | Leucine-rich repeats of kinetochore protein Cenp-F/LEK1 |
| CENP-H | 1 | Centromere protein H (CENP-H) |
| CENP-I | 1 | Mis6 |
| CENP-K | 1 | Centromere-associated protein K |
| CENP-L | 1 | Kinetochore complex Sim4 subunit Fta1 |
| CENP-M | 1 | Centromere protein M (CENP-M) |
| CENP-N | 1 | Kinetochore protein CHL4 like |
| CENP-O | 1 | Cenp-O kinetochore centromere component |
| CENP-P | 1 | CENP-A-nucleosome distal (CAD) centromere subunit, CENP-P |
| CENP-Q | 1 | CENP-Q, a CENPA-CAD centromere complex subunit |
| CENP-R | 1 | Kinetochore component, CENP-R |
| CENP-T_N | 1 | Centromere kinetochore component CENP-T N-terminus |
| CENP-U | 1 | CENP-A nucleosome associated complex (NAC) subunit |
| CENP-W | 1 | CENP-W protein |
| CENP-X | 1 | CENP-S associating Centromere protein X |
| CENP_C_N | 1 | Kinetochore assembly subunit CENP-C N-terminal |
| CEP104_N | 1 | Centrosomal protein CEP104, N-terminal domain |
| CEP104_ZnF | 1 | Centrosomal protein CEP104, ZnF |
| CEP15-like | 1 | Centrosomal protein 15-like |
| CEP19 | 1 | CEP19-like protein |
| CEP209_CC5 | 1 | Coiled-coil region of centrosome protein CE290 |
| CEP44 | 1 | Centrosomal spindle body, CEP44 |
| CEP76-C2 | 1 | CEP76 C2 domain |
| CF222 | 1 | C6orf222, uncharacterised family |
| CFA69_ARM_rpt | 1 | Cilia- and flagella-associated protein 69, ARM repeat |
| CFAI_FIMAC_N | 1 | Complement factor I, FIMAC N-terminal domain |
| CFAI_KAZAL | 1 | Complement factor I, KAZAL domain |
| CFAP141 | 1 | Cilia- and flagella-associated protein 141 |
| CFAP300 | 1 | Cilia- and flagella-associated protein 300 |
| CFAP54_N | 1 | Cilia- and flagella-associated protein 54 |
| CFAP58_CC | 1 | CFAP58 central coiled coil |
| CFAP61_N | 1 | Cilia- and flagella-associated protein 61, N-terminal domain |
| CFAP68 | 1 | Cilia- and flagella-associated protein 68 |
| CFAP77 | 1 | Cilia- and flagella-associated protein 77 |
| CFAP90 | 1 | Cilia and flagella associated protein 90 |
| CFAP91 | 1 | Cilia- and flagella-associated protein 91 |
| CFAP95 | 1 | Protein CFAP95 |
| CFAP96-like | 1 | Cilia-and flagella-associated protein 96 |
| CFTR_R | 1 | Cystic fibrosis TM conductance regulator (CFTR), regulator domain |
| CGI-121 | 1 | Kinase binding protein CGI-121 |
| CHAT | 1 | CHAT domain |
| CHIP_TPR_N | 1 | CHIP N-terminal tetratricopeptide repeat domain |
| CHRD | 1 | CHRD domain |
| CHZ | 1 | Histone chaperone domain CHZ |
| CI-B14_5a | 1 | NADH:ubiquinone oxidoreductase subunit B14.5a (Complex I-B14.5a) |
| CIA30 | 1 | Complex I intermediate-associated protein 30 (CIA30) |
| CIAPIN1 | 1 | Cytokine-induced anti-apoptosis inhibitor 1, Fe-S biogenesis |
| CID_GANP | 1 | Binding region of GANP to ENY2 |
| CIMR | 1 | Cation-independent mannose-6-phosphate receptor repeat |
| CIP2A_N | 1 | CIP2A, N-terminal |
| CK_II_beta | 1 | Casein kinase II regulatory subunit |
| CLEC16A_C | 1 | CLEC16A C-terminal |
| CLIP1_ZNF | 1 | CLIP1 zinc knuckle |
| CLLAC | 1 | CLLAC-motif containing domain |
| CLN3 | 1 | CLN3 protein |
| CLN5 | 1 | Ceroid-lipofuscinosis neuronal protein 5 |
| CLN6 | 1 | Ceroid-lipofuscinosis neuronal protein 6 |
| CLP1_N | 1 | N-terminal beta-sandwich domain of polyadenylation factor |
| CLP_protease | 1 | Clp protease |
| CLU | 1 | Clustered mitochondria |
| CLU_N | 1 | Mitochondrial function, CLU-N-term |
| CMS1 | 1 | U3-containing 90S pre-ribosomal complex subunit |
| CNDH2_C | 1 | Condensin II complex subunit CAP-H2 or CNDH2, C-terminal wHTH domain |
| CNDH2_M | 1 | Condensin II complex subunit CAP-H2 or CNDH2, mid domain |
| CNDH2_N | 1 | Condensin II complex subunit CAP-H2 or CNDH2, N-terminal |
| CNOT11 | 1 | CCR4-NOT transcription complex subunit 11 |
| CNOT1_CAF1_bind | 1 | CCR4-NOT transcription complex subunit 1 CAF1-binding domain |
| CNOT1_HEAT | 1 | CCR4-NOT transcription complex subunit 1 HEAT repeat |
| CNOT1_TTP_bind | 1 | CCR4-NOT transcription complex subunit 1 TTP binding domain |
| CNPase | 1 | 2',3'-cyclic nucleotide 3'-phosphodiesterase (CNP or CNPase) |
| CNRIP1 | 1 | CB1 cannabinoid receptor-interacting protein 1 |
| CNTF | 1 | Ciliary neurotrophic factor |
| COA8 | 1 | Cytochrome c oxidase assembly factor 8 |
| COBRA1 | 1 | Cofactor of BRCA1 (COBRA1) |
| COG2_C | 1 | COG complex component, COG2, C-terminal |
| COG2_N | 1 | COG2 N-terminal |
| COG3_C | 1 | Conserved oligomeric Golgi complex subunit 3, C-terminal |
| COG3_N | 1 | Conserved oligomeric Golgi complex subunit 3, N-terminal |
| COG4_C | 1 | Conserved oligomeric Golgi complex subunit 4, C-terminal |
| COG4_N | 1 | Conserved oligomeric Golgi complex subunit 4, N-terminal |
| COG4_m | 1 | COG4 transport protein, middle alpha-helical bundle |
| COG5_C | 1 | Conserved oligomeric Golgi complex subunit 5, C-terminal |
| COG5_N | 1 | Conserved oligomeric Golgi complex subunit 5, N-terminal |
| COG6_C | 1 | Conserved Oligomeric Golgi complex subunit 6, C-terminal |
| COG6_N | 1 | Conserved oligomeric complex COG6, N-terminal |
| COG7 | 1 | Golgi complex component 7 (COG7) |
| COMMD1_N | 1 | COMMD1 N-terminal domain |
| COMMD9_HN | 1 | COMMD9, helical N-terminal domain |
| COPI_C | 1 | Coatomer (COPI) alpha subunit C-terminus |
| COPR5 | 1 | Cooperator of PRMT5 family |
| COQ7 | 1 | Ubiquinone biosynthesis protein COQ7 |
| COQ9 | 1 | COQ9 |
| COQ9_N | 1 | Ubiquinone biosynthesis protein COQ9, N-terminal domain |
| COX1 | 1 | Cytochrome C and Quinol oxidase polypeptide I |
| COX14 | 1 | Cytochrome oxidase c assembly |
| COX15-CtaA | 1 | Cytochrome oxidase assembly protein |
| COX17 | 1 | Cytochrome C oxidase copper chaperone (COX17) |
| COX2 | 1 | Cytochrome C oxidase subunit II, periplasmic domain |
| COX24_C | 1 | Mitochondrial mRNA-processing protein COX24, C-terminal |
| COX2_TM | 1 | Cytochrome C oxidase subunit II, transmembrane domain |
| COX3 | 1 | Cytochrome c oxidase subunit III |
| COX5A | 1 | Cytochrome c oxidase subunit Va |
| COX5B | 1 | Cytochrome c oxidase subunit Vb |
| COX6C | 1 | Cytochrome c oxidase subunit VIc |
| COX7C | 1 | Cytochrome c oxidase subunit VIIc |
| CPL | 1 | CPL (NUC119) domain |
| CPSF100_C | 1 | Cleavage and polyadenylation factor 2 C-terminal |
| CPSF73-100_C | 1 | Pre-mRNA 3'-end-processing endonuclease polyadenylation factor C-term |
| CR6_interact | 1 | Growth arrest and DNA-damage-inducible proteins-interacting protein 1 |
| CRF-BP | 1 | Corticotropin-releasing factor binding protein (CRF-BP) |
| CRIM | 1 | SAPK-interacting protein 1 (Sin1), middle CRIM domain |
| CRIM1_C | 1 | Cysteine-rich motor neuron 1 protein C-terminal |
| CRM1_repeat | 1 | Chromosome region maintenance or exportin repeat |
| CRM1_repeat_2 | 1 | CRM1 / Exportin repeat 2 |
| CRM1_repeat_3 | 1 | CRM1 / Exportin repeat 3 |
| CReP_N | 1 | eIF2-alpha phosphatase phosphorylation constitutive repressor |
| CSF-1 | 1 | Macrophage colony stimulating factor-1 (CSF-1) |
| CSN1_C | 1 | COP9 signalosome complex subunit 1, C-terminal helix |
| CSN3-like_C | 1 | COP9 signalosome complex subunit 3-like, C-terminal helix |
| CSN4_RPN5_eIF3a | 1 | CSN4/RPN5/eIF3a helix turn helix domain |
| CSN5_C | 1 | Cop9 signalosome subunit 5 C-terminal domain |
| CSTF1_dimer | 1 | Cleavage stimulation factor subunit 1, dimerisation domain |
| CTC1 | 1 | CST, telomere maintenance, complex subunit CTC1 |
| CTNNBL | 1 | Catenin-beta-like, Arm-motif containing nuclear |
| CTP_transf_3 | 1 | Cytidylyltransferase |
| CTSRT | 1 | Cation channel sperm-associated targeting subunit tau |
| CTU2 | 1 | Cytoplasmic tRNA 2-thiolation protein 2 |
| CU062 | 1 | Polycystin-1 Interacting Protein-1 |
| CWC25 | 1 | Pre-mRNA splicing factor |
| CX9C | 1 | CHCH-CHCH-like Cx9C, IMS import disulfide relay-system, |
| CXCL16 | 1 | C-X-C motif chemokine 16 domain |
| CXCL17 | 1 | VEGF co-regulated chemokine 1 |
| CXCR4_N | 1 | CXCR4 Chemokine receptor N terminal |
| CXXC_Zn-b_euk | 1 | CXXC motif containing zinc binding protein, eukaryotic |
| CYLD_phos_site | 1 | Phosphorylation region of CYLD, unstructured |
| CYTH | 1 | CYTH domain |
| CYTL1 | 1 | Cytokine-like protein 1 |
| CYYR1 | 1 | Cysteine and tyrosine-rich protein 1 |
| CaM_bind | 1 | Calcium-dependent calmodulin binding |
| CactinC_cactus | 1 | Cactus-binding C-terminus of cactin protein |
| Cactin_mid | 1 | Conserved mid region of cactin |
| Cadherin-like | 1 | Cadherin-like beta sandwich domain |
| Calpain_inhib | 1 | Calpain inhibitor |
| Calpain_u2 | 1 | Unstructured region on Calpain-3 |
| Carb_kinase | 1 | Carbohydrate kinase |
| Cas1_AcylT | 1 | 10 TM Acyl Transferase domain found in Cas1p |
| Casc1_C | 1 | Cancer susceptibility candidate 1 C-terminal |
| Casc1_N | 1 | Cancer susceptibility candidate 1 N-terminus |
| Casein | 1 | Casein |
| Casein_kappa | 1 | Kappa casein |
| Catalase | 1 | Catalase |
| Catalase-rel | 1 | Catalase-related immune-responsive |
| CathepsinC_exc | 1 | Cathepsin C exclusion domain |
| Ccdc124 | 1 | Coiled-coil domain-containing protein 124 /Oxs1 |
| Cementoin | 1 | Trappin protein transglutaminase binding domain |
| Centro_C10orf90 | 1 | Centrosomal C10orf90 |
| Ceramidase_alk | 1 | Neutral/alkaline non-lysosomal ceramidase, N-terminal |
| Cg6151-P | 1 | Uncharacterized conserved protein CG6151-P |
| Cgr1 | 1 | Cgr1 family |
| ChAPs | 1 | ChAPs (Chs5p-Arf1p-binding proteins) |
| Chisel | 1 | Stretch-responsive small skeletal muscle X protein, Chisel |
| CholecysA-Rec_N | 1 | Cholecystokinin A receptor, N-terminal |
| Chon_Sulph_att | 1 | Chondroitin sulphate attachment domain |
| Chromo_2 | 1 | Chromatin organization modifier domain 2 |
| CiPC | 1 | Clock interacting protein circadian |
| Ciart | 1 | Circadian-associated transcriptional repressor |
| CitMHS | 1 | Citrate transporter |
| Citrate_bind | 1 | ATP citrate lyase citrate-binding |
| Clp1 | 1 | Pre-mRNA cleavage complex II protein Clp1 |
| Cluap1 | 1 | Clusterin-associated protein-1 |
| Cnd1_N | 1 | non-SMC mitotic condensation complex subunit 1, N-term |
| Cnd2 | 1 | Condensin complex subunit 2 |
| Cnd3 | 1 | Nuclear condensing complex subunits, C-term domain |
| Coa1 | 1 | Cytochrome oxidase complex assembly protein 1 |
| Coa3_cc | 1 | Cytochrome c oxidase assembly factor 3 |
| Coatamer_beta_C | 1 | Coatomer beta C-terminal region |
| Coatomer_E | 1 | Coatomer epsilon subunit |
| Coatomer_b_Cpla | 1 | Coatomer beta subunit appendage platform |
| Cob_adeno_trans | 1 | Cobalamin adenosyltransferase |
| Codanin-1_C | 1 | Codanin-1 C-terminus |
| Cohesin_HEAT | 1 | HEAT repeat associated with sister chromatid cohesion |
| Cohesin_load | 1 | Cohesin loading factor |
| Coilin_N | 1 | Coilin N-terminus |
| Colipase | 1 | Colipase, N-terminal domain |
| Colipase-like | 1 | Colipase-like |
| Colipase_C | 1 | Colipase, C-terminal domain |
| Complex1_30kDa | 1 | Respiratory-chain NADH dehydrogenase, 30 Kd subunit |
| Complex1_49kDa | 1 | Respiratory-chain NADH dehydrogenase, 49 Kd subunit |
| Complex1_51K | 1 | Respiratory-chain NADH dehydrogenase 51 Kd subunit |
| Condensin2nSMC | 1 | Condensin II non structural maintenance of chromosomes subunit |
| Connexin40_C | 1 | Connexin 40 C-terminal domain |
| Connexin43 | 1 | Gap junction alpha-1 protein (Cx43) |
| Connexin50 | 1 | Gap junction alpha-8 protein (Cx50) |
| Consortin_C | 1 | Consortin C-terminus |
| Coprogen_oxidas | 1 | Coproporphyrinogen III oxidase |
| Coq4 | 1 | Coenzyme Q (ubiquinone) biosynthesis protein Coq4 |
| Cox20 | 1 | Cytochrome c oxidase assembly protein COX20 |
| CpG_bind_C | 1 | CpG binding protein C-terminal domain |
| Cript | 1 | Microtubule-associated protein CRIPT |
| CtaG_Cox11 | 1 | Cytochrome c oxidase assembly protein CtaG/Cox11 |
| Ctf4_C | 1 | DNA polymerase alpha-binding protein Ctf4, C-terminal domain |
| Ctf8 | 1 | Ctf8 |
| Cu-oxidase_4 | 1 | Multi-copper polyphenol oxidoreductase laccase |
| CutA1 | 1 | CutA1 divalent ion tolerance protein |
| CutC | 1 | CutC family |
| Cwf_Cwc_15 | 1 | Cwf15/Cwc15 cell cycle control protein |
| CxC4 | 1 | CxC4 like cysteine cluster associated with KDZ transposases |
| CybS | 1 | CybS, succinate dehydrogenase cytochrome B small subunit |
| Cybc1_Eros | 1 | Cytochrome b-245 chaperone 1 / Eros |
| Cyclin | 1 | Cyclin |
| Cyclin_D1_bind | 1 | Cyclin D1 binding domain |
| Cys_Met_Meta_PP | 1 | Cys/Met metabolism PLP-dependent enzyme |
| Cys_box | 1 | Anosmin cysteine rich domain |
| Cys_rich_FGFR | 1 | Cysteine rich repeat |
| Cyto_heme_lyase | 1 | Cytochrome c/c1 heme lyase |
| CytochromB561_N | 1 | Cytochrome B561, N terminal |
| Cytochrom_B558a | 1 | Cytochrome Cytochrome b558 alpha-subunit |
| Cytochrom_B_C | 1 | Cytochrome b(C-terminal)/b6/petD |
| Cytochrom_C | 1 | Cytochrome c |
| Cytochrom_C1 | 1 | Cytochrome C1 family |
| Cytochrome_B | 1 | Cytochrome b/b6/petB |
| D-Glu_cyclase | 1 | D-glutamate cyclase |
| D123 | 1 | D123 |
| DAAP1 | 1 | Dynein axonemal-associated protein 1 |
| DAD | 1 | DAD family |
| DAG1 | 1 | Dystroglycan (Dystrophin-associated glycoprotein 1) |
| DAOA | 1 | D-amino acid oxidase activator |
| DAO_C | 1 | C-terminal domain of alpha-glycerophosphate oxidase |
| DAP10 | 1 | DAP10 membrane protein |
| DAP3 | 1 | Mitochondrial ribosomal death-associated protein 3 |
| DASH_Hsk3 | 1 | DASH complex subunit Hsk3 like |
| DAXX_hist_bd | 1 | Death domain-associated protein 6, histone binding domain |
| DAZAP2 | 1 | DAZ associated protein 2 (DAZAP2) |
| DBINO | 1 | DNA-binding domain |
| DBP10CT | 1 | DBP10CT (NUC160) domain |
| DBR1 | 1 | Lariat debranching enzyme, C-terminal domain |
| DCA16 | 1 | DDB1- and CUL4-associated factor 16 |
| DCAF15_WD40 | 1 | DDB1-and CUL4-substrate receptor 15, WD repeat |
| DCAF17 | 1 | DDB1- and CUL4-associated factor 17 |
| DCP2 | 1 | Dcp2, box A domain |
| DCTN5 | 1 | Dynactin subunit 5 |
| DDA1 | 1 | Det1 complexing ubiquitin ligase |
| DDAH_eukar | 1 | N,N dimethylarginine dimethylhydrolase, eukaryotic |
| DDDD | 1 | Putative mitochondrial precursor protein |
| DDE_Tnp_4 | 1 | DDE superfamily endonuclease |
| DDOST_48kD | 1 | Oligosaccharyltransferase 48 kDa subunit beta |
| DDRGK | 1 | DDRGK domain |
| DEFB136 | 1 | Beta defensin 136 |
| DENND11 | 1 | DENN domain-containing protein 11 |
| DENR_N | 1 | DENR, N-terminal |
| DEPDC5_CTD | 1 | DEPDC5 protein C-terminal region |
| DEPP | 1 | Decidual protein induced by progesterone family |
| DERM | 1 | Dermatopontin |
| DFF-C | 1 | DNA Fragmentation factor 45kDa, C terminal domain |
| DFF40 | 1 | DNA fragmentation factor 40 kDa |
| DFP | 1 | DNA / pantothenate metabolism flavoprotein |
| DHH | 1 | DHH family |
| DHHA1 | 1 | DHHA1 domain |
| DIRP | 1 | DIRP |
| DJ-1_PfpI | 1 | DJ-1/PfpI family |
| DJC28_CD | 1 | DnaJ homologue, subfamily C, member 28, conserved domain |
| DKCLD | 1 | DKCLD (NUC011) domain |
| DLEU7 | 1 | Leukemia-associated protein 7 |
| DLH | 1 | Dienelactone hydrolase family |
| DMAP1 | 1 | DNA methyltransferase 1-associated protein 1 (DMAP1) |
| DMP1 | 1 | Dentin matrix protein 1 (DMP1) |
| DMTF1_N | 1 | Cyclin-D-binding Myb-like transcription factor 1 N-terminal |
| DNA-PKcs_N | 1 | DNA-PKcs, N-terminal |
| DNAPKcs_CC1-2 | 1 | DNA-dependent protein kinase catalytic subunit, CC1/2 |
| DNAPKcs_CC3 | 1 | DNA-dependent protein kinase catalytic subunit, CC3 |
| DNAPKcs_CC5 | 1 | DNA-PKcs, CC5 |
| DNA_RNApol_7kD | 1 | DNA directed RNA polymerase, 7 kDa subunit |
| DNA_binding_1 | 1 | 6-O-methylguanine DNA methyltransferase, DNA binding domain |
| DNA_ligase_IV | 1 | DNA ligase IV |
| DNA_pol_D_N | 1 | DNA polymerase delta subunit OB-fold domain |
| DNA_pol_P_Exo | 1 | DNA polymerase nu pseudo-exo |
| DNA_pol_alpha_N | 1 | DNA polymerase alpha subunit p180 N terminal |
| DNA_pol_delta_4 | 1 | DNA polymerase delta, subunit 4 |
| DNA_pol_phi | 1 | DNA polymerase phi |
| DNA_primase_lrg | 1 | Eukaryotic and archaeal DNA primase, large subunit |
| DNA_repr_REX1B | 1 | DNA repair REX1-B |
| DNApol_Exo | 1 | DNA mitochondrial polymerase exonuclease domain |
| DNER_C | 1 | Delta and Notch-like EGF-related receptor C-terminus |
| DNMT1-RFD | 1 | Cytosine specific DNA methyltransferase replication foci domain |
| DNTTIP1_dimer | 1 | DNTTIP1 dimerisation domain |
| DOT1 | 1 | Histone methylation protein DOT1 |
| DPAGT1_ins | 1 | DPAGT1 insertion domain |
| DPCD | 1 | DPCD protein family |
| DPM2 | 1 | Dolichol phosphate-mannose biosynthesis regulatory protein (DPM2) |
| DPM3 | 1 | Dolichol-phosphate mannosyltransferase subunit 3 (DPM3) |
| DREV | 1 | DREV methyltransferase |
| DS | 1 | Deoxyhypusine synthase |
| DSBA | 1 | DSBA-like thioredoxin domain |
| DSS1_SEM1 | 1 | DSS1/SEM1 family |
| DTX3L_KH-like | 1 | DTX3L, KH-like domain |
| DTX3L_a-b | 1 | DTX3L, alpha/beta domain |
| DUF1077 | 1 | Protein of unknown function (DUF1077) |
| DUF1081 | 1 | Domain of Unknown Function (DUF1081) |
| DUF1168 | 1 | Protein of unknown function (DUF1168) |
| DUF1232 | 1 | Protein of unknown function (DUF1232) |
| DUF1308 | 1 | Protein of unknown function (DUF1308) |
| DUF1358 | 1 | Protein of unknown function (DUF1358) |
| DUF1388 | 1 | Repeat of unknown function (DUF1388) |
| DUF155 | 1 | RMND1/Sif2-Sif3/Mrx10, DUF155 |
| DUF1604 | 1 | Protein of unknown function (DUF1604) |
| DUF167 | 1 | Uncharacterised ACR, YggU family COG1872 |
| DUF1674 | 1 | Protein of unknown function (DUF1674) |
| DUF1731 | 1 | Domain of unknown function (DUF1731) |
| DUF1744 | 1 | Domain of unknown function (DUF1744) |
| DUF1751 | 1 | Eukaryotic integral membrane protein (DUF1751) |
| DUF1771 | 1 | Domain of unknown function (DUF1771) |
| DUF1907 | 1 | Domain of Unknown Function (DUF1907) |
| DUF1986 | 1 | Domain of unknown function (DUF1986) |
| DUF2028 | 1 | Domain of unknown function (DUF2028) |
| DUF2039 | 1 | Uncharacterized conserved protein (DUF2039) |
| DUF2043 | 1 | Uncharacterized conserved protein (DUF2043) |
| DUF2045 | 1 | Uncharacterized conserved protein (DUF2045) |
| DUF2046 | 1 | Uncharacterized conserved protein H4 (DUF2046) |
| DUF2053 | 1 | Predicted membrane protein (DUF2053) |
| DUF2152 | 1 | Uncharacterized conserved protein (DUF2152) |
| DUF2205 | 1 | Short coiled-coil protein |
| DUF2340 | 1 | Uncharacterized conserved protein (DUF2340) |
| DUF2358 | 1 | Uncharacterized conserved protein (DUF2358) |
| DUF2362 | 1 | Uncharacterised conserved protein (DUF2362) |
| DUF2368 | 1 | Uncharacterised conserved protein (DUF2368) |
| DUF2428 | 1 | THADA/TRM732, DUF2428 |
| DUF2448 | 1 | Protein of unknown function C-terminus (DUF2448) |
| DUF2452 | 1 | Protein of unknown function (DUF2452) |
| DUF2462 | 1 | Protein of unknown function (DUF2462) |
| DUF2477 | 1 | Protein of unknown function (DUF2477) |
| DUF2615 | 1 | Protein of unknown function (DUF2615) |
| DUF2678 | 1 | Protein of unknown function (DUF2678) |
| DUF3314 | 1 | Protein of unknown function (DUF3314) |
| DUF3337 | 1 | Domain of unknown function (DUF3337) |
| DUF3381 | 1 | Ribosomal RNA methyltransferase Spb1, DUF3381 |
| DUF3384 | 1 | Domain of unknown function (DUF3384) |
| DUF3429 | 1 | Protein of unknown function (DUF3429) |
| DUF3432 | 1 | Domain of unknown function (DUF3432) |
| DUF3437 | 1 | Domain of unknown function (DUF3437) |
| DUF34_NIF3 | 1 | Duf34/NIF3 (NGG1p interacting factor 3) |
| DUF3524 | 1 | Domain of unknown function (DUF3524) |
| DUF3535 | 1 | Domain of unknown function (DUF3535) |
| DUF3544 | 1 | Protein kinase C-binding protein 1 |
| DUF3668 | 1 | Cep120 protein |
| DUF3689 | 1 | Protein of unknown function (DUF3689) |
| DUF3695 | 1 | Protein of unknown function (DUF3695) |
| DUF3730 | 1 | Focadhesin/RST1, DUF3730 |
| DUF3736 | 1 | Protein of unknown function (DUF3736) |
| DUF3752 | 1 | Protein of unknown function (DUF3752) |
| DUF3754 | 1 | Protein of unknown function (DUF3754) |
| DUF3819 | 1 | CCR4-Not complex, Not1 subunit, domain of unknown function DUF3819 |
| DUF382 | 1 | Domain of unknown function (DUF382) |
| DUF383 | 1 | Domain of unknown function (DUF383) |
| DUF384 | 1 | Domain of unknown function (DUF384) |
| DUF4042 | 1 | Domain of unknown function (DUF4042) |
| DUF4062 | 1 | Domain of unknown function (DUF4062) |
| DUF4078 | 1 | Domain of unknown function (DUF4078) |
| DUF4097 | 1 | Toastrack DUF4097 |
| DUF4147 | 1 | Domain of unknown function (DUF4147) |
| DUF4149 | 1 | Domain of unknown function (DUF4149) |
| DUF4171 | 1 | Domain of unknown function (DUF4171) |
| DUF4187 | 1 | Domain of unknown function (DUF4187) |
| DUF4196 | 1 | Domain of unknown function (DUF4196) |
| DUF4209 | 1 | Domain of unknown function (DUF4209) |
| DUF423 | 1 | Protein of unknown function (DUF423) |
| DUF4347 | 1 | Domain of unknown function (DUF4347) |
| DUF4378 | 1 | Domain of unknown function (DUF4378) |
| DUF4455 | 1 | Domain of unknown function (DUF4455) |
| DUF4456 | 1 | Domain of unknown function (DUF4456) |
| DUF4457 | 1 | Domain of unknown function (DUF4457) |
| DUF4460 | 1 | Domain of unknown function (DUF4460) |
| DUF4461 | 1 | Domain of unknown function (DUF4461) |
| DUF4464 | 1 | Domain of unknown function (DUF4464) |
| DUF4470 | 1 | Domain of unknown function (DUF4470) |
| DUF4471 | 1 | Domain of unknown function (DUF4471) |
| DUF4472 | 1 | Domain of unknown function (DUF4472) |
| DUF4476 | 1 | Domain of unknown function (DUF4476) |
| DUF4481 | 1 | Domain of unknown function (DUF4481) |
| DUF4485 | 1 | Domain of unknown function (DUF4485) |
| DUF4487 | 1 | Domain of unknown function (DUF4487) |
| DUF4495 | 1 | Domain of unknown function (DUF4495) |
| DUF4500 | 1 | Domain of unknown function (DUF4500) |
| DUF4501 | 1 | Domain of unknown function (DUF4501) |
| DUF4502 | 1 | Domain of unknown function (DUF4502) |
| DUF4503 | 1 | Domain of unknown function (DUF4503) |
| DUF4504 | 1 | Domain of unknown function (DUF4504) |
| DUF4505 | 1 | Domain of unknown function (DUF4505) |
| DUF4508 | 1 | Domain of unknown function (DUF4508) |
| DUF4512 | 1 | Domain of unknown function (DUF4512) |
| DUF4514 | 1 | Domain of unknown function (DUF4514) |
| DUF4516 | 1 | Domain of unknown function (DUF4516) |
| DUF4517 | 1 | Domain of unknown function (DUF4517) |
| DUF4518 | 1 | Domain of unknown function (DUF4518) |
| DUF4519 | 1 | Domain of unknown function (DUF4519) |
| DUF4520 | 1 | Domain of unknown function (DUF4520) |
| DUF4522 | 1 | Protein of unknown function (DUF4522) |
| DUF4524_N | 1 | Domain of unknown function (DUF4524), N-terminal domain |
| DUF4527 | 1 | Protein of unknown function (DUF4527) |
| DUF4528 | 1 | Domain of unknown function (DUF4528) |
| DUF4529 | 1 | Protein of unknown function (DUF4529) |
| DUF4530 | 1 | Domain of unknown function (DUF4530) |
| DUF4532 | 1 | Protein of unknown function (DUF4532) |
| DUF4535 | 1 | Domain of unknown function (DUF4535) |
| DUF4536 | 1 | Domain of unknown function (DUF4536) |
| DUF4538 | 1 | Domain of unknown function (DUF4538) |
| DUF4543 | 1 | Domain of unknown function (DUF4543) |
| DUF4545 | 1 | Domain of unknown function (DUF4545) |
| DUF4547 | 1 | Domain of unknown function (DUF4547) |
| DUF4548 | 1 | Domain of unknown function (DUF4548) |
| DUF4550 | 1 | Domain of unknown function (DUF4550) |
| DUF4551 | 1 | Protein of unknown function (DUF4551) |
| DUF4554 | 1 | Domain of unknown function (DUF4554) |
| DUF4556 | 1 | Domain of unknown function (DUF4556), Ig-like |
| DUF4559 | 1 | Domain of unknown function (DUF4559) |
| DUF4562 | 1 | Domain of unknown function (DUF4562) |
| DUF4566 | 1 | Domain of unknown function (DUF4566) |
| DUF4568 | 1 | Domain of unknown function (DUF4568) |
| DUF4573 | 1 | Domain of unknown function (DUF4573) |
| DUF4576 | 1 | Domain of unknown function (DUF4576) |
| DUF4577 | 1 | Domain of unknown function (DUF4577) |
| DUF4578 | 1 | Domain of unknown function (DUF4578) |
| DUF4581 | 1 | Domain of unknown function (DUF4581) |
| DUF4590 | 1 | Domain of unknown function (DUF4590) |
| DUF4597 | 1 | Domain of unknown function (DUF4597) |
| DUF4602 | 1 | Domain of unknown function (DUF4602) |
| DUF4603 | 1 | Domain of unknown function (DUF4603) |
| DUF4604 | 1 | Domain of unknown function (DUF4604) |
| DUF4606 | 1 | Domain of unknown function (DUF4606) |
| DUF4607 | 1 | Domain of unknown function (DUF4607) |
| DUF4609 | 1 | Domain of unknown function (DUF4609) |
| DUF4611 | 1 | Domain of unknown function (DUF4611) |
| DUF4612 | 1 | Domain of unknown function (DUF4612) |
| DUF4614 | 1 | Domain of unknown function (DUF4614) |
| DUF4615 | 1 | Domain of unknown function (DUF4615) |
| DUF4616 | 1 | Domain of unknown function (DUF4616) |
| DUF4617 | 1 | Domain of unknown function (DUF4617) |
| DUF4618 | 1 | Domain of unknown function (DUF4618) |
| DUF4619 | 1 | Domain of unknown function (DUF4619) |
| DUF4620 | 1 | Domain of unknown function (DUF4620) |
| DUF4628 | 1 | Domain of unknown function (DUF4628) |
| DUF4629 | 1 | Domain of unknown function (DUF4629) |
| DUF4630 | 1 | Domain of unknown function (DUF4630) |
| DUF4632 | 1 | Domain of unknown function (DUF4632) |
| DUF4633 | 1 | Domain of unknown function (DUF4633) |
| DUF4634 | 1 | Domain of unknown function (DUF4634) |
| DUF4635 | 1 | Domain of unknown function (DUF4635) |
| DUF4636 | 1 | Domain of unknown function (DUF4636) |
| DUF4637 | 1 | Domain of unknown function (DUF4637) |
| DUF4638 | 1 | Domain of unknown function (DUF4638) |
| DUF4639 | 1 | Domain of unknown function (DUF4639) |
| DUF4640 | 1 | Domain of unknown function (DUF4640) |
| DUF4642 | 1 | Domain of unknown function (DUF4642) |
| DUF4643 | 1 | Domain of unknown function (DUF4643) |
| DUF4644 | 1 | Domain of unknown function (DUF4644) |
| DUF4645 | 1 | Domain of unknown function (DUF4645) |
| DUF4647 | 1 | Domain of unknown function (DUF4647) |
| DUF4650 | 1 | Domain of unknown function (DUF4650) |
| DUF4653 | 1 | Domain of unknown function (DUF4653) |
| DUF4655 | 1 | Domain of unknown function (DUF4655) |
| DUF4656 | 1 | Domain of unknown function (DUF4656) |
| DUF4657 | 1 | Domain of unknown function (DUF4657) |
| DUF4658 | 1 | Domain of unknown function (DUF4658) |
| DUF4660 | 1 | Domain of unknown function (DUF4660) |
| DUF4661 | 1 | Domain of unknown function (DUF4661) |
| DUF4662 | 1 | Domain of unknown function (DUF4662) |
| DUF4663 | 1 | Domain of unknown function (DUF4663) |
| DUF4665 | 1 | Domain of unknown function (DUF4665) |
| DUF4670 | 1 | Domain of unknown function (DUF4670) |
| DUF4674 | 1 | Domain of unknown function (DUF4674) |
| DUF4675 | 1 | Domain of unknown function (DUF4675) |
| DUF4677 | 1 | Domain of unknown function (DUF4677) |
| DUF4678 | 1 | Domain of unknown function (DUF4678) |
| DUF4679 | 1 | Domain of unknown function (DUF4679) |
| DUF4680 | 1 | Domain of unknown function (DUF4680) |
| DUF4681 | 1 | Domain of unknown function (DUF4681) |
| DUF4684 | 1 | Domain of unknown function (DUF4684) |
| DUF4686 | 1 | Domain of unknown function (DUF4686) |
| DUF4687 | 1 | Domain of unknown function (DUF4687) |
| DUF4688 | 1 | Domain of unknown function (DUF4688) |
| DUF4689 | 1 | Domain of unknown function (DUF4689) |
| DUF4690 | 1 | Small Novel Rich in Cartilage |
| DUF4691 | 1 | Domain of unknown function (DUF4691) |
| DUF4692 | 1 | Regulator of human erythroid cell expansion (RHEX) |
| DUF4693 | 1 | Domain of unknown function (DUF4693) |
| DUF4694 | 1 | Domain of unknown function (DUF4694) |
| DUF4695 | 1 | Domain of unknown function (DUF4695) |
| DUF4698 | 1 | Domain of unknown function (DUF4698) |
| DUF4699 | 1 | Domain of unknown function (DUF4699) |
| DUF4702 | 1 | Domain of unknown function (DUF4702) |
| DUF4703 | 1 | Domain of unknown function (DUF4703) |
| DUF4706 | 1 | Domain of unknown function (DUF4706) |
| DUF4707 | 1 | Domain of unknown function (DUF4707) |
| DUF4708 | 1 | Domain of unknown function (DUF4708) |
| DUF4709 | 1 | Domain of unknown function (DUF4709) |
| DUF4711 | 1 | Domain of unknown function (DUF4711) |
| DUF4712 | 1 | Domain of unknown function (DUF4712) |
| DUF4714 | 1 | Domain of unknown function (DUF4714) |
| DUF4715 | 1 | Domain of unknown function (DUF4715) |
| DUF4718 | 1 | Domain of unknown function (DUF4718) |
| DUF4719 | 1 | Domain of unknown function (DUF4719) |
| DUF4720 | 1 | Domain of unknown function (DUF4720) |
| DUF4722 | 1 | Domain of unknown function (DUF4722) |
| DUF4723 | 1 | Domain of unknown function (DUF4723) |
| DUF4724 | 1 | Domain of unknown function (DUF4724) |
| DUF4726 | 1 | Domain of unknown function (DUF4726) |
| DUF4730 | 1 | Domain of unknown function (DUF4730) |
| DUF4731 | 1 | Domain of unknown function (DUF4731) |
| DUF4732 | 1 | Domain of unknown function (DUF4732) |
| DUF4735 | 1 | Domain of unknown function (DUF4735) |
| DUF4745 | 1 | Domain of unknown function (DUF4745) |
| DUF4748 | 1 | Domain of unknown function (DUF4748) |
| DUF4750 | 1 | Domain of unknown function (DUF4750) |
| DUF4764 | 1 | Domain of unknown function (DUF4764) |
| DUF4772 | 1 | Domain of unknown function (DUF4772) |
| DUF4796_C | 1 | DUF4796 C-terminal |
| DUF4808 | 1 | Domain of unknown function (DUF4808) |
| DUF4819 | 1 | Domain of unknown function (DUF4819) |
| DUF4962 | 1 | Domain of unknown function (DUF4962) |
| DUF5009 | 1 | Domain of unknown function (DUF5009) |
| DUF543 | 1 | Domain of unknown function (DUF543) |
| DUF5520 | 1 | Family of unknown function (DUF5520) |
| DUF5521 | 1 | Family of unknown function (DUF5521) |
| DUF5522 | 1 | Family of unknown function (DUF5522) |
| DUF5524 | 1 | Family of unknown function (DUF5524) |
| DUF5525 | 1 | Family of unknown function (DUF5525) |
| DUF5527 | 1 | Family of unknown function (DUF5527) |
| DUF5528 | 1 | Family of unknown function (DUF5528) |
| DUF5529 | 1 | Family of unknown function (DUF5529) |
| DUF5530 | 1 | Family of unknown function (DUF5530) |
| DUF5531 | 1 | Family of unknown function (DUF5531) |
| DUF5532 | 1 | Family of unknown function (DUF5532) |
| DUF5533 | 1 | Family of unknown function (DUF5533) |
| DUF5534 | 1 | Family of unknown function (DUF5534) |
| DUF5536 | 1 | Family of unknown function (DUF5536) |
| DUF5537 | 1 | Family of unknown function (DUF5537) |
| DUF5538 | 1 | Family of unknown function (DUF5538) |
| DUF5542 | 1 | Family of unknown function (DUF5542) |
| DUF5543 | 1 | Family of unknown function (DUF5543) |
| DUF5546 | 1 | Family of unknown function (DUF5546) |
| DUF5547 | 1 | Family of unknown function (DUF5547) |
| DUF5548 | 1 | Family of unknown function (DUF5548) |
| DUF5549 | 1 | Family of unknown function (DUF5549) |
| DUF5554 | 1 | Family of unknown function (DUF5554) |
| DUF5559 | 1 | Family of unknown function (DUF5559) |
| DUF5562 | 1 | Family of unknown function (DUF5562) |
| DUF5563 | 1 | Family of unknown function (DUF5563) |
| DUF5564 | 1 | Family of unknown function (DUF5564) |
| DUF5565 | 1 | Family of unknown function (DUF5565) |
| DUF5568 | 1 | Family of unknown function (DUF5568) |
| DUF5569 | 1 | Family of unknown function (DUF5569) |
| DUF5571 | 1 | Family of unknown function (DUF5571) |
| DUF5572 | 1 | Family of unknown function (DUF5572) |
| DUF5575 | 1 | DUF5575 central RNaseH like domain |
| DUF5575_C | 1 | DUF5575 C-terminal domain |
| DUF5575_N | 1 | DUF5575 N-terminal domain |
| DUF5577 | 1 | Family of unknown function (DUF5577) |
| DUF5578 | 1 | Family of unknown function (DUF5578) |
| DUF5580 | 1 | Family of unknown function (DUF5580) N-terminal domain |
| DUF5580_C | 1 | Family of unknown function (DUF5580) C-terminal domain |
| DUF5580_M | 1 | Family of unknown function (DUF5580) middle domain |
| DUF5581 | 1 | Family of unknown function (DUF5581) Fn3-like domain |
| DUF5581_N | 1 | Family of unknown function (DUF5581) N-terminal domain |
| DUF5585 | 1 | Family of unknown function (DUF5585) |
| DUF5586 | 1 | Family of unknown function (DUF5586) |
| DUF5587 | 1 | Family of unknown function (DUF5587) |
| DUF5595 | 1 | Domain of unknown function (DUF5595) |
| DUF5604 | 1 | Domain of unknown function (DUF5604) |
| DUF5614 | 1 | Family of unknown function (DUF5614) |
| DUF5745 | 1 | Domain of unknown function (DUF5745) |
| DUF5915 | 1 | Domain of unknown function (DUF5915) |
| DUF5918 | 1 | Family of unknown function (DUF5918) |
| DUF5920 | 1 | Domain of unknown function (DUF5920) |
| DUF5921 | 1 | Domain of unknown function (DUF5921) |
| DUF608 | 1 | Glycosyl-hydrolase family 116, catalytic region |
| DUF726 | 1 | Protein of unknown function (DUF726) |
| DUF747 | 1 | Eukaryotic membrane protein family |
| DUF776 | 1 | Protein of unknown function (DUF776) |
| DUF778 | 1 | Domain of unknown function (DUF778) |
| DUF829 | 1 | Eukaryotic protein of unknown function (DUF829) |
| DUF842 | 1 | Eukaryotic protein of unknown function (DUF842) |
| DUF908 | 1 | Domain of Unknown Function (DUF908) |
| DUF913 | 1 | Domain of Unknown Function (DUF913) |
| DUF92 | 1 | Integral membrane protein DUF92 |
| DUF938 | 1 | Protein of unknown function (DUF938) |
| DUF959 | 1 | Domain of Unknown Function (DUF959) |
| DUF974 | 1 | Protein of unknown function (DUF974) |
| DWNN | 1 | DWNN domain |
| DYNC2H1_AAA_dom | 1 | Cytoplasmic dynein 2 heavy chain 1, AAA+ ATPase domain |
| DZR | 1 | Double zinc ribbon |
| Dak1 | 1 | Dak1 domain |
| Dak2 | 1 | DAK2 domain |
| Daxx | 1 | Daxx N-terminal Rassf1C-interacting domain |
| Dcc1 | 1 | Sister chromatid cohesion protein Dcc1 |
| DcpS | 1 | Scavenger mRNA decapping enzyme (DcpS) N-terminal |
| Defensin_big | 1 | Big defensin |
| Dendrin | 1 | Nephrin and CD2AP-binding protein, Dendrin |
| DeoC | 1 | DeoC/LacD family aldolase |
| Dermcidin | 1 | Dermcidin, antibiotic peptide |
| Det1 | 1 | De-etiolated protein 1 Det1 |
| Dexa_ind | 1 | Dexamethasone-induced |
| Dicer_PBD | 1 | Dicer, partner-binding domain |
| Dicer_dimer | 1 | Dicer dimerisation domain |
| Dicer_dsRBD | 1 | Dicer, dsRNA-binding domain |
| Dicer_platform | 1 | Dicer, platform domain |
| Diphthami_syn_2 | 1 | Diphthamide synthase |
| Dis3l2_C_term | 1 | DIS3-like exonuclease 2 C terminal |
| Dmrt1 | 1 | Double-sex mab3 related transcription factor 1 |
| Dna2 | 1 | DNA replication factor Dna2 |
| Dna2_Rift | 1 | Dna2 Rift barrel domain |
| DnaJ-like_C11_C | 1 | DnaJ-like protein C11, C-terminal |
| Domain | 1 | Domain |
| Doppel | 1 | Prion-like protein Doppel |
| Dor1 | 1 | Dor1-like family |
| DoxX | 1 | DoxX |
| Dpoe2NT | 1 | DNA polymerases epsilon N terminal |
| Draxin | 1 | Draxin |
| Drc1-Sld2 | 1 | DNA replication and checkpoint protein |
| Drf_FH1 | 1 | Formin Homology Region 1 |
| Dymeclin | 1 | Dyggve-Melchior-Clausen syndrome protein |
| Dynactin | 1 | Dynein associated protein |
| Dynactin_p22 | 1 | Dynactin subunit p22 |
| Dynactin_p62 | 1 | Dynactin p62 family |
| Dynamitin | 1 | Dynamitin |
| Dynein_attach_N | 1 | Dynein attachment factor N-terminus |
| E2_bind | 1 | E2 binding domain |
| E3_UBR4_N | 1 | E3 ubiquitin-protein ligase UBR4 N-terminal |
| E3_UFM1_ligase | 1 | E3 UFM1-protein ligase 1 |
| E3_UbLigase_EDD | 1 | E3 ubiquitin ligase EDD |
| E3_UbLigase_R4 | 1 | E3 ubiquitin-protein ligase UBR4 |
| E3_UbLigase_RBR | 1 | E3 Ubiquitin Ligase RBR C-terminal domain |
| EB1_binding | 1 | EB-1 Binding Domain |
| ECD | 1 | Extracellular Cadherin domain |
| ECH_2 | 1 | Enoyl-CoA hydratase/isomerase |
| ECM1 | 1 | Extracellular matrix protein 1 (ECM1) |
| ECM10 | 1 | ER membrane protein complex subunit 10 |
| ECSCR | 1 | Endothelial cell-specific chemotaxis regulator |
| ECSIT_C | 1 | C-terminal domain of the ECSIT protein |
| ECSIT_N | 1 | Evolutionarily conserved signalling intermediate in Toll pathway |
| ECT2_BRCT0 | 1 | ECT2, BRCT0 domain |
| ECT2_PH | 1 | ECT2, PH domain |
| EDC4_C | 1 | Enhancer of mRNA-decapping protein 4, C-terminal |
| EDR1 | 1 | Ethylene-responsive protein kinase Le-CTR1 |
| EF-hand_10 | 1 | EF hand |
| EF1G | 1 | Elongation factor 1 gamma, conserved domain |
| EFTUD2 | 1 | 116 kDa U5 small nuclear ribonucleoprotein component N-terminus |
| EF_TS | 1 | Elongation factor TS |
| EHN | 1 | Epoxide hydrolase N terminus |
| EI24 | 1 | Sulfate transporter CysZ/Etoposide-induced protein 2.4 |
| EIF3E_C | 1 | Eukaryotic translation initiation factor 3 subunit E, C-terminal |
| EIF4E-T | 1 | Nucleocytoplasmic shuttling protein for mRNA cap-binding EIF4E |
| EIF_2_alpha | 1 | Eukaryotic translation initiation factor 2 alpha subunit |
| EKLF_TAD1 | 1 | Erythroid krueppel-like transcription factor, transactivation 1 |
| EKLF_TAD2 | 1 | Erythroid krueppel-like transcription factor, transactivation 2 |
| ELP6 | 1 | Elongation complex protein 6 |
| ELYS | 1 | Nuclear pore complex assembly |
| ELYS-bb | 1 | beta-propeller of ELYS nucleoporin |
| EMC1_C | 1 | ER membrane protein complex subunit 1, C-terminal |
| EMC7_beta-sandw | 1 | ER membrane protein complex subunit 7, beta-sandwich domain |
| EMG1 | 1 | EMG1/NEP1 methyltransferase |
| ENT | 1 | ENT domain |
| EP400_N | 1 | E1A-binding protein p400, N-terminal |
| ER | 1 | Enhancer of rudimentary |
| ERCC3_RAD25_C | 1 | ERCC3/RAD25/XPB C-terminal helicase |
| ERG2_Sigma1R | 1 | ERG2 and Sigma1 receptor like protein |
| ERK-JNK_inhib | 1 | ERK and JNK pathways, inhibitor |
| ERbeta_N | 1 | Estrogen receptor beta |
| ERp29 | 1 | Endoplasmic reticulum protein ERp29, C-terminal domain |
| ERp29_N | 1 | ERp29, N-terminal domain |
| ESCRT-II | 1 | ESCRT-II complex subunit |
| ESR1_C | 1 | Oestrogen-type nuclear receptor final C-terminal |
| ESSS | 1 | ESSS subunit of NADH:ubiquinone oxidoreductase (complex I) |
| ETAA1 | 1 | Ewing's tumour-associated antigen 1 homologue |
| ETC_C1_NDUFA5 | 1 | ETC complex I subunit conserved region |
| ETFQO_UQ-bd | 1 | ETF-QO, ubiquinone-binding |
| ETF_QO | 1 | Electron transfer flavoprotein-ubiquinone oxidoreductase, 4Fe-4S |
| ETF_alpha | 1 | Electron transfer flavoprotein FAD-binding domain |
| EURL | 1 | EURL protein |
| EVC2_like | 1 | Ellis van Creveld protein 2 like protein |
| EVE | 1 | EVE domain |
| EVI2A | 1 | Ectropic viral integration site 2A protein (EVI2A) |
| EXOSC1 | 1 | Exosome component EXOSC1/CSL4 |
| EXS | 1 | EXS family |
| Eaf7 | 1 | Chromatin modification-related protein EAF7 |
| Eapp_C | 1 | E2F-associated phosphoprotein |
| Ebp2 | 1 | Eukaryotic rRNA processing protein EBP2 |
| Ecm29 | 1 | Proteasome stabiliser |
| Edc3_linker | 1 | Linker region of enhancer of mRNA-decapping protein 3 |
| Elf1 | 1 | Transcription elongation factor Elf1 like |
| EloA-BP1 | 1 | ElonginA binding-protein 1 |
| Elong_Iki1 | 1 | Elongator subunit Iki1 |
| EnY2 | 1 | Transcription factor e(y)2 |
| Enamelin | 1 | Enamelin |
| Endomucin | 1 | Endomucin |
| Endonuclease_5 | 1 | Endonuclease V |
| Ependymin | 1 | Ependymin |
| Epiglycanin_TR | 1 | Tandem-repeating region of mucin, epiglycanin-like |
| Epimerase_2 | 1 | UDP-N-acetylglucosamine 2-epimerase |
| Erg28 | 1 | Erg28 like protein |
| Ermin | 1 | Ermin |
| Erv26 | 1 | Transmembrane adaptor Erv26 |
| Es2 | 1 | Nuclear protein Es2 |
| Esterase | 1 | Putative esterase |
| Exo5 | 1 | Exonuclease V - a 5' deoxyribonuclease |
| Exo70_C | 1 | Exo70 exocyst complex subunit C-terminal |
| Exo70_N | 1 | Exocyst complex component Exo70 N-terminal |
| Exo84_C | 1 | Exocyst component 84 C-terminal |
| Exog_C | 1 | Endo/exonuclease (EXOG) C-terminal domain |
| Exportin-5 | 1 | Exportin-5 family |
| Exportin-T | 1 | Exportin-T |
| FAA_hydrolase_N | 1 | Fumarylacetoacetase N-terminal |
| FACT-Spt16_Nlob | 1 | FACT complex subunit SPT16 N-terminal lobe domain |
| FAD_binding_2 | 1 | FAD binding domain |
| FAIM1 | 1 | Fas apoptotic inhibitory molecule (FAIM1) |
| FAM165 | 1 | FAM165 family |
| FAM183 | 1 | FAM183A and FAM183B related |
| FAM186A_PQQAQ | 1 | FAM186A, PQQAQ repeat |
| FAM192A_Fyv6_N | 1 | FAM192A/Fyv6, N-terminal domain |
| FAM199X | 1 | Protein family FAM199X |
| FAM220 | 1 | FAM220 family |
| FAM32A | 1 | FAM32A |
| FAM60A | 1 | Protein Family FAM60A |
| FAM91_C | 1 | FAM91 C-terminus |
| FAM91_N | 1 | FAM91 N-terminus |
| FAN1_HTH | 1 | FAN1, HTH domain |
| FAN1_TPR | 1 | Fanconi-associated nuclease 1, TPR domain |
| FANCAA | 1 | Fanconi anemia-associated |
| FANCA_interact | 1 | FAAP20 FANCA interaction domain |
| FANCD2OS | 1 | FANCD2 opposite strand protein |
| FANCF | 1 | Fanconi anemia group F protein (FANCF) |
| FANCI_HD1 | 1 | FANCI helical domain 1 |
| FANCI_HD2 | 1 | FANCI helical domain 2 |
| FANCI_S1 | 1 | FANCI solenoid 1 |
| FANCI_S1-cap | 1 | FANCI solenoid 1 cap |
| FANCI_S2 | 1 | FANCI solenoid 2 |
| FANCI_S3 | 1 | FANCI solenoid 3 |
| FANCI_S4 | 1 | FANCI solenoid 4 |
| FANCL_C | 1 | FANCL C-terminal domain |
| FANCL_d1 | 1 | FANCL UBC-like domain 1 |
| FANCL_d2 | 1 | FANCL UBC-like domain 2 |
| FANCL_d3 | 1 | FANCL UBC-like domain 3 |
| FANCM-MHF_bd | 1 | FANCM to MHF binding domain |
| FAP206 | 1 | Domain of unknown function |
| FAS_pseudo-KR | 1 | Fatty acid synthase, pseudo-KR domain |
| FAXC_N | 1 | Failed axon connections homolog N-terminus |
| FA_FANCE | 1 | Fanconi Anaemia group E protein FANCE |
| FBF1 | 1 | Fas-binding factor 1 |
| FBO_C | 1 | F-box only protein C-terminal region |
| FBXL18_LRR | 1 | F-box/LRR-repeat protein 18, LRR |
| FCP1_C | 1 | FCP1, C-terminal |
| FDC-SP | 1 | Follicular dendritic cell secreted peptide |
| FGAR-AT_N | 1 | Formylglycinamide ribonucleotide amidotransferase N-terminal |
| FGAR-AT_linker | 1 | Formylglycinamide ribonucleotide amidotransferase linker domain |
| FGFR3_TM | 1 | Fibroblast growth factor receptor 3 transmembrane domain |
| FIBP | 1 | Acidic fibroblast growth factor binding (FIBP) |
| FIMP | 1 | Fertilisation-influencing membrane protein |
| FIST_C | 1 | FIST C domain |
| FKTN_N | 1 | Fukutin N-terminal |
| FLYWCH | 1 | FLYWCH zinc finger domain |
| FLYWCH_u | 1 | FLYWCH-type zinc finger-containing protein 1 |
| FMP27-BLTP2_C | 1 | BLTP2/FMP27/Hobbit, C-terminal |
| FMP27-BLTP2_N | 1 | FMP27/BTLP1, N-terminal |
| FN3_7 | 1 | Fibronectin type III domain |
| FPL | 1 | Uncharacterised conserved protein |
| FPN1 | 1 | Ferroportin1 (FPN1) |
| FRB_dom | 1 | FKBP12-rapamycin binding domain |
| FRG1 | 1 | FRG1-like domain |
| FSH1 | 1 | Serine hydrolase (FSH1) |
| FSIP1 | 1 | FSIP1 family |
| FSIP2 | 1 | Fibrous sheath-interacting protein 2 |
| FTCD | 1 | Formiminotransferase domain |
| FTCD_C | 1 | Formiminotransferase-cyclodeaminase |
| FTO_CTD | 1 | FTO C-terminal domain |
| FTO_NTD | 1 | FTO catalytic domain |
| FUT8_N_cat | 1 | Alpha-(1,6)-fucosyltransferase N- and catalytic domains |
| FXMR_C2 | 1 | Fragile X messenger ribonucleoprotein 1, C-terminal region 2 |
| FXR_C3 | 1 | Fragile X-related 1 protein C-terminal region 3 |
| FYTT | 1 | Forty-two-three protein |
| F_actin_cap_B | 1 | F-actin capping protein, beta subunit |
| Fan1_SAP | 1 | Fanconi-associated nuclease 1, SAP subdomain |
| FancD2 | 1 | Fanconi anaemia protein FancD2 nuclease |
| Fanconi_A | 1 | Fanconi anaemia group A protein |
| Fanconi_A_N | 1 | Fanconi anaemia group A protein N terminus |
| Fanconi_C | 1 | Fanconi anaemia group C protein |
| Fapy_DNA_glyco | 1 | Formamidopyrimidine-DNA glycosylase N-terminal domain |
| Fcf2 | 1 | Fcf2 pre-rRNA processing |
| Fe-ADH | 1 | Iron-containing alcohol dehydrogenase |
| Fer2_3 | 1 | 2Fe-2S iron-sulfur cluster binding domain |
| Fer2_4 | 1 | 2Fe-2S iron-sulfur cluster binding domain |
| Fer4_17 | 1 | 4Fe-4S dicluster domain |
| Fer4_20 | 1 | Dihydroprymidine dehydrogenase domain II, 4Fe-4S cluster |
| Fer4_21 | 1 | 4Fe-4S dicluster domain |
| Fer4_7 | 1 | 4Fe-4S dicluster domain |
| Ferrochelatase | 1 | Ferrochelatase |
| Fibin | 1 | Fin bud initiation factor homologue |
| Fibrinogen_aC | 1 | Fibrinogen alpha C domain |
| Fic | 1 | Fic/DOC family |
| Filaggrin | 1 | Filaggrin |
| Fip1 | 1 | Fip1 motif |
| Fis1_TPR_C | 1 | Fis1 C-terminal tetratricopeptide repeat |
| Fis1_TPR_N | 1 | Fis1 N-terminal tetratricopeptide repeat |
| Flavokinase | 1 | Riboflavin kinase |
| Flavoprotein | 1 | Flavoprotein |
| Flt3_lig | 1 | flt3 ligand |
| FmiP_Thoc5 | 1 | Fms-interacting protein/Thoc5 |
| Fmp27_GFWDK | 1 | Fmp27/BLTP2/Hobbit, GFWDK motif-containg RBG unit |
| Fn3_assoc | 1 | Fn3 associated |
| Fndc10 | 1 | Fibronectin type III domain-containing protein 10 |
| Focadhesin | 1 | Focadhesin |
| Foie-gras_1 | 1 | Foie gras liver health family 1 |
| Folliculin_C | 1 | Folliculin C-terminal domain |
| Fra10Ac1 | 1 | Folate-sensitive fragile site protein Fra10Ac1 |
| Frataxin_Cyay | 1 | Frataxin-like domain |
| Frey | 1 | Protein Frey |
| Frtz | 1 | WD repeat-containing and planar cell polarity effector protein Fritz |
| FumaraseC_C | 1 | Fumarase C C-terminus |
| G-patch_2 | 1 | G-patch domain |
| G0-G1_switch_2 | 1 | G0/G1 switch protein 2 |
| G2BR | 1 | E3 gp78 Ube2g2-binding region (G2BR) |
| G6B | 1 | G6b-B extracellular V-set Ig-like domain |
| GABP-alpha | 1 | GA-binding protein alpha chain |
| GAD | 1 | GAD domain |
| GAIN_A | 1 | GPCR-Autoproteolysis-INducing (GAIN) subdomain A |
| GAPT | 1 | GRB2-binding adapter (GAPT) |
| GARS_A | 1 | Phosphoribosylglycinamide synthetase, ATP-grasp (A) domain |
| GARS_C | 1 | Phosphoribosylglycinamide synthetase, C domain |
| GARS_N | 1 | Phosphoribosylglycinamide synthetase, N domain |
| GAS | 1 | Growth-arrest specific micro-tubule binding |
| GATase_5 | 1 | CobB/CobQ-like glutamine amidotransferase domain |
| GBR2_CC | 1 | Gamma-aminobutyric acid type B receptor subunit 2 coiled-coil domain |
| GCIP_C | 1 | Grap2 and cyclin-D-interacting, C-terminal |
| GCIP_N | 1 | Grap2 and cyclin-D-interacting, N-terminal |
| GCN5L1 | 1 | GCN5-like protein 1 (GCN5L1) |
| GCP6_N | 1 | Gamma-tubulin complex component 6 N-terminus |
| GCR | 1 | Glucocorticoid receptor |
| GCS | 1 | Glutamate-cysteine ligase |
| GCSF | 1 | Granulocyte colony-stimulating factor |
| GDC-P | 1 | Glycine cleavage system P-protein |
| GDE_C | 1 | Amylo-alpha-1,6-glucosidase |
| GDWWSH | 1 | Protein of unknown function with motif GDWWSH |
| GEMIN8 | 1 | Gemini of Cajal bodies-associated protein 8 |
| GET4 | 1 | Golgi to ER traffic protein 4 |
| GFRP | 1 | GTP cyclohydrolase I feedback regulatory protein (GFRP) |
| GGACT | 1 | Gamma-glutamyl cyclotransferase, AIG2-like |
| GGN | 1 | Gametogenetin |
| GH3 | 1 | GH3 auxin-responsive promoter |
| GHBP | 1 | Growth hormone receptor binding |
| GIDA | 1 | Glucose inhibited division protein A |
| GIDA_C | 1 | tRNA modifying enzyme MnmG/GidA C-terminal helical bundle |
| GIDA_C_1st | 1 | tRNA modifying enzyme MnmG/GidA C-terminal helical domain |
| GIDE | 1 | E3 Ubiquitin ligase |
| GILT | 1 | Gamma interferon inducible lysosomal thiol reductase (GILT) |
| GKRP-like_C | 1 | C-terminal lid domain of glucokinase regulatory protein |
| GLE1 | 1 | GLE1-like protein |
| GLOD4_C | 1 | Glyoxalasedomain-containing protein 4-like, C-terminal domain |
| GLOD4_N | 1 | Glyoxalase domain-containing protein 4, N-terminal |
| GLUCM-like_C | 1 | D-glutamate cyclase-like, C-terminal |
| GMAP | 1 | Galanin message associated peptide (GMAP) |
| GMC_oxred_C | 1 | GMC oxidoreductase |
| GMC_oxred_N | 1 | GMC oxidoreductase |
| GMP_synt_C | 1 | GMP synthase C terminal domain |
| GM_CSF | 1 | Granulocyte-macrophage colony-stimulating factor |
| GN3L_Grn1 | 1 | GNL3L/Grn1 putative GTPase |
| GNAT_acetyltr_2 | 1 | GNAT acetyltransferase 2 |
| GPI2 | 1 | Phosphatidylinositol N-acetylglucosaminyltransferase |
| GPR15L | 1 | G-protein coupled receptor ligand 15 |
| GRIM-19 | 1 | GRIM-19 protein |
| GSAP-16 | 1 | gamma-Secretase-activating protein C-term |
| GSH_synth_ATP | 1 | Eukaryotic glutathione synthase, ATP binding domain |
| GSH_synthase | 1 | Eukaryotic glutathione synthase |
| GSKIP_dom | 1 | GSKIP domain |
| GST_N_5 | 1 | Glutathione S-transferase, N-terminal domain |
| GTP-bdg_M | 1 | GTP-binding GTPase Middle Region |
| GTP-bdg_N | 1 | GTP-binding GTPase N-terminal |
| GTP_EFTU_D4 | 1 | Elongation factor Tu domain 4 |
| GTP_cyclohydroI | 1 | GTP cyclohydrolase I |
| GWT1 | 1 | GWT1 |
| GYF_2 | 1 | GYF domain 2 |
| G_path_suppress | 1 | G-protein pathway suppressor |
| Gaa1 | 1 | Gaa1-like, GPI transamidase component |
| GalP_UDP_tr_C | 1 | Galactose-1-phosphate uridyl transferase, C-terminal domain |
| GalP_UDP_transf | 1 | Galactose-1-phosphate uridyl transferase, N-terminal domain |
| GatB_N | 1 | GatB/GatE catalytic domain |
| GatB_Yqey | 1 | GatB domain |
| GatC | 1 | Glu-tRNAGln amidotransferase C subunit |
| Gcd10p | 1 | Gcd10p family |
| GcvP2_C | 1 | Glycine dehydrogenase, C-terminal domain |
| Ge1_WD40 | 1 | WD40 region of Ge1, enhancer of mRNA-decapping protein |
| Gemin6 | 1 | Gemin6 Sm-like domain |
| Gemin6_C | 1 | Gemin6 C-terminal domain |
| Gemin7 | 1 | Gem-associated protein 7 (Gemin7) |
| Get2_like | 1 | Get2-like |
| GlcNAc-1_reg | 1 | Putative GlcNAc-1 phosphotransferase regulatory domain |
| GlcNAc_2-epim | 1 | N-acylglucosamine 2-epimerase (GlcNAc 2-epimerase) |
| Glce_b_sandwich | 1 | D-glucuronyl C5-epimerase, beta-sandwich domain |
| Gln-synt_N | 1 | Glutamine synthetase, beta-Grasp domain |
| Glrx-like | 1 | Glutaredoxin-like domain (DUF836) |
| Glyco_hydr_116N | 1 | beta-glucosidase 2, glycosyl-hydrolase family 116 N-term |
| Glyco_hydro38C2 | 1 | Glycosyl hydrolases family 38 C-terminal beta sandwich domain |
| Glyco_hydro_2 | 1 | Glycosyl hydrolases family 2 |
| Glyco_hydro_2_N | 1 | Glycosyl hydrolases family 2, sugar binding domain |
| Glyco_hydro_30 | 1 | Glycosyl hydrolase family 30 TIM-barrel domain |
| Glyco_hydro_30C | 1 | Glycosyl hydrolase family 30 beta sandwich domain |
| Glyco_hydro_39 | 1 | Glycosyl hydrolases family 39 |
| Glyco_hydro_39_C | 1 | Alpha-L-iduronidase C-terminal domain |
| Glyco_hydro_59 | 1 | Glycosyl hydrolase family 59 |
| Glyco_hydro_59M | 1 | Glycosyl hydrolase family 59 central domain |
| Glyco_hydro_59_C | 1 | Galactocerebrosidase, C-terminal lectin domain |
| Glyco_hydro_63 | 1 | Glycosyl hydrolase family 63 C-terminal domain |
| Glyco_hydro_63N | 1 | Glycosyl hydrolase family 63 N-terminal domain |
| Glyco_hydro_65m | 1 | Glycosyl hydrolase family 65 central catalytic domain |
| Glyco_hydro_85 | 1 | Glycosyl hydrolase family 85 |
| Glyco_tran_28_C | 1 | Glycosyltransferase family 28 C-terminal domain |
| Glyco_trans_4_4 | 1 | Glycosyl transferase 4-like domain |
| Glyco_transf_17 | 1 | Glycosyltransferase family 17 |
| Glyco_transf_21 | 1 | Glycosyl transferase family 21 |
| Glyco_transf_41 | 1 | Glycosyl transferase family 41 |
| Glycos_trans_3N | 1 | Glycosyl transferase family, helical bundle domain |
| Glycos_transf_3 | 1 | Glycosyl transferase family, a/b domain |
| Glycos_transf_4 | 1 | Glycosyl transferase family 4 |
| Glyoxalase_4 | 1 | Glyoxalase/Bleomycin resistance protein/Dioxygenase superfamily |
| GnHR_trans | 1 | Gonadotropin hormone receptor transmembrane region |
| Golgin_A5 | 1 | Golgin subfamily A member 5 |
| Gpi1 | 1 | N-acetylglucosaminyl transferase component (Gpi1) |
| Gpi16 | 1 | Gpi16 subunit, GPI transamidase component |
| Granulin | 1 | Granulin |
| Gryzun-like | 1 | Gryzun, putative Golgi trafficking |
| Guanylate_cyc_2 | 1 | Guanylylate cyclase |
| H-K_ATPase_N | 1 | Gastric H+/K+-ATPase, N terminal domain |
| HAD_2 | 1 | Haloacid dehalogenase-like hydrolase |
| HARP | 1 | HepA-related protein (HARP) |
| HAT | 1 | HAT (Half-A-TPR) repeat |
| HAT1_C | 1 | Histone acetyltransferase type B catalytic subunit, C-terminal |
| HAUS-augmin3 | 1 | HAUS augmin-like complex subunit 3 |
| HAUS2 | 1 | HAUS augmin-like complex subunit 2 |
| HAUS4 | 1 | HAUS augmin-like complex subunit 4 |
| HAUS5 | 1 | HAUS augmin-like complex subunit 5 |
| HAUS6_N | 1 | HAUS augmin-like complex subunit 6 N-terminus |
| HBB | 1 | Helical and beta-bridge domain |
| HBS1_N | 1 | HBS1 N-terminus |
| HCNGP | 1 | HCNGP-like protein |
| HCR | 1 | Alpha helical coiled-coil rod protein (HCR) |
| HD | 1 | HD domain |
| HD_3 | 1 | HD domain |
| HD_4 | 1 | HD domain |
| HEAT_PBS | 1 | PBS lyase HEAT-like repeat |
| HECA | 1 | Headcase protein family homologue |
| HECT_2 | 1 | HECT-like Ubiquitin-conjugating enzyme (E2)-binding |
| HEM4 | 1 | Uroporphyrinogen-III synthase HemD |
| HEPN | 1 | HEPN domain |
| HEPN_DZIP3 | 1 | DZIP3/ hRUL138-like HEPN |
| HERV-K_REC | 1 | Rec (regulator of expression encoded by corf) of HERV-K-113 |
| HGSNAT_cat | 1 | Heparan-alpha-glucosaminide N-acetyltransferase, catalytic |
| HGTP_anticodon2 | 1 | Anticodon binding domain of tRNAs |
| HHH_2 | 1 | Helix-hairpin-helix motif |
| HHH_7 | 1 | Helix-hairpin-helix motif |
| HILPDA | 1 | Hypoxia-inducible lipid droplet-associated |
| HIND | 1 | HIND motif |
| HIRAN | 1 | HIRAN domain |
| HJURP_mid | 1 | Holliday junction recognition protein-associated repeat |
| HMG-CoA_red | 1 | Hydroxymethylglutaryl-coenzyme A reductase |
| HMG_box_5 | 1 | HMG (high mobility group) box 5 |
| HMMR_N | 1 | Hyaluronan mediated motility receptor N-terminal |
| HNF-1A_C | 1 | Hepatocyte nuclear factor 1 (HNF-1), alpha isoform C terminus |
| HNH | 1 | HNH endonuclease |
| HNH_3 | 1 | HNH endonuclease |
| HOATZ-like | 1 | Cilia- and flagella-associated protein HOATZ-like |
| HOIP-UBA | 1 | HOIP UBA domain pair |
| HOOK_N_NuMA | 1 | NuMA N-terminal hook domain |
| HPF1 | 1 | Histone PARylation factor 1 |
| HPHLAWLY | 1 | Domain of unknown function |
| HPIP | 1 | HCF-1 beta-propeller-interacting protein family |
| HPS3_C | 1 | Hermansky-Pudlak syndrome 3 protein, C-terminal region |
| HPS3_N | 1 | Hermansky-Pudlak syndrome 3 protein, N-terminal |
| HPS6 | 1 | Hermansky-Pudlak syndrome 6 protein N-terminal domain |
| HPS6_C | 1 | Hermansky-Pudlak syndrome 6 protein C-terminal domain |
| HRCT1 | 1 | Histidine-rich carboxyl terminus protein 1 |
| HRG | 1 | Haem-transporter, endosomal/lysosomal, haem-responsive gene |
| HROB | 1 | Homologous recombination OB-fold protein |
| HSCB_C | 1 | HSCB C-terminal oligomerisation domain |
| HSD3 | 1 | Spermatogenesis-associated protein 7, or HSD3 |
| HSL_N | 1 | Hormone-sensitive lipase (HSL) N-terminus |
| HTH_3 | 1 | Helix-turn-helix |
| HTH_40 | 1 | Helix-turn-helix domain |
| HTH_44 | 1 | Helix-turn-helix DNA-binding domain of SPT6 |
| HTH_9 | 1 | RNA polymerase III subunit RPC82 helix-turn-helix domain |
| HTTM | 1 | HTTM domain |
| HU-CCDC81_euk_1 | 1 | CCDC81 eukaryotic HU domain 1 |
| HU-CCDC81_euk_2 | 1 | CCDC81 eukaryotic HU domain 2 |
| HVSL | 1 | Uncharacterised conserved protein |
| HYLS1_C | 1 | Hydrolethalus syndrome protein 1 C-terminus |
| Ham1p_like | 1 | Ham1 family |
| Hamartin | 1 | Hamartin protein |
| Harakiri | 1 | Activator of apoptosis harakiri |
| Hat1_N | 1 | Histone acetyl transferase HAT1 N-terminus |
| He_PIG | 1 | Putative Ig domain |
| Headcase | 1 | Headcase protein |
| Helicase_C_3 | 1 | Helicase conserved C-terminal domain |
| Helicase_PWI | 1 | N-terminal helicase PWI domain |
| Helicase_RecD | 1 | Helicase |
| HemN_C | 1 | HemN C-terminal domain |
| Hemerythrin | 1 | Hemerythrin HHE cation binding domain |
| Hep_59 | 1 | Hepatocellular carcinoma-associated antigen 59 |
| Hepcidin | 1 | Hepcidin |
| Hepsin-SRCR | 1 | Hepsin, SRCR domain |
| Herpes_UL52 | 1 | Herpesviridae UL52/UL70 DNA primase |
| HgmA_C | 1 | Homogentisate 1,2-dioxygenase C-terminal |
| HgmA_N | 1 | Homogentisate 1,2-dioxygenase N-terminal |
| Hid1 | 1 | High-temperature-induced dauer-formation protein |
| Hikeshi-like_C | 1 | Hikeshi-like, C-terminal domain |
| Hikeshi-like_N | 1 | Hikeshi-like, N-terminal domain |
| HipN | 1 | Hsp70-interacting protein N N-terminal domain |
| Hira | 1 | TUP1-like enhancer of split |
| Hist_rich_Ca-bd | 1 | Histidine-rich Calcium-binding repeat region |
| HnRNP_M_NLS | 1 | HnRNP M nuclear localisation signal |
| Hormone_6 | 1 | Glycoprotein hormone |
| HpcH_HpaI | 1 | HpcH/HpaI aldolase/citrate lyase family |
| Hrs_helical | 1 | Hepatocyte growth factor-regulated tyrosine kinase substrate |
| HscB_4_cys | 1 | Co-chaperone HscB tetracysteine metal binding motif |
| Htt_C-HEAT | 1 | Huntingtin, C-terminal HEAT |
| Htt_N-HEAT | 1 | Huntingtin, N-terminal HEAT |
| Htt_N-HEAT_1 | 1 | Huntingtin, N-terminal HEAT 1 |
| Htt_bridge | 1 | Huntingtin, bridge |
| Hydant_A_C | 1 | Hydantoinase/oxoprolinase C-terminal domain |
| Hydant_A_N | 1 | Hydantoinase/oxoprolinase N-terminal region |
| Hydantoinase_A | 1 | Hydantoinase/oxoprolinase |
| Hydantoinase_B | 1 | Hydantoinase B/oxoprolinase |
| Hydin_ADK | 1 | Hydin Adenylate kinase-like domain |
| Hydrolase_3 | 1 | haloacid dehalogenase-like hydrolase |
| IATP | 1 | Mitochondrial ATPase inhibitor, IATP |
| ICAP-1_inte_bdg | 1 | Beta-1 integrin binding protein |
| ICMT | 1 | Isoprenylcysteine carboxyl methyltransferase (ICMT) family |
| IF3_N | 1 | Translation initiation factor IF-3, N-terminal domain |
| IFN-gamma | 1 | Interferon gamma |
| IFNGR1_D2 | 1 | Interferon gamma receptor (IFNGR1), D2 domain |
| IFNGR1_transm | 1 | Interferon gamma receptor 1, transmembrane region |
| IFT20 | 1 | Intraflagellar transport complex B, subunit 20 |
| IFT43 | 1 | Intraflagellar transport protein 43 |
| IFT46_B_C | 1 | Intraflagellar transport complex B protein 46 C terminal |
| IFT57 | 1 | Intra-flagellar transport protein 57 |
| IFT81_CH | 1 | Intraflagellar transport 81 calponin homology domain |
| IFTAP | 1 | Intraflagellar transport-associated protein |
| IGF2_C | 1 | Insulin-like growth factor II E-peptide |
| IHO1 | 1 | Interactor of HORMAD1 protein 1 |
| IKI3 | 1 | IKI3 family |
| IL11 | 1 | Interleukin 11 |
| IL12 | 1 | Interleukin-12 alpha subunit |
| IL12p40_C | 1 | Cytokine interleukin-12p40 C-terminus |
| IL13 | 1 | Interleukin-13 |
| IL17R_D_N | 1 | N-terminus of interleukin 17 receptor D |
| IL2 | 1 | Interleukin 2 |
| IL22 | 1 | Interleukin 22 IL-10-related T-cell-derived-inducible factor |
| IL23 | 1 | Interleukin 23 subunit alpha |
| IL2RB_N1 | 1 | Interleukin-2 receptor subunit beta N-terminal domain 1 |
| IL3 | 1 | Interleukin-3 |
| IL31 | 1 | Interleukin 31 |
| IL32 | 1 | Interleukin 32 |
| IL33_bt | 1 | Interleukin 33 |
| IL34 | 1 | Interleukin 34 |
| IL3Rb_N | 1 | GM-CSF/IL-3/IL-5 receptor common beta subunit, N-terminal |
| IL4 | 1 | Interleukin 4 |
| IL4Ra_N | 1 | Interleukin-4 receptor alpha chain, N-terminal |
| IL5 | 1 | Interleukin 5 |
| IL6 | 1 | Interleukin-6/G-CSF/MGF family |
| IL7 | 1 | Interleukin 7 |
| IML1 | 1 | Vacuolar membrane-associated protein Iml1, N-terminal domain |
| IMUP | 1 | Immortalisation up-regulated protein |
| INCA1 | 1 | INCA1 |
| INCENP_ARK-bind | 1 | Inner centromere protein, ARK binding region |
| INCENP_N | 1 | Chromosome passenger complex (CPC) protein INCENP N terminal |
| INI1_DNA-bd | 1 | SWI/SNF Subunit INI1, DNA binding domain |
| INPP5B_PH | 1 | Type II inositol 1,4,5-trisphosphate 5-phosphatase PH domain |
| INSC_LBD | 1 | Inscuteable LGN-binding domain |
| INT10 | 1 | Integrator subunit 10 |
| INTAP | 1 | Intersectin and clathrin adaptor AP2 binding region |
| INTS15 | 1 | Integrator complex subunit 15 |
| INTS1_RP2B-bd | 1 | Integrator complex subunit 1, RP2B-binding domain |
| INTS2 | 1 | Integrator complex subunit 2 |
| INTS5_C | 1 | Integrator complex subunit 5 C-terminus |
| INTS5_N | 1 | Integrator complex subunit 5 N-terminus |
| IPPT | 1 | IPP transferase |
| IR1-M | 1 | Nup358/RanBP2 E3 ligase domain |
| ISET-FN3_linker | 1 | Unstructured linking region I-set and fnIII on Brother of CDO |
| ISPD_C | 1 | D-ribitol-5-phosphate cytidylyltransferase C-terminal domain |
| IgGFc_binding | 1 | IgGFc binding protein |
| Ig_5 | 1 | Ig-like domain on T-cell surface glycoprotein CD3 epsilon chain |
| Ig_C19orf38 | 1 | Ig domain in C19orf38 (HIDE1) |
| Ig_J_chain | 1 | Immunoglobulin J chain |
| Ig_Tie2_1 | 1 | Tie-2 Ig-like domain 1 |
| Ig_mannosidase | 1 | Ig-fold domain |
| Il13Ra_Ig | 1 | Interleukin-13 receptor subunit alpha Ig-like domain |
| Ima1_N | 1 | Ima1 N-terminal domain |
| Img2 | 1 | Mitochondrial large subunit ribosomal protein (Img2) |
| Importin_rep | 1 | Importin 13 repeat |
| Importin_rep_2 | 1 | Importin 13 repeat |
| Importin_rep_3 | 1 | Importin 13 repeat |
| Importin_rep_5 | 1 | Importin repeat |
| Inhibitor_I9 | 1 | Peptidase inhibitor I9 |
| Inos-1-P_synth | 1 | Myo-inositol-1-phosphate synthase |
| Ins134_P3_kin | 1 | Inositol 1,3,4-trisphosphate 5/6-kinase ATP-grasp domain |
| Ins134_P3_kin_N | 1 | Inositol 1,3,4-trisphosphate 5/6-kinase pre-ATP-grasp domain |
| Ins_P5_2-kin | 1 | Inositol-pentakisphosphate 2-kinase |
| Insc_C | 1 | Protein inscuteable C-terminal |
| Insulin_TMD | 1 | Insulin receptor trans-membrane segment |
| IntS11_C | 1 | Integrator IntS11, C-terminal domain |
| IntS14_C | 1 | Integrator complex subunit 14, alpha-helical domain |
| IntS14_b-barrel | 1 | Integrator complex subunit 14, beta-barrel domain |
| IntS9_C | 1 | Integrator IntS9, C-terminal domain |
| Ints3_N | 1 | Integrator complex subunit 3 N-terminal |
| Involucrin | 1 | Involucrin repeat |
| Involucrin_N | 1 | Involucrin of squamous epithelia N-terminus |
| Ipi1_N | 1 | Rix1 complex component involved in 60S ribosome maturation |
| IspD | 1 | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
| Ist1 | 1 | Regulator of Vps4 activity in the MVB pathway |
| Itf52_C | 1 | Intraflagellar transport protein 52, C-terminal domain |
| Itfg2 | 1 | Integrin-alpha FG-GAP repeat-containing protein 2 |
| Iwr1 | 1 | Transcription factor Iwr1 |
| Izumo-Ig | 1 | Izumo-like Immunoglobulin domain |
| JAMP | 1 | JNK1/MAPK8-associated membrane protein |
| JCAD | 1 | Junctional protein associated with coronary artery disease |
| JHY | 1 | Jhy protein |
| JSRP | 1 | Junctional sarcoplasmic reticulum protein |
| JTB | 1 | Jumping translocation breakpoint protein (JTB) |
| Jagunal | 1 | Jagunal, ER re-organisation during oogenesis |
| Jiraiya | 1 | Jiraiya |
| Jiv90 | 1 | Cleavage inducing molecular chaperone |
| Joubert | 1 | Joubert syndrome-associated |
| K1377 | 1 | Susceptibility to monomelic amyotrophy |
| KAP | 1 | Kinesin-associated protein (KAP) |
| KAsynt_C_assoc | 1 | Ketoacyl-synthetase C-terminal extension |
| KBP_C | 1 | KIF-1 binding protein C terminal |
| KCNQ2_u3 | 1 | Unstructured region on Potassium channel subunit alpha KvLQT2 |
| KCTD18_C | 1 | BTB/POZ domain-containing protein KCTD18 C-terminus |
| KCTD4_C | 1 | BTB/POZ domain-containing protein KCTD4 C-terminus |
| KHA | 1 | KHA, dimerisation domain of potassium ion channel |
| KH_2 | 1 | KH domain |
| KH_8 | 1 | Krr1 KH1 domain |
| KI67R | 1 | KI67R (NUC007) repeat |
| KIAA1328 | 1 | Uncharacterised protein KIAA1328 |
| KKLCAg1 | 1 | Kita-kyushu lung cancer antigen 1 |
| KLRAQ | 1 | Predicted coiled-coil domain-containing protein |
| KN17_SH3 | 1 | KN17 SH3-like C-terminal domain |
| KR | 1 | KR domain |
| KRBA1 | 1 | KRBA1 family repeat |
| KRR1-like_KH2 | 1 | KRR1 small subunit processome component, second KH domain |
| KRTAP7 | 1 | KRTAP type 7 family |
| K_channel_TID | 1 | Potassium channel Kv1.4 tandem inactivation domain |
| Kazal_3 | 1 | Kazal-type serine protease inhibitor domain |
| KcnmB2_inactiv | 1 | KCNMB2, ball and chain domain |
| Kdo | 1 | Lipopolysaccharide kinase (Kdo/WaaP) family |
| Keratin_2_tail | 1 | Keratin type II cytoskeletal 1 tail |
| Keratin_assoc | 1 | Keratinocyte-associated protein 2 |
| Kin17_mid | 1 | Domain of Kin17 curved DNA-binding protein |
| Kinetochor_Ybp2 | 1 | Uncharacterised protein family, YAP/Alf4/glomulin |
| Kinocilin | 1 | Kinocilin protein |
| Kisspeptin | 1 | Kisspeptin |
| Knl1_RWD_C | 1 | Knl1 RWD C-terminal domain |
| Kri1 | 1 | KRI1-like family |
| Kri1_C | 1 | KRI1-like family C-terminal |
| Ku_PK_bind | 1 | Ku C terminal domain like |
| KxDL | 1 | Uncharacterized conserved protein |
| L27_N | 1 | L27_N |
| LAMTOR | 1 | Late endosomal/lysosomal adaptor and MAPK and MTOR activator |
| LAMTOR5 | 1 | Ragulator complex protein LAMTOR5 |
| LAS2 | 1 | Lung adenoma susceptibility protein 2 |
| LAT | 1 | Linker for activation of T-cells |
| LAT2 | 1 | Linker for activation of T-cells family member 2 |
| LAX | 1 | Lymphocyte activation family X |
| LBR_tudor | 1 | Lamin-B receptor of TUDOR domain |
| LCE6A | 1 | Late cornified envelope protein 6A family |
| LEAP-2 | 1 | Liver-expressed antimicrobial peptide 2 precursor (LEAP-2) |
| LELP1 | 1 | Late cornified envelope-like proline-rich protein 1 |
| LEP503 | 1 | Lens epithelial cell protein LEP503 |
| LETM2_N | 1 | LETM1 domain-containing protein LETM2 N-terminus |
| LIAS_N | 1 | N-terminal domain of lipoyl synthase of Radical_SAM family |
| LIDHydrolase | 1 | Lipid-droplet associated hydrolase |
| LIFR_N | 1 | Leukemia inhibitory factor receptor N-terminal domain |
| LIG3_BRCT | 1 | DNA ligase 3 BRCT domain |
| LIME1 | 1 | Lck-interacting transmembrane adapter 1 |
| LIN37 | 1 | LIN37 |
| LIN52 | 1 | Retinal tissue protein |
| LIN9_C | 1 | LIN9 C-terminal |
| LINES_C | 1 | Lines C-terminus |
| LINES_N | 1 | Lines N-terminus |
| LIP1 | 1 | LKB1 serine/threonine kinase interacting protein 1 |
| LKAAEAR | 1 | Family of unknown function with LKAAEAR motif |
| LLC1 | 1 | Normal lung function maintenance, Low in Lung Cancer 1 protein |
| LLCFC1 | 1 | LLLL and CFNLAS motif-containing protein 1 |
| LLPH | 1 | LLP homolog |
| LMWPc | 1 | Low molecular weight phosphotyrosine protein phosphatase |
| LNP1 | 1 | Leukemia NUP98 fusion partner 1 |
| LOH1CR12 | 1 | Tumour suppressor protein |
| LRIF1 | 1 | Ligand-dependent nuclear receptor-interacting factor 1 |
| LRR19-TM | 1 | Leucine-rich repeat family 19 TM domain |
| LRR_11 | 1 | Leucine-rich repeat |
| LRR_12 | 1 | Leucine-rich repeat |
| LRR_RI_capping | 1 | Capping Ribonuclease inhibitor Leucine Rich Repeat |
| LSM12_LSM | 1 | LSM12, LSM domain |
| LSM_int_assoc | 1 | LSM-interacting associated unstructured |
| LST1 | 1 | LST-1 protein |
| LTV | 1 | Low temperature viability protein |
| LYRIC | 1 | Lysine-rich CEACAM1 co-isolated protein family |
| Lactamase_B_4 | 1 | tRNase Z endonuclease |
| Las1 | 1 | Las1-like |
| Leg1 | 1 | Leg1 |
| Legum_prodom | 1 | Legumain, prodomain |
| Leo1 | 1 | Leo1-like protein |
| LepA_C | 1 | GTP-binding protein LepA C-terminus |
| Leptin | 1 | Leptin |
| Leu_zip | 1 | Leucine zipper |
| Lge1 | 1 | Transcriptional regulatory protein LGE1 |
| Lipocalin_2 | 1 | Lipocalin-like domain |
| LisH_2 | 1 | LisH |
| Loricrin | 1 | Major keratinocyte cell envelope protein |
| Lsm_interact | 1 | Lsm interaction motif |
| Lyase_aromatic | 1 | Aromatic amino acid lyase |
| Lycopene_cycl | 1 | Lycopene cyclase protein |
| M16C_assoc | 1 | Peptidase M16C associated |
| MAD | 1 | Mitotic checkpoint protein |
| MAGI_u5 | 1 | Unstructured region on MAGI |
| MAJIN | 1 | Membrane-anchored junction protein |
| MALT1_Ig | 1 | MALT1 Ig-like domain |
| MAM33 | 1 | Mitochondrial glycoprotein |
| MAML2_TAD | 1 | Mastermind-like protein 2, transcriptional activation domain |
| MAP10_N | 1 | Microtubule associated protein 10, N-terminal domain |
| MAP1B_neuraxin | 1 | Neuraxin and MAP1B repeat |
| MAP2_projctn | 1 | MAP2/Tau projection domain |
| MAPKK1_Int | 1 | Mitogen-activated protein kinase kinase 1 interacting |
| MARF1_LOTUS | 1 | MARF1 LOTUS domain |
| MARF1_RRM1 | 1 | MARF1, RNA recognition motif 1 |
| MAT1 | 1 | CDK-activating kinase assembly factor MAT1 |
| MBF1 | 1 | Multiprotein bridging factor 1 |
| MCCD1 | 1 | Mitochondrial coiled-coil domain protein 1 |
| MCD | 1 | Malonyl-CoA decarboxylase C-terminal domain |
| MCD_N | 1 | Malonyl-CoA decarboxylase N-terminal domain |
| MCLC | 1 | Mid-1-related chloride channel (MCLC) |
| MCM2_N | 1 | Mini-chromosome maintenance protein 2 |
| MCM3AP_GANP | 1 | MCM3AP domain of GANP |
| MCM4_WHD | 1 | MCM4, winged helix domain |
| MCM6_C | 1 | MCM6 C-terminal winged-helix domain |
| MCM_bind | 1 | Mini-chromosome maintenance replisome factor |
| MCRS_N | 1 | N-terminal region of micro-spherule protein |
| MDD_C | 1 | Mevalonate 5-diphosphate decarboxylase C-terminal domain |
| MDM1 | 1 | Nuclear protein MDM1 |
| MDN1_4th | 1 | Midasin AAA+ ATPase lid domain |
| MEA1 | 1 | Male enhanced antigen 1 (MEA1) |
| MEF2_binding | 1 | MEF2 binding |
| MEIOC | 1 | Meiosis-specific coiled-coil domain-containing protein MEIOC |
| MEKK4_N | 1 | MEKK4 N-terminal |
| MELK_UBA | 1 | Maternal embryonic leucine zipper kinase, UBA domain |
| MELT | 1 | MELT motif |
| MFAP1 | 1 | Microfibril-associated/Pre-mRNA processing |
| MFS_5 | 1 | Sugar-tranasporters, 12 TM |
| MGAT2 | 1 | N-acetylglucosaminyltransferase II (MGAT2) |
| MGA_dom | 1 | MGA, conserved domain |
| MHC2-interact | 1 | CLIP, MHC2 interacting |
| MHCassoc_trimer | 1 | Class II MHC-associated invariant chain trimerisation domain |
| MIEAP | 1 | Mitochondria-eating protein |
| MIF4G_like | 1 | MIF4G like |
| MIF4G_like_2 | 1 | MIF4G like |
| MIIP | 1 | Migration and invasion-inhibitory |
| MINT_MID | 1 | Msx2-interacting protein, MID domain |
| MINT_RAM7 | 1 | Msx2-interacting protein, RAM7 domain |
| MINT_RID | 1 | Msx2-interacting protein, nuclear receptor-interacting domain |
| MIOS_a-sol | 1 | MIOS, alpha-solenoid |
| MIOX | 1 | Myo-inositol oxygenase |
| MIP-T3 | 1 | Microtubule-binding protein MIP-T3 CH-like domain |
| MIP-T3_C | 1 | Microtubule-binding protein MIP-T3 C-terminal region |
| MIS13 | 1 | Mis12-Mtw1 protein family |
| MISS | 1 | MAPK-interacting and spindle-stabilising protein-like |
| MIT_C | 1 | Phospholipase D-like domain at C-terminus of MIT |
| MIX23 | 1 | Intermembrane space protein MIX23 |
| MJ1316 | 1 | MJ1316 RNA cyclic group end recognition domain |
| MKLP1_Arf_bdg | 1 | Arf6-interacting domain of mitotic kinesin-like protein 1 |
| MLANA | 1 | Protein melan-A |
| MLIP | 1 | Muscular LMNA-interacting protein |
| MLVIN_C | 1 | Murine leukemia virus (MLV) integrase (IN) C-terminal domain |
| MMACHC | 1 | Methylmalonic aciduria and homocystinuria type C family |
| MMADHC | 1 | Methylmalonic aciduria and homocystinuria type D protein |
| MMPL | 1 | MMPL family |
| MMS19_C | 1 | RNAPII transcription regulator C-terminal |
| MMS19_N | 1 | Dos2-interacting transcription regulator of RNA-Pol-II |
| MMS22L_C | 1 | S-phase genomic integrity recombination mediator, C-terminal |
| MMS22L_N | 1 | S-phase genomic integrity recombination mediator, N-terminal |
| MM_CoA_mutase | 1 | Methylmalonyl-CoA mutase |
| MMgT | 1 | Membrane magnesium transporter |
| MMtag | 1 | Multiple myeloma tumor-associated |
| MNR | 1 | Protein moonraker |
| MOFRL | 1 | MOFRL family |
| MOV-10_Ig-like | 1 | Helicase MOV-10, Ig-like domain |
| MOV-10_N | 1 | Helicase MOV-10, N-terminal |
| MOZART1 | 1 | Mitotic-spindle organizing gamma-tubulin ring associated |
| MPDZ_u10 | 1 | Unstructured region 10 on multiple PDZ protein |
| MPLKIP | 1 | M-phase-specific PLK1-interacting protein |
| MPP6 | 1 | M-phase phosphoprotein 6 |
| MREG | 1 | Melanoregulin |
| MRI | 1 | Modulator of retrovirus infection |
| MRNIP | 1 | MRN-interacting protein |
| MRP | 1 | Mitochondrial RNA binding protein MRP |
| MRP-63 | 1 | Mitochondrial ribosome protein 63 |
| MRP-L27 | 1 | Mitochondrial ribosomal protein L27 |
| MRP-L28 | 1 | Mitochondrial ribosomal protein L28 |
| MRP-L46 | 1 | 39S mitochondrial ribosomal protein L46 |
| MRP-L47 | 1 | Mitochondrial 39-S ribosomal protein L47 (MRP-L47) |
| MRP-L51 | 1 | Mitochondrial ribosomal subunit |
| MRP-S22 | 1 | Mitochondrial 28S ribosomal protein S22 |
| MRP-S23 | 1 | Mitochondrial ribosomal protein S23 |
| MRP-S24 | 1 | Mitochondrial ribosome subunit S24 |
| MRP-S26 | 1 | Mitochondrial ribosome subunit S26 |
| MRP-S28 | 1 | Mitochondrial ribosomal subunit protein |
| MRP-S31 | 1 | Mitochondrial 28S ribosomal protein S31 |
| MRP-S32 | 1 | Mitochondrial 28S ribosomal protein S32 |
| MRP-S33 | 1 | Mitochondrial ribosomal subunit S27 |
| MRP-S34 | 1 | Mitochondrial 28S ribosomal protein S34 |
| MRP-S35 | 1 | Mitochondrial ribosomal protein MRP-S35 |
| MRPL52 | 1 | Mitoribosomal protein mL52 |
| MRP_L53 | 1 | 39S ribosomal protein L53/MRP-L53 |
| MR_MLE_C | 1 | Enolase C-terminal domain-like |
| MR_MLE_N | 1 | Mandelate racemase / muconate lactonizing enzyme, N-terminal domain |
| MSL1_dimer | 1 | Dimerisation domain of Male-specific-Lethal 1 |
| MSL2-CXC | 1 | CXC domain of E3 ubiquitin-protein ligase MSL2 |
| MTABC_N | 1 | Mitochondrial ABC-transporter N-terminal five TM region |
| MTBP_C | 1 | MDM2-binding |
| MTBP_N | 1 | MDM2-binding |
| MTBP_mid | 1 | MDM2-binding |
| MTCP1 | 1 | Mature-T-Cell Proliferation I type |
| MTHFR | 1 | Methylenetetrahydrofolate reductase |
| MTP18 | 1 | Mitochondrial 18 KDa protein (MTP18) |
| MTP_lip_bd | 1 | MTP large subunit, lipid-binding domain |
| MUS81-like_WH | 1 | Crossover junction endonuclease MUS81-like, winged helix domain |
| MVP_shoulder | 1 | Shoulder domain |
| MWFE | 1 | NADH-ubiquinone oxidoreductase MWFE subunit |
| MYCBPAP | 1 | MYCBP-associated protein family |
| MYEOV2 | 1 | Myeloma-overexpressed-like |
| MYO10_CC | 1 | Unconventional myosin-X coiled coil domain |
| MYO6_lever | 1 | Myosin VI, lever arm |
| M_domain | 1 | M domain of GW182 |
| Macoilin | 1 | Macoilin family |
| Macscav_rec | 1 | Macrophage scavenger receptor |
| Maelstrom | 1 | piRNA pathway germ-plasm component |
| Maf | 1 | Maf-like protein |
| Maf1 | 1 | Maf1 regulator |
| MafB19-deam | 1 | MafB19-like deaminase |
| Mago-bind | 1 | Mago binding |
| Mak10 | 1 | Mak10 subunit, NatC N(alpha)-terminal acetyltransferase |
| Mak16 | 1 | Mak16 protein C-terminal region |
| Malectin | 1 | Malectin domain |
| Man-6-P_recep | 1 | Mannose-6-phosphate receptor |
| Mannosidase_ig | 1 | Mannosidase Ig/CBM-like domain |
| Mannosyl_trans | 1 | Mannosyltransferase (PIG-M) |
| Mannosyl_trans2 | 1 | Mannosyltransferase (PIG-V) |
| Mat89Bb | 1 | Cell cycle and development regulator Mat89Bb |
| MazG-like | 1 | MazG-like family |
| Mcl1_mid | 1 | Minichromosome loss protein, Mcl1, middle region |
| Mcm10 | 1 | Mcm10 replication factor |
| MeaB | 1 | Methylmalonyl Co-A mutase-associated GTPase MeaB |
| Meckelin | 1 | Meckelin (Transmembrane protein 67) |
| Med1 | 1 | Mediator of RNA polymerase II transcription subunit 1 |
| Med10 | 1 | Transcription factor subunit Med10 of Mediator complex |
| Med11 | 1 | Mediator complex protein |
| Med13_N | 1 | Mediator complex subunit 13 N-terminal |
| Med14 | 1 | Mediator complex subunit MED14 |
| Med15_C | 1 | ARC105/Med15 mediator subunit C-terminal domain |
| Med15_M | 1 | ARC105/Med15 mediator subunit central domain |
| Med15_N | 1 | ARC105/Med15 mediator subunit N-terminal KIX domain |
| Med16_C | 1 | Mediator complex subunit 16, C-terminal |
| Med16_N | 1 | Mediator complex subunit 16, N-terminal |
| Med16_bridge | 1 | Mediator complex subunit 16, central helical bridge |
| Med17 | 1 | Subunit 17 of Mediator complex |
| Med18 | 1 | Med18 protein |
| Med19 | 1 | Mediator of RNA pol II transcription subunit 19 |
| Med20 | 1 | TATA-binding related factor (TRF) of subunit 20 of Mediator complex |
| Med21 | 1 | Subunit 21 of Mediator complex |
| Med22 | 1 | Surfeit locus protein 5 subunit 22 of Mediator complex |
| Med23 | 1 | Mediator complex subunit 23 |
| Med24_N | 1 | Mediator complex subunit 24 N-terminal |
| Med25_SD1 | 1 | Mediator complex subunit 25 synapsin 1 |
| Med25_VWA | 1 | Mediator complex subunit 25 von Willebrand factor type A |
| Med26_C | 1 | Mediator complex subunit 26 C-terminal |
| Med26_M | 1 | Mediator complex subunit 26 middle domain |
| Med27 | 1 | Mediator complex subunit 27 |
| Med28 | 1 | Mediator complex subunit 28 |
| Med29 | 1 | Mediator complex subunit 29 |
| Med30 | 1 | Mediator complex subunit 30 |
| Med31 | 1 | SOH1 |
| Med4 | 1 | Vitamin-D-receptor interacting Mediator subunit 4 |
| Med6 | 1 | MED6 mediator sub complex component |
| Med7 | 1 | MED7 protein |
| Med8 | 1 | Mediator of RNA polymerase II transcription complex subunit 8 |
| Med9 | 1 | RNA polymerase II transcription mediator complex subunit 9 |
| Mei4 | 1 | Meiosis-specific protein Mei4 |
| Mei5 | 1 | Double-strand recombination repair protein |
| Meiosis_expr | 1 | Meiosis-expressed |
| Mem_trans | 1 | Membrane transport protein |
| Membralin | 1 | Tumour-associated protein |
| Memo | 1 | Memo-like protein |
| Menin | 1 | Menin |
| Mesd | 1 | Chaperone for wingless signalling and trafficking of LDL receptor |
| Met_synt_B12 | 1 | Vitamin B12 dependent methionine synthase, activation domain |
| Metallophos_2 | 1 | Calcineurin-like phosphoesterase superfamily domain |
| Metallophos_C | 1 | Iron/zinc purple acid phosphatase-like protein C |
| Methyltr_RsmF_N | 1 | N-terminal domain of 16S rRNA methyltransferase RsmF |
| Methyltransf_10 | 1 | RNA methyltransferase |
| Methyltransf_15 | 1 | RNA cap guanine-N2 methyltransferase |
| Methyltransf_1N | 1 | 6-O-methylguanine DNA methyltransferase, ribonuclease-like domain |
| Methyltransf_22 | 1 | Methyltransferase domain |
| Methyltransf_28 | 1 | Putative S-adenosyl-L-methionine-dependent methyltransferase |
| Methyltransf_4 | 1 | Putative methyltransferase |
| Methyltransf_5 | 1 | MraW methylase family |
| Methyltransf_8 | 1 | Hypothetical methyltransferase |
| Methyltransf_FA | 1 | Farnesoic acid 0-methyl transferase |
| Methyltrn_RNA_3 | 1 | Putative RNA methyltransferase |
| Mg_chelatase | 1 | Magnesium chelatase, subunit ChlI |
| MgsA_C | 1 | MgsA AAA+ ATPase C terminal |
| Microcephalin | 1 | Microcephalin protein |
| Microtub_bind | 1 | Kinesin-associated microtubule-binding |
| Mis12 | 1 | Mis12 protein |
| Mis14 | 1 | Kinetochore protein Mis14 like |
| Misat_Tub_SegII | 1 | Misato Segment II tubulin-like domain |
| Mit_proteolip | 1 | Mitochondrial proteolipid |
| Mito_morph_reg | 1 | Mitochondrial morphogenesis regulator |
| Mitoc_L55 | 1 | Mitochondrial ribosomal protein L55 |
| Mitofilin | 1 | Mitochondrial inner membrane protein |
| Mlh1_C | 1 | DNA mismatch repair protein Mlh1 C-terminus |
| MmgE_PrpD_C | 1 | MmgE/PrpD C-terminal domain |
| MmgE_PrpD_N | 1 | MmgE/PrpD N-terminal domain |
| Mnd1 | 1 | Mnd1 HTH domain |
| MnmE_helical | 1 | MnmE helical domain |
| Mo-co_dimer | 1 | Mo-co oxidoreductase dimerisation domain |
| MoaC | 1 | MoaC family |
| MoaE | 1 | MoaE protein |
| Mob_synth_C | 1 | Molybdenum Cofactor Synthesis C |
| MoeA_C | 1 | MoeA C-terminal region (domain IV) |
| MoeA_N | 1 | MoeA N-terminal region (domain I and II) |
| Mog1 | 1 | Ran-interacting Mog1 protein |
| Molybdopterin | 1 | Molybdopterin oxidoreductase |
| Mon2_C | 1 | C-terminal region of Mon2 protein |
| Mov-10_helical | 1 | Helicase MOV-10, helical domain |
| Mpp10 | 1 | Mpp10 protein |
| Mre11_DNA_bind | 1 | Mre11 DNA-binding presumed domain |
| MreB_Mbl | 1 | MreB/Mbl protein |
| Mss4 | 1 | Mss4 protein |
| MtN3_slv | 1 | Sugar efflux transporter for intercellular exchange |
| Mt_ATP-synt_B | 1 | Mitochondrial ATP synthase B chain precursor (ATP-synt_B) |
| Mt_ATP-synt_D | 1 | ATP synthase D chain, mitochondrial (ATP5H) |
| Mucin-like | 1 | Mucin-like |
| Mucin15 | 1 | Cell-membrane associated Mucin15 |
| Musclin | 1 | Insulin-resistance promoting peptide in skeletal muscle |
| Muskelin_N | 1 | Muskelin N-terminus |
| Mut7-C | 1 | Mut7-C RNAse domain |
| Muted | 1 | Organelle biogenesis, Muted-like protein |
| Myb_Cef | 1 | pre-mRNA splicing factor component |
| Myb_DNA-bind_2 | 1 | Rap1 Myb domain |
| Myb_DNA-bind_7 | 1 | Myb DNA-binding like |
| Myc-LZ | 1 | Myc leucine zipper domain |
| Myc_target_1 | 1 | Myc target protein 1 |
| Myelin-PO_C | 1 | Myelin-PO cytoplasmic C-term p65 binding region |
| Myelin_MBP | 1 | Myelin basic protein |
| Myosin-VI_CBD | 1 | Myosin VI cargo binding domain |
| NADH-G_4Fe-4S_3 | 1 | NADH-ubiquinone oxidoreductase-G iron-sulfur binding region |
| NADH5_C | 1 | NADH dehydrogenase subunit 5 C-terminus |
| NADH_4Fe-4S | 1 | NADH-ubiquinone oxidoreductase-F iron-sulfur binding region |
| NADH_B2 | 1 | NADH dehydrogenase 1 beta subcomplex subunit 2 |
| NADH_dehy_S2_C | 1 | NADH dehydrogenase subunit 2 C-terminus |
| NADH_dh_m_C1 | 1 | NADH dehydrogenase [ubiquinone] 1 subunit C1, mitochondrial |
| NADH_dhqG_C | 1 | NADH-ubiquinone oxidoreductase subunit G, C-terminal |
| NADH_oxidored | 1 | MNLL subunit |
| NADHdh | 1 | NADH dehydrogenase |
| NADHdh_A3 | 1 | NADH dehydrogenase 1 alpha subcomplex subunit 3 |
| NAD_binding_3 | 1 | Homoserine dehydrogenase, NAD binding domain |
| NAD_binding_5 | 1 | Myo-inositol-1-phosphate synthase |
| NAD_kinase_C | 1 | ATP-NAD kinase C-terminal domain |
| NAGLU | 1 | Alpha-N-acetylglucosaminidase (NAGLU) tim-barrel domain |
| NAGLU_C | 1 | Alpha-N-acetylglucosaminidase (NAGLU) C-terminal domain |
| NAGLU_N | 1 | Alpha-N-acetylglucosaminidase (NAGLU) N-terminal domain |
| NAGPA | 1 | Phosphodiester glycosidase |
| NAGidase | 1 | beta-N-acetylglucosaminidase |
| NAMPT_N | 1 | Nicotinamide phosphoribosyltransferase, N-terminal domain |
| NAPRTase_C | 1 | Nicotinate phosphoribosyltransferase C-terminal domain |
| NAPRTase_N | 1 | Nicotinate phosphoribosyltransferase (NAPRTase) N-terminal domain |
| NARG2_C | 1 | NMDA receptor-regulated gene protein 2 C-terminus |
| NAT | 1 | NAT, N-acetyltransferase, of N-acetylglutamate synthase |
| NB-ARC | 1 | NB-ARC domain |
| NCBP3 | 1 | Nuclear cap-binding protein subunit 3 |
| NCF1_PBR_AIR | 1 | Neutrophil cytosol factor 1, PBR/AIR |
| NCOA6_TRADD-N | 1 | Nuclear receptor coactivator 6, TRADD-N domain |
| NCU-G1 | 1 | Lysosomal transcription factor, NCU-G1 |
| NDNF | 1 | Neuron-derived neurotrophic factor, first Fn(III) domain |
| NDNF_C | 1 | Neuron-derived Neurotrophic Factor C-terminal |
| NDUFB10 | 1 | NADH-ubiquinone oxidoreductase subunit 10 |
| NDUFV3 | 1 | NADH dehydrogenase [ubiquinone] flavoprotein 3, mitochondrial |
| NDUF_B12 | 1 | NADH-ubiquinone oxidoreductase B12 subunit family |
| NDUF_B4 | 1 | NADH-ubiquinone oxidoreductase B15 subunit (NDUFB4) |
| NDUF_B5 | 1 | NADH:ubiquinone oxidoreductase, NDUFB5/SGDH subunit |
| NDUF_B6 | 1 | NADH:ubiquinone oxidoreductase, NDUFB6/B17 subunit |
| NDUF_B7 | 1 | NADH-ubiquinone oxidoreductase B18 subunit (NDUFB7) |
| NDUF_B8 | 1 | NADH-ubiquinone oxidoreductase ASHI subunit (CI-ASHI or NDUFB8) |
| NDUS4 | 1 | NADH dehydrogenase ubiquinone Fe-S protein 4 |
| NEPRO_N | 1 | Nucleolus and neural progenitor protein, N-terminal |
| NESP55 | 1 | Neuroendocrine-specific golgi protein P55 (NESP55) |
| NFACT-C | 1 | NFACT protein C-terminal domain |
| NFACT_N | 1 | NFACT N-terminal and middle domains |
| NFRKB_winged | 1 | NFRKB Winged Helix-like |
| NGP1NT | 1 | NGP1NT (NUC091) domain |
| NIBRIN_BRCT_II | 1 | Second BRCT domain on Nijmegen syndrome breakage protein |
| NICE-1 | 1 | Cysteine-rich C-terminal 1 family |
| NICE-3 | 1 | NICE-3 protein |
| NLRX1_C | 1 | NLR family member X1, C-terminal domain |
| NMD3 | 1 | NMD3 family |
| NMD3_OB | 1 | NMD3 OB-fold domain |
| NMD_SH3 | 1 | NMD3 SH3 domain |
| NMU | 1 | Neuromedin U |
| NOA36 | 1 | NOA36 protein |
| NOB1_Zn_bind | 1 | Nin one binding (NOB1) Zn-ribbon like |
| NOC3p | 1 | Nucleolar complex-associated protein |
| NOG1 | 1 | Nucleolar GTP-binding protein 1 (NOG1) |
| NOG1_N | 1 | NOG1 N-terminal helical domain |
| NOGCT | 1 | NOGCT (NUC087) domain |
| NOL11_N | 1 | Nucleolar protein 11 N-terminal domain |
| NOPCHAP1 | 1 | NOP protein chaperone 1 |
| NPAT_C | 1 | NPAT C terminus |
| NPDC1 | 1 | Neural proliferation differentiation control-1 protein (NPDC1) |
| NPF | 1 | Rabosyn-5 repeating NPF sequence-motif |
| NPFF | 1 | Neuropeptide FF |
| NPL4 | 1 | NPL4 family |
| NPM1-C | 1 | Nucleophosmin C-terminal domain |
| NPP | 1 | Pro-opiomelanocortin, N-terminal region |
| NPR2 | 1 | Nitrogen permease regulator 2 |
| NPR3 | 1 | Nitrogen Permease regulator of amino acid transport activity 3 |
| NRBF2 | 1 | Nuclear receptor-binding factor 2, autophagy regulator |
| NRBF2_MIT | 1 | MIT domain of nuclear receptor-binding factor 2 |
| NRDE-2 | 1 | NRDE-2, necessary for RNA interference |
| NRIP1_repr_1 | 1 | Nuclear receptor-interacting protein 1 repression 1 |
| NRIP1_repr_2 | 1 | Nuclear receptor-interacting protein 1 repression 2 |
| NRIP1_repr_3 | 1 | Nuclear receptor-interacting protein 1 repression 3 |
| NRIP1_repr_4 | 1 | Nuclear receptor-interacting protein 1 repression 4 |
| NRP1_C | 1 | Nuclear speckle splicing regulatory protein 1, RS-like domain |
| NR_Repeat | 1 | Nuclear receptor repeat |
| NSRP1_N | 1 | Nuclear speckle splicing regulatory protein 1, N-terminal |
| NSUN5_N | 1 | NOL1/NOP2/Sun domain family member 5, N-terminal domain |
| NTF3_N | 1 | Neutrophin-3 N-terminus |
| NTPase_1 | 1 | NTPase |
| NTPase_I-T | 1 | Protein of unknown function DUF84 |
| NUC129 | 1 | NUC129 domain |
| NUDIX_2 | 1 | Nucleotide hydrolase |
| NUDIX_4 | 1 | NUDIX domain |
| NUDIX_5 | 1 | NUDIX, or N-terminal NPxY motif-rich, region of KRIT |
| NUFIP1 | 1 | FMR1-interacting protein 1 (NUFIP1) |
| NUFIP2 | 1 | FMR1-interacting protein 2 |
| NUP50 | 1 | NUP50 (Nucleoporin 50 kDa) |
| NUSAP | 1 | Nucleolar and spindle-associated protein |
| NYD-SP12_N | 1 | Spermatogenesis-associated, N-terminal |
| NYD-SP28_assoc | 1 | Sperm tail C-terminal domain |
| NYN | 1 | NYN domain |
| N_Asn_amidohyd | 1 | Protein N-terminal asparagine amidohydrolase |
| Nab1 | 1 | Conserved region in Nab1 |
| Nas2_N | 1 | Nas2 N_terminal domain |
| NatB_MDM20 | 1 | N-acetyltransferase B complex (NatB) non catalytic subunit |
| Nbas_N | 1 | Neuroblastoma-amplified sequence, N terminal |
| Nbl1_Borealin_N | 1 | Nbl1 / Borealin N terminal |
| Nbs1_C | 1 | DNA damage repair protein Nbs1 |
| Ncstrn_small | 1 | Nicastrin small lobe |
| Ndc1_Nup | 1 | Nucleoporin protein Ndc1-Nup |
| Ndc80_HEC | 1 | HEC/Ndc80p family |
| Ndufs5 | 1 | NADH:ubiquinone oxidoreductase, NDUFS5-15kDa |
| Neil1-DNA_bind | 1 | Endonuclease VIII-like 1, DNA bind |
| NeuB | 1 | NeuB family |
| Neugrin | 1 | Neugrin |
| Neural_ProG_Cyt | 1 | Neural chondroitin sulphate proteoglycan cytoplasmic domain |
| Neurochondrin | 1 | Neurochondrin |
| Neurokinin_B | 1 | Neurokinin B |
| Neuromodulin | 1 | Neuromodulin |
| Neuromodulin_N | 1 | Gap junction protein N-terminal region |
| Neuropep_like | 1 | Neuropeptide-like |
| Neuropeptide_S | 1 | Neuropeptide S precursor protein |
| Nfu_N | 1 | Scaffold protein Nfu/NifU N terminal |
| Nic96 | 1 | Nup93/Nic96 |
| Nicastrin | 1 | Nicastrin |
| NifU | 1 | NifU-like domain |
| NifU_N | 1 | NifU-like N terminal domain |
| Nipped-B_C | 1 | Sister chromatid cohesion C-terminus |
| Nitroreductase | 1 | Nitroreductase family |
| Njmu-R1 | 1 | Njmu-R1-like protein family |
| NmU-R2_C_term | 1 | Neuromedin-U receptor 2, C-terminal |
| NmrA | 1 | NmrA-like family |
| Noc2 | 1 | Noc2p family |
| Noggin | 1 | Noggin |
| Nol11_C | 1 | Nucleolar protein 11 C-terminal domain |
| Nop10p | 1 | Nucleolar RNA-binding protein, Nop10p family |
| Nop14 | 1 | Nop14-like family |
| Nop16 | 1 | Ribosome biogenesis protein Nop16 |
| Nop25 | 1 | Nucleolar protein 12 (25kDa) |
| Nop53 | 1 | Nop53 (60S ribosomal biogenesis) |
| NopRA1 | 1 | Nucleolar pre-ribosomal-associated protein 1 |
| Not1 | 1 | CCR4-Not complex component, Not1 |
| Not3 | 1 | Not1 N-terminal domain, CCR4-Not complex component |
| Npa1 | 1 | Ribosome 60S biogenesis N-terminal |
| Nrap | 1 | Nrap protein domain 1 |
| Nrap_D2 | 1 | Nrap protein PAP/OAS-like domain |
| Nrap_D3 | 1 | Nrap protein domain 3 |
| Nrap_D4 | 1 | Nrap protein nucleotidyltransferase domain 4 |
| Nrap_D5 | 1 | Nrap protein PAP/OAS1-like domain 5 |
| Nrap_D6 | 1 | Nrap protein domain 6 |
| Nrf1_DNA-bind | 1 | NLS-binding and DNA-binding and dimerisation domains of Nrf1 |
| NtA | 1 | Agrin NtA domain |
| Nt_Gln_amidase | 1 | N-terminal glutamine amidase |
| NuA4 | 1 | Histone acetyltransferase subunit NuA4 |
| NuDC | 1 | Nuclear distribution C domain |
| Nuc_deoxyrib_tr | 1 | Nucleoside 2-deoxyribosyltransferase |
| Nucleolin_bd | 1 | Nucleolin binding domain |
| Nucleopor_Nup85 | 1 | Nup85 Nucleoporin |
| Nucleoporin2 | 1 | Nucleoporin autopeptidase |
| Nucleoporin_C | 1 | Non-repetitive/WGA-negative nucleoporin C-terminal |
| Nucleoporin_FG | 1 | Nucleoporin FG repeat region |
| Nucleoporin_FG2 | 1 | Nucleoporin FG repeated region |
| Nucleoporin_N | 1 | Nup133 N terminal like |
| Nudix_hydro | 1 | Nudix hydrolase domain |
| Nuf2 | 1 | Nuf2 family |
| Nup153 | 1 | Nucleoporin Nup153-like |
| Nup160 | 1 | Nucleoporin Nup120/160 |
| Nup188_N | 1 | Nucleoporin Nup188, N-terminal |
| Nup188_N-subdom_III | 1 | Nucleoporin Nup188, N-terminal subdomain III |
| Nup188_SH3-like | 1 | Nup188 SH3-like domain |
| Nup192 | 1 | Nuclear pore complex scaffold, nucleoporins 186/192/205 |
| Nup214_FG | 1 | Nucleoporin Nup214 phenylalanine-glycine (FG) domain |
| Nup35_RRM | 1 | Nup53/35/40-type RNA recognition motif |
| Nup54 | 1 | Nucleoporin complex subunit 54 |
| Nup54_C | 1 | Nup54 C-terminal interacting domain |
| Nup84_Nup100 | 1 | Nuclear pore protein 84 / 107 |
| Nup88 | 1 | Nuclear pore component |
| Nup96 | 1 | Nuclear protein 96 |
| Nup98_GLEBS | 1 | Nup98, Gle2-binding sequence |
| NupH_GANP | 1 | Nucleoporin homology of Germinal-centre associated nuclear protein |
| Nup_retrotrp_bd | 1 | Retro-transposon transporting motif |
| OAF_N | 1 | Out at first |
| OCC1 | 1 | OCC1 family |
| OCD_Mu_crystall | 1 | Ornithine cyclodeaminase/mu-crystallin family |
| OCRL_clath_bd | 1 | Inositol polyphosphate 5-phosphatase clathrin binding domain |
| ODAM | 1 | Odontogenic ameloblast-associated family |
| ODAPH | 1 | Odontogenesis associated phosphoprotein |
| ODR4-like | 1 | Odorant response abnormal 4-like |
| OGFr_III | 1 | Opioid growth factor receptor repeat |
| OGG_N | 1 | 8-oxoguanine DNA glycosylase, N-terminal domain |
| OHCU_decarbox | 1 | OHCU decarboxylase |
| OMPdecase | 1 | Orotidine 5'-phosphate decarboxylase / HUMPS family |
| OPA1_C | 1 | Dynamin-like GTPase OPA1 C-terminal |
| OPA3 | 1 | Optic atrophy 3 protein (OPA3) |
| ORC2 | 1 | Origin recognition complex subunit 2 |
| ORC3_N | 1 | Origin recognition complex (ORC) subunit 3 N-terminus |
| ORC3_ins | 1 | Origin recognition complex subunit 3 insertion domain |
| ORC4_C | 1 | Origin recognition complex (ORC) subunit 4 C-terminus |
| ORC5_C | 1 | Origin recognition complex (ORC) subunit 5 C-terminus |
| ORC5_lid | 1 | ORC5, lid domain |
| ORC6 | 1 | Origin recognition complex subunit 6 (ORC6) |
| ORC_WH_C | 1 | Origin recognition complex winged helix C-terminal |
| OSCP | 1 | ATP synthase delta (OSCP) subunit |
| OSTMP1 | 1 | Osteopetrosis-associated transmembrane protein 1 precursor |
| OSTbeta | 1 | Organic solute transporter subunit beta protein |
| OTOS | 1 | Otospiralin |
| ObR_Ig | 1 | Obesity receptor immunoglobulin like domain |
| Ocnus | 1 | Janus/Ocnus family (Ocnus) |
| Ocular_alb | 1 | Ocular albinism type 1 protein |
| Oest_recep | 1 | Oestrogen receptor |
| Ofd1_CTDD | 1 | Oxoglutarate and iron-dependent oxygenase degradation C-term |
| Olfactory_mark | 1 | Olfactory marker protein |
| Omp85 | 1 | Omp85 superfamily domain |
| Op_neuropeptide | 1 | Opioids neuropeptide |
| Orexin | 1 | Prepro-orexin |
| Orexin_rec2 | 1 | Orexin receptor type 2 |
| Oscp1 | 1 | Organic solute transport protein 1 |
| Ost4 | 1 | Oligosaccaryltransferase |
| Ost5 | 1 | Oligosaccharyltransferase subunit 5 |
| Osteopontin | 1 | Osteopontin |
| Osteoregulin | 1 | Osteoregulin |
| Oxidored_molyb | 1 | Oxidoreductase molybdopterin binding domain |
| Oxidored_q2 | 1 | NADH-ubiquinone/plastoquinone oxidoreductase chain 4L |
| Oxidored_q3 | 1 | NADH-ubiquinone/plastoquinone oxidoreductase chain 6 |
| Oxidored_q4 | 1 | NADH-ubiquinone/plastoquinone oxidoreductase, chain 3 |
| Oxidored_q5_N | 1 | NADH-ubiquinone oxidoreductase chain 4, amino terminus |
| Oxidored_q6 | 1 | NADH ubiquinone oxidoreductase, 20 Kd subunit |
| P-mevalo_kinase | 1 | Phosphomevalonate kinase |
| P120R | 1 | P120R (NUC006) repeat |
| P19Arf_N | 1 | Cyclin-dependent kinase inhibitor 2a p19Arf N-terminus |
| P21-Arc | 1 | ARP2/3 complex ARPC3 (21 kDa) subunit |
| P2RX7_C | 1 | P2X purinoreceptor 7 intracellular domain |
| P33MONOX | 1 | P33 mono-oxygenase |
| P34-Arc | 1 | Arp2/3 complex, 34 kD subunit p34-Arc |
| P53_TAD | 1 | P53 transactivation motif |
| P68HR | 1 | P68HR (NUC004) repeat |
| PAAT-like | 1 | ATPase PAAT-like |
| PAC1 | 1 | Proteasome assembly chaperone 4 |
| PAC2 | 1 | PAC2 family |
| PAC3 | 1 | Proteasome assembly chaperone 3 |
| PAC4 | 1 | Proteasome assembly chaperone 4 |
| PADR1_N | 1 | PADR1, N-terminal helical domain |
| PADR1_Zn_ribbon | 1 | PADR1 domain, zinc ribbon fold |
| PAE | 1 | Pectinacetylesterase |
| PAF | 1 | PCNA-associated factor histone like domain |
| PAG | 1 | Phosphoprotein associated with glycosphingolipid-enriched |
| PALB2_WD40 | 1 | Partner and localizer of BRCA2 WD40 domain |
| PAN2_N | 1 | PAN2 N-terminal |
| PANTS-like | 1 | Synaptic plasticity regulator PANTS-like |
| PAPA-1 | 1 | PAPA-1-like conserved region |
| PAPS_reduct | 1 | Phosphoadenosine phosphosulfate reductase family |
| PARG_cat_C | 1 | Poly (ADP-ribose) glycohydrolase (PARG), Macro domain fold |
| PARG_cat_N | 1 | Poly (ADP-ribose) glycohydrolase (PARG), helical domain |
| PARM | 1 | PARM |
| PAW | 1 | PNGase C-terminal domain, mannose-binding module PAW |
| PAXIP1_C | 1 | PAXIP1-associated-protein-1 C term PTIP binding protein |
| PAXNEB | 1 | PAXNEB protein |
| PAXX | 1 | PAXX, PAralog of XRCC4 and XLF, also called C9orf142 |
| PBDC1 | 1 | PBDC1 protein |
| PBP_sp32 | 1 | Proacrosin binding protein sp32 |
| PC4 | 1 | Transcriptional Coactivator p15 (PC4) |
| PCC_BT | 1 | Propionyl-coenzyme A carboxylase BT domain |
| PCF11_RFEG_rpt | 1 | Pre-mRNA cleavage complex 2 protein Pcf11, RFEGP repeat |
| PCF11_charged | 1 | PCF11, charged region |
| PCIF1_WW | 1 | Phosphorylated CTD interacting factor 1 WW domain |
| PCM1_C | 1 | Pericentriolar material 1 C terminus |
| PCNA_C | 1 | Proliferating cell nuclear antigen, C-terminal domain |
| PCNA_N | 1 | Proliferating cell nuclear antigen, N-terminal domain |
| PCNP | 1 | PEST, proteolytic signal-containing nuclear protein family |
| PCO_ADO | 1 | PCO_ADO |
| PCSK9_C1 | 1 | Proprotein convertase subtilisin-like/kexin type 9 C-terminal domain |
| PCSK9_C2 | 1 | Proprotein convertase subtilisin-like/kexin type 9 C-terminal domain |
| PCSK9_C3 | 1 | Proprotein convertase subtilisin-like/kexin type 9 C-terminal domain |
| PD-C2-AF1 | 1 | POU domain, class 2, associating factor 1 |
| PD40 | 1 | WD40-like Beta Propeller Repeat |
| PDC10_C | 1 | Programmed cell death protein 10, C-terminal |
| PDCD7 | 1 | Programmed cell death protein 7 |
| PDDEXK_1 | 1 | PD-(D/E)XK nuclease superfamily |
| PDE12-like_N | 1 | 2',5'-phosphodiesterase 12-like, N-terminal domain |
| PDE8 | 1 | PDE8 phosphodiesterase |
| PDZ_2 | 1 | PDZ domain |
| PELP1_HEAT | 1 | PELP1, middle domain |
| PEMT | 1 | Phospholipid methyltransferase |
| PEN-2 | 1 | Presenilin enhancer-2 subunit of gamma secretase |
| PET117 | 1 | PET assembly of cytochrome c oxidase, mitochondrial |
| PEX-1N | 1 | Peroxisome biogenesis factor 1, N-terminal |
| PEX-2N | 1 | Peroxisome biogenesis factor 1, N-terminal |
| PFU | 1 | PFU (PLAA family ubiquitin binding) |
| PGAP1 | 1 | PGAP1-like protein |
| PGDH_inter | 1 | D-3-phosphoglycerate dehydrogenase intervening domain |
| PGI | 1 | Phosphoglucose isomerase |
| PGM_PMM_IV | 1 | Phosphoglucomutase/phosphomannomutase, C-terminal domain |
| PH1_SSRP1-like | 1 | SSRP1 PH domain |
| PHAF1 | 1 | Phagosome assembly factor 1 |
| PHAX_RNA-bd | 1 | Phosphorylated adapter RNA export protein, RNA-binding domain |
| PHD20L1_u1 | 1 | PHD finger protein 20-like protein 1 |
| PHD_ash2p_like | 1 | ASH2, PHD zinc finger |
| PHF12_MRG_bd | 1 | PHD finger protein 12 MRG binding domain |
| PHF20_AT-hook | 1 | PHD finger protein 20, AT-hook |
| PHF5 | 1 | PHF5-like protein |
| PHTB1_C | 1 | PTHB1 C-terminus |
| PHTB1_N | 1 | PTHB1 N-terminus |
| PH_3 | 1 | PH domain |
| PH_TFIIH | 1 | TFIIH p62 subunit, N-terminal domain |
| PI31_Prot_C | 1 | PI31 proteasome regulator |
| PI4KB-PIK1_PIK | 1 | PI4KB/PIK1, accessory (PIK) domain |
| PI4K_N | 1 | PI4-kinase N-terminal region |
| PIF1 | 1 | PIF1-like helicase |
| PIG-F | 1 | GPI biosynthesis protein family Pig-F |
| PIG-H | 1 | GPI-GlcNAc transferase complex, PIG-H component |
| PIG-L | 1 | GlcNAc-PI de-N-acetylase |
| PIG-P | 1 | PIG-P |
| PIG-S | 1 | Phosphatidylinositol-glycan biosynthesis class S protein |
| PIG-U | 1 | GPI transamidase subunit PIG-U |
| PIG-X | 1 | PIG-X / PBN1 |
| PIG-Y | 1 | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Y |
| PIGA | 1 | PIGA (GPI anchor biosynthesis) |
| PIGO_PIGG | 1 | GPI ethanolamine phosphate transferase membrane region |
| PIK3CG_ABD | 1 | PIK3 catalytic subunit gamma adaptor-binding domain |
| PIN_11 | 1 | PIN like domain |
| PIN_6 | 1 | PIN domain of ribonuclease |
| PIRT | 1 | Phosphoinositide-interacting protein family |
| PKS_DH_N | 1 | Polyketide synthase dehydratase domain |
| PKcGMP_CC | 1 | Coiled-coil N-terminus of cGMP-dependent protein kinase |
| PLA2_inh | 1 | Phospholipase A2 inhibitor |
| PLAC9 | 1 | Placenta-specific protein 9 |
| PLL | 1 | PTX/LNS-Like (PLL) domain |
| PMAIP1 | 1 | Phorbol-12-myristate-13-acetate-induced |
| PMC2NT | 1 | PMC2NT (NUC016) domain |
| PMI_typeI_C | 1 | Phosphomannose isomerase type I C-terminal |
| PMI_typeI_cat | 1 | Phosphomannose isomerase type I, catalytic domain |
| PMI_typeI_hel | 1 | Phosphomannose isomerase type I, helical insertion domain |
| PML_CC | 1 | PML-like, coiled-coil |
| PMSI1 | 1 | Protein missing in infertile sperm 1, putative |
| PMSR | 1 | Peptide methionine sulfoxide reductase |
| PND | 1 | FANCM pseudonuclease domain |
| PNISR | 1 | Arginine/serine-rich protein PNISR |
| PNK3P | 1 | Polynucleotide kinase 3 phosphatase |
| PNM8A | 1 | Paraneoplastic antigen 8A protein |
| PNP_phzG_C | 1 | Pyridoxine 5'-phosphate oxidase C-terminal dimerisation region |
| PNPase | 1 | Polyribonucleotide nucleotidyltransferase, RNA binding domain |
| PNTB | 1 | NAD(P) transhydrogenase beta subunit |
| PNTB_4TM | 1 | 4TM region of pyridine nucleotide transhydrogenase, mitoch |
| POB3_N | 1 | POB3-like N-terminal PH domain |
| POP1_N | 1 | Ribonucleases P/MRP protein subunit POP1, N-terminal |
| POPLD | 1 | POPLD (NUC188) domain |
| POT1 | 1 | Telomeric single stranded DNA binding POT1/CDC13 |
| POT1PC | 1 | ssDNA-binding domain of telomere protection protein |
| POT1_C_insert | 1 | Protection of telomeres protein 1 (POT1), C-terminal insertion domain |
| POU2AF2 | 1 | POU domain class 2-associating factor 2 |
| PP28 | 1 | Casein kinase substrate phosphoprotein PP28 |
| PPAK | 1 | PPAK motif |
| PPARgamma_N | 1 | PPAR gamma N-terminal region |
| PPI_Ypi1 | 1 | Protein phosphatase inhibitor |
| PPP1R21_C | 1 | Protein phosphatase 1 regulatory subunit 21, C-terminal |
| PPP1R21_helical | 1 | Protein phosphatase 1 regulatory subunit 21, six-helix bundle |
| PPP1R26_N | 1 | Protein phosphatase 1 regulatory subunit 26 N-terminus |
| PPP1R32 | 1 | Protein phosphatase 1 regulatory subunit 32 |
| PPP1R35_C | 1 | Protein phosphatase 1 regulatory subunit 35 C-terminus |
| PPP4R2 | 1 | PPP4R2 |
| PPPI_inhib | 1 | Protein phosphatase 1 inhibitor |
| PPR | 1 | PPR repeat |
| PPR_2 | 1 | PPR repeat family |
| PQQ | 1 | PQQ enzyme repeat |
| PQQ_3 | 1 | PQQ-like domain |
| PRAS | 1 | Proline-rich AKT1 substrate 1 |
| PRCC | 1 | Mitotic checkpoint regulator, MAD2B-interacting |
| PRF | 1 | Plethodontid receptivity factor PRF |
| PRIMA1 | 1 | Proline-rich membrane anchor 1 |
| PRKCSH-like | 1 | Glucosidase II beta subunit-like |
| PRKCSH_1 | 1 | Glucosidase II beta subunit-like protein |
| PRMT5 | 1 | PRMT5 arginine-N-methyltransferase |
| PRMT5_C | 1 | PRMT5 oligomerisation domain |
| PRMT5_TIM | 1 | PRMT5 TIM barrel domain |
| PRO8NT | 1 | PRO8NT (NUC069), PrP8 N-terminal domain |
| PROCN | 1 | PROCN (NUC071) domain |
| PROCT | 1 | PROCT (NUC072) domain |
| PRORP | 1 | Protein-only RNase P |
| PRP1_N | 1 | PRP1 splicing factor, N-terminal |
| PRP21_like_P | 1 | Pre-mRNA splicing factor PRP21 like protein |
| PRP3 | 1 | pre-mRNA processing factor 3 domain |
| PRP38_assoc | 1 | Pre-mRNA-splicing factor 38-associated hydrophilic C-term |
| PRP8_domainIV | 1 | PRP8 domain IV core |
| PRR18 | 1 | Proline-rich protein family 18 |
| PRR22 | 1 | Proline-rich protein family 22 |
| PRS7_OB | 1 | 26S proteasome regulatory subunit 7, OB domain |
| PS_Dcarbxylase | 1 | Phosphatidylserine decarboxylase |
| PTCRA | 1 | Pre-T-cell antigen receptor |
| PTE | 1 | Phosphotriesterase family |
| PTN13_u3 | 1 | Unstructured linker region on PTN13 protein between PDZ |
| PTPA | 1 | Phosphotyrosyl phosphate activator (PTPA) protein |
| PTPRCAP | 1 | Protein tyrosine phosphatase receptor type C-associated |
| PTPS | 1 | 6-pyruvoyl tetrahydropterin synthase |
| PTP_N | 1 | Protein tyrosine phosphatase N terminal |
| PTPlike_phytase | 1 | Inositol hexakisphosphate |
| PTS_2-RNA | 1 | RNA 2'-phosphotransferase, Tpt1 / KptA family |
| PUB_1 | 1 | PNGase/UBA- or UBX-containing domain |
| PUL | 1 | PUL domain |
| PUMA | 1 | Bcl-2-binding component 3, p53 upregulated modulator of apoptosis |
| PV-1 | 1 | PV-1 protein (PLVAP) |
| PXT1 | 1 | Peroxisomal testis-specific protein 1 |
| PYC_OADA | 1 | Conserved carboxylase domain |
| PYNP_C | 1 | Pyrimidine nucleoside phosphorylase C-terminal domain |
| P_C10 | 1 | Protein C10 |
| Paf1 | 1 | Paf1 |
| Paf67 | 1 | RNA polymerase I-associated factor PAF67 |
| Pan3_PK | 1 | Pan3 Pseudokinase domain |
| Papilin_u7 | 1 | Linking region between Kunitz_BPTI and I-set on papilin |
| Pcf11_helical | 1 | Pre-mRNA cleavage complex 2 protein Pcf11, alpha-helical domain |
| Pdase_M17_N2 | 1 | M17 aminopeptidase N-terminal domain 2 |
| Pentapeptide | 1 | Pentapeptide repeats (8 copies) |
| Pep3_Vps18 | 1 | Pep3/Vps18/deep orange beta-propeller domain |
| Pep_deformylase | 1 | Polypeptide deformylase |
| Pept_tRNA_hydro | 1 | Peptidyl-tRNA hydrolase |
| Peptidase_C1_2 | 1 | Peptidase C1-like family |
| Peptidase_C26 | 1 | Peptidase C26 |
| Peptidase_C98 | 1 | Ubiquitin-specific peptidase-like, SUMO isopeptidase |
| Peptidase_M17_N | 1 | Cytosol aminopeptidase family, N-terminal domain |
| Peptidase_M18 | 1 | Aminopeptidase I zinc metalloprotease (M18) |
| Peptidase_M23 | 1 | Peptidase family M23 |
| Peptidase_M48_N | 1 | CAAX prenyl protease N-terminal, five membrane helices |
| Peptidase_M49 | 1 | Peptidase family M49 |
| Peptidase_M50 | 1 | Peptidase family M50 |
| Peptidase_M54 | 1 | Peptidase family M54 |
| Peptidase_M76 | 1 | Peptidase M76 family |
| Peptidase_S41 | 1 | Peptidase family S41 |
| Peptidase_S41_N | 1 | N-terminal domain of Peptidase_S41 in eukaryotic IRBP |
| Peptidase_S68 | 1 | Peptidase S68 |
| Per1 | 1 | Per1-like family |
| Peroxin-13_N | 1 | Peroxin 13, N-terminal region |
| Peroxin-3 | 1 | Peroxin-3 |
| Pescadillo_N | 1 | Pescadillo N-terminus |
| Pet100 | 1 | Pet100 |
| Pet191_N | 1 | Cytochrome c oxidase assembly protein PET191 |
| Pex14_N | 1 | Pex14 N-terminal domain |
| Pex16 | 1 | Peroxisomal membrane protein (Pex16) |
| Pex19 | 1 | Pex19 protein family |
| Pex24p | 1 | Integral peroxisomal membrane peroxin |
| Pex26 | 1 | Pex26 protein |
| Pfc11_Clp1_ID | 1 | Pcf11, Clp1-interaction domain |
| PheRS_DBD1 | 1 | PheRS DNA binding domain 1 |
| PheRS_DBD2 | 1 | PheRS DNA binding domain 2 |
| PheRS_DBD3 | 1 | PheRS DNA binding domain 3 |
| PhetRS_B1 | 1 | Phe-tRNA synthetase beta subunit B1 domain |
| Phos_pyr_kin | 1 | Phosphomethylpyrimidine kinase |
| Phospholamban | 1 | Phospholamban |
| PhzC-PhzF | 1 | Phenazine biosynthesis-like protein |
| Pif1_2B_dom | 1 | DNA helicase Pif1, 2B domain |
| PigN | 1 | Phosphatidylinositolglycan class N (PIG-N) |
| Pilt | 1 | Protein incorporated later into Tight Junctions |
| Pinin_SDK_N | 1 | pinin/SDK conserved region |
| Pinin_SDK_memA | 1 | pinin/SDK/memA/ protein conserved region |
| Pirin | 1 | Pirin |
| Pirin_C | 1 | Pirin C-terminal cupin domain |
| Plk4_PB1 | 1 | Polo-like Kinase 4 Polo Box 1 |
| Plk4_PB2 | 1 | Polo-like Kinase 4 Polo Box 2 |
| Plus-3 | 1 | Plus-3 domain |
| Podoplanin | 1 | Podoplanin |
| Pol_alpha_B_N | 1 | DNA polymerase alpha subunit B N-terminal |
| PolyA_pol | 1 | Poly A polymerase head domain |
| PolyA_pol_RNAbd | 1 | Probable RNA and SrmB- binding site of polymerase A |
| Porphobil_deam | 1 | Porphobilinogen deaminase, dipyromethane cofactor binding domain |
| Porphobil_deamC | 1 | Porphobilinogen deaminase, C-terminal domain |
| Prion_bPrPp | 1 | Major prion protein bPrPp - N terminal |
| PrmC_N | 1 | PrmC N-terminal domain |
| Pro-MCH | 1 | Pro-melanin-concentrating hormone (Pro-MCH) |
| Pro-NT_NN | 1 | Neurotensin/neuromedin N precursor |
| Pro-kuma_activ | 1 | Pro-kumamolisin, activation domain |
| Pro-rich_19 | 1 | Proline-rich 19 |
| ProRS-C_1 | 1 | Prolyl-tRNA synthetase, C-terminal |
| ProSAAS | 1 | ProSAAS precursor |
| Pro_racemase | 1 | Proline racemase |
| Prog_receptor | 1 | Progesterone receptor |
| Proho_convert | 1 | Prohormone convertase enzyme |
| Prok-JAB | 1 | Prokaryotic homologs of the JAB domain |
| Prolactin_RP | 1 | Prolactin-releasing peptide |
| Promethin | 1 | Promethin |
| Propeptide_C1 | 1 | Peptidase family C1 propeptide |
| Protamine_P1 | 1 | Protamine P1 |
| Protamine_P2 | 1 | Sperm histone P2 |
| Proteasom_PSMB | 1 | Proteasome non-ATPase 26S subunit |
| Proton_antipo_N | 1 | NADH-Ubiquinone oxidoreductase (complex I), chain 5 N-terminus |
| Prp18 | 1 | Prp18 domain |
| Prp19 | 1 | Prp19/Pso4-like |
| Prp31_C | 1 | Prp31 C terminal domain |
| Pterin_bind | 1 | Pterin binding enzyme |
| Pur_DNA_glyco | 1 | Methylpurine-DNA glycosylase (MPG) |
| Pur_ac_phosph_N | 1 | Purple acid Phosphatase, N-terminal domain |
| Pus10_C | 1 | Pus10, C-terminal |
| Pus10_N_euk | 1 | Eukaryotic Pus10, N-terminal |
| Putative_PNPOx | 1 | Pyridoxamine 5'-phosphate oxidase |
| Pyr_redox_3 | 1 | Pyridine nucleotide-disulphide oxidoreductase |
| QIL1 | 1 | MICOS complex subunit MIC13, QIL1 |
| QRPTase_C | 1 | Quinolinate phosphoribosyl transferase, C-terminal domain |
| QRPTase_N | 1 | Quinolinate phosphoribosyl transferase, N-terminal domain |
| Q_salvage | 1 | Queuosine salvage protein |
| Quaking_NLS | 1 | Putative nuclear localisation signal of quaking |
| R3H-assoc | 1 | R3H-associated N-terminal domain |
| RAB3GAP2_C | 1 | Rab3 GTPase-activating protein regulatory subunit C-terminus |
| RAB3GAP2_N | 1 | Rab3 GTPase-activating protein regulatory subunit N-terminus |
| RAC_head | 1 | Ribosome-associated complex head domain |
| RAD51D_N | 1 | RAD51D, N-terminal domain |
| RAG1 | 1 | Recombination-activation protein 1 (RAG1), recombinase |
| RAG1_imp_bd | 1 | RAG1 importin binding |
| RAG2 | 1 | Recombination activating protein 2 |
| RAG2_PHD | 1 | RAG2 PHD domain |
| RAI1 | 1 | RAI1 like PD-(D/E)XK nuclease |
| RANK_CRD_2 | 1 | Receptor activator of the NF-KB cysteine-rich repeat domain 2 |
| RAP80_UIM | 1 | RAP80 N-terminal ubiquitin interaction motif |
| RBFA | 1 | Ribosome-binding factor A |
| RBP_receptor | 1 | Retinol binding protein receptor |
| RCDG1 | 1 | Renal cancer differentiation gene 1 protein |
| RCS1 | 1 | Regulator of chromosome segregation 1 |
| RCSD | 1 | RCSD region |
| RDD | 1 | RDD family |
| RDD1 | 1 | Required for drug-induced death protein 1 |
| REC114-like | 1 | Meiotic recombination protein REC114-like |
| RED_C | 1 | RED-like protein C-terminal region |
| RED_N | 1 | RED-like protein N-terminal region |
| REPA_OB_2 | 1 | Replication protein A OB domain |
| RET_CLD1 | 1 | RET Cadherin like domain 1 |
| RET_CLD3 | 1 | RET Cadherin like domain 3 |
| RET_CLD4 | 1 | RET Cadherin like domain 4 |
| REV1_C | 1 | DNA repair protein REV1 C-terminal domain |
| RFC1 | 1 | Replication factor RFC1 C terminal domain |
| RFX5_DNA_bdg | 1 | RFX5 DNA-binding domain |
| RFXA_RFXANK_bdg | 1 | Regulatory factor X-associated C-terminal binding domain |
| RFamide_26RFa | 1 | Orexigenic neuropeptide Qrfp/P518 |
| RGCC | 1 | Response gene to complement 32 protein family |
| RGS12_us1 | 1 | Unstructured region of RGS12 |
| RGS12_us2 | 1 | Unstructured region between RBD and GoLoco |
| RGS12_usC | 1 | C-terminal unstructured region of RGS12 |
| RHINO | 1 | RAD9, RAD1, HUS1-interacting nuclear orphan protein |
| RIC1 | 1 | RIC1 |
| RIC3 | 1 | Resistance to inhibitors of cholinesterase homologue 3 |
| RICTOR_M | 1 | Rapamycin-insensitive companion of mTOR, middle domain |
| RICTOR_N | 1 | Rapamycin-insensitive companion of mTOR, N-term |
| RICTOR_V | 1 | Rapamycin-insensitive companion of mTOR, domain 5 |
| RICTOR_phospho | 1 | Rapamycin-insensitive companion of mTOR, phosphorylation-site |
| RIMC1 | 1 | RAB7A-interacting MON1-CCZ1 complex subunit 1 |
| RINT1_TIP1 | 1 | RINT-1/TIP-1 family |
| RIOX1_C_WH | 1 | Ribosomal oxygenase 1, C-terminal winged helix domain |
| RITA | 1 | RBPJ-interacting and tubulin associated protein |
| RIX1 | 1 | rRNA processing/ribosome biogenesis |
| RL | 1 | RL domain |
| RLIP76_Ral-bd | 1 | RLIP76, Ral binding domain |
| RLL | 1 | RNA transcription, translation and transport factor protein |
| RMC1_C | 1 | Regulator of MON1-CCZ1 complex, C-terminal |
| RMC1_N | 1 | Regulator of MON1-CCZ1 complex, N-terminal |
| RME-8_N | 1 | DNAJ protein RME-8 N-terminal |
| RMI1_C | 1 | Recq-mediated genome instability protein 1, C-terminal OB-fold |
| RMI1_N_N | 1 | RMI1, N-terminal helical domain |
| RMI2 | 1 | RecQ-mediated genome instability protein 2 |
| RMP | 1 | Retinal Maintenance |
| RNA_bind | 1 | RNA binding domain |
| RNA_pol | 1 | DNA-dependent RNA polymerase |
| RNA_polI_A34 | 1 | DNA-directed RNA polymerase I subunit RPA34.5 |
| RNA_pol_I_A49 | 1 | A49-like RNA polymerase I associated factor |
| RNA_pol_N | 1 | RNA polymerases N / 8 kDa subunit |
| RNA_pol_Rbc25 | 1 | RNA polymerase III subunit Rpc25 |
| RNA_pol_Rpa2_4 | 1 | RNA polymerase I, Rpa2 specific domain |
| RNA_pol_Rpb1_6 | 1 | RNA polymerase Rpb1, domain 6 |
| RNA_pol_Rpb1_7 | 1 | RNA polymerase Rpb1, domain 7 |
| RNA_pol_Rpb5_C | 1 | RNA polymerase Rpb5, C-terminal domain |
| RNA_pol_Rpb5_N | 1 | RNA polymerase Rpb5, N-terminal domain |
| RNA_pol_Rpb6 | 1 | RNA polymerase Rpb6 |
| RNA_pol_Rpb8 | 1 | RNA polymerase Rpb8 |
| RNA_pol_Rpc34 | 1 | RNA polymerase Rpc34 subunit |
| RNA_pol_Rpc4 | 1 | RNA polymerase III RPC4 |
| RNA_pol_Rpc82 | 1 | RNA polymerase III subunit RPC82 |
| RNF152_C | 1 | E3 ubiquitin-protein ligase RNF152 C-terminus |
| RNF180_C | 1 | E3 ubiquitin-protein ligase RNF180 C-terminus |
| RNF220 | 1 | E3 ubiquitin-protein ligase RNF220 |
| RNase_H2-Ydr279 | 1 | Ydr279p protein family (RNase H2 complex component) wHTH domain |
| RNase_H2_suC | 1 | Ribonuclease H2 non-catalytic subunit (Ylr154p-like) |
| RNase_HII | 1 | Ribonuclease HII |
| RNase_P-MRP_p29 | 1 | Ribonuclease P/MRP, subunit p29 |
| RNase_P_p30 | 1 | RNase P subunit p30 |
| ROK | 1 | ROK family |
| ROKNT | 1 | ROKNT (NUC014) domain |
| ROXA-like_wH | 1 | ROXA-like winged helix |
| RPA43_OB | 1 | RPA43 OB domain in RNA Pol I |
| RPAP1_C | 1 | RPAP1-like, C-terminal |
| RPAP1_N | 1 | RPAP1-like, N-terminal |
| RPAP2_Rtr1 | 1 | Rtr1/RPAP2 family |
| RPA_interact_C | 1 | Replication protein A interacting C-terminal |
| RPA_interact_M | 1 | Replication protein A interacting middle |
| RPA_interact_N | 1 | Replication protein A interacting N-terminal |
| RPC3_helical | 1 | DNA-directed RNA polymerase III subunit RPC3, helical hairpin domain |
| RPC5 | 1 | RPC5 protein |
| RPC5_C | 1 | DNA-directed RNA polymerase III subunit RPC5 C-terminal |
| RPN13_C | 1 | UCH-binding domain |
| RPN1_C | 1 | 26S proteasome non-ATPase regulatory subunit RPN1 C-terminal |
| RPN1_RPN2_N | 1 | RPN1 N-terminal domain |
| RPN2_C | 1 | 26S proteasome regulatory subunit RPN2 C-terminal domain |
| RPN2_N | 1 | 26S proteasome subunit RPN2, N-terminal domain |
| RPN5_C | 1 | 26S proteasome regulatory subunit RPN5 C-terminal domain |
| RPN6_C_helix | 1 | 26S proteasome subunit RPN6 C-terminal helix domain |
| RPN6_N | 1 | 26S proteasome regulatory subunit RPN6 N-terminal domain |
| RPN7_PSMD6_C | 1 | 26S proteasome regulatory subunit RPN7/PSMD6 C-terminal helix |
| RPOL_N | 1 | DNA-directed RNA polymerase N-terminal |
| RRF | 1 | Ribosome recycling factor |
| RRM_4 | 1 | RNA recognition motif of the spliceosomal PrP8 |
| RRM_Rrp7 | 1 | Rrp7 RRM-like N-terminal domain |
| RRN3 | 1 | RNA polymerase I specific transcription initiation factor RRN3 |
| RRP12_HEAT | 1 | Ribosomal RNA-processing protein 12, HEAT repeats |
| RRP36 | 1 | rRNA biogenesis protein RRP36 |
| RRP40_N_mamm | 1 | Mammalian exosome complex component RRP40, N-terminal |
| RRP40_S1 | 1 | Exosome complex component RRP40, S1 domain |
| RRP4_S1 | 1 | RRP4, S1 domain |
| RRP7 | 1 | Ribosomal RNA-processing protein 7 (RRP7) C-terminal domain |
| RRS1 | 1 | Ribosome biogenesis regulatory protein (RRS1) |
| RSB_motif | 1 | RNSP1-SAP18 binding (RSB) motif |
| RTP1_C1 | 1 | Required for nuclear transport of RNA pol II C-terminus 1 |
| RTP1_C2 | 1 | Required for nuclear transport of RNA pol II C-terminus 2 |
| RTTN_N | 1 | Rotatin, an armadillo repeat protein, centriole functioning |
| RT_RNaseH | 1 | RNase H-like domain found in reverse transcriptase |
| RVT_1 | 1 | Reverse transcriptase (RNA-dependent DNA polymerase) |
| Rab15_effector | 1 | Rab15 effector |
| Rab3-GAP_cat_C | 1 | Rab3 GTPase-activating protein catalytic subunit C-terminal |
| Rab3-GTPase_cat | 1 | Rab3 GTPase-activating protein catalytic subunit |
| RabGGT_insert | 1 | Rab geranylgeranyl transferase alpha-subunit, insert domain |
| Rad1 | 1 | Repair protein Rad1/Rec1/Rad17 |
| Rad10 | 1 | Binding domain of DNA repair protein Ercc1 (rad10/Swi10) |
| Rad50_zn_hook | 1 | Rad50 zinc hook motif |
| Rad52_Rad22 | 1 | Rad52/22 family double-strand break repair protein |
| Rad60-SLD_2 | 1 | Ubiquitin-2 like Rad60 SUMO-like |
| Radial_spoke_3 | 1 | Radial spoke protein 3 |
| Radical_SAM_C | 1 | Radical_SAM C-terminal domain |
| RanGAP1_C | 1 | RanGAP1 C-terminal domain |
| Rap1_C | 1 | TRF2-interacting telomeric protein/Rap1 - C terminal domain |
| Rapsyn_N | 1 | Rapsyn N-terminal myristoylation and linker region |
| Raptor_N | 1 | Raptor N-terminal CASPase like domain |
| RasGEF_N_2 | 1 | Rapamycin-insensitive companion of mTOR RasGEF_N domain |
| RbsD_FucU | 1 | RbsD / FucU transport protein family |
| Rbsn | 1 | Rabenosyn Rab binding domain |
| Rcd1 | 1 | Cell differentiation family, Rcd1-like |
| Rce1-like | 1 | Type II CAAX prenyl endopeptidase Rce1-like |
| RecQ5 | 1 | RecQ helicase protein-like 5 (RecQ5) |
| Receptor_2B4 | 1 | Natural killer cell receptor 2B4 |
| Redic1-like | 1 | Regulator of DNA class I crossover intermediates 1-like |
| Redoxin | 1 | Redoxin |
| Reductase_C | 1 | Reductase C-terminal |
| Reelin_subrepeat-B | 1 | Reelin subrepeat B |
| RelB_leu_zip | 1 | RelB leucine zipper |
| RelB_transactiv | 1 | RelB transactivation domain |
| Renin_r | 1 | Renin receptor-like protein |
| Rep-A_N | 1 | Replication factor-A protein 1, N-terminal domain |
| Rep_fac-A_3 | 1 | Replication factor A protein 3 |
| Reprolysin_4 | 1 | Metallo-peptidase family M12B Reprolysin-like |
| Rer1 | 1 | Rer1 family |
| Retinal | 1 | Retinal protein |
| Rft-1 | 1 | Rft protein |
| Rgp1 | 1 | Rgp1 |
| Rhodanese_C | 1 | Rhodanase C-terminal |
| Rhodopsin_N | 1 | Amino terminal of the G-protein receptor rhodopsin |
| Rib_5-P_isom_A | 1 | Ribose 5-phosphate isomerase A (phosphoriboisomerase A) |
| Ribo_biogen_C | 1 | Ribosome biogenesis protein, C-terminal |
| Ribonuc_L-PSP | 1 | Endoribonuclease L-PSP |
| Ribonuc_P_40 | 1 | Ribonuclease P 40kDa (Rpp40) subunit |
| Ribonuc_red_lgC | 1 | Ribonucleotide reductase, barrel domain |
| Ribonuc_red_lgN | 1 | Ribonucleotide reductase, all-alpha domain |
| Ribonucleas_3_3 | 1 | Ribonuclease-III-like |
| Ribonuclease_T2 | 1 | Ribonuclease T2 family |
| Ribophorin_I | 1 | Ribophorin I |
| Ribophorin_II | 1 | Oligosaccharyltransferase subunit Ribophorin II |
| Ribos_L4_asso_C | 1 | 60S ribosomal protein L4 C-terminal domain |
| Ribosomal_L12 | 1 | Ribosomal protein L7/L12 C-terminal domain |
| Ribosomal_L12_N | 1 | Ribosomal protein L7/L12 dimerisation domain |
| Ribosomal_L13e | 1 | Ribosomal protein L13e |
| Ribosomal_L14e | 1 | Ribosomal protein L14 |
| Ribosomal_L15e | 1 | Ribosomal L15 |
| Ribosomal_L17 | 1 | Ribosomal protein L17 |
| Ribosomal_L18 | 1 | Ribosomal protein 60S L18 and 50S L18e |
| Ribosomal_L18A | 1 | Ribosomal proteins 50S-L18Ae/60S-L20/60S-L18A |
| Ribosomal_L18_c | 1 | Ribosomal L18 C-terminal region |
| Ribosomal_L18p | 1 | Ribosomal L18 of archaea, bacteria, mitoch. and chloroplast |
| Ribosomal_L19 | 1 | Ribosomal protein L19 |
| Ribosomal_L19e | 1 | Ribosomal protein L19e |
| Ribosomal_L20 | 1 | Ribosomal protein L20 |
| Ribosomal_L21e | 1 | Ribosomal protein L21e |
| Ribosomal_L21p | 1 | Ribosomal prokaryotic L21 protein |
| Ribosomal_L23eN | 1 | Ribosomal protein L23, N-terminal domain |
| Ribosomal_L27 | 1 | Ribosomal L27 protein |
| Ribosomal_L27e | 1 | Ribosomal L27e protein family |
| Ribosomal_L28 | 1 | Ribosomal L28 family |
| Ribosomal_L29 | 1 | Ribosomal L29 protein |
| Ribosomal_L29e | 1 | Ribosomal L29e protein family |
| Ribosomal_L31e | 1 | Ribosomal protein L31e |
| Ribosomal_L32e | 1 | Ribosomal protein L32 |
| Ribosomal_L32p | 1 | Ribosomal L32p protein family |
| Ribosomal_L33 | 1 | Ribosomal protein L33 |
| Ribosomal_L34 | 1 | Ribosomal protein L34 |
| Ribosomal_L34e | 1 | Ribosomal protein L34e |
| Ribosomal_L35Ae | 1 | Ribosomal protein L35Ae |
| Ribosomal_L35p | 1 | Ribosomal protein L35 |
| Ribosomal_L36 | 1 | Ribosomal protein L36 |
| Ribosomal_L36e | 1 | Ribosomal protein L36e |
| Ribosomal_L37 | 1 | Mitochondrial ribosomal protein L37 |
| Ribosomal_L37ae | 1 | Ribosomal L37ae protein family |
| Ribosomal_L37e | 1 | Ribosomal protein L37e |
| Ribosomal_L38e | 1 | Ribosomal L38e protein family |
| Ribosomal_L40e | 1 | Ribosomal L40e family |
| Ribosomal_L41 | 1 | Ribosomal protein L41 |
| Ribosomal_L5 | 1 | Ribosomal protein L5 |
| Ribosomal_L50 | 1 | Ribosomal subunit 39S |
| Ribosomal_L5_C | 1 | ribosomal L5P family C-terminus |
| Ribosomal_L5e | 1 | Ribosomal large subunit proteins 60S L5, and 50S L18 |
| Ribosomal_L6 | 1 | Ribosomal protein L6 |
| Ribosomal_L6e | 1 | Ribosomal protein L6e |
| Ribosomal_L6e_N | 1 | Ribosomal protein L6, N-terminal domain |
| Ribosomal_L9_N | 1 | Ribosomal protein L9, N-terminal domain |
| Ribosomal_S13 | 1 | Ribosomal protein S13/S18 |
| Ribosomal_S13_N | 1 | Ribosomal S13/S15 N-terminal domain |
| Ribosomal_S16 | 1 | Ribosomal protein S16 |
| Ribosomal_S17_N | 1 | Ribosomal_S17 N-terminal |
| Ribosomal_S17e | 1 | Ribosomal S17 |
| Ribosomal_S19 | 1 | Ribosomal protein S19 |
| Ribosomal_S19e | 1 | Ribosomal protein S19e |
| Ribosomal_S21 | 1 | Ribosomal protein S21 |
| Ribosomal_S24e | 1 | Ribosomal protein S24e |
| Ribosomal_S25 | 1 | S25 ribosomal protein |
| Ribosomal_S26e | 1 | Ribosomal protein S26e |
| Ribosomal_S27 | 1 | Ribosomal protein S27a |
| Ribosomal_S28e | 1 | Ribosomal protein S28e |
| Ribosomal_S30 | 1 | Ribosomal protein S30 |
| Ribosomal_S3Ae | 1 | Ribosomal S3Ae family |
| Ribosomal_S3_C | 1 | Ribosomal protein S3, C-terminal domain |
| Ribosomal_S6 | 1 | Ribosomal protein S6 |
| Ribosomal_S6e | 1 | Ribosomal protein S6e |
| Ribosomal_S7e | 1 | Ribosomal protein S7e |
| Ribosomal_S8 | 1 | Ribosomal protein S8 |
| Ribosomal_uS5m_N | 1 | Small ribosomal subunit protein uS5m, N-terminal |
| Rif1_N | 1 | Rap1-interacting factor 1 N terminal |
| Rio2_N | 1 | Rio2, N-terminal |
| RmlD_sub_bind | 1 | RmlD substrate binding domain |
| Rod_C | 1 | Rough deal protein C-terminal region |
| Rod_cone_degen | 1 | Progressive rod-cone degeneration |
| Rogdi_lz | 1 | Rogdi leucine zipper containing protein |
| Romo1 | 1 | Reactive mitochondrial oxygen species modulator 1 |
| Rotamase | 1 | PPIC-type PPIASE domain |
| Rotamase_3 | 1 | PPIC-type PPIASE domain |
| Rpn13_ADRM1_Pru | 1 | Proteasome complex subunit Rpn13, Pru domain |
| Rpn3_C | 1 | Proteasome regulatory subunit C-terminal |
| Rpp20 | 1 | Rpp20 subunit of nuclear RNase MRP and P |
| Rrn7_cyclin_C | 1 | Rrn7/TAF1B, C-terminal cyclin domain |
| Rrn7_cyclin_N | 1 | Rrn7/TAF1B, N-terminal cyclin domain |
| Rrp15p | 1 | Rrp15p |
| RsfS | 1 | Ribosomal silencing factor during starvation |
| Rsm1 | 1 | Rsm1-like |
| Rsm22 | 1 | Mitochondrial small ribosomal subunit Rsm22 |
| RtcB | 1 | tRNA-splicing ligase RtcB |
| Rtf2 | 1 | Rtf2 RING-finger |
| Rubredoxin_C | 1 | Rubredoxin NAD+ reductase C-terminal domain |
| S100PBPR | 1 | S100P-binding protein |
| S6OS1 | 1 | Six6 opposite strand transcript 1 family |
| SAE2 | 1 | DNA endonuclease activator SAE2/CtIP C-terminus |
| SAF | 1 | SAF domain |
| SAGA-Tad1 | 1 | Transcriptional regulator of RNA polII, SAGA, subunit |
| SAICAR_synt | 1 | SAICAR synthetase |
| SAM_4 | 1 | SAM domain (Sterile alpha motif) |
| SAM_KSR1 | 1 | SAM like domain present in kinase suppressor RAS 1 |
| SAM_decarbox | 1 | Adenosylmethionine decarboxylase |
| SANBR_BTB | 1 | SANT and BTB domain regulator of CSR, BTB domain |
| SANTA | 1 | SANTA (SANT Associated) |
| SANT_DAMP1_like | 1 | SANT/Myb-like domain of DAMP1 |
| SAP130_C | 1 | Histone deacetylase complex subunit SAP130 C-terminus |
| SAP18 | 1 | Sin3 associated polypeptide p18 (SAP18) |
| SAP25 | 1 | Histone deacetylase complex subunit SAP25 |
| SARA | 1 | Smad anchor for receptor activation (SARA) |
| SARAF | 1 | SOCE-associated regulatory factor of calcium homoeostasis |
| SART-1 | 1 | SART-1 family |
| SAS-6_N | 1 | Centriolar protein SAS N-terminal domain |
| SASA | 1 | Carbohydrate esterase, sialic acid-specific acetylesterase |
| SASRP1 | 1 | Spermatogenesis-associated serine-rich protein 1 |
| SAXO5-like | 1 | Stabilizer of axonemal microtubules 5 |
| SAYSvFN | 1 | Uncharacterized conserved domain (SAYSvFN) |
| SBDS | 1 | Shwachman-Bodian-Diamond syndrome (SBDS) protein |
| SBDS_C | 1 | SBDS protein, C-terminal domain |
| SBDS_domain_II | 1 | SBDS protein, domain II |
| SBF_like | 1 | SBF-like CPA transporter family (DUF4137) |
| SBP56 | 1 | 56kDa selenium binding protein (SBP56) |
| SBSN_GxHH_rpt | 1 | Suprabasin, GxHH repeat |
| SCAI | 1 | Protein SCAI |
| SCAPER_N | 1 | S phase cyclin A-associated protein in the endoplasmic reticulum |
| SCF | 1 | Stem cell factor |
| SCIMP | 1 | SCIMP protein |
| SCNM1_acidic | 1 | Acidic C-terminal region of sodium channel modifier 1 SCNM1 |
| SCP-1 | 1 | Synaptonemal complex protein 1 (SCP-1) |
| SCRE | 1 | Protein SPO16 homolog |
| SCRG1 | 1 | Scrapie-responsive protein 1 |
| SDA1_C | 1 | SDA1, C-terminal |
| SDA1_HEAT | 1 | SDA1, HEAT repeat |
| SDA1_dom | 1 | SDA1, conserved domain |
| SE | 1 | Squalene epoxidase |
| SEEEED | 1 | Serine-rich region of AP3B1, clathrin-adaptor complex |
| SEEK1 | 1 | Psoriasis susceptibility 1 candidate 1 |
| SERRATE_Ars2_N | 1 | SERRATE/Ars2, N-terminal domain |
| SF1-HH | 1 | Splicing factor 1 helix-hairpin domain |
| SF3A2 | 1 | Pre-mRNA-splicing factor SF3a complex subunit 2 (Prp11) |
| SF3A3 | 1 | Pre-mRNA-splicing factor SF3A3, of SF3a complex, Prp9 |
| SF3a60_Prp9_C | 1 | SF3a60/Prp9 C-terminal |
| SF3a60_bindingd | 1 | Splicing factor SF3a60 binding domain |
| SF3b1 | 1 | Splicing factor 3B subunit 1 |
| SF3b10 | 1 | Splicing factor 3B subunit 10 (SF3b10) |
| SFTA2 | 1 | Surfactant-associated protein 2 |
| SGF29_Tudor | 1 | SGF29 tudor-like domain |
| SGIII | 1 | Secretogranin-3 |
| SGL | 1 | SMP-30/Gluconolactonase/LRE-like region |
| SGT1 | 1 | SGT1 protein |
| SH2_2 | 1 | SH2 domain |
| SH3-WW_linker | 1 | Linker region between SH3 and WW domains on ARHGAP12 |
| SH3_12 | 1 | Xrn1 SH3-like domain |
| SH3_19 | 1 | Myosin X N-terminal SH3 domain |
| SHLD1_C | 1 | Shieldin complex subunit 1, C-terminal domain |
| SHLD2_C | 1 | Shieldin complex subunit 2, C-terminal |
| SHLD2_OB1 | 1 | Shieldin complex subunit 2, first OB fold domain |
| SHNi-TPR | 1 | SHNi-TPR |
| SHPRH_helical-1st | 1 | E3 ubiquitin-protein ligase SHPRH, first helical domain |
| SHPRH_helical-2nd | 1 | E3 ubiquitin-protein ligase SHPRH, second helical domain |
| SHQ1 | 1 | SHQ1 protein, Shq1 domain |
| SHQ1-like_CS | 1 | SHQ1-like, CS domain |
| SIMPL | 1 | Protein of unknown function (DUF541) |
| SIN1 | 1 | Stress-activated map kinase interacting protein 1 (SIN1) |
| SIN1_PH | 1 | SAPK-interacting protein 1 (Sin1), Pleckstrin-homology |
| SIP1 | 1 | Survival motor neuron (SMN) interacting protein 1 (SIP1) |
| SKA1 | 1 | Spindle and kinetochore-associated protein 1 |
| SKA2 | 1 | Spindle and kinetochore-associated protein 2 |
| SKIP_SNW | 1 | SKIP/SNW domain |
| SLAM | 1 | Signaling lymphocytic activation molecule (SLAM) protein |
| SLBP_RNA_bind | 1 | Histone RNA hairpin-binding protein RNA-binding domain |
| SLC3A2_N | 1 | Solute carrier family 3 member 2 N-terminus |
| SLD5_C | 1 | DNA replication complex GINS protein SLD5 C-terminus |
| SLX9 | 1 | Ribosome biogenesis protein SLX9 |
| SMC_Nse1 | 1 | Nse1 non-SMC component of SMC5-6 complex |
| SMG1 | 1 | Serine/threonine-protein kinase smg-1 |
| SMG1_N | 1 | Serine/threonine-protein kinase SMG1 N-terminal |
| SMRP1 | 1 | Spermatid-specific manchette-related protein 1 |
| SMUBP-2_HCS1_1B | 1 | Helicase SMUBP-2/HCS1, 1B domain |
| SMYLE_N | 1 | Short myomegalin-like EB1 binding proteins, N-terminal domain |
| SNAPC1 | 1 | Small nuclear RNA activating complex (SNAPc), subunit 1 |
| SNAPC2 | 1 | Small nuclear RNA activating complex (SNAPc), subunit 2 |
| SNAPC3 | 1 | snRNA-activating protein complex (SNAPc), subunit 3 |
| SNAPC5 | 1 | snRNA-activating protein complex subunit 19, SNAPc subunit 19 |
| SNCAIP_SNCA_bd | 1 | Synphilin-1 alpha-Synuclein-binding domain |
| SNF5 | 1 | SNF5 / SMARCB1 / INI1 |
| SNN_cytoplasm | 1 | Stannin cytoplasmic |
| SNN_linker | 1 | Stannin unstructured linker |
| SNN_transmemb | 1 | Stannin transmembrane |
| SNRNP27 | 1 | U4/U6.U5 small nuclear ribonucleoproteins |
| SNURF | 1 | SNURF/RPN4 protein |
| SNase | 1 | Staphylococcal nuclease homologue |
| SOSSC | 1 | SOSS complex subunit C |
| SPACA7 | 1 | Sperm acrosome-associated protein 7 |
| SPACA9 | 1 | Sperm acrosome-associated protein 9 |
| SPATA19 | 1 | Spermatogenesis-associated protein 19, mitochondrial |
| SPATA1_C | 1 | Spermatogenesis-associated C-terminus |
| SPATA24 | 1 | Spermatogenesis-associated protein 24 |
| SPATA25 | 1 | Spermatogenesis-associated protein 25 |
| SPATA3 | 1 | Spermatogenesis-associated protein 3 family |
| SPATA48 | 1 | Spermatogenesis-associated protein 48 |
| SPATA9 | 1 | Spermatogenesis-associated protein 9 |
| SPC12 | 1 | Microsomal signal peptidase 12 kDa subunit (SPC12) |
| SPC22 | 1 | Signal peptidase subunit |
| SPC25 | 1 | Microsomal signal peptidase 25 kDa subunit (SPC25) |
| SPEG_u2 | 1 | Unstructured region on SPEG complex protein |
| SPESP1 | 1 | Sperm equatorial segment protein 1 |
| SPG48 | 1 | AP-5 complex subunit, vesicle trafficking |
| SPICE | 1 | Centriole duplication and mitotic chromosome congression |
| SPIN90_LRD | 1 | SPIN90/Ldb17, leucine-rich domain |
| SPMAP1-like | 1 | Sperm microtubule associated protein 1 |
| SPMIP10-like | 1 | Sperm-associated microtubule inner protein 10 |
| SPMIP4-like | 1 | Sperm-associated microtubule inner protein 4 |
| SPO11_like | 1 | SPO11 homologue |
| SPO22 | 1 | Meiosis protein SPO22/ZIP4 like |
| SPR1 | 1 | Psoriasis susceptibility locus 2 |
| SPRING1 | 1 | SREBP regulating gene protein |
| SPT16 | 1 | FACT complex subunit (SPT16/CDC68) |
| SPT16_C | 1 | FACT complex subunit SPT16, C-terminal domain |
| SPT2 | 1 | SPT2 chromatin protein |
| SPT6_acidic | 1 | Acidic N-terminal SPT6 |
| SPX | 1 | SPX domain |
| SP_C-Propep | 1 | Surfactant protein C, N terminal propeptide |
| SQHop_cyclase_C | 1 | Squalene-hopene cyclase C-terminal domain |
| SQHop_cyclase_N | 1 | Squalene-hopene cyclase N-terminal domain |
| SR-25 | 1 | Nuclear RNA-splicing-associated protein |
| SRA1 | 1 | Steroid receptor RNA activator (SRA1) |
| SRAP | 1 | SOS response associated peptidase (SRAP) |
| SRI | 1 | SRI (Set2 Rpb1 interacting) domain |
| SRP-alpha_N | 1 | Signal recognition particle, alpha subunit, N-terminal |
| SRP14 | 1 | Signal recognition particle 14kD protein |
| SRP19 | 1 | SRP19 protein |
| SRP40_C | 1 | SRP40, C-terminal domain |
| SRP68 | 1 | RNA-binding signal recognition particle 68 |
| SRP72 | 1 | SRP72 RNA-binding domain |
| SRP9-21 | 1 | Signal recognition particle 9 kDa protein (SRP9) |
| SRPRB | 1 | Signal recognition particle receptor beta subunit |
| SRP_SPB | 1 | Signal peptide binding domain |
| SRP_TPR_like | 1 | Putative TPR-like repeat |
| SRR1 | 1 | SRR1 domain |
| SSB | 1 | Single-strand binding protein family |
| SSL_N | 1 | Strictosidine synthase-like, N-terminal |
| SSRP1_C | 1 | FACT complex subunit SSRP1, C-terminal domain |
| SSTK-IP | 1 | SSTK-interacting protein, TSSK6-activating co-chaperone protein |
| SSrecog | 1 | Structure-specific recognition protein (SSRP1) |
| STAR_dimer | 1 | Homodimerisation region of STAR domain protein |
| STAT1_TAZ2bind | 1 | STAT1 TAZ2 binding domain |
| STAT2_C | 1 | Signal transducer and activator of transcription 2 C terminal |
| STAT6_C | 1 | STAT6 C-terminal |
| STG | 1 | Simian taste bud-specific gene product family |
| STIL_N | 1 | SCL-interrupting locus protein N-terminus |
| STL11_N | 1 | STL11, N-terminal |
| STN1_2 | 1 | CST, complex subunit STN1, C terminal |
| SUFU | 1 | Suppressor of fused protein (SUFU) |
| SUFU_C | 1 | Suppressor of Fused Gli/Ci N terminal binding domain |
| SURF1 | 1 | SURF1 family |
| SURF2 | 1 | Surfeit locus protein 2 (SURF2) |
| SURF4 | 1 | SURF4 family |
| SURF6 | 1 | Surfeit locus protein 6 |
| SUV3_C | 1 | Mitochondrial degradasome RNA helicase subunit C terminal |
| SVA | 1 | Seminal vesicle autoantigen (SVA) |
| SVIP | 1 | Small VCP/p97-interacting protein |
| SYF2 | 1 | SYF2 splicing factor |
| SYMPK_PTA1_N | 1 | Symplekin/PTA1 N-terminal |
| SYS1 | 1 | Integral membrane protein S linking to the trans Golgi network |
| Sacchrp_dh_C | 1 | Saccharopine dehydrogenase C-terminal domain |
| Saposin | 1 | Saposin-like domain |
| Sarcolipin | 1 | Sarcolipin |
| Sas10 | 1 | Sas10 C-terminal domain |
| Sas6_CC | 1 | Sas6/XLF/XRCC4 coiled-coil domain |
| SbcC_Walker_B | 1 | SbcC/RAD50-like, Walker B motif |
| Scm3 | 1 | Centromere protein Scm3 |
| Sde2_N_Ubi_yeast | 1 | Yeast Splicing regulator sde2, N-terminal ubiquitin domain |
| Sdh5 | 1 | Flavinator of succinate dehydrogenase |
| Sdh_cyt | 1 | Succinate dehydrogenase/Fumarate reductase transmembrane subunit |
| Sec10_HB | 1 | Exocyst complex component Sec10-like, alpha-helical bundle |
| Sec10_N | 1 | Exocyst complex component Sec10, N-terminal |
| Sec20 | 1 | Sec20 |
| Sec39 | 1 | Secretory pathway protein Sec39 |
| Sec3_C-term | 1 | Exocyst complex component Sec3, C-terminal |
| Sec3_CC | 1 | Exocyst complex component Sec3, coiled-coil |
| Sec61_beta | 1 | Sec61beta family |
| Sec62 | 1 | Translocation protein Sec62 |
| Sec8_C | 1 | Exocyst complex component Sec8 C-terminal |
| Sec8_N | 1 | Exocyst complex component Sec8 N-terminal |
| SecE | 1 | SecE/Sec61-gamma subunits of protein translocation complex |
| Seipin | 1 | Putative adipose-regulatory protein (Seipin) |
| SelK_SelG | 1 | Selenoprotein SelK_SelG |
| SelO | 1 | Protein adenylyltransferase SelO |
| SelP_C | 1 | Selenoprotein P, C terminal region |
| SelP_N | 1 | Selenoprotein P, N terminal region |
| Selenoprotein_S | 1 | Selenoprotein S (SelS) |
| Sema4F_C | 1 | Semaphorin 4F C-terminal |
| Sen15 | 1 | Sen15 protein |
| Senescence | 1 | Senescence domain |
| SepSecS | 1 | O-phosphoseryl-tRNA(Sec) selenium transferase, SepSecS |
| Ser_hydrolase | 1 | Serine hydrolase |
| Serglycin | 1 | Serglycin |
| Seryl_tRNA_N | 1 | Seryl-tRNA synthetase N-terminal domain |
| Sgf11 | 1 | Sgf11 (transcriptional regulation protein) |
| ShK | 1 | ShK domain-like |
| Sharpin_PH | 1 | Sharpin PH domain |
| Shugoshin_C | 1 | Shugoshin C terminus |
| Shugoshin_N | 1 | Shugoshin N-terminal coiled-coil region |
| Siah-Interact_N | 1 | Siah interacting protein, N terminal |
| Siva | 1 | Cd27 binding protein (Siva) |
| Ski2_N | 1 | Ski2 N-terminal region |
| Skp1 | 1 | Skp1 family, dimerisation domain |
| Slu7 | 1 | Pre-mRNA splicing Prp18-interacting factor |
| Slx4 | 1 | Slx4 endonuclease |
| SmAKAP | 1 | Small membrane A-kinase anchor protein |
| Smac_DIABLO | 1 | Second Mitochondria-derived Activator of Caspases |
| Smg8_Smg9 | 1 | Smg8_Smg9 |
| Smim3 | 1 | Small integral membrane protein 3 |
| Smoothelin | 1 | Smoothelin cytoskeleton protein |
| Smr | 1 | Smr domain |
| Snurportin1 | 1 | Snurportin1 |
| Snx8_BAR_dom | 1 | Sorting nexin 8/Mvp1 BAR domain |
| Sod_Fe_C | 1 | Iron/manganese superoxide dismutases, C-terminal domain |
| Sod_Fe_N | 1 | Iron/manganese superoxide dismutases, alpha-hairpin domain |
| Sof1 | 1 | Sof1-like domain |
| Sororin | 1 | Sororin protein |
| Spatacsin_C | 1 | Spatacsin C-terminus |
| Spb1_C | 1 | Spb1 C-terminal domain |
| Spc24 | 1 | Spc24 subunit of Ndc80 |
| Spec3 | 1 | Ectodermal ciliogenesis protein |
| Spexin | 1 | Neuropeptide secretory protein family, NPQ, spexin |
| Spindle_Spc25 | 1 | Chromosome segregation protein Spc25 |
| SpmSyn_N | 1 | S-adenosylmethionine decarboxylase N -terminal |
| SpoIIE | 1 | Stage II sporulation protein E (SpoIIE) |
| SpoU_sub_bind | 1 | RNA 2'-O ribose methyltransferase substrate binding |
| Spp-24 | 1 | Secreted phosphoprotein 24 (Spp-24) cystatin-like domain |
| Spt4 | 1 | Spt4/RpoE2 zinc finger |
| Spt5-NGN | 1 | Early transcription elongation factor of RNA pol II, NGN section |
| Spt5_N | 1 | Spt5 transcription elongation factor, acidic N-terminal |
| Statherin | 1 | Statherin |
| Stealth_CR1 | 1 | Stealth protein CR1, conserved region 1 |
| Stealth_CR2 | 1 | Stealth protein CR2, conserved region 2 |
| Stealth_CR3 | 1 | Stealth protein CR3, conserved region 3 |
| Stealth_CR4 | 1 | Stealth protein CR4, conserved region 4 |
| Stereocilin | 1 | Stereocilin |
| Stk19 | 1 | Serine-threonine protein kinase 19 |
| Stonin2_N | 1 | Stonin 2 |
| Str_synth | 1 | Strictosidine synthase |
| Strumpellin | 1 | Hereditary spastic paraplegia protein strumpellin |
| Sua5_yciO_yrdC | 1 | Telomere recombination |
| Succ_DH_flav_C | 1 | Fumarate reductase flavoprotein C-term |
| Surfac_D-trimer | 1 | Lung surfactant protein D coiled-coil trimerisation |
| Sushi_2 | 1 | Beta-2-glycoprotein-1 fifth domain |
| Suv3_C_1 | 1 | Suv3 C-terminal domain 1 |
| Suv3_N | 1 | Suv3 helical N-terminal domain |
| Swi3 | 1 | Replication Fork Protection Component Swi3 |
| Swi5 | 1 | Swi5 |
| Symplekin_C | 1 | Symplekin tight junction protein C terminal |
| Synaptonemal_3 | 1 | Synaptonemal complex central element protein 3 |
| Syncollin | 1 | Syncollin |
| Syndetin_C | 1 | Syndetin, C-terminal |
| Syntaxin-18_N | 1 | SNARE-complex protein Syntaxin-18 N-terminus |
| Syntaxin-5_N | 1 | Syntaxin-5 N-terminal, Sly1p-binding domain |
| TACI-CRD2 | 1 | TACI, cysteine-rich domain |
| TACO1_YebC_N | 1 | TACO1/YebC protein N-terminal domain |
| TAD2 | 1 | Transactivation domain 2 |
| TAF1C_C | 1 | TAF1C, C-terminal domain |
| TAF1C_HB | 1 | TAF1C, helical bundle domain |
| TAF1C_beta-prop | 1 | TAF1C, beta-propeller |
| TAF1D | 1 | TATA box-binding protein-associated factor 1D |
| TAF1_subA | 1 | TAF RNA Polymerase I subunit A |
| TAF8_C | 1 | Transcription factor TFIID complex subunit 8 C-term |
| TALPID3 | 1 | Hedgehog signalling target |
| TAL_FSA | 1 | Transaldolase/Fructose-6-phosphate aldolase |
| TAN | 1 | Telomere-length maintenance and DNA damage repair |
| TANGO2 | 1 | Transport and Golgi organisation 2 |
| TASL | 1 | TLR adaptor interacting with SLC15A4 on the lysosome |
| TAT_ubiq | 1 | Aminotransferase ubiquitination site |
| TBC1D23_C | 1 | TBC1 domain family member 23 C-terminal |
| TBCA | 1 | Tubulin binding cofactor A |
| TBCC_N | 1 | Tubulin-specific chaperone C N-terminal domain |
| TBPIP | 1 | TBPIP/Hop2 winged helix domain |
| TCTP | 1 | Translationally controlled tumour protein |
| TDP-43_C | 1 | TAR DNA-binding protein 43, C-terminal |
| TDP43_N | 1 | TAR DNA-binding protein 43, N-terminal domain |
| TDRP | 1 | Testis development-related protein |
| TEDC1 | 1 | Tubulin epsilon and delta complex protein 1 |
| TEP1_N | 1 | TEP1 N-terminal domain |
| TERB2 | 1 | Telomere-associated protein TERB2 |
| TERF2_RBM | 1 | Telomeric repeat-binding factor 2 Rap1-binding motif |
| TERT_C | 1 | Telomerase reverse transcriptase, C-terminal extension |
| TEX12 | 1 | Testis-expressed 12 |
| TEX15 | 1 | Testis expressed sequence 15 |
| TEX19 | 1 | Testis-expressed protein 19 |
| TEX29 | 1 | Testis-expressed sequence 29 protein |
| TEX33 | 1 | Testis-expressed sequence 33 protein family |
| TFA2_Winged_2 | 1 | TFA2 Winged helix domain 2 |
| TFCD_C | 1 | Tubulin folding cofactor D C terminal |
| TFIIA_gamma_C | 1 | Transcription initiation factor IIA, gamma subunit |
| TFIIA_gamma_N | 1 | Transcription initiation factor IIA, gamma subunit, helical domain |
| TFIID_20kDa | 1 | Transcription initiation factor TFIID subunit A |
| TFIID_30kDa | 1 | Transcription initiation factor TFIID 23-30kDa subunit |
| TFIIE-A_C | 1 | C-terminal general transcription factor TFIIE alpha |
| TFIIE_alpha | 1 | TFIIE alpha subunit |
| TFIIE_beta | 1 | TFIIE beta subunit core domain |
| TFIIF_alpha | 1 | Transcription initiation factor IIF, alpha subunit (TFIIF-alpha) |
| TFIIF_beta | 1 | TFIIF, beta subunit HTH domain |
| TFIIF_beta_N | 1 | TFIIF, beta subunit N-terminus |
| TFIIIC_delta | 1 | Transcription factor IIIC subunit delta bet-propeller domain |
| TFIIIC_sub6 | 1 | TFIIIC subunit triple barrel domain |
| TH1 | 1 | TH1 protein |
| THAP4_heme-bd | 1 | THAP4-like, heme-binding beta-barrel domain |
| THEG4 | 1 | Testis highly expressed protein 4 |
| THOC2_N | 1 | THO complex subunit 2 N-terminus |
| THOC7 | 1 | Tho complex subunit 7 |
| TH_ACT | 1 | Tyrosine 3-monooxygenase-like, ACT domain |
| TIM | 1 | Triosephosphate isomerase |
| TIM21 | 1 | TIM21 |
| TIMELESS | 1 | Timeless protein |
| TIMELESS_C | 1 | Timeless PAB domain |
| TINF2_N | 1 | TERF1-interacting nuclear factor 2 N-terminus |
| TIP39 | 1 | TIP39 peptide |
| TIP41 | 1 | TIP41-like family |
| TIP_N | 1 | Tuftelin interacting protein N terminal |
| TK | 1 | Thymidine kinase |
| TKTI1 | 1 | Tektin bundle interacting protein 1 |
| TM140 | 1 | TM140 protein family |
| TM231 | 1 | Transmembrane protein 231 |
| TMCCDC2 | 1 | Transmembrane and coiled-coil domain-containing protein 2 |
| TMEM100 | 1 | Transmembrane protein 100 |
| TMEM101 | 1 | TMEM101 protein family |
| TMEM107 | 1 | Transmembrane protein |
| TMEM108 | 1 | TMEM108 family |
| TMEM117 | 1 | TMEM117 protein family |
| TMEM119 | 1 | TMEM119 family |
| TMEM125 | 1 | TMEM125 protein family |
| TMEM127 | 1 | Transmembrane protein 127 |
| TMEM128 | 1 | TMEM128 protein |
| TMEM135_C_rich | 1 | N-terminal cysteine-rich region of Transmembrane protein 135 |
| TMEM138 | 1 | Transmembrane protein 138 |
| TMEM141 | 1 | TMEM141 protein family |
| TMEM144 | 1 | Transmembrane family, TMEM144 of transporters |
| TMEM154 | 1 | TMEM154 protein family |
| TMEM156 | 1 | TMEM156 protein family |
| TMEM164 | 1 | TMEM164 family |
| TMEM169 | 1 | TMEM169 protein family |
| TMEM171 | 1 | Transmembrane protein family 171 |
| TMEM173 | 1 | Transmembrane protein 173 |
| TMEM174 | 1 | Transmembrane protein 174 |
| TMEM175 | 1 | Endosomal/lysosomal potassium channel TMEM175 |
| TMEM18 | 1 | Transmembrane protein 18 |
| TMEM187 | 1 | TMEM187 protein family |
| TMEM190 | 1 | Transmembrane protein 190 |
| TMEM192 | 1 | TMEM192 family |
| TMEM206 | 1 | TMEM206 protein family |
| TMEM208_SND2 | 1 | SRP-independent targeting protein 2/TMEM208 |
| TMEM210 | 1 | TMEM210 family |
| TMEM213 | 1 | TMEM213 family |
| TMEM214 | 1 | TMEM214, C-terminal, caspase 4 activator |
| TMEM215 | 1 | TMEM215 family |
| TMEM220 | 1 | Transmembrane family 220, helix |
| TMEM232 | 1 | Transmembrane protein family 232 |
| TMEM234 | 1 | Putative transmembrane family 234 |
| TMEM237 | 1 | Transmembrane protein 237 |
| TMEM239 | 1 | Transmembrane protein 239 family |
| TMEM240 | 1 | TMEM240 family |
| TMEM247 | 1 | Transmembrane protein 247 |
| TMEM249 | 1 | Cation channel sperm-associated auxiliary subunit TMEM249 |
| TMEM251 | 1 | Transmembrane protein 251 |
| TMEM252 | 1 | Transmembrane protein 252 family |
| TMEM254 | 1 | Transmembrane protein 254 |
| TMEM260-like | 1 | Protein O-mannosyl-transferase TMEM260-like |
| TMEM33_Pom33 | 1 | Transmembrane protein 33/Nucleoporin POM33 |
| TMEM37 | 1 | Voltage-dependent calcium channel gamma-like subunit protein family |
| TMEM40 | 1 | Transmembrane protein 40 family |
| TMEM43 | 1 | Transmembrane protein 43 |
| TMEM51 | 1 | Transmembrane protein 51 |
| TMEM61 | 1 | TMEM61 protein family |
| TMEM65 | 1 | Transmembrane protein 65 |
| TMEM71 | 1 | TMEM71 protein family |
| TMEM72 | 1 | Transmembrane protein family 72 |
| TMEM82 | 1 | Transmembrane protein 82 |
| TMEM89 | 1 | TMEM89 protein family |
| TMEM95 | 1 | TMEM95 family |
| TMF_DNA_bd | 1 | TATA element modulatory factor 1 DNA binding |
| TMF_TATA_bd | 1 | TATA element modulatory factor 1 TATA binding |
| TMIE | 1 | TMIE protein |
| TNFR_16_TM | 1 | Tumor necrosis factor receptor member 16 trans-membrane domain |
| TNP-like_GBD | 1 | TNP-like, GTP-binding insertion domain |
| TNP-like_RNaseH_C | 1 | TNP-like, RNase H C-terminal domain |
| TNP-like_RNaseH_N | 1 | TNP-like, RNase H N-terminal domain |
| TOH_N | 1 | Tyrosine hydroxylase N terminal |
| TOM6p | 1 | Mitochondrial import receptor subunit TOM6 homolog |
| TOP6A-Spo11_Toprim | 1 | Topoisomerase 6 subunit A/Spo11, Toprim domain |
| TP1 | 1 | Nuclear transition protein 1 |
| TP2 | 1 | Nuclear transition protein 2 |
| TP53IP5 | 1 | Cellular tumour antigen p53-inducible 5 |
| TP6A_N | 1 | Type IIB DNA topoisomerase |
| TPD | 1 | Protein of unknown function TPD sequence-motif |
| TPIP1 | 1 | p53-regulated apoptosis-inducing protein 1 |
| TPK_B1_binding | 1 | Thiamin pyrophosphokinase, vitamin B1 binding domain |
| TPK_catalytic | 1 | Thiamin pyrophosphokinase, catalytic domain |
| TPMT | 1 | Thiopurine S-methyltransferase (TPMT) |
| TPP1 | 1 | Shelterin complex subunit, TPP1/ACD |
| TPPII | 1 | Tripeptidyl peptidase II |
| TPPII_C | 1 | Tripeptidyl peptidase II, C-terminal |
| TPPII_GBD | 1 | TPPII, galactose-binding domain-like |
| TPPII_Ig-like-1 | 1 | Tripeptidyl-peptidase II, first Ig-like domain |
| TPR_14 | 1 | Tetratricopeptide repeat |
| TPR_MLP1_2 | 1 | TPR/MLP1/MLP2-like protein |
| TPR_MalT | 1 | MalT-like TPR region |
| TPX2 | 1 | Targeting protein for Xklp2 (TPX2) domain |
| TPX2_importin | 1 | Cell cycle regulated microtubule associated protein |
| TP_methylase | 1 | Tetrapyrrole (Corrin/Porphyrin) Methylases |
| TRADD_N | 1 | TRADD, N-terminal domain |
| TRAF2_zf | 1 | TRAF2, second zinc finger |
| TRAF6_Z2 | 1 | TNF receptor-associated factor 6 zinc finger 2 |
| TRAP-delta | 1 | Translocon-associated protein, delta subunit precursor (TRAP-delta) |
| TRAP-gamma | 1 | Translocon-associated protein, gamma subunit (TRAP-gamma) |
| TRAPPC-Trs85 | 1 | ER-Golgi trafficking TRAPP I complex 85 kDa subunit |
| TRAPPC10 | 1 | Trafficking protein particle complex subunit 10, TRAPPC10 |
| TRAPPC9-Trs120 | 1 | Transport protein Trs120 or TRAPPC9, TRAPP II complex subunit |
| TRAP_alpha | 1 | Translocon-associated protein (TRAP), alpha subunit |
| TRAP_beta | 1 | Translocon-associated protein beta (TRAPB) |
| TRAUB | 1 | Apoptosis-antagonizing transcription factor, C-terminal |
| TRH | 1 | Thyrotropin-releasing hormone (TRH) |
| TRIF-NTD | 1 | TRIF N-terminal domain |
| TRIQK | 1 | Triple QxxK/R motif-containing protein family |
| TRM13 | 1 | Methyltransferase TRM13 |
| TSC21 | 1 | TSC21 family |
| TSGA13 | 1 | Testis-specific gene 13 protein |
| TSKS | 1 | Testis-specific serine kinase substrate |
| TSLP | 1 | Thymic stromal lymphopoietin |
| TSR | 1 | Thrombospondin type 1 repeat |
| TSSC4 | 1 | Tumour suppressing sub-chromosomal transferable candidate 4 |
| TTC5_OB | 1 | Tetratricopeptide repeat protein 5 OB fold domain |
| TTI1 | 1 | TELO2-interacting protein 1 |
| TUG-UBL1 | 1 | TUG ubiquitin-like domain |
| TUSC2 | 1 | Tumour suppressor candidate 2 |
| TUTF7_u4 | 1 | Unstructured region 4 on terminal uridylyltransferase 7 |
| TXD17-like_Trx | 1 | Thioredoxin domain-containing protein 17-like, thioredoxin domain |
| TYW3 | 1 | Methyltransferase TYW3 |
| T_cell_tran_alt | 1 | T-cell leukemia translocation-altered |
| Tachykinin | 1 | Tachykinin family |
| Takusan | 1 | Takusan |
| Tam41_Mmp37 | 1 | Phosphatidate cytidylyltransferase, mitochondrial |
| Tau95 | 1 | RNA polymerase III transcription factor (TF)IIIC subunit HTH domain |
| Tau95_N | 1 | Tau95 Triple barrel domain |
| Tcell_CD4_C | 1 | T cell CD4 receptor C terminal region |
| Tcf25 | 1 | Transcriptional repressor TCF25 |
| Tcp10_C | 1 | T-complex protein 10 C-terminus |
| TdIF1_2nd | 1 | TdIF1, C-terminal |
| Telethonin | 1 | Telethonin protein |
| Telomerase_RBD | 1 | Telomerase ribonucleoprotein complex - RNA binding domain |
| Telomere_reg-2 | 1 | Telomere length regulation protein |
| Ten1_2 | 1 | Telomere-capping, CST complex subunit |
| Tex55 | 1 | Testis-specific expressed protein 55 |
| Tex_N | 1 | Tex-like protein N-terminal domain |
| Tex_YqgF | 1 | Tex protein YqgF-like domain |
| Tfb2 | 1 | Transcription factor Tfb2 |
| Tfb2_C | 1 | Transcription factor Tfb2 (p52) C-terminal domain |
| Tfb4 | 1 | Transcription factor Tfb4 |
| Tfb5 | 1 | Transcription factor TFIIH complex subunit Tfb5 |
| Thg1 | 1 | tRNAHis guanylyltransferase |
| Thg1C | 1 | Thg1 C terminal domain |
| Thi4 | 1 | Thi4 family |
| ThiS | 1 | ThiS family |
| Thioredox_DsbH | 1 | Protein of unknown function, DUF255 |
| Thioredoxin_16 | 1 | Thioredoxin-like domain |
| Tho2 | 1 | Transcription factor/nuclear export subunit protein 2 |
| Thoc2 | 1 | Transcription- and export-related complex subunit |
| Thrombin_light | 1 | Thrombin light chain |
| Thymidylat_synt | 1 | Thymidylate synthase |
| Thymopoietin | 1 | Thymopoietin protein |
| Tim29 | 1 | Translocase of the Inner Mitochondrial membrane 29 |
| Titin_Z | 1 | Titin Z |
| Tlr3_TMD | 1 | Toll-like receptor 3 trans-membrane domain |
| Tma16 | 1 | Translation machinery-associated protein 16 |
| TmcA_N | 1 | tRNA(Met) cytidine acetyltransferase TmcA, N-terminal |
| Tme5_EGF_like | 1 | Thrombomodulin like fifth domain, EGF-like |
| Tmem26 | 1 | Transmembrane protein 26 |
| Tmemb_18A | 1 | Transmembrane protein 188 |
| Tmemb_40 | 1 | Predicted membrane protein |
| Tmpp129 | 1 | Putative transmembrane protein precursor |
| Tnp_22_dsRBD | 1 | L1 transposable element dsRBD-like domain |
| Tnp_P_element | 1 | Transposable element P transposase-like, HTH domain |
| Tnp_zf-ribbon_2 | 1 | DDE_Tnp_1-like zinc-ribbon |
| Tom22 | 1 | Mitochondrial import receptor subunit Tom22 |
| Tom5 | 1 | Mitochondrial import receptor subunit or translocase |
| Tom7 | 1 | TOM7 family |
| TopBP1_BRCT0 | 1 | TopBP1, BRCT0 domain |
| Tower | 1 | Tower |
| Tra1_central | 1 | Tra1 HEAT repeat central region |
| Tra1_ring | 1 | Tra1 HEAT repeat ring region |
| Transcrip_act | 1 | Transcriptional activator |
| Transcrip_reg | 1 | TACO1/YebC second and third domain |
| Transglut_core2 | 1 | Transglutaminase-like superfamily |
| Transposase_1 | 1 | Transposase (partial DDE domain) |
| Transposase_22 | 1 | L1 transposable element RBD-like domain |
| Transthyretin | 1 | HIUase/Transthyretin family |
| Treacle | 1 | Treacher Collins syndrome protein Treacle |
| Trehalase | 1 | Trehalase |
| Treslin_M | 1 | Treslin, M domain |
| Trimer_CC | 1 | Trimerisation motif |
| TrkA_TMD | 1 | Tyrosine kinase receptor A trans-membrane domain |
| TrmE_N | 1 | GTP-binding protein TrmE N-terminus |
| TrmO | 1 | tRNA-methyltransferase O |
| Troponin-I_N | 1 | Troponin I residues 1-32 |
| Trp_dioxygenase | 1 | Tryptophan 2,3-dioxygenase |
| TruB_C_2 | 1 | tRNA pseudouridylate synthase B C-terminal domain |
| Tsc35 | 1 | Testis-specific protein 35 |
| Tsg | 1 | Twisted gastrulation (Tsg) protein conserved region |
| Tti2 | 1 | Tti2 family |
| Tuberin | 1 | Tuberin |
| Tubulin_3 | 1 | Tubulin domain |
| Tudor_3 | 1 | DNA repair protein Crb2 Tudor domain |
| Tudor_4 | 1 | Histone methyltransferase Tudor domain |
| Tudor_5 | 1 | Histone methyltransferase Tudor domain 1 |
| Tyr-DNA_phospho | 1 | Tyrosyl-DNA phosphodiesterase |
| U1snRNP70_N | 1 | U1 small nuclear ribonucleoprotein of 70kDa MW N terminal |
| U3_assoc_6 | 1 | U3 small nucleolar RNA-associated protein 6 |
| U3snoRNP10 | 1 | U3 small nucleolar RNA-associated protein 10 |
| U5_2-snRNA_bdg | 1 | U5-snRNA binding site 2 of PrP8 |
| U6-snRNA_bdg | 1 | U6-snRNA interacting domain of PrP8 |
| UAE_UbL | 1 | Ubiquitin/SUMO-activating enzyme ubiquitin-like domain |
| UBA2_C | 1 | SUMO-activating enzyme subunit 2 C-terminus |
| UBA_5 | 1 | UBA domain |
| UBZ_FAAP20 | 1 | Ubiquitin-binding zinc-finger |
| UCH_1 | 1 | Ubiquitin carboxyl-terminal hydrolase |
| UCMA | 1 | Unique cartilage matrix associated protein |
| UCR_14kD | 1 | Ubiquinol-cytochrome C reductase complex 14kD subunit |
| UCR_6-4kD | 1 | Ubiquinol-cytochrome C reductase complex, 6.4kD protein |
| UCR_TM | 1 | Ubiquinol cytochrome reductase transmembrane region |
| UCR_UQCRX_QCR9 | 1 | Ubiquinol-cytochrome C reductase, UQCRX/QCR9 like |
| UDPG_MGDP_dh | 1 | UDP-glucose/GDP-mannose dehydrogenase family, central domain |
| UDPG_MGDP_dh_C | 1 | UDP-glucose/GDP-mannose dehydrogenase family, UDP binding domain |
| UDPG_MGDP_dh_N | 1 | UDP-glucose/GDP-mannose dehydrogenase family, NAD binding domain |
| UFC1 | 1 | Ubiquitin-fold modifier-conjugating enzyme 1 |
| UFD1 | 1 | Ubiquitin fusion degradation protein UFD1 |
| UME | 1 | UME (NUC010) domain |
| UMP1 | 1 | Proteasome maturation factor UMP1 |
| UNC-50 | 1 | UNC-50 family |
| UNC-79 | 1 | Cation-channel complex subunit UNC-79 |
| UNC119_bdg | 1 | UNC119-binding protein C5orf30 homologue |
| UNC80 | 1 | Protein UNC80 central region |
| UNC80_C | 1 | Protein UNC80 C-terminal region |
| UNC80_N | 1 | UNC80 N-terminal |
| UN_NPL4 | 1 | Nuclear pore localisation protein NPL4 |
| UPF0016 | 1 | Uncharacterized protein family UPF0016 |
| UPF0029 | 1 | Uncharacterized protein family UPF0029 |
| UPF0113 | 1 | UPF0113 PUA domain |
| UPF0113_N | 1 | UPF0113 Pre-PUA domain |
| UPF0160 | 1 | Uncharacterised protein family (UPF0160) |
| UPF0176_N | 1 | UPF0176 acylphosphatase like domain |
| UPF0184 | 1 | Uncharacterised protein family (UPF0184) |
| UPF0193 | 1 | Uncharacterised protein family (UPF0193) |
| UPF0203 | 1 | Uncharacterised protein family (UPF0203) |
| UPF0239 | 1 | Uncharacterised protein family (UPF0239) |
| UPF0240 | 1 | Uncharacterised protein family (UPF0240) |
| UPF0444 | 1 | Transmembrane protein C12orf23, UPF0444 |
| UPF0449 | 1 | Uncharacterised protein family UPF0449 |
| UPF0489 | 1 | UPF0489 domain |
| UPF0492 | 1 | Uncharacterized protein family UPF0492 |
| UPF0515 | 1 | Uncharacterised protein UPF0515 |
| UPF0524 | 1 | UPF0524 of C3orf70 |
| UPF0542 | 1 | Uncharacterised protein family UPF0542 |
| UPF0547 | 1 | Uncharacterised protein family UPF0547 |
| UPF0556 | 1 | UPF0556 domain |
| UPF0561 | 1 | Uncharacterised protein family UPF0561 |
| UPF0640 | 1 | Uncharacterised protein family UPF0640 |
| UPF0669 | 1 | Putative cytokine, C6ORF120 |
| UPF0688 | 1 | UPF0688 family |
| UPF0697 | 1 | Uncharacterised protein family UPF0697 |
| UPF0728 | 1 | Uncharacterised protein family UPF0728 |
| UPF0767 | 1 | UPF0767 family |
| UPF1_1B_dom | 1 | RNA helicase UPF1, 1B domain |
| UPF1_Zn_bind | 1 | RNA helicase (UPF2 interacting domain) |
| UQCC2_CBP6 | 1 | Complex III assembly factor UQCC2/CBP6 |
| UQCC3 | 1 | Ubiquinol-cytochrome-c reductase complex assembly factor 3 |
| URO-D | 1 | Uroporphyrinogen decarboxylase (URO-D) |
| USH1C_N | 1 | Harmonin, N-terminal domain |
| USP19_linker | 1 | Linker region of USP19 deubiquitinase |
| USP47_C | 1 | Ubiquitin carboxyl-terminal hydrolase 47 C-terminal |
| USP7_ICP0_bdg | 1 | ICP0-binding domain of Ubiquitin-specific protease 7 |
| USP8_interact | 1 | USP8 interacting |
| UTP15_C | 1 | UTP15 C terminal |
| UTP20_C | 1 | U3 small nucleolar RNA-associated protein 20, C-terminal |
| UTP20_N | 1 | U3 small nucleolar RNA-associated protein 20, N-terminal |
| UVB_sens_prot | 1 | Vitamin B6 photo-protection and homoeostasis |
| UVSSA_N | 1 | UVSSA N-terminal domain |
| UXS1_N | 1 | UDP-glucuronate decarboxylase N-terminal |
| Ubie_methyltran | 1 | ubiE/COQ5 methyltransferase family |
| Ubiq-Cytc-red_N | 1 | Ubiquinol-cytochrome c reductase 8 kDa, N-terminal |
| Ubiq_cyt_C_chap | 1 | Ubiquinol-cytochrome C chaperone |
| Ubiquitin_4 | 1 | Ubiquitin-like domain |
| Ubiquitin_5 | 1 | Ubiquitin-like domain |
| UcrQ | 1 | UcrQ family |
| UfSP2_N | 1 | Ubiquitin-fold modifier 1 specific protease 2, N-terminal |
| Ufm1 | 1 | Ubiquitin fold modifier 1 protein |
| UnbV_ASPIC | 1 | ASPIC and UnbV |
| Upf2 | 1 | Up-frameshift suppressor 2 |
| Urb2 | 1 | Urb2/Npa2 family |
| Urm1 | 1 | Urm1 (Ubiquitin related modifier) |
| Urocanase | 1 | Urocanase Rossmann-like domain |
| Urocanase_C | 1 | Urocanase C-terminal domain |
| Urocanase_N | 1 | Urocanase N-terminal domain |
| Uroplakin_II | 1 | Uroplakin II |
| Urotensin_II | 1 | Urotensin II |
| Use1 | 1 | Membrane fusion protein Use1 |
| Uso1_p115_C | 1 | Uso1 / p115 like vesicle tethering protein, C terminal region |
| Uso1_p115_head | 1 | Uso1 / p115 like vesicle tethering protein, head region |
| Utp11 | 1 | Utp11 protein |
| Utp13 | 1 | Utp13 specific WD40 associated domain |
| Utp21 | 1 | Utp21 specific WD40 associated putative domain |
| Utp25_C | 1 | Utp25, C-terminal domain |
| UvrD_C | 1 | UvrD-like helicase C-terminal domain |
| UvrD_C_2 | 1 | UvrD-like helicase C-terminal domain |
| V-ATPase_H_C | 1 | V-ATPase subunit H |
| V-ATPase_H_N | 1 | V-ATPase subunit H |
| V-set_CD47 | 1 | CD47 immunoglobulin-like domain |
| V1R_C | 1 | Vasopressin V1 receptor, C-terminal |
| VAD1-2 | 1 | Vitamin A-deficiency (VAD) rat model signalling |
| VAS1_LD | 1 | V-type proton ATPase subunit S1, luminal domain |
| VATC | 1 | Vms1-associating treble clef domain |
| VCIP135_N | 1 | VCIP135 N-terminal |
| VEFS-Box | 1 | VEFS-Box of polycomb protein |
| VEGF_C | 1 | VEGF heparin-binding domain |
| VGLL4 | 1 | Transcription cofactor vestigial-like protein 4 |
| VGPC1_C | 1 | C-terminal membrane-localisation domain of ion-channel, VCN1 |
| VHL_C | 1 | VHL box domain |
| VIGSSK | 1 | Helicase-associated putative binding domain, C-terminal |
| VIR_N | 1 | Virilizer, N-terminal |
| VOMI | 1 | Vitelline membrane outer layer protein I (VOMI) |
| VPS11_C | 1 | Vacuolar protein sorting protein 11 C terminal |
| VPS28 | 1 | VPS28 protein |
| VPS53_C | 1 | Vacuolar protein sorting-associated protein 53 C-terminus |
| VRR_NUC | 1 | VRR-NUC domain |
| VWA_N2 | 1 | VWA N-terminal |
| Vac14_Fig4_bd | 1 | Vacuolar protein 14 C-terminal Fig4p binding |
| Vac_ImportDeg | 1 | Vacuolar import and degradation protein |
| Vault | 1 | Major Vault Protein repeat domain |
| Vault_2 | 1 | Major Vault Protein repeat domain |
| Vault_3 | 1 | Major Vault Protein Repeat domain |
| Vault_4 | 1 | Major Vault Protein repeat domain |
| Vert_IL3-reg_TF | 1 | Vertebrate interleukin-3 regulated transcription factor |
| Vexin | 1 | Vexin domain |
| Vezatin | 1 | Mysoin-binding motif of peroxisomes |
| VitD-bind_III | 1 | Vitamin D binding protein, domain III |
| Vit_open_b-sht | 1 | Vitellinogen, open beta-sheet |
| Vma12 | 1 | Endoplasmic reticulum-based factor for assembly of V-ATPase |
| Vma22_CCDC115 | 1 | Vma22/CCDC115 |
| Voldacs | 1 | Regulator of volume decrease after cellular swelling |
| Vps16_N | 1 | Vps16, N-terminal region |
| Vps23_core | 1 | Vps23 core domain |
| Vps35 | 1 | Vacuolar protein sorting-associated protein 35 |
| Vps36_ESCRT-II | 1 | Vacuolar protein sorting protein 36 Vps36 |
| Vps52_C | 1 | Vps52, C-terminal |
| Vps52_CC | 1 | Vps52/Sac2 family, coiled-coil |
| Vps53_N | 1 | Vps53, N-terminal |
| Vps54 | 1 | Vps54-like protein |
| Vps8 | 1 | Golgi CORVET complex core vacuolar protein 8 |
| Vta1 | 1 | Vta1 like |
| Vta1_C | 1 | Vta1 C-terminal domain |
| WAPL | 1 | Wings apart-like protein regulation of heterochromatin |
| WASH-4_N | 1 | WASH complex subunit 4, N-terminal |
| WASH-7_C | 1 | WASH complex subunit 7, C-terminal |
| WASH-7_mid | 1 | WASH complex subunit 7 |
| WBS28 | 1 | Williams-Beuren syndrome chromosomal region 28 protein homologue |
| WBS_methylT | 1 | Methyltransferase involved in Williams-Beuren syndrome |
| WC-rich | 1 | Wolframin cysteine-rich domain |
| WCOB | 1 | Wolframin C-terminal OB-fold domain |
| WD40_3 | 1 | WD domain, G-beta repeat |
| WD40_alt | 1 | Alternative WD40 repeat motif |
| WDCP | 1 | WD repeat and coiled-coil-containing protein family |
| WDR54 | 1 | WDR54 |
| WDR93 | 1 | WD repeat-containing protein 93 |
| WEF-hand | 1 | Wolframin EF-hand domain |
| WGG | 1 | Pre-rRNA-processing protein TSR2 |
| WKF | 1 | WKF domain |
| WRW | 1 | Mitochondrial F1F0-ATP synthase, subunit f |
| WSLR | 1 | Wolframin Sel1-like repeat |
| WT1 | 1 | Wilm's tumour protein |
| WW_FCH_linker | 1 | Unstructured linker region between on GAS7 protein |
| WW_USP8 | 1 | USP8 WW domain |
| Wbp11 | 1 | WW domain binding protein 11 |
| Wheel | 1 | Cns1/TTC4 Wheel domain |
| Wtap | 1 | WTAP/Mum2p family |
| XAF1_C | 1 | XIAP-associated factor 1 C-terminal domain |
| XLF | 1 | XLF N-terminal domain |
| XPA_C | 1 | XPA protein C-terminus |
| XPA_N | 1 | XPA protein N-terminal |
| XRCC4 | 1 | XRCC4 N-terminal domain |
| XRN1_D1 | 1 | Exoribonuclease Xrn1 D1 domain |
| XRN1_D2_D3 | 1 | Exoribonuclease Xrn1 D2/D3 domain |
| XendoU | 1 | Endoribonuclease XendoU |
| YL1 | 1 | YL1 nuclear protein |
| YbeY | 1 | Endoribonuclease YbeY |
| YchF-GTPase_C | 1 | Protein of unknown function (DUF933) |
| YdjC | 1 | YdjC-like protein |
| Ydr279_N | 1 | Ydr279p protein triple barrel domain |
| YegS_C | 1 | YegS C-terminal NAD kinase beta sandwich-like domain |
| Yos1 | 1 | Yos1-like |
| YqeH-like_C | 1 | YqeH-like, C-terminal |
| YqgF | 1 | Holliday-junction resolvase-like of SPT6 |
| Ysc84 | 1 | Las17-binding protein actin regulator |
| ZFYVE21_C | 1 | Zinc finger FYVE domain-containing protein 21 C-terminus |
| ZGRF1-like_N | 1 | DNA helicase ZGRF1-like, N-terminal domain |
| ZNF512_zf-C2H2 | 1 | Zinc finger 512, zinc fingers |
| ZNHIT3_C | 1 | Zinc finger HIT domain-containing protein 3, C-terminal domain |
| ZSWIM3_N | 1 | Zinc finger SWIM domain-containing protein 3, N-terminal |
| ZW10_C | 1 | Centromere/kinetochore protein zw10, C-terminal |
| Zmiz1_N | 1 | Zmiz1 N-terminal tetratricopeptide repeat domain |
| Zn_dep_PLPC | 1 | Zinc dependent phospholipase C |
| Zw10_N | 1 | Centromere/kinetochore Zw10 N-terminal |
| Zw10_middle | 1 | Centromere/kinetochore protein zw10, middle domain |
| Zwilch | 1 | RZZ complex, subunit zwilch |
| Zwint | 1 | ZW10 interactor |
| a_DG1_N2 | 1 | Alpha-Dystroglycan N-terminal domain 2 |
| bVLRF1 | 1 | Bacteroidetes VLRF1 release factor |
| bcl-2I13 | 1 | Bcl2-interacting killer, BH3-domain containing |
| cwf18 | 1 | cwf18 pre-mRNA splicing factor |
| dUTPase | 1 | dUTPase |
| dbPDZ_assoc | 1 | Unstructured region between two PDZ domains on Dlg5 |
| dsDNA_bind | 1 | Double-stranded DNA-binding domain |
| eEFSec_4th | 1 | eEFSec C-terminal RIFT domain |
| eIF-3_zeta | 1 | Eukaryotic translation initiation factor 3 subunit 7 (eIF-3) |
| eIF-6 | 1 | eIF-6 family |
| eIF3_N | 1 | eIF3 subunit 6 N terminal domain |
| eIF3_p135 | 1 | Translation initiation factor eIF3 subunit 135 |
| eIF3_subunit | 1 | Translation initiation factor eIF3 subunit |
| eIF3g | 1 | Eukaryotic translation initiation factor 3 subunit G |
| eIF3h_C | 1 | C-terminal region of eIF3h |
| eIF3m_C_helix | 1 | eIF3 subunit M, C-terminal helix |
| ecTbetaR2 | 1 | Transforming growth factor beta receptor 2 ectodomain |
| efThoc1 | 1 | THO complex subunit 1 transcription elongation factor |
| euk_SelB_III | 1 | Selenocysteine-specific elongation factor, eukaryotes, 3rd domain |
| gag-asp_proteas | 1 | gag-polyprotein putative aspartyl protease |
| hDGE_amylase | 1 | Glycogen debranching enzyme, glucanotransferase domain |
| hGDE_N | 1 | N-terminal domain from the human glycogen debranching enzyme |
| hGDE_central | 1 | Central domain of human glycogen debranching enzyme |
| hNIFK_binding | 1 | FHA Ki67 binding domain of hNIFK |
| mRNA_G-N7_MeTrfase | 1 | mRNA (guanine-N(7))-methyltransferase domain |
| mRNA_cap_C | 1 | mRNA capping enzyme, C-terminal domain |
| mRNA_cap_enzyme | 1 | mRNA capping enzyme, catalytic domain |
| mTOR_dom | 1 | Serine/threonine-protein kinase mTOR domain |
| mit_SMPDase | 1 | Mitochondrial-associated sphingomyelin phosphodiesterase |
| name | 1 | name |
| names | 1 | names |
| p31comet | 1 | Mad1 and Cdc20-bound-Mad2 binding |
| p47_phox_C | 1 | NADPH oxidase subunit p47Phox, C terminal domain |
| p53-inducible11 | 1 | Tumour protein p53-inducible protein 11 |
| rRNA_processing | 1 | rRNA processing |
| ribosomal_L24 | 1 | Ribosomal proteins 50S L24/mitochondrial 39S L24 |
| rve | 1 | Integrase core domain |
| stn_TNFRSF12A | 1 | Tumour necrosis factor receptor stn_TNFRSF12A_TNFR domain |
| tRNA-synt_1_2 | 1 | Leucyl-tRNA synthetase, editing domain |
| tRNA_Me_trans | 1 | tRNA methyl transferase HUP domain |
| tRNA_Me_trans_C | 1 | Aminomethyltransferase beta-barrel domain |
| tRNA_Me_trans_M | 1 | tRNA methyl transferase PRC-barrel domain |
| tRNA_bind_2 | 1 | Possible tRNA binding domain |
| tRNA_int_end_N2 | 1 | tRNA-splicing endonuclease subunit sen54 N-term |
| tRNA_int_endo_N | 1 | tRNA intron endonuclease, N-terminal domain |
| tRNA_synt_1c_R1 | 1 | Glutaminyl-tRNA synthetase, non-specific RNA binding region part 1 |
| tRNA_synt_1c_R2 | 1 | Glutaminyl-tRNA synthetase, non-specific RNA binding region part 2 |
| tRNA_synthFbeta | 1 | Phenylalanyl tRNA synthetase beta chain CLM domain |
| zf-3CxxC_2 | 1 | Zinc-binding domain |
| zf-ACC | 1 | Acetyl-CoA carboxylase zinc finger domain |
| zf-C2H2_10 | 1 | C2H2 zinc-finger |
| zf-C2H2_8 | 1 | C2H2-type zinc ribbon |
| zf-C2H2_assoc | 1 | Unstructured conserved, between two C2H2-type zinc-fingers |
| zf-C2H2_assoc2 | 1 | Unstructured region upstream of a zinc-finger |
| zf-C2H2_assoc3 | 1 | Putative zinc-finger between two C2H2 zinc-fingers on Patz |
| zf-C2HC5 | 1 | Putative zinc finger motif, C2HC5-type |
| zf-C2HE | 1 | C2HE / C2H2 / C2HC zinc-binding finger |
| zf-C3H1 | 1 | Putative zinc-finger domain |
| zf-C3HC | 1 | C3HC zinc finger-like |
| zf-C3HC4_5 | 1 | Zinc finger, C3HC4 type (RING finger) |
| zf-C4H2 | 1 | Zinc finger-containing protein |
| zf-C4_Topoisom | 1 | Topoisomerase DNA binding C4 zinc finger |
| zf-C6H2 | 1 | zf-MYND-like zinc finger, mRNA-binding |
| zf-CCCH_8 | 1 | Zinc-finger antiviral protein (ZAP) zinc finger domain 3 |
| zf-CCHH | 1 | PBZ domain |
| zf-CHCC | 1 | Zinc-finger domain |
| zf-CHY | 1 | CHY zinc finger |
| zf-CRD | 1 | Cysteine rich domain with multizinc binding regions |
| zf-DNA_Pol | 1 | DNA Polymerase alpha zinc finger |
| zf-DNL | 1 | DNL zinc finger |
| zf-FPG_IleRS | 1 | Zinc finger found in FPG and IleRS |
| zf-LYAR | 1 | LYAR-type C2HC zinc finger |
| zf-NOSIP | 1 | Zinc-finger of nitric oxide synthase-interacting protein |
| zf-NPL4 | 1 | NPL4 family, putative zinc binding region |
| zf-Nse | 1 | Zinc-finger of the MIZ type in Nse subunit |
| zf-RAG1 | 1 | Recombination-activating protein 1 zinc-finger domain |
| zf-RING-like | 1 | RING-like domain |
| zf-RING_10 | 1 | zinc RING finger of MSL2 |
| zf-RING_12 | 1 | RING/Ubox like zinc-binding domain |
| zf-RING_14 | 1 | RING/Ubox like zinc-binding domain |
| zf-RING_15 | 1 | KIAA1045 RING finger |
| zf-RING_16 | 1 | RING/Ubox like zinc-binding domain |
| zf-RING_4 | 1 | RING/Ubox like zinc-binding domain |
| zf-RING_6 | 1 | zf-RING of BARD1-type protein |
| zf-RRN7 | 1 | Zinc-finger of RNA-polymerase I-specific TFIIB, Rrn7 |
| zf-SCNM1 | 1 | Zinc-finger of sodium channel modifier 1 |
| zf-TFIIIC | 1 | Putative zinc-finger of transcription factor IIIC complex |
| zf-TRM13_CCCH | 1 | CCCH zinc finger in TRM13 protein |
| zf-WRNIP1_ubi | 1 | Werner helicase-interacting protein 1 ubiquitin-binding domain |
| zf-ZPR1 | 1 | ZPR1 zinc-finger domain |
| zf-primase | 1 | Primase zinc finger |
| zf-tcix | 1 | Putative treble-clef, zinc-finger, Zn-binding |
| zf_C2H2_6 | 1 | Zinc Finger domain |
| zf_PR_Knuckle | 1 | PR zinc knuckle motif |
| zf_UBZ | 1 | Ubiquitin-Binding Zinc Finger |
| zinc_ribbon_10 | 1 | Predicted integral membrane zinc-ribbon metal-binding protein |
| zinc_ribbon_16 | 1 | Zinc-ribbon like family |
| zinc_ribbon_2 | 1 | zinc-ribbon domain |
| zinc_ribbon_6 | 1 | Zinc-ribbon |
| zinc_ribbon_9 | 1 | zinc-ribbon |
| zn-ribbon_14 | 1 | Zinc-ribbon |
Format: map
Web label: Protein domains
Web label: key| (|count|): |desc
| Filename | Size | md5 |
|---|---|---|
| PFAMdataprep.py | 1.24 KB | 877182d5106bab82fd361d9112cac832 |
| Pfam-A.clans.xlsx | 977.25 KB | a788714d5cf6b12ca575c78db5c248a1 |
| domain-map.txt | 388.88 KB | 5a6886188467d28a85d4b5a91633169e |
| domain-mapnames.txt | 934.78 KB | 61147b89e31e0b0dbf06da8fa99f6b75 |
| genomic_resource.yaml | 1.93 KB | 282d4ac6c357238ef8800ff6d74ff02b |
| mart_export.xlsx | 603.91 KB | a28ced2cbc46699dbf66499fee45f534 |
| statistics/ |