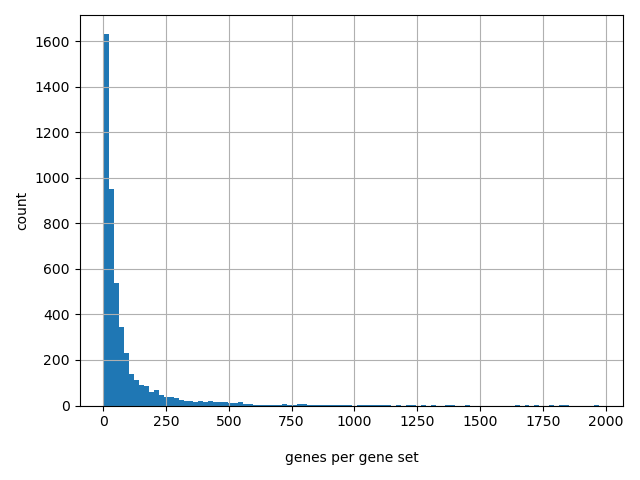
Count of genes per gene set

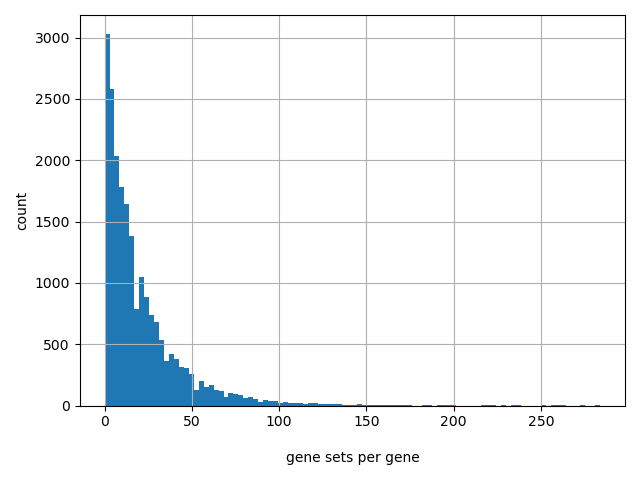
Count of gene sets per gene

| Id: | gene_properties/gene_sets/MSigDB_curated |
| Type: | gene_set_collection |
| Version: | 0 |
| Summary: |
MSigDB (Molecular Signatures Database) gene sets derived from a variety of curated sources |
| Description: |
N/A
|
| Labels: |
Number of gene sets: 4722
Number of unique genes: 21047


| Gene Set | Gene Count | Description |
|---|---|---|
| PILON_KLF1_TARGETS_DN | 1972 | http://www.broadinstitute.org/gsea/msigdb/cards/PILON_KLF1_TARGETS_DN |
| ZWANG_TRANSIENTLY_UP_BY_1ST_EGF_PULSE_ONLY | 1839 | http://www.broadinstitute.org/gsea/msigdb/cards/ZWANG_TRANSIENTLY_UP_BY_1ST_EGF_PULSE_ONLY |
| DODD_NASOPHARYNGEAL_CARCINOMA_UP | 1821 | http://www.broadinstitute.org/gsea/msigdb/cards/DODD_NASOPHARYNGEAL_CARCINOMA_UP |
| GRAESSMANN_APOPTOSIS_BY_DOXORUBICIN_DN | 1781 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAESSMANN_APOPTOSIS_BY_DOXORUBICIN_DN |
| ZWANG_TRANSIENTLY_UP_BY_2ND_EGF_PULSE_ONLY | 1725 | http://www.broadinstitute.org/gsea/msigdb/cards/ZWANG_TRANSIENTLY_UP_BY_2ND_EGF_PULSE_ONLY |
| BLALOCK_ALZHEIMERS_DISEASE_UP | 1691 | http://www.broadinstitute.org/gsea/msigdb/cards/BLALOCK_ALZHEIMERS_DISEASE_UP |
| PUJANA_BRCA1_PCC_NETWORK | 1652 | http://www.broadinstitute.org/gsea/msigdb/cards/PUJANA_BRCA1_PCC_NETWORK |
| PUJANA_ATM_PCC_NETWORK | 1442 | http://www.broadinstitute.org/gsea/msigdb/cards/PUJANA_ATM_PCC_NETWORK |
| DIAZ_CHRONIC_MEYLOGENOUS_LEUKEMIA_UP | 1382 | http://www.broadinstitute.org/gsea/msigdb/cards/DIAZ_CHRONIC_MEYLOGENOUS_LEUKEMIA_UP |
| DODD_NASOPHARYNGEAL_CARCINOMA_DN | 1375 | http://www.broadinstitute.org/gsea/msigdb/cards/DODD_NASOPHARYNGEAL_CARCINOMA_DN |
| YOSHIMURA_MAPK8_TARGETS_UP | 1305 | http://www.broadinstitute.org/gsea/msigdb/cards/YOSHIMURA_MAPK8_TARGETS_UP |
| KINSEY_TARGETS_OF_EWSR1_FLII_FUSION_UP | 1278 | http://www.broadinstitute.org/gsea/msigdb/cards/KINSEY_TARGETS_OF_EWSR1_FLII_FUSION_UP |
| BLALOCK_ALZHEIMERS_DISEASE_DN | 1237 | http://www.broadinstitute.org/gsea/msigdb/cards/BLALOCK_ALZHEIMERS_DISEASE_DN |
| MARSON_BOUND_BY_FOXP3_UNSTIMULATED | 1229 | http://www.broadinstitute.org/gsea/msigdb/cards/MARSON_BOUND_BY_FOXP3_UNSTIMULATED |
| CHEN_METABOLIC_SYNDROM_NETWORK | 1210 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEN_METABOLIC_SYNDROM_NETWORK |
| PEREZ_TP53_TARGETS | 1174 | http://www.broadinstitute.org/gsea/msigdb/cards/PEREZ_TP53_TARGETS |
| GRAESSMANN_APOPTOSIS_BY_DOXORUBICIN_UP | 1142 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAESSMANN_APOPTOSIS_BY_DOXORUBICIN_UP |
| BRUINS_UVC_RESPONSE_LATE | 1137 | http://www.broadinstitute.org/gsea/msigdb/cards/BRUINS_UVC_RESPONSE_LATE |
| BENPORATH_ES_WITH_H3K27ME3 | 1118 | http://www.broadinstitute.org/gsea/msigdb/cards/BENPORATH_ES_WITH_H3K27ME3 |
| DANG_BOUND_BY_MYC | 1103 | http://www.broadinstitute.org/gsea/msigdb/cards/DANG_BOUND_BY_MYC |
| GOBERT_OLIGODENDROCYTE_DIFFERENTIATION_DN | 1080 | http://www.broadinstitute.org/gsea/msigdb/cards/GOBERT_OLIGODENDROCYTE_DIFFERENTIATION_DN |
| MEISSNER_BRAIN_HCP_WITH_H3K4ME3_AND_H3K27ME3 | 1069 | http://www.broadinstitute.org/gsea/msigdb/cards/MEISSNER_BRAIN_HCP_WITH_H3K4ME3_AND_H3K27ME3 |
| BENPORATH_EED_TARGETS | 1062 | http://www.broadinstitute.org/gsea/msigdb/cards/BENPORATH_EED_TARGETS |
| BENPORATH_SUZ12_TARGETS | 1038 | http://www.broadinstitute.org/gsea/msigdb/cards/BENPORATH_SUZ12_TARGETS |
| NUYTTEN_EZH2_TARGETS_UP | 1037 | http://www.broadinstitute.org/gsea/msigdb/cards/NUYTTEN_EZH2_TARGETS_UP |
| NUYTTEN_EZH2_TARGETS_DN | 1024 | http://www.broadinstitute.org/gsea/msigdb/cards/NUYTTEN_EZH2_TARGETS_DN |
| MARSON_BOUND_BY_FOXP3_STIMULATED | 1022 | http://www.broadinstitute.org/gsea/msigdb/cards/MARSON_BOUND_BY_FOXP3_STIMULATED |
| KRIGE_RESPONSE_TO_TOSEDOSTAT_24HR_DN | 1011 | http://www.broadinstitute.org/gsea/msigdb/cards/KRIGE_RESPONSE_TO_TOSEDOSTAT_24HR_DN |
| BENPORATH_NANOG_TARGETS | 988 | http://www.broadinstitute.org/gsea/msigdb/cards/BENPORATH_NANOG_TARGETS |
| ACEVEDO_LIVER_CANCER_UP | 973 | http://www.broadinstitute.org/gsea/msigdb/cards/ACEVEDO_LIVER_CANCER_UP |
| LOPEZ_MBD_TARGETS | 957 | http://www.broadinstitute.org/gsea/msigdb/cards/LOPEZ_MBD_TARGETS |
| KRIGE_RESPONSE_TO_TOSEDOSTAT_6HR_UP | 953 | http://www.broadinstitute.org/gsea/msigdb/cards/KRIGE_RESPONSE_TO_TOSEDOSTAT_6HR_UP |
| ACEVEDO_METHYLATED_IN_LIVER_CANCER_DN | 940 | http://www.broadinstitute.org/gsea/msigdb/cards/ACEVEDO_METHYLATED_IN_LIVER_CANCER_DN |
| REACTOME_IMMUNE_SYSTEM | 933 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_IMMUNE_SYSTEM |
| REACTOME_SIGNALING_BY_GPCR | 920 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_GPCR |
| JOHNSTONE_PARVB_TARGETS_3_DN | 918 | http://www.broadinstitute.org/gsea/msigdb/cards/JOHNSTONE_PARVB_TARGETS_3_DN |
| KRIGE_RESPONSE_TO_TOSEDOSTAT_6HR_DN | 911 | http://www.broadinstitute.org/gsea/msigdb/cards/KRIGE_RESPONSE_TO_TOSEDOSTAT_6HR_DN |
| BRUINS_UVC_RESPONSE_VIA_TP53_GROUP_A | 898 | http://www.broadinstitute.org/gsea/msigdb/cards/BRUINS_UVC_RESPONSE_VIA_TP53_GROUP_A |
| GEORGES_TARGETS_OF_MIR192_AND_MIR215 | 893 | http://www.broadinstitute.org/gsea/msigdb/cards/GEORGES_TARGETS_OF_MIR192_AND_MIR215 |
| BYSTRYKH_HEMATOPOIESIS_STEM_CELL_QTL_TRANS | 882 | http://www.broadinstitute.org/gsea/msigdb/cards/BYSTRYKH_HEMATOPOIESIS_STEM_CELL_QTL_TRANS |
| LEE_BMP2_TARGETS_DN | 882 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_BMP2_TARGETS_DN |
| WAKABAYASHI_ADIPOGENESIS_PPARG_RXRA_BOUND_8D | 882 | http://www.broadinstitute.org/gsea/msigdb/cards/WAKABAYASHI_ADIPOGENESIS_PPARG_RXRA_BOUND_8D |
| GRADE_COLON_CANCER_UP | 871 | http://www.broadinstitute.org/gsea/msigdb/cards/GRADE_COLON_CANCER_UP |
| ACEVEDO_LIVER_TUMOR_VS_NORMAL_ADJACENT_TISSUE_UP | 863 | http://www.broadinstitute.org/gsea/msigdb/cards/ACEVEDO_LIVER_TUMOR_VS_NORMAL_ADJACENT_TISSUE_UP |
| MARTENS_TRETINOIN_RESPONSE_UP | 857 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTENS_TRETINOIN_RESPONSE_UP |
| DACOSTA_UV_RESPONSE_VIA_ERCC3_DN | 855 | http://www.broadinstitute.org/gsea/msigdb/cards/DACOSTA_UV_RESPONSE_VIA_ERCC3_DN |
| NUYTTEN_NIPP1_TARGETS_DN | 848 | http://www.broadinstitute.org/gsea/msigdb/cards/NUYTTEN_NIPP1_TARGETS_DN |
| MARTENS_TRETINOIN_RESPONSE_DN | 841 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTENS_TRETINOIN_RESPONSE_DN |
| CUI_TCF21_TARGETS_2_DN | 830 | http://www.broadinstitute.org/gsea/msigdb/cards/CUI_TCF21_TARGETS_2_DN |
| KOINUMA_TARGETS_OF_SMAD2_OR_SMAD3 | 824 | http://www.broadinstitute.org/gsea/msigdb/cards/KOINUMA_TARGETS_OF_SMAD2_OR_SMAD3 |
| BUYTAERT_PHOTODYNAMIC_THERAPY_STRESS_UP | 811 | http://www.broadinstitute.org/gsea/msigdb/cards/BUYTAERT_PHOTODYNAMIC_THERAPY_STRESS_UP |
| REACTOME_GPCR_DOWNSTREAM_SIGNALING | 805 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GPCR_DOWNSTREAM_SIGNALING |
| RODRIGUES_THYROID_CARCINOMA_POORLY_DIFFERENTIATED_DN | 805 | http://www.broadinstitute.org/gsea/msigdb/cards/RODRIGUES_THYROID_CARCINOMA_POORLY_DIFFERENTIATED_DN |
| LASTOWSKA_NEUROBLASTOMA_COPY_NUMBER_DN | 800 | http://www.broadinstitute.org/gsea/msigdb/cards/LASTOWSKA_NEUROBLASTOMA_COPY_NUMBER_DN |
| WEI_MYCN_TARGETS_WITH_E_BOX | 795 | http://www.broadinstitute.org/gsea/msigdb/cards/WEI_MYCN_TARGETS_WITH_E_BOX |
| FORTSCHEGGER_PHF8_TARGETS_DN | 784 | http://www.broadinstitute.org/gsea/msigdb/cards/FORTSCHEGGER_PHF8_TARGETS_DN |
| KRIGE_RESPONSE_TO_TOSEDOSTAT_24HR_UP | 783 | http://www.broadinstitute.org/gsea/msigdb/cards/KRIGE_RESPONSE_TO_TOSEDOSTAT_24HR_UP |
| ONKEN_UVEAL_MELANOMA_UP | 783 | http://www.broadinstitute.org/gsea/msigdb/cards/ONKEN_UVEAL_MELANOMA_UP |
| GOZGIT_ESR1_TARGETS_DN | 781 | http://www.broadinstitute.org/gsea/msigdb/cards/GOZGIT_ESR1_TARGETS_DN |
| PUJANA_CHEK2_PCC_NETWORK | 779 | http://www.broadinstitute.org/gsea/msigdb/cards/PUJANA_CHEK2_PCC_NETWORK |
| BENPORATH_MYC_MAX_TARGETS | 775 | http://www.broadinstitute.org/gsea/msigdb/cards/BENPORATH_MYC_MAX_TARGETS |
| GRAESSMANN_RESPONSE_TO_MC_AND_DOXORUBICIN_DN | 770 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAESSMANN_RESPONSE_TO_MC_AND_DOXORUBICIN_DN |
| KRIEG_HYPOXIA_NOT_VIA_KDM3A | 770 | http://www.broadinstitute.org/gsea/msigdb/cards/KRIEG_HYPOXIA_NOT_VIA_KDM3A |
| NUYTTEN_NIPP1_TARGETS_UP | 769 | http://www.broadinstitute.org/gsea/msigdb/cards/NUYTTEN_NIPP1_TARGETS_UP |
| KIM_ALL_DISORDERS_OLIGODENDROCYTE_NUMBER_CORR_UP | 756 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_ALL_DISORDERS_OLIGODENDROCYTE_NUMBER_CORR_UP |
| PATIL_LIVER_CANCER | 747 | http://www.broadinstitute.org/gsea/msigdb/cards/PATIL_LIVER_CANCER |
| LEE_BMP2_TARGETS_UP | 745 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_BMP2_TARGETS_UP |
| BENPORATH_SOX2_TARGETS | 734 | http://www.broadinstitute.org/gsea/msigdb/cards/BENPORATH_SOX2_TARGETS |
| MARSON_BOUND_BY_E2F4_UNSTIMULATED | 728 | http://www.broadinstitute.org/gsea/msigdb/cards/MARSON_BOUND_BY_E2F4_UNSTIMULATED |
| RODRIGUES_THYROID_CARCINOMA_ANAPLASTIC_UP | 722 | http://www.broadinstitute.org/gsea/msigdb/cards/RODRIGUES_THYROID_CARCINOMA_ANAPLASTIC_UP |
| WONG_ADULT_TISSUE_STEM_MODULE | 721 | http://www.broadinstitute.org/gsea/msigdb/cards/WONG_ADULT_TISSUE_STEM_MODULE |
| CREIGHTON_ENDOCRINE_THERAPY_RESISTANCE_3 | 720 | http://www.broadinstitute.org/gsea/msigdb/cards/CREIGHTON_ENDOCRINE_THERAPY_RESISTANCE_3 |
| MIKKELSEN_ES_ICP_WITH_H3K4ME3 | 718 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_ES_ICP_WITH_H3K4ME3 |
| SCHLOSSER_SERUM_RESPONSE_DN | 712 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHLOSSER_SERUM_RESPONSE_DN |
| SMID_BREAST_CANCER_BASAL_DN | 701 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_BASAL_DN |
| FEVR_CTNNB1_TARGETS_UP | 682 | http://www.broadinstitute.org/gsea/msigdb/cards/FEVR_CTNNB1_TARGETS_UP |
| KIM_BIPOLAR_DISORDER_OLIGODENDROCYTE_DENSITY_CORR_UP | 682 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_BIPOLAR_DISORDER_OLIGODENDROCYTE_DENSITY_CORR_UP |
| IVANOVA_HEMATOPOIESIS_STEM_CELL_AND_PROGENITOR | 681 | http://www.broadinstitute.org/gsea/msigdb/cards/IVANOVA_HEMATOPOIESIS_STEM_CELL_AND_PROGENITOR |
| MARTINEZ_RB1_TARGETS_UP | 673 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTINEZ_RB1_TARGETS_UP |
| MILI_PSEUDOPODIA_HAPTOTAXIS_DN | 668 | http://www.broadinstitute.org/gsea/msigdb/cards/MILI_PSEUDOPODIA_HAPTOTAXIS_DN |
| CASORELLI_ACUTE_PROMYELOCYTIC_LEUKEMIA_DN | 663 | http://www.broadinstitute.org/gsea/msigdb/cards/CASORELLI_ACUTE_PROMYELOCYTIC_LEUKEMIA_DN |
| WAKABAYASHI_ADIPOGENESIS_PPARG_BOUND_8D | 658 | http://www.broadinstitute.org/gsea/msigdb/cards/WAKABAYASHI_ADIPOGENESIS_PPARG_BOUND_8D |
| BENPORATH_PRC2_TARGETS | 652 | http://www.broadinstitute.org/gsea/msigdb/cards/BENPORATH_PRC2_TARGETS |
| BENPORATH_CYCLING_GENES | 648 | http://www.broadinstitute.org/gsea/msigdb/cards/BENPORATH_CYCLING_GENES |
| SMID_BREAST_CANCER_BASAL_UP | 648 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_BASAL_UP |
| BUYTAERT_PHOTODYNAMIC_THERAPY_STRESS_DN | 637 | http://www.broadinstitute.org/gsea/msigdb/cards/BUYTAERT_PHOTODYNAMIC_THERAPY_STRESS_DN |
| NAKAMURA_TUMOR_ZONE_PERIPHERAL_VS_CENTRAL_DN | 634 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAMURA_TUMOR_ZONE_PERIPHERAL_VS_CENTRAL_DN |
| RODRIGUES_THYROID_CARCINOMA_POORLY_DIFFERENTIATED_UP | 633 | http://www.broadinstitute.org/gsea/msigdb/cards/RODRIGUES_THYROID_CARCINOMA_POORLY_DIFFERENTIATED_UP |
| GRAESSMANN_RESPONSE_TO_MC_AND_DOXORUBICIN_UP | 612 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAESSMANN_RESPONSE_TO_MC_AND_DOXORUBICIN_UP |
| CAIRO_HEPATOBLASTOMA_CLASSES_UP | 605 | http://www.broadinstitute.org/gsea/msigdb/cards/CAIRO_HEPATOBLASTOMA_CLASSES_UP |
| MARTINEZ_TP53_TARGETS_UP | 602 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTINEZ_TP53_TARGETS_UP |
| MARTINEZ_RB1_AND_TP53_TARGETS_UP | 601 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTINEZ_RB1_AND_TP53_TARGETS_UP |
| MARTINEZ_TP53_TARGETS_DN | 593 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTINEZ_TP53_TARGETS_DN |
| MARTINEZ_RB1_AND_TP53_TARGETS_DN | 591 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTINEZ_RB1_AND_TP53_TARGETS_DN |
| MIKKELSEN_MEF_HCP_WITH_H3K27ME3 | 590 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_MEF_HCP_WITH_H3K27ME3 |
| HAMAI_APOPTOSIS_VIA_TRAIL_UP | 584 | http://www.broadinstitute.org/gsea/msigdb/cards/HAMAI_APOPTOSIS_VIA_TRAIL_UP |
| SPIELMAN_LYMPHOBLAST_EUROPEAN_VS_ASIAN_DN | 584 | http://www.broadinstitute.org/gsea/msigdb/cards/SPIELMAN_LYMPHOBLAST_EUROPEAN_VS_ASIAN_DN |
| FULCHER_INFLAMMATORY_RESPONSE_LECTIN_VS_LPS_UP | 579 | http://www.broadinstitute.org/gsea/msigdb/cards/FULCHER_INFLAMMATORY_RESPONSE_LECTIN_VS_LPS_UP |
| MASSARWEH_TAMOXIFEN_RESISTANCE_UP | 578 | http://www.broadinstitute.org/gsea/msigdb/cards/MASSARWEH_TAMOXIFEN_RESISTANCE_UP |
| CHICAS_RB1_TARGETS_SENESCENT | 572 | http://www.broadinstitute.org/gsea/msigdb/cards/CHICAS_RB1_TARGETS_SENESCENT |
| GOBERT_OLIGODENDROCYTE_DIFFERENTIATION_UP | 570 | http://www.broadinstitute.org/gsea/msigdb/cards/GOBERT_OLIGODENDROCYTE_DIFFERENTIATION_UP |
| CHICAS_RB1_TARGETS_CONFLUENT | 567 | http://www.broadinstitute.org/gsea/msigdb/cards/CHICAS_RB1_TARGETS_CONFLUENT |
| PARENT_MTOR_SIGNALING_UP | 567 | http://www.broadinstitute.org/gsea/msigdb/cards/PARENT_MTOR_SIGNALING_UP |
| DOUGLAS_BMI1_TARGETS_UP | 566 | http://www.broadinstitute.org/gsea/msigdb/cards/DOUGLAS_BMI1_TARGETS_UP |
| SMID_BREAST_CANCER_LUMINAL_B_DN | 564 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_LUMINAL_B_DN |
| TIEN_INTESTINE_PROBIOTICS_24HR_UP | 557 | http://www.broadinstitute.org/gsea/msigdb/cards/TIEN_INTESTINE_PROBIOTICS_24HR_UP |
| FLECHNER_BIOPSY_KIDNEY_TRANSPLANT_OK_VS_DONOR_UP | 555 | http://www.broadinstitute.org/gsea/msigdb/cards/FLECHNER_BIOPSY_KIDNEY_TRANSPLANT_OK_VS_DONOR_UP |
| FEVR_CTNNB1_TARGETS_DN | 553 | http://www.broadinstitute.org/gsea/msigdb/cards/FEVR_CTNNB1_TARGETS_DN |
| GRAESSMANN_APOPTOSIS_BY_SERUM_DEPRIVATION_UP | 552 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAESSMANN_APOPTOSIS_BY_SERUM_DEPRIVATION_UP |
| BRUINS_UVC_RESPONSE_VIA_TP53_GROUP_B | 549 | http://www.broadinstitute.org/gsea/msigdb/cards/BRUINS_UVC_RESPONSE_VIA_TP53_GROUP_B |
| KIM_ALL_DISORDERS_CALB1_CORR_UP | 548 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_ALL_DISORDERS_CALB1_CORR_UP |
| FLECHNER_BIOPSY_KIDNEY_TRANSPLANT_REJECTED_VS_OK_DN | 546 | http://www.broadinstitute.org/gsea/msigdb/cards/FLECHNER_BIOPSY_KIDNEY_TRANSPLANT_REJECTED_VS_OK_DN |
| WEST_ADRENOCORTICAL_TUMOR_DN | 546 | http://www.broadinstitute.org/gsea/msigdb/cards/WEST_ADRENOCORTICAL_TUMOR_DN |
| IVANOVA_HEMATOPOIESIS_LATE_PROGENITOR | 544 | http://www.broadinstitute.org/gsea/msigdb/cards/IVANOVA_HEMATOPOIESIS_LATE_PROGENITOR |
| ZHOU_INFLAMMATORY_RESPONSE_FIMA_UP | 544 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHOU_INFLAMMATORY_RESPONSE_FIMA_UP |
| MARTINEZ_RB1_TARGETS_DN | 543 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTINEZ_RB1_TARGETS_DN |
| ACEVEDO_LIVER_CANCER_DN | 540 | http://www.broadinstitute.org/gsea/msigdb/cards/ACEVEDO_LIVER_CANCER_DN |
| LIAO_METASTASIS | 539 | http://www.broadinstitute.org/gsea/msigdb/cards/LIAO_METASTASIS |
| REACTOME_ADAPTIVE_IMMUNE_SYSTEM | 539 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ADAPTIVE_IMMUNE_SYSTEM |
| RODRIGUES_THYROID_CARCINOMA_ANAPLASTIC_DN | 537 | http://www.broadinstitute.org/gsea/msigdb/cards/RODRIGUES_THYROID_CARCINOMA_ANAPLASTIC_DN |
| BERENJENO_TRANSFORMED_BY_RHOA_UP | 536 | http://www.broadinstitute.org/gsea/msigdb/cards/BERENJENO_TRANSFORMED_BY_RHOA_UP |
| MEISSNER_NPC_HCP_WITH_H3_UNMETHYLATED | 536 | http://www.broadinstitute.org/gsea/msigdb/cards/MEISSNER_NPC_HCP_WITH_H3_UNMETHYLATED |
| SENESE_HDAC3_TARGETS_DN | 536 | http://www.broadinstitute.org/gsea/msigdb/cards/SENESE_HDAC3_TARGETS_DN |
| STEIN_ESRRA_TARGETS | 535 | http://www.broadinstitute.org/gsea/msigdb/cards/STEIN_ESRRA_TARGETS |
| IVANOVA_HEMATOPOIESIS_EARLY_PROGENITOR | 532 | http://www.broadinstitute.org/gsea/msigdb/cards/IVANOVA_HEMATOPOIESIS_EARLY_PROGENITOR |
| ENK_UV_RESPONSE_KERATINOCYTE_UP | 530 | http://www.broadinstitute.org/gsea/msigdb/cards/ENK_UV_RESPONSE_KERATINOCYTE_UP |
| CREIGHTON_ENDOCRINE_THERAPY_RESISTANCE_1 | 528 | http://www.broadinstitute.org/gsea/msigdb/cards/CREIGHTON_ENDOCRINE_THERAPY_RESISTANCE_1 |
| ONKEN_UVEAL_MELANOMA_DN | 526 | http://www.broadinstitute.org/gsea/msigdb/cards/ONKEN_UVEAL_MELANOMA_DN |
| MILI_PSEUDOPODIA_HAPTOTAXIS_UP | 518 | http://www.broadinstitute.org/gsea/msigdb/cards/MILI_PSEUDOPODIA_HAPTOTAXIS_UP |
| REACTOME_METABOLISM_OF_PROTEINS | 518 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_METABOLISM_OF_PROTEINS |
| RAO_BOUND_BY_SALL4_ISOFORM_B | 517 | http://www.broadinstitute.org/gsea/msigdb/cards/RAO_BOUND_BY_SALL4_ISOFORM_B |
| STARK_PREFRONTAL_CORTEX_22Q11_DELETION_DN | 517 | http://www.broadinstitute.org/gsea/msigdb/cards/STARK_PREFRONTAL_CORTEX_22Q11_DELETION_DN |
| ZWANG_CLASS_1_TRANSIENTLY_INDUCED_BY_EGF | 516 | http://www.broadinstitute.org/gsea/msigdb/cards/ZWANG_CLASS_1_TRANSIENTLY_INDUCED_BY_EGF |
| MARTORIATI_MDM4_TARGETS_FETAL_LIVER_DN | 514 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTORIATI_MDM4_TARGETS_FETAL_LIVER_DN |
| SCHAEFFER_PROSTATE_DEVELOPMENT_6HR_DN | 514 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_PROSTATE_DEVELOPMENT_6HR_DN |
| BONOME_OVARIAN_CANCER_SURVIVAL_SUBOPTIMAL_DEBULKING | 510 | http://www.broadinstitute.org/gsea/msigdb/cards/BONOME_OVARIAN_CANCER_SURVIVAL_SUBOPTIMAL_DEBULKING |
| ENK_UV_RESPONSE_EPIDERMIS_DN | 508 | http://www.broadinstitute.org/gsea/msigdb/cards/ENK_UV_RESPONSE_EPIDERMIS_DN |
| DUTERTRE_ESTRADIOL_RESPONSE_24HR_DN | 505 | http://www.broadinstitute.org/gsea/msigdb/cards/DUTERTRE_ESTRADIOL_RESPONSE_24HR_DN |
| BROWNE_HCMV_INFECTION_48HR_DN | 504 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_48HR_DN |
| PILON_KLF1_TARGETS_UP | 504 | http://www.broadinstitute.org/gsea/msigdb/cards/PILON_KLF1_TARGETS_UP |
| SENESE_HDAC3_TARGETS_UP | 501 | http://www.broadinstitute.org/gsea/msigdb/cards/SENESE_HDAC3_TARGETS_UP |
| GAZDA_DIAMOND_BLACKFAN_ANEMIA_ERYTHROID_DN | 493 | http://www.broadinstitute.org/gsea/msigdb/cards/GAZDA_DIAMOND_BLACKFAN_ANEMIA_ERYTHROID_DN |
| MEISSNER_NPC_HCP_WITH_H3K4ME2 | 491 | http://www.broadinstitute.org/gsea/msigdb/cards/MEISSNER_NPC_HCP_WITH_H3K4ME2 |
| SWEET_LUNG_CANCER_KRAS_UP | 491 | http://www.broadinstitute.org/gsea/msigdb/cards/SWEET_LUNG_CANCER_KRAS_UP |
| ZHENG_BOUND_BY_FOXP3 | 491 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHENG_BOUND_BY_FOXP3 |
| LIM_MAMMARY_STEM_CELL_UP | 489 | http://www.broadinstitute.org/gsea/msigdb/cards/LIM_MAMMARY_STEM_CELL_UP |
| RODWELL_AGING_KIDNEY_UP | 487 | http://www.broadinstitute.org/gsea/msigdb/cards/RODWELL_AGING_KIDNEY_UP |
| SCHAEFFER_PROSTATE_DEVELOPMENT_48HR_UP | 487 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_PROSTATE_DEVELOPMENT_48HR_UP |
| ENK_UV_RESPONSE_KERATINOCYTE_DN | 485 | http://www.broadinstitute.org/gsea/msigdb/cards/ENK_UV_RESPONSE_KERATINOCYTE_DN |
| ZHOU_INFLAMMATORY_RESPONSE_LIVE_UP | 485 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHOU_INFLAMMATORY_RESPONSE_LIVE_UP |
| DACOSTA_UV_RESPONSE_VIA_ERCC3_COMMON_DN | 483 | http://www.broadinstitute.org/gsea/msigdb/cards/DACOSTA_UV_RESPONSE_VIA_ERCC3_COMMON_DN |
| CREIGHTON_ENDOCRINE_THERAPY_RESISTANCE_5 | 482 | http://www.broadinstitute.org/gsea/msigdb/cards/CREIGHTON_ENDOCRINE_THERAPY_RESISTANCE_5 |
| LIU_PROSTATE_CANCER_DN | 481 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_PROSTATE_CANCER_DN |
| SPIELMAN_LYMPHOBLAST_EUROPEAN_VS_ASIAN_UP | 479 | http://www.broadinstitute.org/gsea/msigdb/cards/SPIELMAN_LYMPHOBLAST_EUROPEAN_VS_ASIAN_UP |
| REACTOME_METABOLISM_OF_LIPIDS_AND_LIPOPROTEINS | 478 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_METABOLISM_OF_LIPIDS_AND_LIPOPROTEINS |
| SMID_BREAST_CANCER_NORMAL_LIKE_UP | 476 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_NORMAL_LIKE_UP |
| MATSUDA_NATURAL_KILLER_DIFFERENTIATION | 475 | http://www.broadinstitute.org/gsea/msigdb/cards/MATSUDA_NATURAL_KILLER_DIFFERENTIATION |
| CREIGHTON_ENDOCRINE_THERAPY_RESISTANCE_2 | 473 | http://www.broadinstitute.org/gsea/msigdb/cards/CREIGHTON_ENDOCRINE_THERAPY_RESISTANCE_2 |
| GARY_CD5_TARGETS_UP | 473 | http://www.broadinstitute.org/gsea/msigdb/cards/GARY_CD5_TARGETS_UP |
| MCCABE_BOUND_BY_HOXC6 | 469 | http://www.broadinstitute.org/gsea/msigdb/cards/MCCABE_BOUND_BY_HOXC6 |
| RATTENBACHER_BOUND_BY_CELF1 | 467 | http://www.broadinstitute.org/gsea/msigdb/cards/RATTENBACHER_BOUND_BY_CELF1 |
| REACTOME_HEMOSTASIS | 466 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_HEMOSTASIS |
| ONDER_CDH1_TARGETS_2_DN | 464 | http://www.broadinstitute.org/gsea/msigdb/cards/ONDER_CDH1_TARGETS_2_DN |
| FULCHER_INFLAMMATORY_RESPONSE_LECTIN_VS_LPS_DN | 463 | http://www.broadinstitute.org/gsea/msigdb/cards/FULCHER_INFLAMMATORY_RESPONSE_LECTIN_VS_LPS_DN |
| TOYOTA_TARGETS_OF_MIR34B_AND_MIR34C | 463 | http://www.broadinstitute.org/gsea/msigdb/cards/TOYOTA_TARGETS_OF_MIR34B_AND_MIR34C |
| DELACROIX_RAR_BOUND_ES | 462 | http://www.broadinstitute.org/gsea/msigdb/cards/DELACROIX_RAR_BOUND_ES |
| HELLER_HDAC_TARGETS_SILENCED_BY_METHYLATION_UP | 461 | http://www.broadinstitute.org/gsea/msigdb/cards/HELLER_HDAC_TARGETS_SILENCED_BY_METHYLATION_UP |
| CHARAFE_BREAST_CANCER_LUMINAL_VS_MESENCHYMAL_DN | 460 | http://www.broadinstitute.org/gsea/msigdb/cards/CHARAFE_BREAST_CANCER_LUMINAL_VS_MESENCHYMAL_DN |
| KIM_WT1_TARGETS_DN | 459 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_WT1_TARGETS_DN |
| MILI_PSEUDOPODIA_CHEMOTAXIS_DN | 457 | http://www.broadinstitute.org/gsea/msigdb/cards/MILI_PSEUDOPODIA_CHEMOTAXIS_DN |
| SENESE_HDAC1_TARGETS_UP | 457 | http://www.broadinstitute.org/gsea/msigdb/cards/SENESE_HDAC1_TARGETS_UP |
| MARTENS_BOUND_BY_PML_RARA_FUSION | 456 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTENS_BOUND_BY_PML_RARA_FUSION |
| ODONNELL_TFRC_TARGETS_UP | 456 | http://www.broadinstitute.org/gsea/msigdb/cards/ODONNELL_TFRC_TARGETS_UP |
| SHEDDEN_LUNG_CANCER_POOR_SURVIVAL_A6 | 456 | http://www.broadinstitute.org/gsea/msigdb/cards/SHEDDEN_LUNG_CANCER_POOR_SURVIVAL_A6 |
| CHARAFE_BREAST_CANCER_LUMINAL_VS_BASAL_DN | 455 | http://www.broadinstitute.org/gsea/msigdb/cards/CHARAFE_BREAST_CANCER_LUMINAL_VS_BASAL_DN |
| CHARAFE_BREAST_CANCER_LUMINAL_VS_MESENCHYMAL_UP | 450 | http://www.broadinstitute.org/gsea/msigdb/cards/CHARAFE_BREAST_CANCER_LUMINAL_VS_MESENCHYMAL_UP |
| PURBEY_TARGETS_OF_CTBP1_NOT_SATB1_DN | 448 | http://www.broadinstitute.org/gsea/msigdb/cards/PURBEY_TARGETS_OF_CTBP1_NOT_SATB1_DN |
| MOOTHA_MITOCHONDRIA | 447 | http://www.broadinstitute.org/gsea/msigdb/cards/MOOTHA_MITOCHONDRIA |
| MIKKELSEN_NPC_ICP_WITH_H3K4ME3 | 445 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_NPC_ICP_WITH_H3K4ME3 |
| DELYS_THYROID_CANCER_UP | 443 | http://www.broadinstitute.org/gsea/msigdb/cards/DELYS_THYROID_CANCER_UP |
| HAN_SATB1_TARGETS_DN | 442 | http://www.broadinstitute.org/gsea/msigdb/cards/HAN_SATB1_TARGETS_DN |
| ROME_INSULIN_TARGETS_IN_MUSCLE_UP | 442 | http://www.broadinstitute.org/gsea/msigdb/cards/ROME_INSULIN_TARGETS_IN_MUSCLE_UP |
| MITSIADES_RESPONSE_TO_APLIDIN_UP | 439 | http://www.broadinstitute.org/gsea/msigdb/cards/MITSIADES_RESPONSE_TO_APLIDIN_UP |
| MIKKELSEN_MCV6_HCP_WITH_H3K27ME3 | 435 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_MCV6_HCP_WITH_H3K27ME3 |
| SWEET_LUNG_CANCER_KRAS_DN | 435 | http://www.broadinstitute.org/gsea/msigdb/cards/SWEET_LUNG_CANCER_KRAS_DN |
| BAELDE_DIABETIC_NEPHROPATHY_DN | 434 | http://www.broadinstitute.org/gsea/msigdb/cards/BAELDE_DIABETIC_NEPHROPATHY_DN |
| GARY_CD5_TARGETS_DN | 431 | http://www.broadinstitute.org/gsea/msigdb/cards/GARY_CD5_TARGETS_DN |
| ZHOU_INFLAMMATORY_RESPONSE_LPS_UP | 431 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHOU_INFLAMMATORY_RESPONSE_LPS_UP |
| JOHNSTONE_PARVB_TARGETS_3_UP | 430 | http://www.broadinstitute.org/gsea/msigdb/cards/JOHNSTONE_PARVB_TARGETS_3_UP |
| RIGGI_EWING_SARCOMA_PROGENITOR_UP | 430 | http://www.broadinstitute.org/gsea/msigdb/cards/RIGGI_EWING_SARCOMA_PROGENITOR_UP |
| VECCHI_GASTRIC_CANCER_EARLY_UP | 430 | http://www.broadinstitute.org/gsea/msigdb/cards/VECCHI_GASTRIC_CANCER_EARLY_UP |
| MOOTHA_HUMAN_MITODB_6_2002 | 429 | http://www.broadinstitute.org/gsea/msigdb/cards/MOOTHA_HUMAN_MITODB_6_2002 |
| CUI_TCF21_TARGETS_2_UP | 428 | http://www.broadinstitute.org/gsea/msigdb/cards/CUI_TCF21_TARGETS_2_UP |
| LIM_MAMMARY_STEM_CELL_DN | 428 | http://www.broadinstitute.org/gsea/msigdb/cards/LIM_MAMMARY_STEM_CELL_DN |
| SCHAEFFER_PROSTATE_DEVELOPMENT_48HR_DN | 428 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_PROSTATE_DEVELOPMENT_48HR_DN |
| BOCHKIS_FOXA2_TARGETS | 425 | http://www.broadinstitute.org/gsea/msigdb/cards/BOCHKIS_FOXA2_TARGETS |
| BOQUEST_STEM_CELL_CULTURED_VS_FRESH_UP | 425 | http://www.broadinstitute.org/gsea/msigdb/cards/BOQUEST_STEM_CELL_CULTURED_VS_FRESH_UP |
| ZHANG_BREAST_CANCER_PROGENITORS_UP | 425 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_BREAST_CANCER_PROGENITORS_UP |
| SHEN_SMARCA2_TARGETS_UP | 424 | http://www.broadinstitute.org/gsea/msigdb/cards/SHEN_SMARCA2_TARGETS_UP |
| PUJANA_BRCA2_PCC_NETWORK | 423 | http://www.broadinstitute.org/gsea/msigdb/cards/PUJANA_BRCA2_PCC_NETWORK |
| THUM_SYSTOLIC_HEART_FAILURE_UP | 423 | http://www.broadinstitute.org/gsea/msigdb/cards/THUM_SYSTOLIC_HEART_FAILURE_UP |
| YAGI_AML_WITH_INV_16_TRANSLOCATION | 422 | http://www.broadinstitute.org/gsea/msigdb/cards/YAGI_AML_WITH_INV_16_TRANSLOCATION |
| REACTOME_CELL_CYCLE | 421 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CELL_CYCLE |
| MOOTHA_PGC | 420 | http://www.broadinstitute.org/gsea/msigdb/cards/MOOTHA_PGC |
| SATO_SILENCED_BY_METHYLATION_IN_PANCREATIC_CANCER_1 | 419 | http://www.broadinstitute.org/gsea/msigdb/cards/SATO_SILENCED_BY_METHYLATION_IN_PANCREATIC_CANCER_1 |
| MULLIGHAN_MLL_SIGNATURE_2_UP | 418 | http://www.broadinstitute.org/gsea/msigdb/cards/MULLIGHAN_MLL_SIGNATURE_2_UP |
| PEDRIOLI_MIR31_TARGETS_DN | 418 | http://www.broadinstitute.org/gsea/msigdb/cards/PEDRIOLI_MIR31_TARGETS_DN |
| RUTELLA_RESPONSE_TO_HGF_UP | 418 | http://www.broadinstitute.org/gsea/msigdb/cards/RUTELLA_RESPONSE_TO_HGF_UP |
| LU_EZH2_TARGETS_DN | 414 | http://www.broadinstitute.org/gsea/msigdb/cards/LU_EZH2_TARGETS_DN |
| MOHANKUMAR_TLX1_TARGETS_UP | 414 | http://www.broadinstitute.org/gsea/msigdb/cards/MOHANKUMAR_TLX1_TARGETS_UP |
| REACTOME_TRANSMEMBRANE_TRANSPORT_OF_SMALL_MOLECULES | 413 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANSMEMBRANE_TRANSPORT_OF_SMALL_MOLECULES |
| MOREAUX_MULTIPLE_MYELOMA_BY_TACI_UP | 412 | http://www.broadinstitute.org/gsea/msigdb/cards/MOREAUX_MULTIPLE_MYELOMA_BY_TACI_UP |
| KOYAMA_SEMA3B_TARGETS_DN | 411 | http://www.broadinstitute.org/gsea/msigdb/cards/KOYAMA_SEMA3B_TARGETS_DN |
| FOSTER_TOLERANT_MACROPHAGE_DN | 409 | http://www.broadinstitute.org/gsea/msigdb/cards/FOSTER_TOLERANT_MACROPHAGE_DN |
| REACTOME_GPCR_LIGAND_BINDING | 408 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GPCR_LIGAND_BINDING |
| REN_ALVEOLAR_RHABDOMYOSARCOMA_DN | 408 | http://www.broadinstitute.org/gsea/msigdb/cards/REN_ALVEOLAR_RHABDOMYOSARCOMA_DN |
| RUTELLA_RESPONSE_TO_HGF_VS_CSF2RB_AND_IL4_UP | 408 | http://www.broadinstitute.org/gsea/msigdb/cards/RUTELLA_RESPONSE_TO_HGF_VS_CSF2RB_AND_IL4_UP |
| SENGUPTA_NASOPHARYNGEAL_CARCINOMA_WITH_LMP1_UP | 408 | http://www.broadinstitute.org/gsea/msigdb/cards/SENGUPTA_NASOPHARYNGEAL_CARCINOMA_WITH_LMP1_UP |
| OSMAN_BLADDER_CANCER_DN | 406 | http://www.broadinstitute.org/gsea/msigdb/cards/OSMAN_BLADDER_CANCER_DN |
| KUMAR_TARGETS_OF_MLL_AF9_FUSION | 405 | http://www.broadinstitute.org/gsea/msigdb/cards/KUMAR_TARGETS_OF_MLL_AF9_FUSION |
| OSMAN_BLADDER_CANCER_UP | 404 | http://www.broadinstitute.org/gsea/msigdb/cards/OSMAN_BLADDER_CANCER_UP |
| PEDERSEN_METASTASIS_BY_ERBB2_ISOFORM_7 | 403 | http://www.broadinstitute.org/gsea/msigdb/cards/PEDERSEN_METASTASIS_BY_ERBB2_ISOFORM_7 |
| TARTE_PLASMA_CELL_VS_PLASMABLAST_UP | 398 | http://www.broadinstitute.org/gsea/msigdb/cards/TARTE_PLASMA_CELL_VS_PLASMABLAST_UP |
| WANG_RESPONSE_TO_GSK3_INHIBITOR_SB216763_UP | 397 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_RESPONSE_TO_GSK3_INHIBITOR_SB216763_UP |
| REACTOME_DEVELOPMENTAL_BIOLOGY | 396 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DEVELOPMENTAL_BIOLOGY |
| HAN_SATB1_TARGETS_UP | 395 | http://www.broadinstitute.org/gsea/msigdb/cards/HAN_SATB1_TARGETS_UP |
| BERENJENO_TRANSFORMED_BY_RHOA_DN | 394 | http://www.broadinstitute.org/gsea/msigdb/cards/BERENJENO_TRANSFORMED_BY_RHOA_DN |
| MONNIER_POSTRADIATION_TUMOR_ESCAPE_UP | 393 | http://www.broadinstitute.org/gsea/msigdb/cards/MONNIER_POSTRADIATION_TUMOR_ESCAPE_UP |
| LINDGREN_BLADDER_CANCER_CLUSTER_2B | 392 | http://www.broadinstitute.org/gsea/msigdb/cards/LINDGREN_BLADDER_CANCER_CLUSTER_2B |
| CHEMNITZ_RESPONSE_TO_PROSTAGLANDIN_E2_DN | 391 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEMNITZ_RESPONSE_TO_PROSTAGLANDIN_E2_DN |
| BLALOCK_ALZHEIMERS_DISEASE_INCIPIENT_UP | 390 | http://www.broadinstitute.org/gsea/msigdb/cards/BLALOCK_ALZHEIMERS_DISEASE_INCIPIENT_UP |
| HATADA_METHYLATED_IN_LUNG_CANCER_UP | 390 | http://www.broadinstitute.org/gsea/msigdb/cards/HATADA_METHYLATED_IN_LUNG_CANCER_UP |
| HSIAO_HOUSEKEEPING_GENES | 389 | http://www.broadinstitute.org/gsea/msigdb/cards/HSIAO_HOUSEKEEPING_GENES |
| KEGG_OLFACTORY_TRANSDUCTION | 389 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_OLFACTORY_TRANSDUCTION |
| STEIN_ESRRA_TARGETS_UP | 388 | http://www.broadinstitute.org/gsea/msigdb/cards/STEIN_ESRRA_TARGETS_UP |
| CHYLA_CBFA2T3_TARGETS_UP | 387 | http://www.broadinstitute.org/gsea/msigdb/cards/CHYLA_CBFA2T3_TARGETS_UP |
| BASAKI_YBX1_TARGETS_DN | 384 | http://www.broadinstitute.org/gsea/msigdb/cards/BASAKI_YBX1_TARGETS_DN |
| ZHOU_INFLAMMATORY_RESPONSE_LIVE_DN | 384 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHOU_INFLAMMATORY_RESPONSE_LIVE_DN |
| RICKMAN_TUMOR_DIFFERENTIATED_WELL_VS_POORLY_DN | 382 | http://www.broadinstitute.org/gsea/msigdb/cards/RICKMAN_TUMOR_DIFFERENTIATED_WELL_VS_POORLY_DN |
| CHARAFE_BREAST_CANCER_LUMINAL_VS_BASAL_UP | 380 | http://www.broadinstitute.org/gsea/msigdb/cards/CHARAFE_BREAST_CANCER_LUMINAL_VS_BASAL_UP |
| DAIRKEE_TERT_TARGETS_UP | 380 | http://www.broadinstitute.org/gsea/msigdb/cards/DAIRKEE_TERT_TARGETS_UP |
| MULLIGHAN_MLL_SIGNATURE_1_UP | 380 | http://www.broadinstitute.org/gsea/msigdb/cards/MULLIGHAN_MLL_SIGNATURE_1_UP |
| BENPORATH_ES_1 | 379 | http://www.broadinstitute.org/gsea/msigdb/cards/BENPORATH_ES_1 |
| LINDGREN_BLADDER_CANCER_CLUSTER_1_DN | 378 | http://www.broadinstitute.org/gsea/msigdb/cards/LINDGREN_BLADDER_CANCER_CLUSTER_1_DN |
| WANG_RESPONSE_TO_GSK3_INHIBITOR_SB216763_DN | 374 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_RESPONSE_TO_GSK3_INHIBITOR_SB216763_DN |
| WANG_TUMOR_INVASIVENESS_UP | 374 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_TUMOR_INVASIVENESS_UP |
| MONNIER_POSTRADIATION_TUMOR_ESCAPE_DN | 373 | http://www.broadinstitute.org/gsea/msigdb/cards/MONNIER_POSTRADIATION_TUMOR_ESCAPE_DN |
| WANG_LMO4_TARGETS_UP | 372 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_LMO4_TARGETS_UP |
| WANG_SMARCE1_TARGETS_DN | 371 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_SMARCE1_TARGETS_DN |
| DEURIG_T_CELL_PROLYMPHOCYTIC_LEUKEMIA_UP | 368 | http://www.broadinstitute.org/gsea/msigdb/cards/DEURIG_T_CELL_PROLYMPHOCYTIC_LEUKEMIA_UP |
| YAGI_AML_WITH_T_8_21_TRANSLOCATION | 368 | http://www.broadinstitute.org/gsea/msigdb/cards/YAGI_AML_WITH_T_8_21_TRANSLOCATION |
| DELACROIX_RARG_BOUND_MEF | 367 | http://www.broadinstitute.org/gsea/msigdb/cards/DELACROIX_RARG_BOUND_MEF |
| VECCHI_GASTRIC_CANCER_EARLY_DN | 367 | http://www.broadinstitute.org/gsea/msigdb/cards/VECCHI_GASTRIC_CANCER_EARLY_DN |
| SANSOM_APC_TARGETS_DN | 366 | http://www.broadinstitute.org/gsea/msigdb/cards/SANSOM_APC_TARGETS_DN |
| YOSHIMURA_MAPK8_TARGETS_DN | 366 | http://www.broadinstitute.org/gsea/msigdb/cards/YOSHIMURA_MAPK8_TARGETS_DN |
| YANG_BCL3_TARGETS_UP | 364 | http://www.broadinstitute.org/gsea/msigdb/cards/YANG_BCL3_TARGETS_UP |
| RHEIN_ALL_GLUCOCORTICOID_THERAPY_DN | 362 | http://www.broadinstitute.org/gsea/msigdb/cards/RHEIN_ALL_GLUCOCORTICOID_THERAPY_DN |
| GRUETZMANN_PANCREATIC_CANCER_UP | 358 | http://www.broadinstitute.org/gsea/msigdb/cards/GRUETZMANN_PANCREATIC_CANCER_UP |
| SHEN_SMARCA2_TARGETS_DN | 357 | http://www.broadinstitute.org/gsea/msigdb/cards/SHEN_SMARCA2_TARGETS_DN |
| COLINA_TARGETS_OF_4EBP1_AND_4EBP2 | 356 | http://www.broadinstitute.org/gsea/msigdb/cards/COLINA_TARGETS_OF_4EBP1_AND_4EBP2 |
| PEREZ_TP63_TARGETS | 355 | http://www.broadinstitute.org/gsea/msigdb/cards/PEREZ_TP63_TARGETS |
| ACEVEDO_NORMAL_TISSUE_ADJACENT_TO_LIVER_TUMOR_DN | 354 | http://www.broadinstitute.org/gsea/msigdb/cards/ACEVEDO_NORMAL_TISSUE_ADJACENT_TO_LIVER_TUMOR_DN |
| IWANAGA_CARCINOGENESIS_BY_KRAS_PTEN_DN | 353 | http://www.broadinstitute.org/gsea/msigdb/cards/IWANAGA_CARCINOGENESIS_BY_KRAS_PTEN_DN |
| REACTOME_GENERIC_TRANSCRIPTION_PATHWAY | 352 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GENERIC_TRANSCRIPTION_PATHWAY |
| WANG_LMO4_TARGETS_DN | 352 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_LMO4_TARGETS_DN |
| SCHUETZ_BREAST_CANCER_DUCTAL_INVASIVE_UP | 351 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHUETZ_BREAST_CANCER_DUCTAL_INVASIVE_UP |
| YAGI_AML_WITH_11Q23_REARRANGED | 351 | http://www.broadinstitute.org/gsea/msigdb/cards/YAGI_AML_WITH_11Q23_REARRANGED |
| MEISSNER_NPC_HCP_WITH_H3K4ME2_AND_H3K27ME3 | 349 | http://www.broadinstitute.org/gsea/msigdb/cards/MEISSNER_NPC_HCP_WITH_H3K4ME2_AND_H3K27ME3 |
| SENGUPTA_NASOPHARYNGEAL_CARCINOMA_DN | 349 | http://www.broadinstitute.org/gsea/msigdb/cards/SENGUPTA_NASOPHARYNGEAL_CARCINOMA_DN |
| GOLDRATH_ANTIGEN_RESPONSE | 346 | http://www.broadinstitute.org/gsea/msigdb/cards/GOLDRATH_ANTIGEN_RESPONSE |
| PURBEY_TARGETS_OF_CTBP1_NOT_SATB1_UP | 344 | http://www.broadinstitute.org/gsea/msigdb/cards/PURBEY_TARGETS_OF_CTBP1_NOT_SATB1_UP |
| RICKMAN_METASTASIS_UP | 344 | http://www.broadinstitute.org/gsea/msigdb/cards/RICKMAN_METASTASIS_UP |
| BLUM_RESPONSE_TO_SALIRASIB_DN | 342 | http://www.broadinstitute.org/gsea/msigdb/cards/BLUM_RESPONSE_TO_SALIRASIB_DN |
| MIKKELSEN_NPC_HCP_WITH_H3K27ME3 | 341 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_NPC_HCP_WITH_H3K27ME3 |
| MULLIGHAN_NPM1_SIGNATURE_3_UP | 341 | http://www.broadinstitute.org/gsea/msigdb/cards/MULLIGHAN_NPM1_SIGNATURE_3_UP |
| RUTELLA_RESPONSE_TO_CSF2RB_AND_IL4_UP | 338 | http://www.broadinstitute.org/gsea/msigdb/cards/RUTELLA_RESPONSE_TO_CSF2RB_AND_IL4_UP |
| PENG_GLUTAMINE_DEPRIVATION_DN | 337 | http://www.broadinstitute.org/gsea/msigdb/cards/PENG_GLUTAMINE_DEPRIVATION_DN |
| PHONG_TNF_RESPONSE_NOT_VIA_P38 | 337 | http://www.broadinstitute.org/gsea/msigdb/cards/PHONG_TNF_RESPONSE_NOT_VIA_P38 |
| JOHNSTONE_PARVB_TARGETS_2_DN | 336 | http://www.broadinstitute.org/gsea/msigdb/cards/JOHNSTONE_PARVB_TARGETS_2_DN |
| GINESTIER_BREAST_CANCER_ZNF217_AMPLIFIED_DN | 335 | http://www.broadinstitute.org/gsea/msigdb/cards/GINESTIER_BREAST_CANCER_ZNF217_AMPLIFIED_DN |
| NIKOLSKY_BREAST_CANCER_17Q21_Q25_AMPLICON | 335 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_17Q21_Q25_AMPLICON |
| WONG_EMBRYONIC_STEM_CELL_CORE | 335 | http://www.broadinstitute.org/gsea/msigdb/cards/WONG_EMBRYONIC_STEM_CELL_CORE |
| DAVICIONI_MOLECULAR_ARMS_VS_ERMS_UP | 332 | http://www.broadinstitute.org/gsea/msigdb/cards/DAVICIONI_MOLECULAR_ARMS_VS_ERMS_UP |
| FARMER_BREAST_CANCER_APOCRINE_VS_BASAL | 330 | http://www.broadinstitute.org/gsea/msigdb/cards/FARMER_BREAST_CANCER_APOCRINE_VS_BASAL |
| FARMER_BREAST_CANCER_BASAL_VS_LULMINAL | 330 | http://www.broadinstitute.org/gsea/msigdb/cards/FARMER_BREAST_CANCER_BASAL_VS_LULMINAL |
| REACTOME_METABOLISM_OF_RNA | 330 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_METABOLISM_OF_RNA |
| KINSEY_TARGETS_OF_EWSR1_FLII_FUSION_DN | 329 | http://www.broadinstitute.org/gsea/msigdb/cards/KINSEY_TARGETS_OF_EWSR1_FLII_FUSION_DN |
| LINDGREN_BLADDER_CANCER_CLUSTER_3_UP | 329 | http://www.broadinstitute.org/gsea/msigdb/cards/LINDGREN_BLADDER_CANCER_CLUSTER_3_UP |
| KEGG_PATHWAYS_IN_CANCER | 328 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PATHWAYS_IN_CANCER |
| REACTOME_OLFACTORY_SIGNALING_PATHWAY | 328 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_OLFACTORY_SIGNALING_PATHWAY |
| FARMER_BREAST_CANCER_APOCRINE_VS_LUMINAL | 326 | http://www.broadinstitute.org/gsea/msigdb/cards/FARMER_BREAST_CANCER_APOCRINE_VS_LUMINAL |
| REACTOME_CELL_CYCLE_MITOTIC | 325 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CELL_CYCLE_MITOTIC |
| DUTERTRE_ESTRADIOL_RESPONSE_24HR_UP | 324 | http://www.broadinstitute.org/gsea/msigdb/cards/DUTERTRE_ESTRADIOL_RESPONSE_24HR_UP |
| GRESHOCK_CANCER_COPY_NUMBER_UP | 323 | http://www.broadinstitute.org/gsea/msigdb/cards/GRESHOCK_CANCER_COPY_NUMBER_UP |
| TORCHIA_TARGETS_OF_EWSR1_FLI1_FUSION_DN | 321 | http://www.broadinstitute.org/gsea/msigdb/cards/TORCHIA_TARGETS_OF_EWSR1_FLI1_FUSION_DN |
| DEURIG_T_CELL_PROLYMPHOCYTIC_LEUKEMIA_DN | 320 | http://www.broadinstitute.org/gsea/msigdb/cards/DEURIG_T_CELL_PROLYMPHOCYTIC_LEUKEMIA_DN |
| DAZARD_RESPONSE_TO_UV_NHEK_DN | 318 | http://www.broadinstitute.org/gsea/msigdb/cards/DAZARD_RESPONSE_TO_UV_NHEK_DN |
| LEI_MYB_TARGETS | 318 | http://www.broadinstitute.org/gsea/msigdb/cards/LEI_MYB_TARGETS |
| BRUINS_UVC_RESPONSE_EARLY_LATE | 317 | http://www.broadinstitute.org/gsea/msigdb/cards/BRUINS_UVC_RESPONSE_EARLY_LATE |
| HELLER_HDAC_TARGETS_UP | 317 | http://www.broadinstitute.org/gsea/msigdb/cards/HELLER_HDAC_TARGETS_UP |
| SHEDDEN_LUNG_CANCER_GOOD_SURVIVAL_A12 | 317 | http://www.broadinstitute.org/gsea/msigdb/cards/SHEDDEN_LUNG_CANCER_GOOD_SURVIVAL_A12 |
| JAATINEN_HEMATOPOIETIC_STEM_CELL_UP | 316 | http://www.broadinstitute.org/gsea/msigdb/cards/JAATINEN_HEMATOPOIETIC_STEM_CELL_UP |
| PASINI_SUZ12_TARGETS_DN | 315 | http://www.broadinstitute.org/gsea/msigdb/cards/PASINI_SUZ12_TARGETS_DN |
| RUTELLA_RESPONSE_TO_CSF2RB_AND_IL4_DN | 315 | http://www.broadinstitute.org/gsea/msigdb/cards/RUTELLA_RESPONSE_TO_CSF2RB_AND_IL4_DN |
| SMID_BREAST_CANCER_RELAPSE_IN_BONE_DN | 315 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_RELAPSE_IN_BONE_DN |
| DEBIASI_APOPTOSIS_BY_REOVIRUS_INFECTION_UP | 314 | http://www.broadinstitute.org/gsea/msigdb/cards/DEBIASI_APOPTOSIS_BY_REOVIRUS_INFECTION_UP |
| DOUGLAS_BMI1_TARGETS_DN | 314 | http://www.broadinstitute.org/gsea/msigdb/cards/DOUGLAS_BMI1_TARGETS_DN |
| GENTILE_UV_HIGH_DOSE_DN | 312 | http://www.broadinstitute.org/gsea/msigdb/cards/GENTILE_UV_HIGH_DOSE_DN |
| JIANG_HYPOXIA_NORMAL | 311 | http://www.broadinstitute.org/gsea/msigdb/cards/JIANG_HYPOXIA_NORMAL |
| HORIUCHI_WTAP_TARGETS_DN | 310 | http://www.broadinstitute.org/gsea/msigdb/cards/HORIUCHI_WTAP_TARGETS_DN |
| DACOSTA_UV_RESPONSE_VIA_ERCC3_UP | 309 | http://www.broadinstitute.org/gsea/msigdb/cards/DACOSTA_UV_RESPONSE_VIA_ERCC3_UP |
| LIU_SOX4_TARGETS_DN | 309 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_SOX4_TARGETS_DN |
| TARTE_PLASMA_CELL_VS_PLASMABLAST_DN | 309 | http://www.broadinstitute.org/gsea/msigdb/cards/TARTE_PLASMA_CELL_VS_PLASMABLAST_DN |
| ACEVEDO_FGFR1_TARGETS_IN_PROSTATE_CANCER_MODEL_DN | 308 | http://www.broadinstitute.org/gsea/msigdb/cards/ACEVEDO_FGFR1_TARGETS_IN_PROSTATE_CANCER_MODEL_DN |
| CREIGHTON_ENDOCRINE_THERAPY_RESISTANCE_4 | 307 | http://www.broadinstitute.org/gsea/msigdb/cards/CREIGHTON_ENDOCRINE_THERAPY_RESISTANCE_4 |
| LI_INDUCED_T_TO_NATURAL_KILLER_UP | 307 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_INDUCED_T_TO_NATURAL_KILLER_UP |
| CHANDRAN_METASTASIS_DN | 306 | http://www.broadinstitute.org/gsea/msigdb/cards/CHANDRAN_METASTASIS_DN |
| HORIUCHI_WTAP_TARGETS_UP | 306 | http://www.broadinstitute.org/gsea/msigdb/cards/HORIUCHI_WTAP_TARGETS_UP |
| BORCZUK_MALIGNANT_MESOTHELIOMA_UP | 305 | http://www.broadinstitute.org/gsea/msigdb/cards/BORCZUK_MALIGNANT_MESOTHELIOMA_UP |
| REACTOME_CLASS_A1_RHODOPSIN_LIKE_RECEPTORS | 305 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CLASS_A1_RHODOPSIN_LIKE_RECEPTORS |
| IVANOVA_HEMATOPOIESIS_STEM_CELL_LONG_TERM | 302 | http://www.broadinstitute.org/gsea/msigdb/cards/IVANOVA_HEMATOPOIESIS_STEM_CELL_LONG_TERM |
| MUELLER_PLURINET | 299 | http://www.broadinstitute.org/gsea/msigdb/cards/MUELLER_PLURINET |
| WALLACE_PROSTATE_CANCER_RACE_UP | 299 | http://www.broadinstitute.org/gsea/msigdb/cards/WALLACE_PROSTATE_CANCER_RACE_UP |
| AGUIRRE_PANCREATIC_CANCER_COPY_NUMBER_UP | 298 | http://www.broadinstitute.org/gsea/msigdb/cards/AGUIRRE_PANCREATIC_CANCER_COPY_NUMBER_UP |
| BROWNE_HCMV_INFECTION_14HR_DN | 298 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_14HR_DN |
| DURAND_STROMA_MAX_UP | 296 | http://www.broadinstitute.org/gsea/msigdb/cards/DURAND_STROMA_MAX_UP |
| ACEVEDO_LIVER_CANCER_WITH_H3K27ME3_UP | 295 | http://www.broadinstitute.org/gsea/msigdb/cards/ACEVEDO_LIVER_CANCER_WITH_H3K27ME3_UP |
| LU_EZH2_TARGETS_UP | 295 | http://www.broadinstitute.org/gsea/msigdb/cards/LU_EZH2_TARGETS_UP |
| SENGUPTA_NASOPHARYNGEAL_CARCINOMA_UP | 294 | http://www.broadinstitute.org/gsea/msigdb/cards/SENGUPTA_NASOPHARYNGEAL_CARCINOMA_UP |
| WEST_ADRENOCORTICAL_TUMOR_UP | 294 | http://www.broadinstitute.org/gsea/msigdb/cards/WEST_ADRENOCORTICAL_TUMOR_UP |
| ENK_UV_RESPONSE_EPIDERMIS_UP | 293 | http://www.broadinstitute.org/gsea/msigdb/cards/ENK_UV_RESPONSE_EPIDERMIS_UP |
| HADDAD_B_LYMPHOCYTE_PROGENITOR | 293 | http://www.broadinstitute.org/gsea/msigdb/cards/HADDAD_B_LYMPHOCYTE_PROGENITOR |
| IVANOVA_HEMATOPOIESIS_MATURE_CELL | 293 | http://www.broadinstitute.org/gsea/msigdb/cards/IVANOVA_HEMATOPOIESIS_MATURE_CELL |
| ZHANG_TLX_TARGETS_60HR_UP | 293 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_TLX_TARGETS_60HR_UP |
| ZWANG_DOWN_BY_2ND_EGF_PULSE | 293 | http://www.broadinstitute.org/gsea/msigdb/cards/ZWANG_DOWN_BY_2ND_EGF_PULSE |
| HELLER_HDAC_TARGETS_DN | 292 | http://www.broadinstitute.org/gsea/msigdb/cards/HELLER_HDAC_TARGETS_DN |
| KOYAMA_SEMA3B_TARGETS_UP | 292 | http://www.broadinstitute.org/gsea/msigdb/cards/KOYAMA_SEMA3B_TARGETS_UP |
| SABATES_COLORECTAL_ADENOMA_DN | 291 | http://www.broadinstitute.org/gsea/msigdb/cards/SABATES_COLORECTAL_ADENOMA_DN |
| BASAKI_YBX1_TARGETS_UP | 290 | http://www.broadinstitute.org/gsea/msigdb/cards/BASAKI_YBX1_TARGETS_UP |
| BENPORATH_OCT4_TARGETS | 290 | http://www.broadinstitute.org/gsea/msigdb/cards/BENPORATH_OCT4_TARGETS |
| ACEVEDO_FGFR1_TARGETS_IN_PROSTATE_CANCER_MODEL_UP | 289 | http://www.broadinstitute.org/gsea/msigdb/cards/ACEVEDO_FGFR1_TARGETS_IN_PROSTATE_CANCER_MODEL_UP |
| GABRIELY_MIR21_TARGETS | 289 | http://www.broadinstitute.org/gsea/msigdb/cards/GABRIELY_MIR21_TARGETS |
| MANALO_HYPOXIA_DN | 289 | http://www.broadinstitute.org/gsea/msigdb/cards/MANALO_HYPOXIA_DN |
| WANG_MLL_TARGETS | 289 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_MLL_TARGETS |
| POOLA_INVASIVE_BREAST_CANCER_UP | 288 | http://www.broadinstitute.org/gsea/msigdb/cards/POOLA_INVASIVE_BREAST_CANCER_UP |
| DEBIASI_APOPTOSIS_BY_REOVIRUS_INFECTION_DN | 287 | http://www.broadinstitute.org/gsea/msigdb/cards/DEBIASI_APOPTOSIS_BY_REOVIRUS_INFECTION_DN |
| GRADE_COLON_AND_RECTAL_CANCER_UP | 285 | http://www.broadinstitute.org/gsea/msigdb/cards/GRADE_COLON_AND_RECTAL_CANCER_UP |
| NAKAMURA_TUMOR_ZONE_PERIPHERAL_VS_CENTRAL_UP | 285 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAMURA_TUMOR_ZONE_PERIPHERAL_VS_CENTRAL_UP |
| REACTOME_METABOLISM_OF_MRNA | 284 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_METABOLISM_OF_MRNA |
| ZHOU_INFLAMMATORY_RESPONSE_FIMA_DN | 284 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHOU_INFLAMMATORY_RESPONSE_FIMA_DN |
| PASQUALUCCI_LYMPHOMA_BY_GC_STAGE_UP | 283 | http://www.broadinstitute.org/gsea/msigdb/cards/PASQUALUCCI_LYMPHOMA_BY_GC_STAGE_UP |
| HELLER_SILENCED_BY_METHYLATION_UP | 282 | http://www.broadinstitute.org/gsea/msigdb/cards/HELLER_SILENCED_BY_METHYLATION_UP |
| BHAT_ESR1_TARGETS_VIA_AKT1_UP | 281 | http://www.broadinstitute.org/gsea/msigdb/cards/BHAT_ESR1_TARGETS_VIA_AKT1_UP |
| HELLER_HDAC_TARGETS_SILENCED_BY_METHYLATION_DN | 281 | http://www.broadinstitute.org/gsea/msigdb/cards/HELLER_HDAC_TARGETS_SILENCED_BY_METHYLATION_DN |
| MULLIGHAN_MLL_SIGNATURE_2_DN | 281 | http://www.broadinstitute.org/gsea/msigdb/cards/MULLIGHAN_MLL_SIGNATURE_2_DN |
| BRUINS_UVC_RESPONSE_VIA_TP53_GROUP_D | 280 | http://www.broadinstitute.org/gsea/msigdb/cards/BRUINS_UVC_RESPONSE_VIA_TP53_GROUP_D |
| WANG_SMARCE1_TARGETS_UP | 280 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_SMARCE1_TARGETS_UP |
| FORTSCHEGGER_PHF8_TARGETS_UP | 279 | http://www.broadinstitute.org/gsea/msigdb/cards/FORTSCHEGGER_PHF8_TARGETS_UP |
| REACTOME_INNATE_IMMUNE_SYSTEM | 279 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INNATE_IMMUNE_SYSTEM |
| REACTOME_NEURONAL_SYSTEM | 279 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NEURONAL_SYSTEM |
| FOURNIER_ACINAR_DEVELOPMENT_LATE_2 | 277 | http://www.broadinstitute.org/gsea/msigdb/cards/FOURNIER_ACINAR_DEVELOPMENT_LATE_2 |
| ZHANG_TLX_TARGETS_60HR_DN | 277 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_TLX_TARGETS_60HR_DN |
| HUTTMANN_B_CLL_POOR_SURVIVAL_UP | 276 | http://www.broadinstitute.org/gsea/msigdb/cards/HUTTMANN_B_CLL_POOR_SURVIVAL_UP |
| MULLIGHAN_NPM1_MUTATED_SIGNATURE_1_UP | 276 | http://www.broadinstitute.org/gsea/msigdb/cards/MULLIGHAN_NPM1_MUTATED_SIGNATURE_1_UP |
| KUMAR_PATHOGEN_LOAD_BY_MACROPHAGES | 275 | http://www.broadinstitute.org/gsea/msigdb/cards/KUMAR_PATHOGEN_LOAD_BY_MACROPHAGES |
| OSWALD_HEMATOPOIETIC_STEM_CELL_IN_COLLAGEN_GEL_DN | 275 | http://www.broadinstitute.org/gsea/msigdb/cards/OSWALD_HEMATOPOIETIC_STEM_CELL_IN_COLLAGEN_GEL_DN |
| ACEVEDO_LIVER_TUMOR_VS_NORMAL_ADJACENT_TISSUE_DN | 274 | http://www.broadinstitute.org/gsea/msigdb/cards/ACEVEDO_LIVER_TUMOR_VS_NORMAL_ADJACENT_TISSUE_DN |
| KEGG_NEUROACTIVE_LIGAND_RECEPTOR_INTERACTION | 272 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_NEUROACTIVE_LIGAND_RECEPTOR_INTERACTION |
| MARTINEZ_RESPONSE_TO_TRABECTEDIN_DN | 271 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTINEZ_RESPONSE_TO_TRABECTEDIN_DN |
| MCBRYAN_PUBERTAL_BREAST_4_5WK_UP | 271 | http://www.broadinstitute.org/gsea/msigdb/cards/MCBRYAN_PUBERTAL_BREAST_4_5WK_UP |
| TORCHIA_TARGETS_OF_EWSR1_FLI1_FUSION_UP | 271 | http://www.broadinstitute.org/gsea/msigdb/cards/TORCHIA_TARGETS_OF_EWSR1_FLI1_FUSION_UP |
| REACTOME_CYTOKINE_SIGNALING_IN_IMMUNE_SYSTEM | 270 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CYTOKINE_SIGNALING_IN_IMMUNE_SYSTEM |
| RIZKI_TUMOR_INVASIVENESS_3D_DN | 270 | http://www.broadinstitute.org/gsea/msigdb/cards/RIZKI_TUMOR_INVASIVENESS_3D_DN |
| MEISSNER_BRAIN_HCP_WITH_H3K27ME3 | 269 | http://www.broadinstitute.org/gsea/msigdb/cards/MEISSNER_BRAIN_HCP_WITH_H3K27ME3 |
| WANG_CLIM2_TARGETS_UP | 269 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_CLIM2_TARGETS_UP |
| XU_GH1_AUTOCRINE_TARGETS_UP | 268 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_GH1_AUTOCRINE_TARGETS_UP |
| CAIRO_HEPATOBLASTOMA_DN | 267 | http://www.broadinstitute.org/gsea/msigdb/cards/CAIRO_HEPATOBLASTOMA_DN |
| HOLLMANN_APOPTOSIS_VIA_CD40_DN | 267 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLMANN_APOPTOSIS_VIA_CD40_DN |
| KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION | 267 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION |
| KEGG_MAPK_SIGNALING_PATHWAY | 267 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_MAPK_SIGNALING_PATHWAY |
| FOSTER_KDM1A_TARGETS_UP | 266 | http://www.broadinstitute.org/gsea/msigdb/cards/FOSTER_KDM1A_TARGETS_UP |
| HOSHIDA_LIVER_CANCER_SUBCLASS_S3 | 266 | http://www.broadinstitute.org/gsea/msigdb/cards/HOSHIDA_LIVER_CANCER_SUBCLASS_S3 |
| ROZANOV_MMP14_TARGETS_UP | 266 | http://www.broadinstitute.org/gsea/msigdb/cards/ROZANOV_MMP14_TARGETS_UP |
| WAMUNYOKOLI_OVARIAN_CANCER_LMP_UP | 265 | http://www.broadinstitute.org/gsea/msigdb/cards/WAMUNYOKOLI_OVARIAN_CANCER_LMP_UP |
| DURCHDEWALD_SKIN_CARCINOGENESIS_DN | 264 | http://www.broadinstitute.org/gsea/msigdb/cards/DURCHDEWALD_SKIN_CARCINOGENESIS_DN |
| YAUCH_HEDGEHOG_SIGNALING_PARACRINE_DN | 264 | http://www.broadinstitute.org/gsea/msigdb/cards/YAUCH_HEDGEHOG_SIGNALING_PARACRINE_DN |
| LU_AGING_BRAIN_UP | 262 | http://www.broadinstitute.org/gsea/msigdb/cards/LU_AGING_BRAIN_UP |
| BILD_HRAS_ONCOGENIC_SIGNATURE | 261 | http://www.broadinstitute.org/gsea/msigdb/cards/BILD_HRAS_ONCOGENIC_SIGNATURE |
| KAAB_HEART_ATRIUM_VS_VENTRICLE_DN | 261 | http://www.broadinstitute.org/gsea/msigdb/cards/KAAB_HEART_ATRIUM_VS_VENTRICLE_DN |
| RICKMAN_METASTASIS_DN | 261 | http://www.broadinstitute.org/gsea/msigdb/cards/RICKMAN_METASTASIS_DN |
| SHETH_LIVER_CANCER_VS_TXNIP_LOSS_PAM4 | 261 | http://www.broadinstitute.org/gsea/msigdb/cards/SHETH_LIVER_CANCER_VS_TXNIP_LOSS_PAM4 |
| BOQUEST_STEM_CELL_UP | 260 | http://www.broadinstitute.org/gsea/msigdb/cards/BOQUEST_STEM_CELL_UP |
| SENESE_HDAC1_TARGETS_DN | 260 | http://www.broadinstitute.org/gsea/msigdb/cards/SENESE_HDAC1_TARGETS_DN |
| MIYAGAWA_TARGETS_OF_EWSR1_ETS_FUSIONS_UP | 259 | http://www.broadinstitute.org/gsea/msigdb/cards/MIYAGAWA_TARGETS_OF_EWSR1_ETS_FUSIONS_UP |
| QI_PLASMACYTOMA_UP | 259 | http://www.broadinstitute.org/gsea/msigdb/cards/QI_PLASMACYTOMA_UP |
| JAEGER_METASTASIS_DN | 258 | http://www.broadinstitute.org/gsea/msigdb/cards/JAEGER_METASTASIS_DN |
| MASSARWEH_TAMOXIFEN_RESISTANCE_DN | 258 | http://www.broadinstitute.org/gsea/msigdb/cards/MASSARWEH_TAMOXIFEN_RESISTANCE_DN |
| ONDER_CDH1_TARGETS_2_UP | 256 | http://www.broadinstitute.org/gsea/msigdb/cards/ONDER_CDH1_TARGETS_2_UP |
| DAVICIONI_TARGETS_OF_PAX_FOXO1_FUSIONS_UP | 255 | http://www.broadinstitute.org/gsea/msigdb/cards/DAVICIONI_TARGETS_OF_PAX_FOXO1_FUSIONS_UP |
| BROWNE_HCMV_INFECTION_4HR_DN | 254 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_4HR_DN |
| IVANOVA_HEMATOPOIESIS_STEM_CELL | 254 | http://www.broadinstitute.org/gsea/msigdb/cards/IVANOVA_HEMATOPOIESIS_STEM_CELL |
| DANG_REGULATED_BY_MYC_DN | 253 | http://www.broadinstitute.org/gsea/msigdb/cards/DANG_REGULATED_BY_MYC_DN |
| MCLACHLAN_DENTAL_CARIES_UP | 253 | http://www.broadinstitute.org/gsea/msigdb/cards/MCLACHLAN_DENTAL_CARIES_UP |
| BOYLAN_MULTIPLE_MYELOMA_C_D_DN | 252 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYLAN_MULTIPLE_MYELOMA_C_D_DN |
| KOBAYASHI_EGFR_SIGNALING_24HR_DN | 251 | http://www.broadinstitute.org/gsea/msigdb/cards/KOBAYASHI_EGFR_SIGNALING_24HR_DN |
| REACTOME_AXON_GUIDANCE | 251 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_AXON_GUIDANCE |
| REACTOME_CLASS_I_MHC_MEDIATED_ANTIGEN_PROCESSING_PRESENTATION | 251 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CLASS_I_MHC_MEDIATED_ANTIGEN_PROCESSING_PRESENTATION |
| ZHENG_GLIOBLASTOMA_PLASTICITY_UP | 250 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHENG_GLIOBLASTOMA_PLASTICITY_UP |
| KAAB_HEART_ATRIUM_VS_VENTRICLE_UP | 249 | http://www.broadinstitute.org/gsea/msigdb/cards/KAAB_HEART_ATRIUM_VS_VENTRICLE_UP |
| MITSIADES_RESPONSE_TO_APLIDIN_DN | 249 | http://www.broadinstitute.org/gsea/msigdb/cards/MITSIADES_RESPONSE_TO_APLIDIN_DN |
| REACTOME_METABOLISM_OF_CARBOHYDRATES | 247 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_METABOLISM_OF_CARBOHYDRATES |
| BILD_E2F3_ONCOGENIC_SIGNATURE | 246 | http://www.broadinstitute.org/gsea/msigdb/cards/BILD_E2F3_ONCOGENIC_SIGNATURE |
| BONOME_OVARIAN_CANCER_SURVIVAL_OPTIMAL_DEBULKING | 246 | http://www.broadinstitute.org/gsea/msigdb/cards/BONOME_OVARIAN_CANCER_SURVIVAL_OPTIMAL_DEBULKING |
| VERHAAK_AML_WITH_NPM1_MUTATED_DN | 246 | http://www.broadinstitute.org/gsea/msigdb/cards/VERHAAK_AML_WITH_NPM1_MUTATED_DN |
| BLUM_RESPONSE_TO_SALIRASIB_UP | 245 | http://www.broadinstitute.org/gsea/msigdb/cards/BLUM_RESPONSE_TO_SALIRASIB_UP |
| KONDO_EZH2_TARGETS | 245 | http://www.broadinstitute.org/gsea/msigdb/cards/KONDO_EZH2_TARGETS |
| MCLACHLAN_DENTAL_CARIES_DN | 245 | http://www.broadinstitute.org/gsea/msigdb/cards/MCLACHLAN_DENTAL_CARIES_DN |
| PENG_RAPAMYCIN_RESPONSE_DN | 245 | http://www.broadinstitute.org/gsea/msigdb/cards/PENG_RAPAMYCIN_RESPONSE_DN |
| RUTELLA_RESPONSE_TO_HGF_VS_CSF2RB_AND_IL4_DN | 245 | http://www.broadinstitute.org/gsea/msigdb/cards/RUTELLA_RESPONSE_TO_HGF_VS_CSF2RB_AND_IL4_DN |
| DAZARD_RESPONSE_TO_UV_NHEK_UP | 244 | http://www.broadinstitute.org/gsea/msigdb/cards/DAZARD_RESPONSE_TO_UV_NHEK_UP |
| GAL_LEUKEMIC_STEM_CELL_DN | 244 | http://www.broadinstitute.org/gsea/msigdb/cards/GAL_LEUKEMIC_STEM_CELL_DN |
| HSIAO_LIVER_SPECIFIC_GENES | 244 | http://www.broadinstitute.org/gsea/msigdb/cards/HSIAO_LIVER_SPECIFIC_GENES |
| KAYO_AGING_MUSCLE_UP | 244 | http://www.broadinstitute.org/gsea/msigdb/cards/KAYO_AGING_MUSCLE_UP |
| PLASARI_TGFB1_TARGETS_10HR_DN | 244 | http://www.broadinstitute.org/gsea/msigdb/cards/PLASARI_TGFB1_TARGETS_10HR_DN |
| THUM_SYSTOLIC_HEART_FAILURE_DN | 244 | http://www.broadinstitute.org/gsea/msigdb/cards/THUM_SYSTOLIC_HEART_FAILURE_DN |
| CHICAS_RB1_TARGETS_GROWING | 243 | http://www.broadinstitute.org/gsea/msigdb/cards/CHICAS_RB1_TARGETS_GROWING |
| CHYLA_CBFA2T3_TARGETS_DN | 242 | http://www.broadinstitute.org/gsea/msigdb/cards/CHYLA_CBFA2T3_TARGETS_DN |
| HIRSCH_CELLULAR_TRANSFORMATION_SIGNATURE_UP | 242 | http://www.broadinstitute.org/gsea/msigdb/cards/HIRSCH_CELLULAR_TRANSFORMATION_SIGNATURE_UP |
| MULLIGHAN_MLL_SIGNATURE_1_DN | 242 | http://www.broadinstitute.org/gsea/msigdb/cards/MULLIGHAN_MLL_SIGNATURE_1_DN |
| WINTER_HYPOXIA_METAGENE | 242 | http://www.broadinstitute.org/gsea/msigdb/cards/WINTER_HYPOXIA_METAGENE |
| DOANE_RESPONSE_TO_ANDROGEN_DN | 241 | http://www.broadinstitute.org/gsea/msigdb/cards/DOANE_RESPONSE_TO_ANDROGEN_DN |
| REACTOME_SLC_MEDIATED_TRANSMEMBRANE_TRANSPORT | 241 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SLC_MEDIATED_TRANSMEMBRANE_TRANSPORT |
| BROWNE_HCMV_INFECTION_20HR_UP | 240 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_20HR_UP |
| UDAYAKUMAR_MED1_TARGETS_DN | 240 | http://www.broadinstitute.org/gsea/msigdb/cards/UDAYAKUMAR_MED1_TARGETS_DN |
| VANTVEER_BREAST_CANCER_ESR1_DN | 240 | http://www.broadinstitute.org/gsea/msigdb/cards/VANTVEER_BREAST_CANCER_ESR1_DN |
| ALFANO_MYC_TARGETS | 239 | http://www.broadinstitute.org/gsea/msigdb/cards/ALFANO_MYC_TARGETS |
| CONCANNON_APOPTOSIS_BY_EPOXOMICIN_UP | 239 | http://www.broadinstitute.org/gsea/msigdb/cards/CONCANNON_APOPTOSIS_BY_EPOXOMICIN_UP |
| AGUIRRE_PANCREATIC_CANCER_COPY_NUMBER_DN | 238 | http://www.broadinstitute.org/gsea/msigdb/cards/AGUIRRE_PANCREATIC_CANCER_COPY_NUMBER_DN |
| GAUSSMANN_MLL_AF4_FUSION_TARGETS_G_UP | 238 | http://www.broadinstitute.org/gsea/msigdb/cards/GAUSSMANN_MLL_AF4_FUSION_TARGETS_G_UP |
| SENESE_HDAC1_AND_HDAC2_TARGETS_UP | 238 | http://www.broadinstitute.org/gsea/msigdb/cards/SENESE_HDAC1_AND_HDAC2_TARGETS_UP |
| HOSHIDA_LIVER_CANCER_SUBCLASS_S1 | 237 | http://www.broadinstitute.org/gsea/msigdb/cards/HOSHIDA_LIVER_CANCER_SUBCLASS_S1 |
| RICKMAN_TUMOR_DIFFERENTIATED_WELL_VS_POORLY_UP | 236 | http://www.broadinstitute.org/gsea/msigdb/cards/RICKMAN_TUMOR_DIFFERENTIATED_WELL_VS_POORLY_UP |
| RUTELLA_RESPONSE_TO_HGF_DN | 235 | http://www.broadinstitute.org/gsea/msigdb/cards/RUTELLA_RESPONSE_TO_HGF_DN |
| AFFAR_YY1_TARGETS_DN | 234 | http://www.broadinstitute.org/gsea/msigdb/cards/AFFAR_YY1_TARGETS_DN |
| GRAESSMANN_APOPTOSIS_BY_SERUM_DEPRIVATION_DN | 234 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAESSMANN_APOPTOSIS_BY_SERUM_DEPRIVATION_DN |
| OSWALD_HEMATOPOIETIC_STEM_CELL_IN_COLLAGEN_GEL_UP | 233 | http://www.broadinstitute.org/gsea/msigdb/cards/OSWALD_HEMATOPOIETIC_STEM_CELL_IN_COLLAGEN_GEL_UP |
| DELYS_THYROID_CANCER_DN | 232 | http://www.broadinstitute.org/gsea/msigdb/cards/DELYS_THYROID_CANCER_DN |
| SENESE_HDAC1_AND_HDAC2_TARGETS_DN | 232 | http://www.broadinstitute.org/gsea/msigdb/cards/SENESE_HDAC1_AND_HDAC2_TARGETS_DN |
| BENPORATH_MYC_TARGETS_WITH_EBOX | 230 | http://www.broadinstitute.org/gsea/msigdb/cards/BENPORATH_MYC_TARGETS_WITH_EBOX |
| COLDREN_GEFITINIB_RESISTANCE_DN | 230 | http://www.broadinstitute.org/gsea/msigdb/cards/COLDREN_GEFITINIB_RESISTANCE_DN |
| KAUFFMANN_DNA_REPAIR_GENES | 230 | http://www.broadinstitute.org/gsea/msigdb/cards/KAUFFMANN_DNA_REPAIR_GENES |
| DUTERTRE_ESTRADIOL_RESPONSE_6HR_UP | 229 | http://www.broadinstitute.org/gsea/msigdb/cards/DUTERTRE_ESTRADIOL_RESPONSE_6HR_UP |
| LINDGREN_BLADDER_CANCER_CLUSTER_3_DN | 229 | http://www.broadinstitute.org/gsea/msigdb/cards/LINDGREN_BLADDER_CANCER_CLUSTER_3_DN |
| MIYAGAWA_TARGETS_OF_EWSR1_ETS_FUSIONS_DN | 229 | http://www.broadinstitute.org/gsea/msigdb/cards/MIYAGAWA_TARGETS_OF_EWSR1_ETS_FUSIONS_DN |
| SHETH_LIVER_CANCER_VS_TXNIP_LOSS_PAM1 | 229 | http://www.broadinstitute.org/gsea/msigdb/cards/SHETH_LIVER_CANCER_VS_TXNIP_LOSS_PAM1 |
| ACEVEDO_LIVER_CANCER_WITH_H3K27ME3_DN | 228 | http://www.broadinstitute.org/gsea/msigdb/cards/ACEVEDO_LIVER_CANCER_WITH_H3K27ME3_DN |
| MARKEY_RB1_ACUTE_LOF_DN | 228 | http://www.broadinstitute.org/gsea/msigdb/cards/MARKEY_RB1_ACUTE_LOF_DN |
| MIKKELSEN_MEF_HCP_WITH_H3_UNMETHYLATED | 228 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_MEF_HCP_WITH_H3_UNMETHYLATED |
| WANG_CISPLATIN_RESPONSE_AND_XPC_DN | 228 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_CISPLATIN_RESPONSE_AND_XPC_DN |
| LEE_RECENT_THYMIC_EMIGRANT | 227 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_RECENT_THYMIC_EMIGRANT |
| MARTORIATI_MDM4_TARGETS_FETAL_LIVER_UP | 227 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTORIATI_MDM4_TARGETS_FETAL_LIVER_UP |
| PHONG_TNF_RESPONSE_VIA_P38_COMPLETE | 227 | http://www.broadinstitute.org/gsea/msigdb/cards/PHONG_TNF_RESPONSE_VIA_P38_COMPLETE |
| RAO_BOUND_BY_SALL4 | 227 | http://www.broadinstitute.org/gsea/msigdb/cards/RAO_BOUND_BY_SALL4 |
| JAATINEN_HEMATOPOIETIC_STEM_CELL_DN | 226 | http://www.broadinstitute.org/gsea/msigdb/cards/JAATINEN_HEMATOPOIETIC_STEM_CELL_DN |
| KYNG_DNA_DAMAGE_UP | 226 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_DNA_DAMAGE_UP |
| BROWNE_HCMV_INFECTION_16HR_UP | 225 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_16HR_UP |
| KYNG_WERNER_SYNDROM_AND_NORMAL_AGING_DN | 225 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_WERNER_SYNDROM_AND_NORMAL_AGING_DN |
| THEILGAARD_NEUTROPHIL_AT_SKIN_WOUND_DN | 225 | http://www.broadinstitute.org/gsea/msigdb/cards/THEILGAARD_NEUTROPHIL_AT_SKIN_WOUND_DN |
| CHEN_HOXA5_TARGETS_9HR_UP | 223 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEN_HOXA5_TARGETS_9HR_UP |
| ZHANG_RESPONSE_TO_IKK_INHIBITOR_AND_TNF_UP | 223 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_RESPONSE_TO_IKK_INHIBITOR_AND_TNF_UP |
| BROWNE_HCMV_INFECTION_1HR_DN | 222 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_1HR_DN |
| CAIRO_LIVER_DEVELOPMENT_DN | 222 | http://www.broadinstitute.org/gsea/msigdb/cards/CAIRO_LIVER_DEVELOPMENT_DN |
| REACTOME_TRANSLATION | 222 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANSLATION |
| RODWELL_AGING_KIDNEY_NO_BLOOD_UP | 222 | http://www.broadinstitute.org/gsea/msigdb/cards/RODWELL_AGING_KIDNEY_NO_BLOOD_UP |
| ZWANG_CLASS_3_TRANSIENTLY_INDUCED_BY_EGF | 222 | http://www.broadinstitute.org/gsea/msigdb/cards/ZWANG_CLASS_3_TRANSIENTLY_INDUCED_BY_EGF |
| CHANDRAN_METASTASIS_UP | 221 | http://www.broadinstitute.org/gsea/msigdb/cards/CHANDRAN_METASTASIS_UP |
| PEDRIOLI_MIR31_TARGETS_UP | 221 | http://www.broadinstitute.org/gsea/msigdb/cards/PEDRIOLI_MIR31_TARGETS_UP |
| ZHANG_TLX_TARGETS_36HR_UP | 221 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_TLX_TARGETS_36HR_UP |
| RIGGINS_TAMOXIFEN_RESISTANCE_DN | 220 | http://www.broadinstitute.org/gsea/msigdb/cards/RIGGINS_TAMOXIFEN_RESISTANCE_DN |
| VANOEVELEN_MYOGENESIS_SIN3A_TARGETS | 220 | http://www.broadinstitute.org/gsea/msigdb/cards/VANOEVELEN_MYOGENESIS_SIN3A_TARGETS |
| AMUNDSON_RESPONSE_TO_ARSENITE | 217 | http://www.broadinstitute.org/gsea/msigdb/cards/AMUNDSON_RESPONSE_TO_ARSENITE |
| REACTOME_SIGNALLING_BY_NGF | 217 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALLING_BY_NGF |
| SANSOM_APC_MYC_TARGETS | 217 | http://www.broadinstitute.org/gsea/msigdb/cards/SANSOM_APC_MYC_TARGETS |
| WIERENGA_STAT5A_TARGETS_UP | 217 | http://www.broadinstitute.org/gsea/msigdb/cards/WIERENGA_STAT5A_TARGETS_UP |
| WONG_MITOCHONDRIA_GENE_MODULE | 217 | http://www.broadinstitute.org/gsea/msigdb/cards/WONG_MITOCHONDRIA_GENE_MODULE |
| BOQUEST_STEM_CELL_DN | 216 | http://www.broadinstitute.org/gsea/msigdb/cards/BOQUEST_STEM_CELL_DN |
| KEGG_REGULATION_OF_ACTIN_CYTOSKELETON | 216 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_REGULATION_OF_ACTIN_CYTOSKELETON |
| WHITFIELD_CELL_CYCLE_G2_M | 216 | http://www.broadinstitute.org/gsea/msigdb/cards/WHITFIELD_CELL_CYCLE_G2_M |
| MARKEY_RB1_ACUTE_LOF_UP | 215 | http://www.broadinstitute.org/gsea/msigdb/cards/MARKEY_RB1_ACUTE_LOF_UP |
| YAMAZAKI_TCEB3_TARGETS_DN | 215 | http://www.broadinstitute.org/gsea/msigdb/cards/YAMAZAKI_TCEB3_TARGETS_DN |
| AFFAR_YY1_TARGETS_UP | 214 | http://www.broadinstitute.org/gsea/msigdb/cards/AFFAR_YY1_TARGETS_UP |
| BIDUS_METASTASIS_UP | 214 | http://www.broadinstitute.org/gsea/msigdb/cards/BIDUS_METASTASIS_UP |
| KIM_WT1_TARGETS_UP | 214 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_WT1_TARGETS_UP |
| LOCKWOOD_AMPLIFIED_IN_LUNG_CANCER | 214 | http://www.broadinstitute.org/gsea/msigdb/cards/LOCKWOOD_AMPLIFIED_IN_LUNG_CANCER |
| MCBRYAN_PUBERTAL_BREAST_3_4WK_UP | 214 | http://www.broadinstitute.org/gsea/msigdb/cards/MCBRYAN_PUBERTAL_BREAST_3_4WK_UP |
| TIEN_INTESTINE_PROBIOTICS_24HR_DN | 214 | http://www.broadinstitute.org/gsea/msigdb/cards/TIEN_INTESTINE_PROBIOTICS_24HR_DN |
| ZWANG_EGF_INTERVAL_DN | 214 | http://www.broadinstitute.org/gsea/msigdb/cards/ZWANG_EGF_INTERVAL_DN |
| WIERENGA_STAT5A_TARGETS_DN | 213 | http://www.broadinstitute.org/gsea/msigdb/cards/WIERENGA_STAT5A_TARGETS_DN |
| CHANG_CORE_SERUM_RESPONSE_UP | 212 | http://www.broadinstitute.org/gsea/msigdb/cards/CHANG_CORE_SERUM_RESPONSE_UP |
| REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | 212 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION |
| SANSOM_APC_TARGETS | 212 | http://www.broadinstitute.org/gsea/msigdb/cards/SANSOM_APC_TARGETS |
| BHAT_ESR1_TARGETS_NOT_VIA_AKT1_UP | 211 | http://www.broadinstitute.org/gsea/msigdb/cards/BHAT_ESR1_TARGETS_NOT_VIA_AKT1_UP |
| FOSTER_KDM1A_TARGETS_DN | 211 | http://www.broadinstitute.org/gsea/msigdb/cards/FOSTER_KDM1A_TARGETS_DN |
| GRAESSMANN_RESPONSE_TO_MC_AND_SERUM_DEPRIVATION_UP | 211 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAESSMANN_RESPONSE_TO_MC_AND_SERUM_DEPRIVATION_UP |
| CAIRO_HEPATOBLASTOMA_CLASSES_DN | 210 | http://www.broadinstitute.org/gsea/msigdb/cards/CAIRO_HEPATOBLASTOMA_CLASSES_DN |
| MIKKELSEN_NPC_HCP_WITH_H3K4ME3_AND_H3K27ME3 | 210 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_NPC_HCP_WITH_H3K4ME3_AND_H3K27ME3 |
| REACTOME_TRANSCRIPTION | 210 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANSCRIPTION |
| RIZKI_TUMOR_INVASIVENESS_3D_UP | 210 | http://www.broadinstitute.org/gsea/msigdb/cards/RIZKI_TUMOR_INVASIVENESS_3D_UP |
| SANSOM_APC_TARGETS_REQUIRE_MYC | 210 | http://www.broadinstitute.org/gsea/msigdb/cards/SANSOM_APC_TARGETS_REQUIRE_MYC |
| VERHAAK_GLIOBLASTOMA_MESENCHYMAL | 210 | http://www.broadinstitute.org/gsea/msigdb/cards/VERHAAK_GLIOBLASTOMA_MESENCHYMAL |
| VERHAAK_GLIOBLASTOMA_NEURAL | 210 | http://www.broadinstitute.org/gsea/msigdb/cards/VERHAAK_GLIOBLASTOMA_NEURAL |
| VERHAAK_GLIOBLASTOMA_PRONEURAL | 210 | http://www.broadinstitute.org/gsea/msigdb/cards/VERHAAK_GLIOBLASTOMA_PRONEURAL |
| WANG_TUMOR_INVASIVENESS_DN | 210 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_TUMOR_INVASIVENESS_DN |
| CHANG_CORE_SERUM_RESPONSE_DN | 209 | http://www.broadinstitute.org/gsea/msigdb/cards/CHANG_CORE_SERUM_RESPONSE_DN |
| GROSS_HYPOXIA_VIA_ELK3_UP | 209 | http://www.broadinstitute.org/gsea/msigdb/cards/GROSS_HYPOXIA_VIA_ELK3_UP |
| KIM_WT1_TARGETS_12HR_DN | 209 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_WT1_TARGETS_12HR_DN |
| KRIEG_KDM3A_TARGETS_NOT_HYPOXIA | 208 | http://www.broadinstitute.org/gsea/msigdb/cards/KRIEG_KDM3A_TARGETS_NOT_HYPOXIA |
| REACTOME_PLATELET_ACTIVATION_SIGNALING_AND_AGGREGATION | 208 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PLATELET_ACTIVATION_SIGNALING_AND_AGGREGATION |
| CAIRO_HEPATOBLASTOMA_UP | 207 | http://www.broadinstitute.org/gsea/msigdb/cards/CAIRO_HEPATOBLASTOMA_UP |
| EBAUER_TARGETS_OF_PAX3_FOXO1_FUSION_UP | 207 | http://www.broadinstitute.org/gsea/msigdb/cards/EBAUER_TARGETS_OF_PAX3_FOXO1_FUSION_UP |
| KARLSSON_TGFB1_TARGETS_DN | 207 | http://www.broadinstitute.org/gsea/msigdb/cards/KARLSSON_TGFB1_TARGETS_DN |
| MANALO_HYPOXIA_UP | 207 | http://www.broadinstitute.org/gsea/msigdb/cards/MANALO_HYPOXIA_UP |
| REACTOME_HIV_INFECTION | 207 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_HIV_INFECTION |
| VERHAAK_GLIOBLASTOMA_CLASSICAL | 207 | http://www.broadinstitute.org/gsea/msigdb/cards/VERHAAK_GLIOBLASTOMA_CLASSICAL |
| BERTUCCI_MEDULLARY_VS_DUCTAL_BREAST_CANCER_UP | 206 | http://www.broadinstitute.org/gsea/msigdb/cards/BERTUCCI_MEDULLARY_VS_DUCTAL_BREAST_CANCER_UP |
| BILD_MYC_ONCOGENIC_SIGNATURE | 206 | http://www.broadinstitute.org/gsea/msigdb/cards/BILD_MYC_ONCOGENIC_SIGNATURE |
| GOTZMANN_EPITHELIAL_TO_MESENCHYMAL_TRANSITION_DN | 206 | http://www.broadinstitute.org/gsea/msigdb/cards/GOTZMANN_EPITHELIAL_TO_MESENCHYMAL_TRANSITION_DN |
| LINSLEY_MIR16_TARGETS | 206 | http://www.broadinstitute.org/gsea/msigdb/cards/LINSLEY_MIR16_TARGETS |
| MIKKELSEN_MEF_ICP_WITH_H3K27ME3 | 206 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_MEF_ICP_WITH_H3K27ME3 |
| RAMALHO_STEMNESS_UP | 206 | http://www.broadinstitute.org/gsea/msigdb/cards/RAMALHO_STEMNESS_UP |
| PEREZ_TP53_AND_TP63_TARGETS | 205 | http://www.broadinstitute.org/gsea/msigdb/cards/PEREZ_TP53_AND_TP63_TARGETS |
| PICCALUGA_ANGIOIMMUNOBLASTIC_LYMPHOMA_UP | 205 | http://www.broadinstitute.org/gsea/msigdb/cards/PICCALUGA_ANGIOIMMUNOBLASTIC_LYMPHOMA_UP |
| REACTOME_GASTRIN_CREB_SIGNALLING_PATHWAY_VIA_PKC_AND_MAPK | 205 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GASTRIN_CREB_SIGNALLING_PATHWAY_VIA_PKC_AND_MAPK |
| SHEPARD_BMYB_MORPHOLINO_UP | 205 | http://www.broadinstitute.org/gsea/msigdb/cards/SHEPARD_BMYB_MORPHOLINO_UP |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_8D_DN | 205 | http://www.broadinstitute.org/gsea/msigdb/cards/TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_8D_DN |
| ROME_INSULIN_TARGETS_IN_MUSCLE_DN | 204 | http://www.broadinstitute.org/gsea/msigdb/cards/ROME_INSULIN_TARGETS_IN_MUSCLE_DN |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_MONOCYTE_UP | 204 | http://www.broadinstitute.org/gsea/msigdb/cards/TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_MONOCYTE_UP |
| GRUETZMANN_PANCREATIC_CANCER_DN | 203 | http://www.broadinstitute.org/gsea/msigdb/cards/GRUETZMANN_PANCREATIC_CANCER_DN |
| PAL_PRMT5_TARGETS_UP | 203 | http://www.broadinstitute.org/gsea/msigdb/cards/PAL_PRMT5_TARGETS_UP |
| PENG_RAPAMYCIN_RESPONSE_UP | 203 | http://www.broadinstitute.org/gsea/msigdb/cards/PENG_RAPAMYCIN_RESPONSE_UP |
| REACTOME_INFLUENZA_LIFE_CYCLE | 203 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INFLUENZA_LIFE_CYCLE |
| FUJII_YBX1_TARGETS_DN | 202 | http://www.broadinstitute.org/gsea/msigdb/cards/FUJII_YBX1_TARGETS_DN |
| WANG_CISPLATIN_RESPONSE_AND_XPC_UP | 202 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_CISPLATIN_RESPONSE_AND_XPC_UP |
| HOLLMANN_APOPTOSIS_VIA_CD40_UP | 201 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLMANN_APOPTOSIS_VIA_CD40_UP |
| KEGG_FOCAL_ADHESION | 201 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_FOCAL_ADHESION |
| KIM_MYC_AMPLIFICATION_TARGETS_UP | 201 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_MYC_AMPLIFICATION_TARGETS_UP |
| LEE_DIFFERENTIATING_T_LYMPHOCYTE | 200 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_DIFFERENTIATING_T_LYMPHOCYTE |
| REACTOME_METABOLISM_OF_AMINO_ACIDS_AND_DERIVATIVES | 200 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_METABOLISM_OF_AMINO_ACIDS_AND_DERIVATIVES |
| SHEPARD_BMYB_MORPHOLINO_DN | 200 | http://www.broadinstitute.org/gsea/msigdb/cards/SHEPARD_BMYB_MORPHOLINO_DN |
| PLASARI_TGFB1_TARGETS_10HR_UP | 199 | http://www.broadinstitute.org/gsea/msigdb/cards/PLASARI_TGFB1_TARGETS_10HR_UP |
| WAMUNYOKOLI_OVARIAN_CANCER_LMP_DN | 199 | http://www.broadinstitute.org/gsea/msigdb/cards/WAMUNYOKOLI_OVARIAN_CANCER_LMP_DN |
| ALONSO_METASTASIS_UP | 198 | http://www.broadinstitute.org/gsea/msigdb/cards/ALONSO_METASTASIS_UP |
| REACTOME_PHOSPHOLIPID_METABOLISM | 198 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PHOSPHOLIPID_METABOLISM |
| TURASHVILI_BREAST_DUCTAL_CARCINOMA_VS_DUCTAL_NORMAL_DN | 198 | http://www.broadinstitute.org/gsea/msigdb/cards/TURASHVILI_BREAST_DUCTAL_CARCINOMA_VS_DUCTAL_NORMAL_DN |
| WELCSH_BRCA1_TARGETS_UP | 198 | http://www.broadinstitute.org/gsea/msigdb/cards/WELCSH_BRCA1_TARGETS_UP |
| MCBRYAN_PUBERTAL_BREAST_6_7WK_UP | 197 | http://www.broadinstitute.org/gsea/msigdb/cards/MCBRYAN_PUBERTAL_BREAST_6_7WK_UP |
| SHEPARD_CRUSH_AND_BURN_MUTANT_UP | 197 | http://www.broadinstitute.org/gsea/msigdb/cards/SHEPARD_CRUSH_AND_BURN_MUTANT_UP |
| KONDO_PROSTATE_CANCER_WITH_H3K27ME3 | 196 | http://www.broadinstitute.org/gsea/msigdb/cards/KONDO_PROSTATE_CANCER_WITH_H3K27ME3 |
| MCBRYAN_PUBERTAL_BREAST_4_5WK_DN | 196 | http://www.broadinstitute.org/gsea/msigdb/cards/MCBRYAN_PUBERTAL_BREAST_4_5WK_DN |
| SHEDDEN_LUNG_CANCER_GOOD_SURVIVAL_A4 | 196 | http://www.broadinstitute.org/gsea/msigdb/cards/SHEDDEN_LUNG_CANCER_GOOD_SURVIVAL_A4 |
| ZHENG_FOXP3_TARGETS_IN_THYMUS_UP | 196 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHENG_FOXP3_TARGETS_IN_THYMUS_UP |
| KYNG_DNA_DAMAGE_DN | 195 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_DNA_DAMAGE_DN |
| REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | 195 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_G_ALPHA_I_SIGNALLING_EVENTS |
| STARK_PREFRONTAL_CORTEX_22Q11_DELETION_UP | 195 | http://www.broadinstitute.org/gsea/msigdb/cards/STARK_PREFRONTAL_CORTEX_22Q11_DELETION_UP |
| KASLER_HDAC7_TARGETS_1_UP | 194 | http://www.broadinstitute.org/gsea/msigdb/cards/KASLER_HDAC7_TARGETS_1_UP |
| PROVENZANI_METASTASIS_UP | 194 | http://www.broadinstitute.org/gsea/msigdb/cards/PROVENZANI_METASTASIS_UP |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_10D_UP | 194 | http://www.broadinstitute.org/gsea/msigdb/cards/TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_10D_UP |
| CHIANG_LIVER_CANCER_SUBCLASS_UNANNOTATED_DN | 193 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIANG_LIVER_CANCER_SUBCLASS_UNANNOTATED_DN |
| MOHANKUMAR_TLX1_TARGETS_DN | 193 | http://www.broadinstitute.org/gsea/msigdb/cards/MOHANKUMAR_TLX1_TARGETS_DN |
| PYEON_CANCER_HEAD_AND_NECK_VS_CERVICAL_UP | 193 | http://www.broadinstitute.org/gsea/msigdb/cards/PYEON_CANCER_HEAD_AND_NECK_VS_CERVICAL_UP |
| REACTOME_DNA_REPLICATION | 192 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DNA_REPLICATION |
| GAUSSMANN_MLL_AF4_FUSION_TARGETS_A_UP | 191 | http://www.broadinstitute.org/gsea/msigdb/cards/GAUSSMANN_MLL_AF4_FUSION_TARGETS_A_UP |
| RIGGI_EWING_SARCOMA_PROGENITOR_DN | 191 | http://www.broadinstitute.org/gsea/msigdb/cards/RIGGI_EWING_SARCOMA_PROGENITOR_DN |
| YAGI_AML_FAB_MARKERS | 191 | http://www.broadinstitute.org/gsea/msigdb/cards/YAGI_AML_FAB_MARKERS |
| KEGG_CHEMOKINE_SIGNALING_PATHWAY | 190 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_CHEMOKINE_SIGNALING_PATHWAY |
| BURTON_ADIPOGENESIS_6 | 189 | http://www.broadinstitute.org/gsea/msigdb/cards/BURTON_ADIPOGENESIS_6 |
| BOYAULT_LIVER_CANCER_SUBCLASS_G3_UP | 188 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYAULT_LIVER_CANCER_SUBCLASS_G3_UP |
| REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | 188 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS |
| REACTOME_POST_TRANSLATIONAL_PROTEIN_MODIFICATION | 188 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_POST_TRANSLATIONAL_PROTEIN_MODIFICATION |
| BAKKER_FOXO3_TARGETS_DN | 187 | http://www.broadinstitute.org/gsea/msigdb/cards/BAKKER_FOXO3_TARGETS_DN |
| PENG_LEUCINE_DEPRIVATION_DN | 187 | http://www.broadinstitute.org/gsea/msigdb/cards/PENG_LEUCINE_DEPRIVATION_DN |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_HSC_DN | 187 | http://www.broadinstitute.org/gsea/msigdb/cards/TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_HSC_DN |
| HAMAI_APOPTOSIS_VIA_TRAIL_DN | 186 | http://www.broadinstitute.org/gsea/msigdb/cards/HAMAI_APOPTOSIS_VIA_TRAIL_DN |
| REACTOME_TRANSMISSION_ACROSS_CHEMICAL_SYNAPSES | 186 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANSMISSION_ACROSS_CHEMICAL_SYNAPSES |
| WANG_CLIM2_TARGETS_DN | 186 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_CLIM2_TARGETS_DN |
| BYSTRYKH_HEMATOPOIESIS_STEM_CELL_AND_BRAIN_QTL_TRANS | 185 | http://www.broadinstitute.org/gsea/msigdb/cards/BYSTRYKH_HEMATOPOIESIS_STEM_CELL_AND_BRAIN_QTL_TRANS |
| DARWICHE_SKIN_TUMOR_PROMOTER_DN | 185 | http://www.broadinstitute.org/gsea/msigdb/cards/DARWICHE_SKIN_TUMOR_PROMOTER_DN |
| GAUSSMANN_MLL_AF4_FUSION_TARGETS_F_UP | 185 | http://www.broadinstitute.org/gsea/msigdb/cards/GAUSSMANN_MLL_AF4_FUSION_TARGETS_F_UP |
| KEGG_HUNTINGTONS_DISEASE | 185 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_HUNTINGTONS_DISEASE |
| LEE_LIVER_CANCER_SURVIVAL_UP | 185 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_SURVIVAL_UP |
| SHEPARD_CRUSH_AND_BURN_MUTANT_DN | 185 | http://www.broadinstitute.org/gsea/msigdb/cards/SHEPARD_CRUSH_AND_BURN_MUTANT_DN |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_HSC_UP | 185 | http://www.broadinstitute.org/gsea/msigdb/cards/TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_HSC_UP |
| ZHANG_TLX_TARGETS_36HR_DN | 185 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_TLX_TARGETS_36HR_DN |
| ALCALAY_AML_BY_NPM1_LOCALIZATION_DN | 184 | http://www.broadinstitute.org/gsea/msigdb/cards/ALCALAY_AML_BY_NPM1_LOCALIZATION_DN |
| DOANE_RESPONSE_TO_ANDROGEN_UP | 184 | http://www.broadinstitute.org/gsea/msigdb/cards/DOANE_RESPONSE_TO_ANDROGEN_UP |
| REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS | 184 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS |
| WU_CELL_MIGRATION | 184 | http://www.broadinstitute.org/gsea/msigdb/cards/WU_CELL_MIGRATION |
| KEGG_ENDOCYTOSIS | 183 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ENDOCYTOSIS |
| OKUMURA_INFLAMMATORY_RESPONSE_LPS | 183 | http://www.broadinstitute.org/gsea/msigdb/cards/OKUMURA_INFLAMMATORY_RESPONSE_LPS |
| VERHAAK_AML_WITH_NPM1_MUTATED_UP | 183 | http://www.broadinstitute.org/gsea/msigdb/cards/VERHAAK_AML_WITH_NPM1_MUTATED_UP |
| ZHONG_RESPONSE_TO_AZACITIDINE_AND_TSA_UP | 183 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHONG_RESPONSE_TO_AZACITIDINE_AND_TSA_UP |
| DAVICIONI_MOLECULAR_ARMS_VS_ERMS_DN | 182 | http://www.broadinstitute.org/gsea/msigdb/cards/DAVICIONI_MOLECULAR_ARMS_VS_ERMS_DN |
| FERNANDEZ_BOUND_BY_MYC | 182 | http://www.broadinstitute.org/gsea/msigdb/cards/FERNANDEZ_BOUND_BY_MYC |
| LI_WILMS_TUMOR_VS_FETAL_KIDNEY_1_UP | 182 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_WILMS_TUMOR_VS_FETAL_KIDNEY_1_UP |
| RAO_BOUND_BY_SALL4_ISOFORM_A | 182 | http://www.broadinstitute.org/gsea/msigdb/cards/RAO_BOUND_BY_SALL4_ISOFORM_A |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_3D_UP | 182 | http://www.broadinstitute.org/gsea/msigdb/cards/TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_3D_UP |
| WHITFIELD_CELL_CYCLE_G2 | 182 | http://www.broadinstitute.org/gsea/msigdb/cards/WHITFIELD_CELL_CYCLE_G2 |
| DARWICHE_SQUAMOUS_CELL_CARCINOMA_DN | 181 | http://www.broadinstitute.org/gsea/msigdb/cards/DARWICHE_SQUAMOUS_CELL_CARCINOMA_DN |
| GRAHAM_CML_DIVIDING_VS_NORMAL_QUIESCENT_UP | 181 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAHAM_CML_DIVIDING_VS_NORMAL_QUIESCENT_UP |
| IWANAGA_CARCINOGENESIS_BY_KRAS_PTEN_UP | 181 | http://www.broadinstitute.org/gsea/msigdb/cards/IWANAGA_CARCINOGENESIS_BY_KRAS_PTEN_UP |
| JISON_SICKLE_CELL_DISEASE_DN | 181 | http://www.broadinstitute.org/gsea/msigdb/cards/JISON_SICKLE_CELL_DISEASE_DN |
| JISON_SICKLE_CELL_DISEASE_UP | 181 | http://www.broadinstitute.org/gsea/msigdb/cards/JISON_SICKLE_CELL_DISEASE_UP |
| LASTOWSKA_NEUROBLASTOMA_COPY_NUMBER_UP | 181 | http://www.broadinstitute.org/gsea/msigdb/cards/LASTOWSKA_NEUROBLASTOMA_COPY_NUMBER_UP |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_17 | 181 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_17 |
| BROWNE_HCMV_INFECTION_48HR_UP | 180 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_48HR_UP |
| DARWICHE_PAPILLOMA_RISK_HIGH_DN | 180 | http://www.broadinstitute.org/gsea/msigdb/cards/DARWICHE_PAPILLOMA_RISK_HIGH_DN |
| GINESTIER_BREAST_CANCER_20Q13_AMPLIFICATION_DN | 180 | http://www.broadinstitute.org/gsea/msigdb/cards/GINESTIER_BREAST_CANCER_20Q13_AMPLIFICATION_DN |
| PURBEY_TARGETS_OF_CTBP1_AND_SATB1_DN | 180 | http://www.broadinstitute.org/gsea/msigdb/cards/PURBEY_TARGETS_OF_CTBP1_AND_SATB1_DN |
| SARRIO_EPITHELIAL_MESENCHYMAL_TRANSITION_UP | 180 | http://www.broadinstitute.org/gsea/msigdb/cards/SARRIO_EPITHELIAL_MESENCHYMAL_TRANSITION_UP |
| BENPORATH_NOS_TARGETS | 179 | http://www.broadinstitute.org/gsea/msigdb/cards/BENPORATH_NOS_TARGETS |
| CHIANG_LIVER_CANCER_SUBCLASS_PROLIFERATION_DN | 179 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIANG_LIVER_CANCER_SUBCLASS_PROLIFERATION_DN |
| REACTOME_SRP_DEPENDENT_COTRANSLATIONAL_PROTEIN_TARGETING_TO_MEMBRANE | 179 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SRP_DEPENDENT_COTRANSLATIONAL_PROTEIN_TARGETING_TO_MEMBRANE |
| BROWNE_HCMV_INFECTION_18HR_DN | 178 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_18HR_DN |
| BROWNE_HCMV_INFECTION_18HR_UP | 178 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_18HR_UP |
| CHIANG_LIVER_CANCER_SUBCLASS_PROLIFERATION_UP | 178 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIANG_LIVER_CANCER_SUBCLASS_PROLIFERATION_UP |
| KEGG_CALCIUM_SIGNALING_PATHWAY | 178 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_CALCIUM_SIGNALING_PATHWAY |
| LI_AMPLIFIED_IN_LUNG_CANCER | 178 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_AMPLIFIED_IN_LUNG_CANCER |
| CASORELLI_ACUTE_PROMYELOCYTIC_LEUKEMIA_UP | 177 | http://www.broadinstitute.org/gsea/msigdb/cards/CASORELLI_ACUTE_PROMYELOCYTIC_LEUKEMIA_UP |
| HAHTOLA_MYCOSIS_FUNGOIDES_SKIN_UP | 177 | http://www.broadinstitute.org/gsea/msigdb/cards/HAHTOLA_MYCOSIS_FUNGOIDES_SKIN_UP |
| CHIANG_LIVER_CANCER_SUBCLASS_CTNNB1_UP | 176 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIANG_LIVER_CANCER_SUBCLASS_CTNNB1_UP |
| GARCIA_TARGETS_OF_FLI1_AND_DAX1_DN | 176 | http://www.broadinstitute.org/gsea/msigdb/cards/GARCIA_TARGETS_OF_FLI1_AND_DAX1_DN |
| MARTORIATI_MDM4_TARGETS_NEUROEPITHELIUM_UP | 176 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTORIATI_MDM4_TARGETS_NEUROEPITHELIUM_UP |
| NAGASHIMA_NRG1_SIGNALING_UP | 176 | http://www.broadinstitute.org/gsea/msigdb/cards/NAGASHIMA_NRG1_SIGNALING_UP |
| REACTOME_3_UTR_MEDIATED_TRANSLATIONAL_REGULATION | 176 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_3_UTR_MEDIATED_TRANSLATIONAL_REGULATION |
| REACTOME_NONSENSE_MEDIATED_DECAY_ENHANCED_BY_THE_EXON_JUNCTION_COMPLEX | 176 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NONSENSE_MEDIATED_DECAY_ENHANCED_BY_THE_EXON_JUNCTION_COMPLEX |
| SASAKI_ADULT_T_CELL_LEUKEMIA | 176 | http://www.broadinstitute.org/gsea/msigdb/cards/SASAKI_ADULT_T_CELL_LEUKEMIA |
| SCHAEFFER_PROSTATE_DEVELOPMENT_6HR_UP | 176 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_PROSTATE_DEVELOPMENT_6HR_UP |
| FIRESTEIN_PROLIFERATION | 175 | http://www.broadinstitute.org/gsea/msigdb/cards/FIRESTEIN_PROLIFERATION |
| LEE_LIVER_CANCER_SURVIVAL_DN | 175 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_SURVIVAL_DN |
| SENGUPTA_NASOPHARYNGEAL_CARCINOMA_WITH_LMP1_DN | 175 | http://www.broadinstitute.org/gsea/msigdb/cards/SENGUPTA_NASOPHARYNGEAL_CARCINOMA_WITH_LMP1_DN |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_16D_UP | 175 | http://www.broadinstitute.org/gsea/msigdb/cards/TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_16D_UP |
| VECCHI_GASTRIC_CANCER_ADVANCED_VS_EARLY_UP | 175 | http://www.broadinstitute.org/gsea/msigdb/cards/VECCHI_GASTRIC_CANCER_ADVANCED_VS_EARLY_UP |
| YAMAZAKI_TCEB3_TARGETS_UP | 175 | http://www.broadinstitute.org/gsea/msigdb/cards/YAMAZAKI_TCEB3_TARGETS_UP |
| ACEVEDO_NORMAL_TISSUE_ADJACENT_TO_LIVER_TUMOR_UP | 174 | http://www.broadinstitute.org/gsea/msigdb/cards/ACEVEDO_NORMAL_TISSUE_ADJACENT_TO_LIVER_TUMOR_UP |
| MIKKELSEN_IPS_LCP_WITH_H3K4ME3 | 174 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_IPS_LCP_WITH_H3K4ME3 |
| SENGUPTA_EBNA1_ANTICORRELATED | 173 | http://www.broadinstitute.org/gsea/msigdb/cards/SENGUPTA_EBNA1_ANTICORRELATED |
| CONCANNON_APOPTOSIS_BY_EPOXOMICIN_DN | 172 | http://www.broadinstitute.org/gsea/msigdb/cards/CONCANNON_APOPTOSIS_BY_EPOXOMICIN_DN |
| MOREAUX_MULTIPLE_MYELOMA_BY_TACI_DN | 172 | http://www.broadinstitute.org/gsea/msigdb/cards/MOREAUX_MULTIPLE_MYELOMA_BY_TACI_DN |
| REACTOME_MITOTIC_M_M_G1_PHASES | 172 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MITOTIC_M_M_G1_PHASES |
| SMID_BREAST_CANCER_LUMINAL_B_UP | 172 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_LUMINAL_B_UP |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_13 | 172 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_13 |
| AMUNDSON_POOR_SURVIVAL_AFTER_GAMMA_RADIATION_2G | 171 | http://www.broadinstitute.org/gsea/msigdb/cards/AMUNDSON_POOR_SURVIVAL_AFTER_GAMMA_RADIATION_2G |
| ELVIDGE_HYPOXIA_UP | 171 | http://www.broadinstitute.org/gsea/msigdb/cards/ELVIDGE_HYPOXIA_UP |
| GOLDRATH_HOMEOSTATIC_PROLIFERATION | 171 | http://www.broadinstitute.org/gsea/msigdb/cards/GOLDRATH_HOMEOSTATIC_PROLIFERATION |
| CHIANG_LIVER_CANCER_SUBCLASS_CTNNB1_DN | 170 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIANG_LIVER_CANCER_SUBCLASS_CTNNB1_DN |
| FIGUEROA_AML_METHYLATION_CLUSTER_3_UP | 170 | http://www.broadinstitute.org/gsea/msigdb/cards/FIGUEROA_AML_METHYLATION_CLUSTER_3_UP |
| GAUSSMANN_MLL_AF4_FUSION_TARGETS_C_UP | 170 | http://www.broadinstitute.org/gsea/msigdb/cards/GAUSSMANN_MLL_AF4_FUSION_TARGETS_C_UP |
| IWANAGA_CARCINOGENESIS_BY_KRAS_UP | 170 | http://www.broadinstitute.org/gsea/msigdb/cards/IWANAGA_CARCINOGENESIS_BY_KRAS_UP |
| BERTUCCI_MEDULLARY_VS_DUCTAL_BREAST_CANCER_DN | 169 | http://www.broadinstitute.org/gsea/msigdb/cards/BERTUCCI_MEDULLARY_VS_DUCTAL_BREAST_CANCER_DN |
| KATSANOU_ELAVL1_TARGETS_UP | 169 | http://www.broadinstitute.org/gsea/msigdb/cards/KATSANOU_ELAVL1_TARGETS_UP |
| KEGG_ALZHEIMERS_DISEASE | 169 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ALZHEIMERS_DISEASE |
| MCBRYAN_PUBERTAL_TGFB1_TARGETS_UP | 169 | http://www.broadinstitute.org/gsea/msigdb/cards/MCBRYAN_PUBERTAL_TGFB1_TARGETS_UP |
| ONDER_CDH1_TARGETS_1_DN | 169 | http://www.broadinstitute.org/gsea/msigdb/cards/ONDER_CDH1_TARGETS_1_DN |
| PENG_GLUCOSE_DEPRIVATION_DN | 169 | http://www.broadinstitute.org/gsea/msigdb/cards/PENG_GLUCOSE_DEPRIVATION_DN |
| REACTOME_INFLUENZA_VIRAL_RNA_TRANSCRIPTION_AND_REPLICATION | 169 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INFLUENZA_VIRAL_RNA_TRANSCRIPTION_AND_REPLICATION |
| UEDA_PERIFERAL_CLOCK | 169 | http://www.broadinstitute.org/gsea/msigdb/cards/UEDA_PERIFERAL_CLOCK |
| PUJANA_XPRSS_INT_NETWORK | 168 | http://www.broadinstitute.org/gsea/msigdb/cards/PUJANA_XPRSS_INT_NETWORK |
| REACTOME_FATTY_ACID_TRIACYLGLYCEROL_AND_KETONE_BODY_METABOLISM | 168 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FATTY_ACID_TRIACYLGLYCEROL_AND_KETONE_BODY_METABOLISM |
| FERREIRA_EWINGS_SARCOMA_UNSTABLE_VS_STABLE_UP | 167 | http://www.broadinstitute.org/gsea/msigdb/cards/FERREIRA_EWINGS_SARCOMA_UNSTABLE_VS_STABLE_UP |
| TIEN_INTESTINE_PROBIOTICS_6HR_DN | 167 | http://www.broadinstitute.org/gsea/msigdb/cards/TIEN_INTESTINE_PROBIOTICS_6HR_DN |
| VANTVEER_BREAST_CANCER_ESR1_UP | 167 | http://www.broadinstitute.org/gsea/msigdb/cards/VANTVEER_BREAST_CANCER_ESR1_UP |
| CAIRO_LIVER_DEVELOPMENT_UP | 166 | http://www.broadinstitute.org/gsea/msigdb/cards/CAIRO_LIVER_DEVELOPMENT_UP |
| SMIRNOV_RESPONSE_TO_IR_6HR_UP | 166 | http://www.broadinstitute.org/gsea/msigdb/cards/SMIRNOV_RESPONSE_TO_IR_6HR_UP |
| BLALOCK_ALZHEIMERS_DISEASE_INCIPIENT_DN | 165 | http://www.broadinstitute.org/gsea/msigdb/cards/BLALOCK_ALZHEIMERS_DISEASE_INCIPIENT_DN |
| BROWN_MYELOID_CELL_DEVELOPMENT_UP | 165 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWN_MYELOID_CELL_DEVELOPMENT_UP |
| DARWICHE_PAPILLOMA_RISK_LOW_DN | 165 | http://www.broadinstitute.org/gsea/msigdb/cards/DARWICHE_PAPILLOMA_RISK_LOW_DN |
| LIU_CMYB_TARGETS_UP | 165 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_CMYB_TARGETS_UP |
| PASQUALUCCI_LYMPHOMA_BY_GC_STAGE_DN | 165 | http://www.broadinstitute.org/gsea/msigdb/cards/PASQUALUCCI_LYMPHOMA_BY_GC_STAGE_DN |
| VART_KSHV_INFECTION_ANGIOGENIC_MARKERS_UP | 165 | http://www.broadinstitute.org/gsea/msigdb/cards/VART_KSHV_INFECTION_ANGIOGENIC_MARKERS_UP |
| AMIT_EGF_RESPONSE_480_HELA | 164 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_EGF_RESPONSE_480_HELA |
| KIM_WT1_TARGETS_8HR_UP | 164 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_WT1_TARGETS_8HR_UP |
| MARTORIATI_MDM4_TARGETS_NEUROEPITHELIUM_DN | 164 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTORIATI_MDM4_TARGETS_NEUROEPITHELIUM_DN |
| HEDENFALK_BREAST_CANCER_BRCA1_VS_BRCA2 | 163 | http://www.broadinstitute.org/gsea/msigdb/cards/HEDENFALK_BREAST_CANCER_BRCA1_VS_BRCA2 |
| HOFFMANN_LARGE_TO_SMALL_PRE_BII_LYMPHOCYTE_UP | 163 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFFMANN_LARGE_TO_SMALL_PRE_BII_LYMPHOCYTE_UP |
| LIN_NPAS4_TARGETS_UP | 163 | http://www.broadinstitute.org/gsea/msigdb/cards/LIN_NPAS4_TARGETS_UP |
| LI_WILMS_TUMOR_VS_FETAL_KIDNEY_1_DN | 163 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_WILMS_TUMOR_VS_FETAL_KIDNEY_1_DN |
| SERVITJA_ISLET_HNF1A_TARGETS_UP | 163 | http://www.broadinstitute.org/gsea/msigdb/cards/SERVITJA_ISLET_HNF1A_TARGETS_UP |
| DARWICHE_PAPILLOMA_RISK_LOW_UP | 162 | http://www.broadinstitute.org/gsea/msigdb/cards/DARWICHE_PAPILLOMA_RISK_LOW_UP |
| KIM_WT1_TARGETS_12HR_UP | 162 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_WT1_TARGETS_12HR_UP |
| MIKKELSEN_MCV6_LCP_WITH_H3K4ME3 | 162 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_MCV6_LCP_WITH_H3K4ME3 |
| MULLIGHAN_NPM1_SIGNATURE_3_DN | 162 | http://www.broadinstitute.org/gsea/msigdb/cards/MULLIGHAN_NPM1_SIGNATURE_3_DN |
| PETROVA_ENDOTHELIUM_LYMPHATIC_VS_BLOOD_DN | 162 | http://www.broadinstitute.org/gsea/msigdb/cards/PETROVA_ENDOTHELIUM_LYMPHATIC_VS_BLOOD_DN |
| WHITFIELD_CELL_CYCLE_S | 162 | http://www.broadinstitute.org/gsea/msigdb/cards/WHITFIELD_CELL_CYCLE_S |
| WINNEPENNINCKX_MELANOMA_METASTASIS_UP | 162 | http://www.broadinstitute.org/gsea/msigdb/cards/WINNEPENNINCKX_MELANOMA_METASTASIS_UP |
| BANDRES_RESPONSE_TO_CARMUSTIN_MGMT_48HR_DN | 161 | http://www.broadinstitute.org/gsea/msigdb/cards/BANDRES_RESPONSE_TO_CARMUSTIN_MGMT_48HR_DN |
| BIDUS_METASTASIS_DN | 161 | http://www.broadinstitute.org/gsea/msigdb/cards/BIDUS_METASTASIS_DN |
| REACTOME_MRNA_PROCESSING | 161 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MRNA_PROCESSING |
| ASTON_MAJOR_DEPRESSIVE_DISORDER_DN | 160 | http://www.broadinstitute.org/gsea/msigdb/cards/ASTON_MAJOR_DEPRESSIVE_DISORDER_DN |
| BROWNE_HCMV_INFECTION_6HR_DN | 160 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_6HR_DN |
| GAVIN_FOXP3_TARGETS_CLUSTER_P3 | 160 | http://www.broadinstitute.org/gsea/msigdb/cards/GAVIN_FOXP3_TARGETS_CLUSTER_P3 |
| PHONG_TNF_RESPONSE_VIA_P38_PARTIAL | 160 | http://www.broadinstitute.org/gsea/msigdb/cards/PHONG_TNF_RESPONSE_VIA_P38_PARTIAL |
| KEGG_PURINE_METABOLISM | 159 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PURINE_METABOLISM |
| RAY_TUMORIGENESIS_BY_ERBB2_CDC25A_DN | 159 | http://www.broadinstitute.org/gsea/msigdb/cards/RAY_TUMORIGENESIS_BY_ERBB2_CDC25A_DN |
| REACTOME_INTERFERON_SIGNALING | 159 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INTERFERON_SIGNALING |
| SCHLOSSER_MYC_TARGETS_REPRESSED_BY_SERUM | 159 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHLOSSER_MYC_TARGETS_REPRESSED_BY_SERUM |
| NIKOLSKY_BREAST_CANCER_11Q12_Q14_AMPLICON | 158 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_11Q12_Q14_AMPLICON |
| RODRIGUES_NTN1_TARGETS_DN | 158 | http://www.broadinstitute.org/gsea/msigdb/cards/RODRIGUES_NTN1_TARGETS_DN |
| SMIRNOV_CIRCULATING_ENDOTHELIOCYTES_IN_CANCER_UP | 158 | http://www.broadinstitute.org/gsea/msigdb/cards/SMIRNOV_CIRCULATING_ENDOTHELIOCYTES_IN_CANCER_UP |
| NIKOLSKY_BREAST_CANCER_8Q23_Q24_AMPLICON | 157 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_8Q23_Q24_AMPLICON |
| NING_CHRONIC_OBSTRUCTIVE_PULMONARY_DISEASE_UP | 157 | http://www.broadinstitute.org/gsea/msigdb/cards/NING_CHRONIC_OBSTRUCTIVE_PULMONARY_DISEASE_UP |
| PANGAS_TUMOR_SUPPRESSION_BY_SMAD1_AND_SMAD5_DN | 157 | http://www.broadinstitute.org/gsea/msigdb/cards/PANGAS_TUMOR_SUPPRESSION_BY_SMAD1_AND_SMAD5_DN |
| SERVITJA_LIVER_HNF1A_TARGETS_DN | 157 | http://www.broadinstitute.org/gsea/msigdb/cards/SERVITJA_LIVER_HNF1A_TARGETS_DN |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_8D_UP | 157 | http://www.broadinstitute.org/gsea/msigdb/cards/TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_8D_UP |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_ERYTHROCYTE_UP | 157 | http://www.broadinstitute.org/gsea/msigdb/cards/TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_ERYTHROCYTE_UP |
| BROWNE_HCMV_INFECTION_14HR_UP | 156 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_14HR_UP |
| FOSTER_TOLERANT_MACROPHAGE_UP | 156 | http://www.broadinstitute.org/gsea/msigdb/cards/FOSTER_TOLERANT_MACROPHAGE_UP |
| GROSS_HYPOXIA_VIA_ELK3_DN | 156 | http://www.broadinstitute.org/gsea/msigdb/cards/GROSS_HYPOXIA_VIA_ELK3_DN |
| KEGG_JAK_STAT_SIGNALING_PATHWAY | 155 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_JAK_STAT_SIGNALING_PATHWAY |
| SARRIO_EPITHELIAL_MESENCHYMAL_TRANSITION_DN | 154 | http://www.broadinstitute.org/gsea/msigdb/cards/SARRIO_EPITHELIAL_MESENCHYMAL_TRANSITION_DN |
| CHIARADONNA_NEOPLASTIC_TRANSFORMATION_CDC25_DN | 153 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIARADONNA_NEOPLASTIC_TRANSFORMATION_CDC25_DN |
| DAZARD_UV_RESPONSE_CLUSTER_G6 | 153 | http://www.broadinstitute.org/gsea/msigdb/cards/DAZARD_UV_RESPONSE_CLUSTER_G6 |
| DURAND_STROMA_MAX_DN | 153 | http://www.broadinstitute.org/gsea/msigdb/cards/DURAND_STROMA_MAX_DN |
| LU_AGING_BRAIN_DN | 153 | http://www.broadinstitute.org/gsea/msigdb/cards/LU_AGING_BRAIN_DN |
| REACTOME_PEPTIDE_CHAIN_ELONGATION | 153 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PEPTIDE_CHAIN_ELONGATION |
| RUIZ_TNC_TARGETS_UP | 153 | http://www.broadinstitute.org/gsea/msigdb/cards/RUIZ_TNC_TARGETS_UP |
| SHETH_LIVER_CANCER_VS_TXNIP_LOSS_PAM2 | 153 | http://www.broadinstitute.org/gsea/msigdb/cards/SHETH_LIVER_CANCER_VS_TXNIP_LOSS_PAM2 |
| WAKABAYASHI_ADIPOGENESIS_PPARG_RXRA_BOUND_36HR | 152 | http://www.broadinstitute.org/gsea/msigdb/cards/WAKABAYASHI_ADIPOGENESIS_PPARG_RXRA_BOUND_36HR |
| WILCOX_PRESPONSE_TO_ROGESTERONE_UP | 152 | http://www.broadinstitute.org/gsea/msigdb/cards/WILCOX_PRESPONSE_TO_ROGESTERONE_UP |
| BOSCO_ALLERGEN_INDUCED_TH2_ASSOCIATED_MODULE | 151 | http://www.broadinstitute.org/gsea/msigdb/cards/BOSCO_ALLERGEN_INDUCED_TH2_ASSOCIATED_MODULE |
| FLECHNER_PBL_KIDNEY_TRANSPLANT_OK_VS_DONOR_UP | 151 | http://www.broadinstitute.org/gsea/msigdb/cards/FLECHNER_PBL_KIDNEY_TRANSPLANT_OK_VS_DONOR_UP |
| IGLESIAS_E2F_TARGETS_UP | 151 | http://www.broadinstitute.org/gsea/msigdb/cards/IGLESIAS_E2F_TARGETS_UP |
| KEGG_WNT_SIGNALING_PATHWAY | 151 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_WNT_SIGNALING_PATHWAY |
| SOTIRIOU_BREAST_CANCER_GRADE_1_VS_3_UP | 151 | http://www.broadinstitute.org/gsea/msigdb/cards/SOTIRIOU_BREAST_CANCER_GRADE_1_VS_3_UP |
| BROWNE_HCMV_INFECTION_30MIN_DN | 150 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_30MIN_DN |
| LANDIS_ERBB2_BREAST_TUMORS_324_UP | 150 | http://www.broadinstitute.org/gsea/msigdb/cards/LANDIS_ERBB2_BREAST_TUMORS_324_UP |
| RODWELL_AGING_KIDNEY_NO_BLOOD_DN | 150 | http://www.broadinstitute.org/gsea/msigdb/cards/RODWELL_AGING_KIDNEY_NO_BLOOD_DN |
| GOZGIT_ESR1_TARGETS_UP | 149 | http://www.broadinstitute.org/gsea/msigdb/cards/GOZGIT_ESR1_TARGETS_UP |
| HUANG_GATA2_TARGETS_UP | 149 | http://www.broadinstitute.org/gsea/msigdb/cards/HUANG_GATA2_TARGETS_UP |
| IVANOVA_HEMATOPOIESIS_INTERMEDIATE_PROGENITOR | 149 | http://www.broadinstitute.org/gsea/msigdb/cards/IVANOVA_HEMATOPOIESIS_INTERMEDIATE_PROGENITOR |
| LANDIS_ERBB2_BREAST_TUMORS_324_DN | 149 | http://www.broadinstitute.org/gsea/msigdb/cards/LANDIS_ERBB2_BREAST_TUMORS_324_DN |
| NIKOLSKY_BREAST_CANCER_20Q12_Q13_AMPLICON | 149 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_20Q12_Q13_AMPLICON |
| OHGUCHI_LIVER_HNF4A_TARGETS_DN | 149 | http://www.broadinstitute.org/gsea/msigdb/cards/OHGUCHI_LIVER_HNF4A_TARGETS_DN |
| TSAI_RESPONSE_TO_IONIZING_RADIATION | 149 | http://www.broadinstitute.org/gsea/msigdb/cards/TSAI_RESPONSE_TO_IONIZING_RADIATION |
| YAUCH_HEDGEHOG_SIGNALING_PARACRINE_UP | 149 | http://www.broadinstitute.org/gsea/msigdb/cards/YAUCH_HEDGEHOG_SIGNALING_PARACRINE_UP |
| BROWNE_HCMV_INFECTION_24HR_DN | 148 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_24HR_DN |
| BROWNE_HCMV_INFECTION_24HR_UP | 148 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_24HR_UP |
| CHANG_CYCLING_GENES | 148 | http://www.broadinstitute.org/gsea/msigdb/cards/CHANG_CYCLING_GENES |
| KATSANOU_ELAVL1_TARGETS_DN | 148 | http://www.broadinstitute.org/gsea/msigdb/cards/KATSANOU_ELAVL1_TARGETS_DN |
| REACTOME_APOPTOSIS | 148 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_APOPTOSIS |
| WEI_MIR34A_TARGETS | 148 | http://www.broadinstitute.org/gsea/msigdb/cards/WEI_MIR34A_TARGETS |
| WHITFIELD_CELL_CYCLE_M_G1 | 148 | http://www.broadinstitute.org/gsea/msigdb/cards/WHITFIELD_CELL_CYCLE_M_G1 |
| BENPORATH_PROLIFERATION | 147 | http://www.broadinstitute.org/gsea/msigdb/cards/BENPORATH_PROLIFERATION |
| DARWICHE_PAPILLOMA_RISK_HIGH_UP | 147 | http://www.broadinstitute.org/gsea/msigdb/cards/DARWICHE_PAPILLOMA_RISK_HIGH_UP |
| KAUFFMANN_DNA_REPLICATION_GENES | 147 | http://www.broadinstitute.org/gsea/msigdb/cards/KAUFFMANN_DNA_REPLICATION_GENES |
| PODAR_RESPONSE_TO_ADAPHOSTIN_UP | 147 | http://www.broadinstitute.org/gsea/msigdb/cards/PODAR_RESPONSE_TO_ADAPHOSTIN_UP |
| RASHI_RESPONSE_TO_IONIZING_RADIATION_5 | 147 | http://www.broadinstitute.org/gsea/msigdb/cards/RASHI_RESPONSE_TO_IONIZING_RADIATION_5 |
| SMID_BREAST_CANCER_ERBB2_UP | 147 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_ERBB2_UP |
| WHITFIELD_CELL_CYCLE_G1_S | 147 | http://www.broadinstitute.org/gsea/msigdb/cards/WHITFIELD_CELL_CYCLE_G1_S |
| CHEMNITZ_RESPONSE_TO_PROSTAGLANDIN_E2_UP | 146 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEMNITZ_RESPONSE_TO_PROSTAGLANDIN_E2_UP |
| DARWICHE_SQUAMOUS_CELL_CARCINOMA_UP | 146 | http://www.broadinstitute.org/gsea/msigdb/cards/DARWICHE_SQUAMOUS_CELL_CARCINOMA_UP |
| ELVIDGE_HYPOXIA_DN | 146 | http://www.broadinstitute.org/gsea/msigdb/cards/ELVIDGE_HYPOXIA_DN |
| KIM_ALL_DISORDERS_DURATION_CORR_DN | 146 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_ALL_DISORDERS_DURATION_CORR_DN |
| LEE_NEURAL_CREST_STEM_CELL_UP | 146 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_NEURAL_CREST_STEM_CELL_UP |
| PECE_MAMMARY_STEM_CELL_DN | 146 | http://www.broadinstitute.org/gsea/msigdb/cards/PECE_MAMMARY_STEM_CELL_DN |
| PECE_MAMMARY_STEM_CELL_UP | 146 | http://www.broadinstitute.org/gsea/msigdb/cards/PECE_MAMMARY_STEM_CELL_UP |
| GREGORY_SYNTHETIC_LETHAL_WITH_IMATINIB | 145 | http://www.broadinstitute.org/gsea/msigdb/cards/GREGORY_SYNTHETIC_LETHAL_WITH_IMATINIB |
| PUIFFE_INVASION_INHIBITED_BY_ASCITES_DN | 145 | http://www.broadinstitute.org/gsea/msigdb/cards/PUIFFE_INVASION_INHIBITED_BY_ASCITES_DN |
| RODWELL_AGING_KIDNEY_DN | 145 | http://www.broadinstitute.org/gsea/msigdb/cards/RODWELL_AGING_KIDNEY_DN |
| SMITH_TERT_TARGETS_UP | 145 | http://www.broadinstitute.org/gsea/msigdb/cards/SMITH_TERT_TARGETS_UP |
| WAKABAYASHI_ADIPOGENESIS_PPARG_RXRA_BOUND_WITH_H4K20ME1_MARK | 145 | http://www.broadinstitute.org/gsea/msigdb/cards/WAKABAYASHI_ADIPOGENESIS_PPARG_RXRA_BOUND_WITH_H4K20ME1_MARK |
| ZHANG_BREAST_CANCER_PROGENITORS_DN | 145 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_BREAST_CANCER_PROGENITORS_DN |
| BROCKE_APOPTOSIS_REVERSED_BY_IL6 | 144 | http://www.broadinstitute.org/gsea/msigdb/cards/BROCKE_APOPTOSIS_REVERSED_BY_IL6 |
| BASSO_B_LYMPHOCYTE_NETWORK | 143 | http://www.broadinstitute.org/gsea/msigdb/cards/BASSO_B_LYMPHOCYTE_NETWORK |
| DANG_MYC_TARGETS_UP | 143 | http://www.broadinstitute.org/gsea/msigdb/cards/DANG_MYC_TARGETS_UP |
| NOUZOVA_TRETINOIN_AND_H4_ACETYLATION | 143 | http://www.broadinstitute.org/gsea/msigdb/cards/NOUZOVA_TRETINOIN_AND_H4_ACETYLATION |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_16D_DN | 143 | http://www.broadinstitute.org/gsea/msigdb/cards/TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_16D_DN |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_14 | 143 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_14 |
| CHIARADONNA_NEOPLASTIC_TRANSFORMATION_KRAS_DN | 142 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIARADONNA_NEOPLASTIC_TRANSFORMATION_KRAS_DN |
| DARWICHE_SKIN_TUMOR_PROMOTER_UP | 142 | http://www.broadinstitute.org/gsea/msigdb/cards/DARWICHE_SKIN_TUMOR_PROMOTER_UP |
| GROSS_HYPOXIA_VIA_ELK3_AND_HIF1A_UP | 142 | http://www.broadinstitute.org/gsea/msigdb/cards/GROSS_HYPOXIA_VIA_ELK3_AND_HIF1A_UP |
| MEISSNER_NPC_HCP_WITH_H3K4ME3_AND_H3K27ME3 | 142 | http://www.broadinstitute.org/gsea/msigdb/cards/MEISSNER_NPC_HCP_WITH_H3K4ME3_AND_H3K27ME3 |
| MIKKELSEN_ES_LCP_WITH_H3K4ME3 | 142 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_ES_LCP_WITH_H3K4ME3 |
| PENG_LEUCINE_DEPRIVATION_UP | 142 | http://www.broadinstitute.org/gsea/msigdb/cards/PENG_LEUCINE_DEPRIVATION_UP |
| RUIZ_TNC_TARGETS_DN | 142 | http://www.broadinstitute.org/gsea/msigdb/cards/RUIZ_TNC_TARGETS_DN |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_10D_DN | 142 | http://www.broadinstitute.org/gsea/msigdb/cards/TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_10D_DN |
| XU_GH1_AUTOCRINE_TARGETS_DN | 142 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_GH1_AUTOCRINE_TARGETS_DN |
| ACEVEDO_LIVER_CANCER_WITH_H3K9ME3_UP | 141 | http://www.broadinstitute.org/gsea/msigdb/cards/ACEVEDO_LIVER_CANCER_WITH_H3K9ME3_UP |
| KAAB_FAILED_HEART_ATRIUM_DN | 141 | http://www.broadinstitute.org/gsea/msigdb/cards/KAAB_FAILED_HEART_ATRIUM_DN |
| LINDGREN_BLADDER_CANCER_CLUSTER_2A_DN | 141 | http://www.broadinstitute.org/gsea/msigdb/cards/LINDGREN_BLADDER_CANCER_CLUSTER_2A_DN |
| REACTOME_TCA_CYCLE_AND_RESPIRATORY_ELECTRON_TRANSPORT | 141 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TCA_CYCLE_AND_RESPIRATORY_ELECTRON_TRANSPORT |
| SABATES_COLORECTAL_ADENOMA_UP | 141 | http://www.broadinstitute.org/gsea/msigdb/cards/SABATES_COLORECTAL_ADENOMA_UP |
| WELCSH_BRCA1_TARGETS_DN | 141 | http://www.broadinstitute.org/gsea/msigdb/cards/WELCSH_BRCA1_TARGETS_DN |
| ALCALAY_AML_BY_NPM1_LOCALIZATION_UP | 140 | http://www.broadinstitute.org/gsea/msigdb/cards/ALCALAY_AML_BY_NPM1_LOCALIZATION_UP |
| FIGUEROA_AML_METHYLATION_CLUSTER_6_UP | 140 | http://www.broadinstitute.org/gsea/msigdb/cards/FIGUEROA_AML_METHYLATION_CLUSTER_6_UP |
| JOHNSTONE_PARVB_TARGETS_2_UP | 140 | http://www.broadinstitute.org/gsea/msigdb/cards/JOHNSTONE_PARVB_TARGETS_2_UP |
| KEGG_SYSTEMIC_LUPUS_ERYTHEMATOSUS | 140 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_SYSTEMIC_LUPUS_ERYTHEMATOSUS |
| ONDER_CDH1_TARGETS_1_UP | 140 | http://www.broadinstitute.org/gsea/msigdb/cards/ONDER_CDH1_TARGETS_1_UP |
| QI_HYPOXIA | 140 | http://www.broadinstitute.org/gsea/msigdb/cards/QI_HYPOXIA |
| REACTOME_PROCESSING_OF_CAPPED_INTRON_CONTAINING_PRE_MRNA | 140 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PROCESSING_OF_CAPPED_INTRON_CONTAINING_PRE_MRNA |
| ROSTY_CERVICAL_CANCER_PROLIFERATION_CLUSTER | 140 | http://www.broadinstitute.org/gsea/msigdb/cards/ROSTY_CERVICAL_CANCER_PROLIFERATION_CLUSTER |
| BOYLAN_MULTIPLE_MYELOMA_C_D_UP | 139 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYLAN_MULTIPLE_MYELOMA_C_D_UP |
| HOELZEL_NF1_TARGETS_UP | 139 | http://www.broadinstitute.org/gsea/msigdb/cards/HOELZEL_NF1_TARGETS_UP |
| MULLIGHAN_NPM1_MUTATED_SIGNATURE_2_UP | 139 | http://www.broadinstitute.org/gsea/msigdb/cards/MULLIGHAN_NPM1_MUTATED_SIGNATURE_2_UP |
| ODONNELL_TFRC_TARGETS_DN | 139 | http://www.broadinstitute.org/gsea/msigdb/cards/ODONNELL_TFRC_TARGETS_DN |
| REACTOME_BIOLOGICAL_OXIDATIONS | 139 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_BIOLOGICAL_OXIDATIONS |
| COULOUARN_TEMPORAL_TGFB1_SIGNATURE_DN | 138 | http://www.broadinstitute.org/gsea/msigdb/cards/COULOUARN_TEMPORAL_TGFB1_SIGNATURE_DN |
| JIANG_VHL_TARGETS | 138 | http://www.broadinstitute.org/gsea/msigdb/cards/JIANG_VHL_TARGETS |
| KEGG_UBIQUITIN_MEDIATED_PROTEOLYSIS | 138 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_UBIQUITIN_MEDIATED_PROTEOLYSIS |
| VART_KSHV_INFECTION_ANGIOGENIC_MARKERS_DN | 138 | http://www.broadinstitute.org/gsea/msigdb/cards/VART_KSHV_INFECTION_ANGIOGENIC_MARKERS_DN |
| VECCHI_GASTRIC_CANCER_ADVANCED_VS_EARLY_DN | 138 | http://www.broadinstitute.org/gsea/msigdb/cards/VECCHI_GASTRIC_CANCER_ADVANCED_VS_EARLY_DN |
| KEGG_INSULIN_SIGNALING_PATHWAY | 137 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_INSULIN_SIGNALING_PATHWAY |
| KEGG_NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY | 137 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY |
| LIU_SOX4_TARGETS_UP | 137 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_SOX4_TARGETS_UP |
| MCBRYAN_PUBERTAL_BREAST_5_6WK_DN | 137 | http://www.broadinstitute.org/gsea/msigdb/cards/MCBRYAN_PUBERTAL_BREAST_5_6WK_DN |
| MIKKELSEN_ES_ICP_WITH_H3K4ME3_AND_H3K27ME3 | 137 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_ES_ICP_WITH_H3K4ME3_AND_H3K27ME3 |
| PID_P53DOWNSTREAMPATHWAY | 137 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_P53DOWNSTREAMPATHWAY |
| REACTOME_MITOTIC_G1_G1_S_PHASES | 137 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MITOTIC_G1_G1_S_PHASES |
| REACTOME_NEUROTRANSMITTER_RECEPTOR_BINDING_AND_DOWNSTREAM_TRANSMISSION_IN_THE_POSTSYNAPTIC_CELL | 137 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NEUROTRANSMITTER_RECEPTOR_BINDING_AND_DOWNSTREAM_TRANSMISSION_IN_THE_POSTSYNAPTIC_CELL |
| REACTOME_NGF_SIGNALLING_VIA_TRKA_FROM_THE_PLASMA_MEMBRANE | 137 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NGF_SIGNALLING_VIA_TRKA_FROM_THE_PLASMA_MEMBRANE |
| WAMUNYOKOLI_OVARIAN_CANCER_GRADES_1_2_UP | 137 | http://www.broadinstitute.org/gsea/msigdb/cards/WAMUNYOKOLI_OVARIAN_CANCER_GRADES_1_2_UP |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_BLUE_UP | 136 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_BLUE_UP |
| PICCALUGA_ANGIOIMMUNOBLASTIC_LYMPHOMA_DN | 136 | http://www.broadinstitute.org/gsea/msigdb/cards/PICCALUGA_ANGIOIMMUNOBLASTIC_LYMPHOMA_DN |
| PROVENZANI_METASTASIS_DN | 136 | http://www.broadinstitute.org/gsea/msigdb/cards/PROVENZANI_METASTASIS_DN |
| WIERENGA_STAT5A_TARGETS_GROUP1 | 136 | http://www.broadinstitute.org/gsea/msigdb/cards/WIERENGA_STAT5A_TARGETS_GROUP1 |
| KEGG_OXIDATIVE_PHOSPHORYLATION | 135 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_OXIDATIVE_PHOSPHORYLATION |
| SERVITJA_LIVER_HNF1A_TARGETS_UP | 135 | http://www.broadinstitute.org/gsea/msigdb/cards/SERVITJA_LIVER_HNF1A_TARGETS_UP |
| TSENG_IRS1_TARGETS_DN | 135 | http://www.broadinstitute.org/gsea/msigdb/cards/TSENG_IRS1_TARGETS_DN |
| UDAYAKUMAR_MED1_TARGETS_UP | 135 | http://www.broadinstitute.org/gsea/msigdb/cards/UDAYAKUMAR_MED1_TARGETS_UP |
| CHUNG_BLISTER_CYTOTOXICITY_UP | 134 | http://www.broadinstitute.org/gsea/msigdb/cards/CHUNG_BLISTER_CYTOTOXICITY_UP |
| KEGG_CELL_ADHESION_MOLECULES_CAMS | 134 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_CELL_ADHESION_MOLECULES_CAMS |
| KEGG_TIGHT_JUNCTION | 134 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_TIGHT_JUNCTION |
| PANGAS_TUMOR_SUPPRESSION_BY_SMAD1_AND_SMAD5_UP | 134 | http://www.broadinstitute.org/gsea/msigdb/cards/PANGAS_TUMOR_SUPPRESSION_BY_SMAD1_AND_SMAD5_UP |
| POOLA_INVASIVE_BREAST_CANCER_DN | 134 | http://www.broadinstitute.org/gsea/msigdb/cards/POOLA_INVASIVE_BREAST_CANCER_DN |
| SCHLOSSER_SERUM_RESPONSE_UP | 134 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHLOSSER_SERUM_RESPONSE_UP |
| GAL_LEUKEMIC_STEM_CELL_UP | 133 | http://www.broadinstitute.org/gsea/msigdb/cards/GAL_LEUKEMIC_STEM_CELL_UP |
| KEGG_PARKINSONS_DISEASE | 133 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PARKINSONS_DISEASE |
| NIKOLSKY_BREAST_CANCER_17Q11_Q21_AMPLICON | 133 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_17Q11_Q21_AMPLICON |
| REACTOME_DIABETES_PATHWAYS | 133 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DIABETES_PATHWAYS |
| SENESE_HDAC2_TARGETS_DN | 133 | http://www.broadinstitute.org/gsea/msigdb/cards/SENESE_HDAC2_TARGETS_DN |
| NIKOLSKY_BREAST_CANCER_8Q12_Q22_AMPLICON | 132 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_8Q12_Q22_AMPLICON |
| REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | 132 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION |
| REACTOME_HOST_INTERACTIONS_OF_HIV_FACTORS | 132 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_HOST_INTERACTIONS_OF_HIV_FACTORS |
| ZHONG_SECRETOME_OF_LUNG_CANCER_AND_FIBROBLAST | 132 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHONG_SECRETOME_OF_LUNG_CANCER_AND_FIBROBLAST |
| ICHIBA_GRAFT_VERSUS_HOST_DISEASE_35D_UP | 131 | http://www.broadinstitute.org/gsea/msigdb/cards/ICHIBA_GRAFT_VERSUS_HOST_DISEASE_35D_UP |
| PETROVA_ENDOTHELIUM_LYMPHATIC_VS_BLOOD_UP | 131 | http://www.broadinstitute.org/gsea/msigdb/cards/PETROVA_ENDOTHELIUM_LYMPHATIC_VS_BLOOD_UP |
| ELVIDGE_HYPOXIA_BY_DMOG_UP | 130 | http://www.broadinstitute.org/gsea/msigdb/cards/ELVIDGE_HYPOXIA_BY_DMOG_UP |
| YAGI_AML_WITH_T_9_11_TRANSLOCATION | 130 | http://www.broadinstitute.org/gsea/msigdb/cards/YAGI_AML_WITH_T_9_11_TRANSLOCATION |
| BROWN_MYELOID_CELL_DEVELOPMENT_DN | 129 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWN_MYELOID_CELL_DEVELOPMENT_DN |
| KEGG_AXON_GUIDANCE | 129 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_AXON_GUIDANCE |
| KIM_RESPONSE_TO_TSA_AND_DECITABINE_UP | 129 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_RESPONSE_TO_TSA_AND_DECITABINE_UP |
| KIM_WT1_TARGETS_8HR_DN | 129 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_WT1_TARGETS_8HR_DN |
| PID_PDGFRBPATHWAY | 129 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_PDGFRBPATHWAY |
| REACTOME_MEMBRANE_TRAFFICKING | 129 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MEMBRANE_TRAFFICKING |
| YAGI_AML_SURVIVAL | 129 | http://www.broadinstitute.org/gsea/msigdb/cards/YAGI_AML_SURVIVAL |
| ALTEMEIER_RESPONSE_TO_LPS_WITH_MECHANICAL_VENTILATION | 128 | http://www.broadinstitute.org/gsea/msigdb/cards/ALTEMEIER_RESPONSE_TO_LPS_WITH_MECHANICAL_VENTILATION |
| BYSTRYKH_HEMATOPOIESIS_STEM_CELL_QTL_CIS | 128 | http://www.broadinstitute.org/gsea/msigdb/cards/BYSTRYKH_HEMATOPOIESIS_STEM_CELL_QTL_CIS |
| KEGG_CELL_CYCLE | 128 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_CELL_CYCLE |
| KEGG_SPLICEOSOME | 128 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_SPLICEOSOME |
| KOKKINAKIS_METHIONINE_DEPRIVATION_48HR_UP | 128 | http://www.broadinstitute.org/gsea/msigdb/cards/KOKKINAKIS_METHIONINE_DEPRIVATION_48HR_UP |
| LENAOUR_DENDRITIC_CELL_MATURATION_DN | 128 | http://www.broadinstitute.org/gsea/msigdb/cards/LENAOUR_DENDRITIC_CELL_MATURATION_DN |
| MIKKELSEN_MEF_LCP_WITH_H3K4ME3 | 128 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_MEF_LCP_WITH_H3K4ME3 |
| POTTI_TOPOTECAN_SENSITIVITY | 128 | http://www.broadinstitute.org/gsea/msigdb/cards/POTTI_TOPOTECAN_SENSITIVITY |
| YEGNASUBRAMANIAN_PROSTATE_CANCER | 128 | http://www.broadinstitute.org/gsea/msigdb/cards/YEGNASUBRAMANIAN_PROSTATE_CANCER |
| ZHOU_CELL_CYCLE_GENES_IN_IR_RESPONSE_24HR | 128 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHOU_CELL_CYCLE_GENES_IN_IR_RESPONSE_24HR |
| ZHU_CMV_ALL_DN | 128 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHU_CMV_ALL_DN |
| BHATI_G2M_ARREST_BY_2METHOXYESTRADIOL_DN | 127 | http://www.broadinstitute.org/gsea/msigdb/cards/BHATI_G2M_ARREST_BY_2METHOXYESTRADIOL_DN |
| INGRAM_SHH_TARGETS_UP | 127 | http://www.broadinstitute.org/gsea/msigdb/cards/INGRAM_SHH_TARGETS_UP |
| KARLSSON_TGFB1_TARGETS_UP | 127 | http://www.broadinstitute.org/gsea/msigdb/cards/KARLSSON_TGFB1_TARGETS_UP |
| LIU_VMYB_TARGETS_UP | 127 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_VMYB_TARGETS_UP |
| RASHI_RESPONSE_TO_IONIZING_RADIATION_2 | 127 | http://www.broadinstitute.org/gsea/msigdb/cards/RASHI_RESPONSE_TO_IONIZING_RADIATION_2 |
| REACTOME_SIGNALING_BY_FGFR_IN_DISEASE | 127 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_FGFR_IN_DISEASE |
| BURTON_ADIPOGENESIS_5 | 126 | http://www.broadinstitute.org/gsea/msigdb/cards/BURTON_ADIPOGENESIS_5 |
| CHIARADONNA_NEOPLASTIC_TRANSFORMATION_KRAS_UP | 126 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIARADONNA_NEOPLASTIC_TRANSFORMATION_KRAS_UP |
| GERY_CEBP_TARGETS | 126 | http://www.broadinstitute.org/gsea/msigdb/cards/GERY_CEBP_TARGETS |
| IZADPANAH_STEM_CELL_ADIPOSE_VS_BONE_UP | 126 | http://www.broadinstitute.org/gsea/msigdb/cards/IZADPANAH_STEM_CELL_ADIPOSE_VS_BONE_UP |
| KEGG_NEUROTROPHIN_SIGNALING_PATHWAY | 126 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_NEUROTROPHIN_SIGNALING_PATHWAY |
| MIKKELSEN_IPS_ICP_WITH_H3K4ME3_AND_H327ME3 | 126 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_IPS_ICP_WITH_H3K4ME3_AND_H327ME3 |
| MISSIAGLIA_REGULATED_BY_METHYLATION_UP | 126 | http://www.broadinstitute.org/gsea/msigdb/cards/MISSIAGLIA_REGULATED_BY_METHYLATION_UP |
| MULLIGHAN_NPM1_MUTATED_SIGNATURE_1_DN | 126 | http://www.broadinstitute.org/gsea/msigdb/cards/MULLIGHAN_NPM1_MUTATED_SIGNATURE_1_DN |
| REACTOME_SIGNALING_BY_THE_B_CELL_RECEPTOR_BCR | 126 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_THE_B_CELL_RECEPTOR_BCR |
| SANSOM_APC_TARGETS_UP | 126 | http://www.broadinstitute.org/gsea/msigdb/cards/SANSOM_APC_TARGETS_UP |
| BHATI_G2M_ARREST_BY_2METHOXYESTRADIOL_UP | 125 | http://www.broadinstitute.org/gsea/msigdb/cards/BHATI_G2M_ARREST_BY_2METHOXYESTRADIOL_UP |
| FIGUEROA_AML_METHYLATION_CLUSTER_1_UP | 125 | http://www.broadinstitute.org/gsea/msigdb/cards/FIGUEROA_AML_METHYLATION_CLUSTER_1_UP |
| REACTOME_HIV_LIFE_CYCLE | 125 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_HIV_LIFE_CYCLE |
| STEARMAN_LUNG_CANCER_EARLY_VS_LATE_UP | 125 | http://www.broadinstitute.org/gsea/msigdb/cards/STEARMAN_LUNG_CANCER_EARLY_VS_LATE_UP |
| DAIRKEE_TERT_TARGETS_DN | 124 | http://www.broadinstitute.org/gsea/msigdb/cards/DAIRKEE_TERT_TARGETS_DN |
| REACTOME_CELL_CYCLE_CHECKPOINTS | 124 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CELL_CYCLE_CHECKPOINTS |
| DAZARD_RESPONSE_TO_UV_SCC_DN | 123 | http://www.broadinstitute.org/gsea/msigdb/cards/DAZARD_RESPONSE_TO_UV_SCC_DN |
| DAZARD_RESPONSE_TO_UV_SCC_UP | 123 | http://www.broadinstitute.org/gsea/msigdb/cards/DAZARD_RESPONSE_TO_UV_SCC_UP |
| HOOI_ST7_TARGETS_DN | 123 | http://www.broadinstitute.org/gsea/msigdb/cards/HOOI_ST7_TARGETS_DN |
| KAN_RESPONSE_TO_ARSENIC_TRIOXIDE | 123 | http://www.broadinstitute.org/gsea/msigdb/cards/KAN_RESPONSE_TO_ARSENIC_TRIOXIDE |
| KAYO_AGING_MUSCLE_DN | 123 | http://www.broadinstitute.org/gsea/msigdb/cards/KAYO_AGING_MUSCLE_DN |
| MISSIAGLIA_REGULATED_BY_METHYLATION_DN | 122 | http://www.broadinstitute.org/gsea/msigdb/cards/MISSIAGLIA_REGULATED_BY_METHYLATION_DN |
| REACTOME_CHROMOSOME_MAINTENANCE | 122 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CHROMOSOME_MAINTENANCE |
| REACTOME_RNA_POL_I_RNA_POL_III_AND_MITOCHONDRIAL_TRANSCRIPTION | 122 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RNA_POL_I_RNA_POL_III_AND_MITOCHONDRIAL_TRANSCRIPTION |
| REACTOME_SIGNALING_BY_PDGF | 122 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_PDGF |
| TOOKER_GEMCITABINE_RESISTANCE_DN | 122 | http://www.broadinstitute.org/gsea/msigdb/cards/TOOKER_GEMCITABINE_RESISTANCE_DN |
| AZARE_NEOPLASTIC_TRANSFORMATION_BY_STAT3_DN | 121 | http://www.broadinstitute.org/gsea/msigdb/cards/AZARE_NEOPLASTIC_TRANSFORMATION_BY_STAT3_DN |
| CHARAFE_BREAST_CANCER_BASAL_VS_MESENCHYMAL_UP | 121 | http://www.broadinstitute.org/gsea/msigdb/cards/CHARAFE_BREAST_CANCER_BASAL_VS_MESENCHYMAL_UP |
| KEGG_LYSOSOME | 121 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_LYSOSOME |
| LINDGREN_BLADDER_CANCER_CLUSTER_1_UP | 121 | http://www.broadinstitute.org/gsea/msigdb/cards/LINDGREN_BLADDER_CANCER_CLUSTER_1_UP |
| NING_CHRONIC_OBSTRUCTIVE_PULMONARY_DISEASE_DN | 121 | http://www.broadinstitute.org/gsea/msigdb/cards/NING_CHRONIC_OBSTRUCTIVE_PULMONARY_DISEASE_DN |
| REACTOME_G_ALPHA_S_SIGNALLING_EVENTS | 121 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_G_ALPHA_S_SIGNALLING_EVENTS |
| RICKMAN_TUMOR_DIFFERENTIATED_MODERATELY_VS_POORLY_UP | 121 | http://www.broadinstitute.org/gsea/msigdb/cards/RICKMAN_TUMOR_DIFFERENTIATED_MODERATELY_VS_POORLY_UP |
| RODRIGUES_DCC_TARGETS_DN | 121 | http://www.broadinstitute.org/gsea/msigdb/cards/RODRIGUES_DCC_TARGETS_DN |
| VANTVEER_BREAST_CANCER_METASTASIS_DN | 121 | http://www.broadinstitute.org/gsea/msigdb/cards/VANTVEER_BREAST_CANCER_METASTASIS_DN |
| WANG_ESOPHAGUS_CANCER_VS_NORMAL_UP | 121 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_ESOPHAGUS_CANCER_VS_NORMAL_UP |
| ACEVEDO_LIVER_CANCER_WITH_H3K9ME3_DN | 120 | http://www.broadinstitute.org/gsea/msigdb/cards/ACEVEDO_LIVER_CANCER_WITH_H3K9ME3_DN |
| CHEBOTAEV_GR_TARGETS_DN | 120 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEBOTAEV_GR_TARGETS_DN |
| CHIARADONNA_NEOPLASTIC_TRANSFORMATION_CDC25_UP | 120 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIARADONNA_NEOPLASTIC_TRANSFORMATION_CDC25_UP |
| IWANAGA_CARCINOGENESIS_BY_KRAS_DN | 120 | http://www.broadinstitute.org/gsea/msigdb/cards/IWANAGA_CARCINOGENESIS_BY_KRAS_DN |
| NIKOLSKY_BREAST_CANCER_16P13_AMPLICON | 120 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_16P13_AMPLICON |
| REACTOME_CELL_CELL_COMMUNICATION | 120 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CELL_CELL_COMMUNICATION |
| REACTOME_INTEGRATION_OF_ENERGY_METABOLISM | 120 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INTEGRATION_OF_ENERGY_METABOLISM |
| XU_GH1_EXOGENOUS_TARGETS_DN | 120 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_GH1_EXOGENOUS_TARGETS_DN |
| ZHU_CMV_ALL_UP | 120 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHU_CMV_ALL_UP |
| GINESTIER_BREAST_CANCER_20Q13_AMPLIFICATION_UP | 119 | http://www.broadinstitute.org/gsea/msigdb/cards/GINESTIER_BREAST_CANCER_20Q13_AMPLIFICATION_UP |
| CERVERA_SDHB_TARGETS_1_UP | 118 | http://www.broadinstitute.org/gsea/msigdb/cards/CERVERA_SDHB_TARGETS_1_UP |
| DAIRKEE_CANCER_PRONE_RESPONSE_BPA_E2 | 118 | http://www.broadinstitute.org/gsea/msigdb/cards/DAIRKEE_CANCER_PRONE_RESPONSE_BPA_E2 |
| FIGUEROA_AML_METHYLATION_CLUSTER_7_UP | 118 | http://www.broadinstitute.org/gsea/msigdb/cards/FIGUEROA_AML_METHYLATION_CLUSTER_7_UP |
| KEGG_LEUKOCYTE_TRANSENDOTHELIAL_MIGRATION | 118 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_LEUKOCYTE_TRANSENDOTHELIAL_MIGRATION |
| LEE_NEURAL_CREST_STEM_CELL_DN | 118 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_NEURAL_CREST_STEM_CELL_DN |
| MARKEY_RB1_CHRONIC_LOF_DN | 118 | http://www.broadinstitute.org/gsea/msigdb/cards/MARKEY_RB1_CHRONIC_LOF_DN |
| REACTOME_TOLL_RECEPTOR_CASCADES | 118 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TOLL_RECEPTOR_CASCADES |
| KOKKINAKIS_METHIONINE_DEPRIVATION_96HR_UP | 117 | http://www.broadinstitute.org/gsea/msigdb/cards/KOKKINAKIS_METHIONINE_DEPRIVATION_96HR_UP |
| OUELLET_OVARIAN_CANCER_INVASIVE_VS_LMP_UP | 117 | http://www.broadinstitute.org/gsea/msigdb/cards/OUELLET_OVARIAN_CANCER_INVASIVE_VS_LMP_UP |
| PUJANA_BRCA_CENTERED_NETWORK | 117 | http://www.broadinstitute.org/gsea/msigdb/cards/PUJANA_BRCA_CENTERED_NETWORK |
| ACOSTA_PROLIFERATION_INDEPENDENT_MYC_TARGETS_DN | 116 | http://www.broadinstitute.org/gsea/msigdb/cards/ACOSTA_PROLIFERATION_INDEPENDENT_MYC_TARGETS_DN |
| DIAZ_CHRONIC_MEYLOGENOUS_LEUKEMIA_DN | 116 | http://www.broadinstitute.org/gsea/msigdb/cards/DIAZ_CHRONIC_MEYLOGENOUS_LEUKEMIA_DN |
| HAHTOLA_MYCOSIS_FUNGOIDES_CD4_DN | 116 | http://www.broadinstitute.org/gsea/msigdb/cards/HAHTOLA_MYCOSIS_FUNGOIDES_CD4_DN |
| IKEDA_MIR30_TARGETS_UP | 116 | http://www.broadinstitute.org/gsea/msigdb/cards/IKEDA_MIR30_TARGETS_UP |
| LIM_MAMMARY_LUMINAL_MATURE_UP | 116 | http://www.broadinstitute.org/gsea/msigdb/cards/LIM_MAMMARY_LUMINAL_MATURE_UP |
| LINDGREN_BLADDER_CANCER_WITH_LOH_IN_CHR9Q | 116 | http://www.broadinstitute.org/gsea/msigdb/cards/LINDGREN_BLADDER_CANCER_WITH_LOH_IN_CHR9Q |
| LI_INDUCED_T_TO_NATURAL_KILLER_DN | 116 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_INDUCED_T_TO_NATURAL_KILLER_DN |
| MATTIOLI_MGUS_VS_PCL | 116 | http://www.broadinstitute.org/gsea/msigdb/cards/MATTIOLI_MGUS_VS_PCL |
| MCBRYAN_PUBERTAL_BREAST_5_6WK_UP | 116 | http://www.broadinstitute.org/gsea/msigdb/cards/MCBRYAN_PUBERTAL_BREAST_5_6WK_UP |
| REACTOME_MEIOSIS | 116 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MEIOSIS |
| SCHAEFFER_PROSTATE_DEVELOPMENT_12HR_UP | 116 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_PROSTATE_DEVELOPMENT_12HR_UP |
| WHITEFORD_PEDIATRIC_CANCER_MARKERS | 116 | http://www.broadinstitute.org/gsea/msigdb/cards/WHITEFORD_PEDIATRIC_CANCER_MARKERS |
| HOELZEL_NF1_TARGETS_DN | 115 | http://www.broadinstitute.org/gsea/msigdb/cards/HOELZEL_NF1_TARGETS_DN |
| HOSHIDA_LIVER_CANCER_SUBCLASS_S2 | 115 | http://www.broadinstitute.org/gsea/msigdb/cards/HOSHIDA_LIVER_CANCER_SUBCLASS_S2 |
| JUBAN_TARGETS_OF_SPI1_AND_FLI1_UP | 115 | http://www.broadinstitute.org/gsea/msigdb/cards/JUBAN_TARGETS_OF_SPI1_AND_FLI1_UP |
| KEGG_VASCULAR_SMOOTH_MUSCLE_CONTRACTION | 115 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_VASCULAR_SMOOTH_MUSCLE_CONTRACTION |
| MARKEY_RB1_CHRONIC_LOF_UP | 115 | http://www.broadinstitute.org/gsea/msigdb/cards/MARKEY_RB1_CHRONIC_LOF_UP |
| MARZEC_IL2_SIGNALING_UP | 115 | http://www.broadinstitute.org/gsea/msigdb/cards/MARZEC_IL2_SIGNALING_UP |
| BOSCO_TH1_CYTOTOXIC_MODULE | 114 | http://www.broadinstitute.org/gsea/msigdb/cards/BOSCO_TH1_CYTOTOXIC_MODULE |
| CERVERA_SDHB_TARGETS_2 | 114 | http://www.broadinstitute.org/gsea/msigdb/cards/CERVERA_SDHB_TARGETS_2 |
| KEGG_OOCYTE_MEIOSIS | 114 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_OOCYTE_MEIOSIS |
| LENAOUR_DENDRITIC_CELL_MATURATION_UP | 114 | http://www.broadinstitute.org/gsea/msigdb/cards/LENAOUR_DENDRITIC_CELL_MATURATION_UP |
| LIEN_BREAST_CARCINOMA_METAPLASTIC_VS_DUCTAL_DN | 114 | http://www.broadinstitute.org/gsea/msigdb/cards/LIEN_BREAST_CARCINOMA_METAPLASTIC_VS_DUCTAL_DN |
| MATZUK_SPERMATOZOA | 114 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_SPERMATOZOA |
| ROPERO_HDAC2_TARGETS | 114 | http://www.broadinstitute.org/gsea/msigdb/cards/ROPERO_HDAC2_TARGETS |
| SENESE_HDAC2_TARGETS_UP | 114 | http://www.broadinstitute.org/gsea/msigdb/cards/SENESE_HDAC2_TARGETS_UP |
| SMIRNOV_RESPONSE_TO_IR_6HR_DN | 114 | http://www.broadinstitute.org/gsea/msigdb/cards/SMIRNOV_RESPONSE_TO_IR_6HR_DN |
| BOYAULT_LIVER_CANCER_SUBCLASS_G1_UP | 113 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYAULT_LIVER_CANCER_SUBCLASS_G1_UP |
| HOSHIDA_LIVER_CANCER_SURVIVAL_DN | 113 | http://www.broadinstitute.org/gsea/msigdb/cards/HOSHIDA_LIVER_CANCER_SURVIVAL_DN |
| REACTOME_SIGNALING_BY_RHO_GTPASES | 113 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_RHO_GTPASES |
| RICKMAN_HEAD_AND_NECK_CANCER_C | 113 | http://www.broadinstitute.org/gsea/msigdb/cards/RICKMAN_HEAD_AND_NECK_CANCER_C |
| TSENG_IRS1_TARGETS_UP | 113 | http://www.broadinstitute.org/gsea/msigdb/cards/TSENG_IRS1_TARGETS_UP |
| DITTMER_PTHLH_TARGETS_UP | 112 | http://www.broadinstitute.org/gsea/msigdb/cards/DITTMER_PTHLH_TARGETS_UP |
| DOANE_BREAST_CANCER_ESR1_UP | 112 | http://www.broadinstitute.org/gsea/msigdb/cards/DOANE_BREAST_CANCER_ESR1_UP |
| FIGUEROA_AML_METHYLATION_CLUSTER_4_UP | 112 | http://www.broadinstitute.org/gsea/msigdb/cards/FIGUEROA_AML_METHYLATION_CLUSTER_4_UP |
| LABBE_WNT3A_TARGETS_UP | 112 | http://www.broadinstitute.org/gsea/msigdb/cards/LABBE_WNT3A_TARGETS_UP |
| LAIHO_COLORECTAL_CANCER_SERRATED_UP | 112 | http://www.broadinstitute.org/gsea/msigdb/cards/LAIHO_COLORECTAL_CANCER_SERRATED_UP |
| PASINI_SUZ12_TARGETS_UP | 112 | http://www.broadinstitute.org/gsea/msigdb/cards/PASINI_SUZ12_TARGETS_UP |
| REACTOME_DNA_REPAIR | 112 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DNA_REPAIR |
| REACTOME_G1_S_TRANSITION | 112 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_G1_S_TRANSITION |
| REACTOME_SIGNALING_BY_FGFR | 112 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_FGFR |
| BOUDOUKHA_BOUND_BY_IGF2BP2 | 111 | http://www.broadinstitute.org/gsea/msigdb/cards/BOUDOUKHA_BOUND_BY_IGF2BP2 |
| BROWNE_HCMV_INFECTION_12HR_UP | 111 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_12HR_UP |
| BRUECKNER_TARGETS_OF_MIRLET7A3_UP | 111 | http://www.broadinstitute.org/gsea/msigdb/cards/BRUECKNER_TARGETS_OF_MIRLET7A3_UP |
| LABBE_TARGETS_OF_TGFB1_AND_WNT3A_UP | 111 | http://www.broadinstitute.org/gsea/msigdb/cards/LABBE_TARGETS_OF_TGFB1_AND_WNT3A_UP |
| REACTOME_GLYCOSAMINOGLYCAN_METABOLISM | 111 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GLYCOSAMINOGLYCAN_METABOLISM |
| REACTOME_MRNA_SPLICING | 111 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MRNA_SPLICING |
| WANG_HCP_PROSTATE_CANCER | 111 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_HCP_PROSTATE_CANCER |
| CHENG_IMPRINTED_BY_ESTRADIOL | 110 | http://www.broadinstitute.org/gsea/msigdb/cards/CHENG_IMPRINTED_BY_ESTRADIOL |
| GHANDHI_DIRECT_IRRADIATION_UP | 110 | http://www.broadinstitute.org/gsea/msigdb/cards/GHANDHI_DIRECT_IRRADIATION_UP |
| GROSS_HYPOXIA_VIA_HIF1A_DN | 110 | http://www.broadinstitute.org/gsea/msigdb/cards/GROSS_HYPOXIA_VIA_HIF1A_DN |
| MORI_EMU_MYC_LYMPHOMA_BY_ONSET_TIME_UP | 110 | http://www.broadinstitute.org/gsea/msigdb/cards/MORI_EMU_MYC_LYMPHOMA_BY_ONSET_TIME_UP |
| PEDERSEN_METASTASIS_BY_ERBB2_ISOFORM_4 | 110 | http://www.broadinstitute.org/gsea/msigdb/cards/PEDERSEN_METASTASIS_BY_ERBB2_ISOFORM_4 |
| RICKMAN_TUMOR_DIFFERENTIATED_WELL_VS_MODERATELY_DN | 110 | http://www.broadinstitute.org/gsea/msigdb/cards/RICKMAN_TUMOR_DIFFERENTIATED_WELL_VS_MODERATELY_DN |
| SUNG_METASTASIS_STROMA_UP | 110 | http://www.broadinstitute.org/gsea/msigdb/cards/SUNG_METASTASIS_STROMA_UP |
| WOOD_EBV_EBNA1_TARGETS_UP | 110 | http://www.broadinstitute.org/gsea/msigdb/cards/WOOD_EBV_EBNA1_TARGETS_UP |
| AMBROSINI_FLAVOPIRIDOL_TREATMENT_TP53 | 109 | http://www.broadinstitute.org/gsea/msigdb/cards/AMBROSINI_FLAVOPIRIDOL_TREATMENT_TP53 |
| COULOUARN_TEMPORAL_TGFB1_SIGNATURE_UP | 109 | http://www.broadinstitute.org/gsea/msigdb/cards/COULOUARN_TEMPORAL_TGFB1_SIGNATURE_UP |
| REACTOME_SIGNALING_BY_EGFR_IN_CANCER | 109 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_EGFR_IN_CANCER |
| REACTOME_S_PHASE | 109 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_S_PHASE |
| RICKMAN_TUMOR_DIFFERENTIATED_WELL_VS_MODERATELY_UP | 109 | http://www.broadinstitute.org/gsea/msigdb/cards/RICKMAN_TUMOR_DIFFERENTIATED_WELL_VS_MODERATELY_UP |
| SERVITJA_ISLET_HNF1A_TARGETS_DN | 109 | http://www.broadinstitute.org/gsea/msigdb/cards/SERVITJA_ISLET_HNF1A_TARGETS_DN |
| IZADPANAH_STEM_CELL_ADIPOSE_VS_BONE_DN | 108 | http://www.broadinstitute.org/gsea/msigdb/cards/IZADPANAH_STEM_CELL_ADIPOSE_VS_BONE_DN |
| JAZAG_TGFB1_SIGNALING_UP | 108 | http://www.broadinstitute.org/gsea/msigdb/cards/JAZAG_TGFB1_SIGNALING_UP |
| JAZAG_TGFB1_SIGNALING_VIA_SMAD4_UP | 108 | http://www.broadinstitute.org/gsea/msigdb/cards/JAZAG_TGFB1_SIGNALING_VIA_SMAD4_UP |
| KEGG_T_CELL_RECEPTOR_SIGNALING_PATHWAY | 108 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_T_CELL_RECEPTOR_SIGNALING_PATHWAY |
| LABBE_TARGETS_OF_TGFB1_AND_WNT3A_DN | 108 | http://www.broadinstitute.org/gsea/msigdb/cards/LABBE_TARGETS_OF_TGFB1_AND_WNT3A_DN |
| LABBE_TGFB1_TARGETS_DN | 108 | http://www.broadinstitute.org/gsea/msigdb/cards/LABBE_TGFB1_TARGETS_DN |
| LE_EGR2_TARGETS_DN | 108 | http://www.broadinstitute.org/gsea/msigdb/cards/LE_EGR2_TARGETS_DN |
| LE_EGR2_TARGETS_UP | 108 | http://www.broadinstitute.org/gsea/msigdb/cards/LE_EGR2_TARGETS_UP |
| LIN_SILENCED_BY_TUMOR_MICROENVIRONMENT | 108 | http://www.broadinstitute.org/gsea/msigdb/cards/LIN_SILENCED_BY_TUMOR_MICROENVIRONMENT |
| REACTOME_SIGNALING_BY_INSULIN_RECEPTOR | 108 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_INSULIN_RECEPTOR |
| SCHLOSSER_SERUM_RESPONSE_AUGMENTED_BY_MYC | 108 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHLOSSER_SERUM_RESPONSE_AUGMENTED_BY_MYC |
| TAVAZOIE_METASTASIS | 108 | http://www.broadinstitute.org/gsea/msigdb/cards/TAVAZOIE_METASTASIS |
| WATANABE_RECTAL_CANCER_RADIOTHERAPY_RESPONSIVE_UP | 108 | http://www.broadinstitute.org/gsea/msigdb/cards/WATANABE_RECTAL_CANCER_RADIOTHERAPY_RESPONSIVE_UP |
| WESTON_VEGFA_TARGETS | 108 | http://www.broadinstitute.org/gsea/msigdb/cards/WESTON_VEGFA_TARGETS |
| ZHANG_TLX_TARGETS_DN | 108 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_TLX_TARGETS_DN |
| ICHIBA_GRAFT_VERSUS_HOST_DISEASE_D7_UP | 107 | http://www.broadinstitute.org/gsea/msigdb/cards/ICHIBA_GRAFT_VERSUS_HOST_DISEASE_D7_UP |
| LEE_EARLY_T_LYMPHOCYTE_UP | 107 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_EARLY_T_LYMPHOCYTE_UP |
| REACTOME_SIGNALING_BY_ILS | 107 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_ILS |
| ROESSLER_LIVER_CANCER_METASTASIS_UP | 107 | http://www.broadinstitute.org/gsea/msigdb/cards/ROESSLER_LIVER_CANCER_METASTASIS_UP |
| SESTO_RESPONSE_TO_UV_C0 | 107 | http://www.broadinstitute.org/gsea/msigdb/cards/SESTO_RESPONSE_TO_UV_C0 |
| KEEN_RESPONSE_TO_ROSIGLITAZONE_DN | 106 | http://www.broadinstitute.org/gsea/msigdb/cards/KEEN_RESPONSE_TO_ROSIGLITAZONE_DN |
| PLASARI_TGFB1_SIGNALING_VIA_NFIC_1HR_DN | 106 | http://www.broadinstitute.org/gsea/msigdb/cards/PLASARI_TGFB1_SIGNALING_VIA_NFIC_1HR_DN |
| AMUNDSON_GENOTOXIC_SIGNATURE | 105 | http://www.broadinstitute.org/gsea/msigdb/cards/AMUNDSON_GENOTOXIC_SIGNATURE |
| BROWNE_HCMV_INFECTION_8HR_UP | 105 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_8HR_UP |
| HELLER_SILENCED_BY_METHYLATION_DN | 105 | http://www.broadinstitute.org/gsea/msigdb/cards/HELLER_SILENCED_BY_METHYLATION_DN |
| PID_ERBB1_DOWNSTREAM_PATHWAY | 105 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ERBB1_DOWNSTREAM_PATHWAY |
| REACTOME_RNA_POL_II_TRANSCRIPTION | 105 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RNA_POL_II_TRANSCRIPTION |
| STEIN_ESRRA_TARGETS_DN | 105 | http://www.broadinstitute.org/gsea/msigdb/cards/STEIN_ESRRA_TARGETS_DN |
| WOO_LIVER_CANCER_RECURRENCE_UP | 105 | http://www.broadinstitute.org/gsea/msigdb/cards/WOO_LIVER_CANCER_RECURRENCE_UP |
| ZWANG_EGF_INTERVAL_UP | 105 | http://www.broadinstitute.org/gsea/msigdb/cards/ZWANG_EGF_INTERVAL_UP |
| BORCZUK_MALIGNANT_MESOTHELIOMA_DN | 104 | http://www.broadinstitute.org/gsea/msigdb/cards/BORCZUK_MALIGNANT_MESOTHELIOMA_DN |
| ELVIDGE_HIF1A_AND_HIF2A_TARGETS_DN | 104 | http://www.broadinstitute.org/gsea/msigdb/cards/ELVIDGE_HIF1A_AND_HIF2A_TARGETS_DN |
| NAKAMURA_ADIPOGENESIS_LATE_UP | 104 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAMURA_ADIPOGENESIS_LATE_UP |
| RAY_TUMORIGENESIS_BY_ERBB2_CDC25A_UP | 104 | http://www.broadinstitute.org/gsea/msigdb/cards/RAY_TUMORIGENESIS_BY_ERBB2_CDC25A_UP |
| REACTOME_LATE_PHASE_OF_HIV_LIFE_CYCLE | 104 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_LATE_PHASE_OF_HIV_LIFE_CYCLE |
| REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION | 104 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION |
| CHEN_LVAD_SUPPORT_OF_FAILING_HEART_UP | 103 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEN_LVAD_SUPPORT_OF_FAILING_HEART_UP |
| GROSS_HYPOXIA_VIA_ELK3_AND_HIF1A_DN | 103 | http://www.broadinstitute.org/gsea/msigdb/cards/GROSS_HYPOXIA_VIA_ELK3_AND_HIF1A_DN |
| HIRSCH_CELLULAR_TRANSFORMATION_SIGNATURE_DN | 103 | http://www.broadinstitute.org/gsea/msigdb/cards/HIRSCH_CELLULAR_TRANSFORMATION_SIGNATURE_DN |
| KIM_GASTRIC_CANCER_CHEMOSENSITIVITY | 103 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_GASTRIC_CANCER_CHEMOSENSITIVITY |
| KIM_MYCN_AMPLIFICATION_TARGETS_DN | 103 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_MYCN_AMPLIFICATION_TARGETS_DN |
| LEIN_CHOROID_PLEXUS_MARKERS | 103 | http://www.broadinstitute.org/gsea/msigdb/cards/LEIN_CHOROID_PLEXUS_MARKERS |
| REACTOME_SIGNALING_BY_NOTCH | 103 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_NOTCH |
| TANAKA_METHYLATED_IN_ESOPHAGEAL_CARCINOMA | 103 | http://www.broadinstitute.org/gsea/msigdb/cards/TANAKA_METHYLATED_IN_ESOPHAGEAL_CARCINOMA |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_11 | 103 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_11 |
| ZHANG_RESPONSE_TO_IKK_INHIBITOR_AND_TNF_DN | 103 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_RESPONSE_TO_IKK_INHIBITOR_AND_TNF_DN |
| CHAUHAN_RESPONSE_TO_METHOXYESTRADIOL_DN | 102 | http://www.broadinstitute.org/gsea/msigdb/cards/CHAUHAN_RESPONSE_TO_METHOXYESTRADIOL_DN |
| DER_IFN_BETA_RESPONSE_UP | 102 | http://www.broadinstitute.org/gsea/msigdb/cards/DER_IFN_BETA_RESPONSE_UP |
| KANG_IMMORTALIZED_BY_TERT_DN | 102 | http://www.broadinstitute.org/gsea/msigdb/cards/KANG_IMMORTALIZED_BY_TERT_DN |
| KEGG_MELANOGENESIS | 102 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_MELANOGENESIS |
| KEGG_TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY | 102 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY |
| LABBE_TGFB1_TARGETS_UP | 102 | http://www.broadinstitute.org/gsea/msigdb/cards/LABBE_TGFB1_TARGETS_UP |
| MIKKELSEN_IPS_WITH_HCP_H3K27ME3 | 102 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_IPS_WITH_HCP_H3K27ME3 |
| PID_CXCR4_PATHWAY | 102 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_CXCR4_PATHWAY |
| BASSO_CD40_SIGNALING_UP | 101 | http://www.broadinstitute.org/gsea/msigdb/cards/BASSO_CD40_SIGNALING_UP |
| BOYLAN_MULTIPLE_MYELOMA_PCA1_UP | 101 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYLAN_MULTIPLE_MYELOMA_PCA1_UP |
| BROWNE_HCMV_INFECTION_10HR_UP | 101 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_10HR_UP |
| BROWNE_HCMV_INFECTION_12HR_DN | 101 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_12HR_DN |
| BROWNE_HCMV_INFECTION_20HR_DN | 101 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_20HR_DN |
| BURTON_ADIPOGENESIS_3 | 101 | http://www.broadinstitute.org/gsea/msigdb/cards/BURTON_ADIPOGENESIS_3 |
| GRADE_COLON_AND_RECTAL_CANCER_DN | 101 | http://www.broadinstitute.org/gsea/msigdb/cards/GRADE_COLON_AND_RECTAL_CANCER_DN |
| KEGG_GNRH_SIGNALING_PATHWAY | 101 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GNRH_SIGNALING_PATHWAY |
| KOBAYASHI_EGFR_SIGNALING_24HR_UP | 101 | http://www.broadinstitute.org/gsea/msigdb/cards/KOBAYASHI_EGFR_SIGNALING_24HR_UP |
| PUJANA_BREAST_CANCER_LIT_INT_NETWORK | 101 | http://www.broadinstitute.org/gsea/msigdb/cards/PUJANA_BREAST_CANCER_LIT_INT_NETWORK |
| REACTOME_SIGNALING_BY_ERBB2 | 101 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_ERBB2 |
| SHAFFER_IRF4_TARGETS_IN_MYELOMA_VS_MATURE_B_LYMPHOCYTE | 101 | http://www.broadinstitute.org/gsea/msigdb/cards/SHAFFER_IRF4_TARGETS_IN_MYELOMA_VS_MATURE_B_LYMPHOCYTE |
| WANG_ESOPHAGUS_CANCER_VS_NORMAL_DN | 101 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_ESOPHAGUS_CANCER_VS_NORMAL_DN |
| WIELAND_UP_BY_HBV_INFECTION | 101 | http://www.broadinstitute.org/gsea/msigdb/cards/WIELAND_UP_BY_HBV_INFECTION |
| BENPORATH_ES_CORE_NINE_CORRELATED | 100 | http://www.broadinstitute.org/gsea/msigdb/cards/BENPORATH_ES_CORE_NINE_CORRELATED |
| CHICAS_RB1_TARGETS_LOW_SERUM | 100 | http://www.broadinstitute.org/gsea/msigdb/cards/CHICAS_RB1_TARGETS_LOW_SERUM |
| DING_LUNG_CANCER_EXPRESSION_BY_COPY_NUMBER | 100 | http://www.broadinstitute.org/gsea/msigdb/cards/DING_LUNG_CANCER_EXPRESSION_BY_COPY_NUMBER |
| GAVIN_FOXP3_TARGETS_CLUSTER_P4 | 100 | http://www.broadinstitute.org/gsea/msigdb/cards/GAVIN_FOXP3_TARGETS_CLUSTER_P4 |
| HUMMERICH_SKIN_CANCER_PROGRESSION_DN | 100 | http://www.broadinstitute.org/gsea/msigdb/cards/HUMMERICH_SKIN_CANCER_PROGRESSION_DN |
| MCCLUNG_CREB1_TARGETS_UP | 100 | http://www.broadinstitute.org/gsea/msigdb/cards/MCCLUNG_CREB1_TARGETS_UP |
| QI_PLASMACYTOMA_DN | 100 | http://www.broadinstitute.org/gsea/msigdb/cards/QI_PLASMACYTOMA_DN |
| REACTOME_DOWNSTREAM_SIGNALING_OF_ACTIVATED_FGFR | 100 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DOWNSTREAM_SIGNALING_OF_ACTIVATED_FGFR |
| RICKMAN_HEAD_AND_NECK_CANCER_A | 100 | http://www.broadinstitute.org/gsea/msigdb/cards/RICKMAN_HEAD_AND_NECK_CANCER_A |
| ROVERSI_GLIOMA_COPY_NUMBER_UP | 100 | http://www.broadinstitute.org/gsea/msigdb/cards/ROVERSI_GLIOMA_COPY_NUMBER_UP |
| WINZEN_DEGRADED_VIA_KHSRP | 100 | http://www.broadinstitute.org/gsea/msigdb/cards/WINZEN_DEGRADED_VIA_KHSRP |
| CROONQUIST_NRAS_VS_STROMAL_STIMULATION_DN | 99 | http://www.broadinstitute.org/gsea/msigdb/cards/CROONQUIST_NRAS_VS_STROMAL_STIMULATION_DN |
| LIM_MAMMARY_LUMINAL_MATURE_DN | 99 | http://www.broadinstitute.org/gsea/msigdb/cards/LIM_MAMMARY_LUMINAL_MATURE_DN |
| BRUINS_UVC_RESPONSE_MIDDLE | 98 | http://www.broadinstitute.org/gsea/msigdb/cards/BRUINS_UVC_RESPONSE_MIDDLE |
| CROONQUIST_IL6_DEPRIVATION_DN | 98 | http://www.broadinstitute.org/gsea/msigdb/cards/CROONQUIST_IL6_DEPRIVATION_DN |
| FERREIRA_EWINGS_SARCOMA_UNSTABLE_VS_STABLE_DN | 98 | http://www.broadinstitute.org/gsea/msigdb/cards/FERREIRA_EWINGS_SARCOMA_UNSTABLE_VS_STABLE_DN |
| GAVIN_FOXP3_TARGETS_CLUSTER_T7 | 98 | http://www.broadinstitute.org/gsea/msigdb/cards/GAVIN_FOXP3_TARGETS_CLUSTER_T7 |
| GUO_TARGETS_OF_IRS1_AND_IRS2 | 98 | http://www.broadinstitute.org/gsea/msigdb/cards/GUO_TARGETS_OF_IRS1_AND_IRS2 |
| HAHTOLA_SEZARY_SYNDROM_UP | 98 | http://www.broadinstitute.org/gsea/msigdb/cards/HAHTOLA_SEZARY_SYNDROM_UP |
| KEGG_PYRIMIDINE_METABOLISM | 98 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PYRIMIDINE_METABOLISM |
| MENSE_HYPOXIA_UP | 98 | http://www.broadinstitute.org/gsea/msigdb/cards/MENSE_HYPOXIA_UP |
| NIELSEN_GIST | 98 | http://www.broadinstitute.org/gsea/msigdb/cards/NIELSEN_GIST |
| PYEON_HPV_POSITIVE_TUMORS_UP | 98 | http://www.broadinstitute.org/gsea/msigdb/cards/PYEON_HPV_POSITIVE_TUMORS_UP |
| REACTOME_POTASSIUM_CHANNELS | 98 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_POTASSIUM_CHANNELS |
| REACTOME_RESPIRATORY_ELECTRON_TRANSPORT_ATP_SYNTHESIS_BY_CHEMIOSMOTIC_COUPLING_AND_HEAT_PRODUCTION_BY_UNCOUPLING_PROTEINS_ | 98 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RESPIRATORY_ELECTRON_TRANSPORT_ATP_SYNTHESIS_BY_CHEMIOSMOTIC_COUPLING_AND_HEAT_PRODUCTION_BY_UNCOUPLING_PROTEINS_ |
| REN_ALVEOLAR_RHABDOMYOSARCOMA_UP | 98 | http://www.broadinstitute.org/gsea/msigdb/cards/REN_ALVEOLAR_RHABDOMYOSARCOMA_UP |
| TAKAO_RESPONSE_TO_UVB_RADIATION_DN | 98 | http://www.broadinstitute.org/gsea/msigdb/cards/TAKAO_RESPONSE_TO_UVB_RADIATION_DN |
| GAUSSMANN_MLL_AF4_FUSION_TARGETS_E_UP | 97 | http://www.broadinstitute.org/gsea/msigdb/cards/GAUSSMANN_MLL_AF4_FUSION_TARGETS_E_UP |
| KEGG_FC_GAMMA_R_MEDIATED_PHAGOCYTOSIS | 97 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_FC_GAMMA_R_MEDIATED_PHAGOCYTOSIS |
| KIM_MYC_AMPLIFICATION_TARGETS_DN | 97 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_MYC_AMPLIFICATION_TARGETS_DN |
| KONDO_PROSTATE_CANCER_HCP_WITH_H3K27ME3 | 97 | http://www.broadinstitute.org/gsea/msigdb/cards/KONDO_PROSTATE_CANCER_HCP_WITH_H3K27ME3 |
| KONG_E2F3_TARGETS | 97 | http://www.broadinstitute.org/gsea/msigdb/cards/KONG_E2F3_TARGETS |
| LABBE_WNT3A_TARGETS_DN | 97 | http://www.broadinstitute.org/gsea/msigdb/cards/LABBE_WNT3A_TARGETS_DN |
| REACTOME_DOWNSTREAM_SIGNALING_EVENTS_OF_B_CELL_RECEPTOR_BCR | 97 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DOWNSTREAM_SIGNALING_EVENTS_OF_B_CELL_RECEPTOR_BCR |
| SMID_BREAST_CANCER_RELAPSE_IN_BONE_UP | 97 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_RELAPSE_IN_BONE_UP |
| LIU_PROSTATE_CANCER_UP | 96 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_PROSTATE_CANCER_UP |
| ZAMORA_NOS2_TARGETS_DN | 96 | http://www.broadinstitute.org/gsea/msigdb/cards/ZAMORA_NOS2_TARGETS_DN |
| AMUNDSON_POOR_SURVIVAL_AFTER_GAMMA_RADIATION_8G | 95 | http://www.broadinstitute.org/gsea/msigdb/cards/AMUNDSON_POOR_SURVIVAL_AFTER_GAMMA_RADIATION_8G |
| GRAHAM_CML_DIVIDING_VS_NORMAL_QUIESCENT_DN | 95 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAHAM_CML_DIVIDING_VS_NORMAL_QUIESCENT_DN |
| HECKER_IFNB1_TARGETS | 95 | http://www.broadinstitute.org/gsea/msigdb/cards/HECKER_IFNB1_TARGETS |
| HOEBEKE_LYMPHOID_STEM_CELL_UP | 95 | http://www.broadinstitute.org/gsea/msigdb/cards/HOEBEKE_LYMPHOID_STEM_CELL_UP |
| KAYO_CALORIE_RESTRICTION_MUSCLE_UP | 95 | http://www.broadinstitute.org/gsea/msigdb/cards/KAYO_CALORIE_RESTRICTION_MUSCLE_UP |
| REACTOME_DOWNSTREAM_SIGNAL_TRANSDUCTION | 95 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DOWNSTREAM_SIGNAL_TRANSDUCTION |
| XU_HGF_TARGETS_REPRESSED_BY_AKT1_DN | 95 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_HGF_TARGETS_REPRESSED_BY_AKT1_DN |
| GAVIN_FOXP3_TARGETS_CLUSTER_T4 | 94 | http://www.broadinstitute.org/gsea/msigdb/cards/GAVIN_FOXP3_TARGETS_CLUSTER_T4 |
| HOOI_ST7_TARGETS_UP | 94 | http://www.broadinstitute.org/gsea/msigdb/cards/HOOI_ST7_TARGETS_UP |
| LU_IL4_SIGNALING | 94 | http://www.broadinstitute.org/gsea/msigdb/cards/LU_IL4_SIGNALING |
| NIKOLSKY_MUTATED_AND_AMPLIFIED_IN_BREAST_CANCER | 94 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_MUTATED_AND_AMPLIFIED_IN_BREAST_CANCER |
| REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | 94 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES |
| SHETH_LIVER_CANCER_VS_TXNIP_LOSS_PAM5 | 94 | http://www.broadinstitute.org/gsea/msigdb/cards/SHETH_LIVER_CANCER_VS_TXNIP_LOSS_PAM5 |
| TURASHVILI_BREAST_LOBULAR_CARCINOMA_VS_LOBULAR_NORMAL_UP | 94 | http://www.broadinstitute.org/gsea/msigdb/cards/TURASHVILI_BREAST_LOBULAR_CARCINOMA_VS_LOBULAR_NORMAL_UP |
| CADWELL_ATG16L1_TARGETS_UP | 93 | http://www.broadinstitute.org/gsea/msigdb/cards/CADWELL_ATG16L1_TARGETS_UP |
| CHEN_LIVER_METABOLISM_QTL_CIS | 93 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEN_LIVER_METABOLISM_QTL_CIS |
| KYNG_WERNER_SYNDROM_AND_NORMAL_AGING_UP | 93 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_WERNER_SYNDROM_AND_NORMAL_AGING_UP |
| REACTOME_ACTIVATED_TLR4_SIGNALLING | 93 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATED_TLR4_SIGNALLING |
| REACTOME_REGULATION_OF_INSULIN_SECRETION | 93 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_INSULIN_SECRETION |
| ZHU_CMV_24_HR_UP | 93 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHU_CMV_24_HR_UP |
| BRUINS_UVC_RESPONSE_VIA_TP53_GROUP_C | 92 | http://www.broadinstitute.org/gsea/msigdb/cards/BRUINS_UVC_RESPONSE_VIA_TP53_GROUP_C |
| BURTON_ADIPOGENESIS_9 | 92 | http://www.broadinstitute.org/gsea/msigdb/cards/BURTON_ADIPOGENESIS_9 |
| CAMPS_COLON_CANCER_COPY_NUMBER_UP | 92 | http://www.broadinstitute.org/gsea/msigdb/cards/CAMPS_COLON_CANCER_COPY_NUMBER_UP |
| DE_YY1_TARGETS_DN | 92 | http://www.broadinstitute.org/gsea/msigdb/cards/DE_YY1_TARGETS_DN |
| HILLION_HMGA1B_TARGETS | 92 | http://www.broadinstitute.org/gsea/msigdb/cards/HILLION_HMGA1B_TARGETS |
| JUBAN_TARGETS_OF_SPI1_AND_FLI1_DN | 92 | http://www.broadinstitute.org/gsea/msigdb/cards/JUBAN_TARGETS_OF_SPI1_AND_FLI1_DN |
| KEGG_DILATED_CARDIOMYOPATHY | 92 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_DILATED_CARDIOMYOPATHY |
| KIM_MYCN_AMPLIFICATION_TARGETS_UP | 92 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_MYCN_AMPLIFICATION_TARGETS_UP |
| MOREAUX_B_LYMPHOCYTE_MATURATION_BY_TACI_UP | 92 | http://www.broadinstitute.org/gsea/msigdb/cards/MOREAUX_B_LYMPHOCYTE_MATURATION_BY_TACI_UP |
| REACTOME_SYNTHESIS_OF_DNA | 92 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_OF_DNA |
| WATANABE_RECTAL_CANCER_RADIOTHERAPY_RESPONSIVE_DN | 92 | http://www.broadinstitute.org/gsea/msigdb/cards/WATANABE_RECTAL_CANCER_RADIOTHERAPY_RESPONSIVE_DN |
| WINTER_HYPOXIA_UP | 92 | http://www.broadinstitute.org/gsea/msigdb/cards/WINTER_HYPOXIA_UP |
| BONCI_TARGETS_OF_MIR15A_AND_MIR16_1 | 91 | http://www.broadinstitute.org/gsea/msigdb/cards/BONCI_TARGETS_OF_MIR15A_AND_MIR16_1 |
| DUTERTRE_ESTRADIOL_RESPONSE_6HR_DN | 91 | http://www.broadinstitute.org/gsea/msigdb/cards/DUTERTRE_ESTRADIOL_RESPONSE_6HR_DN |
| ELVIDGE_HIF1A_TARGETS_DN | 91 | http://www.broadinstitute.org/gsea/msigdb/cards/ELVIDGE_HIF1A_TARGETS_DN |
| GAVIN_FOXP3_TARGETS_CLUSTER_P6 | 91 | http://www.broadinstitute.org/gsea/msigdb/cards/GAVIN_FOXP3_TARGETS_CLUSTER_P6 |
| HINATA_NFKB_TARGETS_KERATINOCYTE_UP | 91 | http://www.broadinstitute.org/gsea/msigdb/cards/HINATA_NFKB_TARGETS_KERATINOCYTE_UP |
| REACTOME_CELL_SURFACE_INTERACTIONS_AT_THE_VASCULAR_WALL | 91 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CELL_SURFACE_INTERACTIONS_AT_THE_VASCULAR_WALL |
| REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | 91 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION |
| SASSON_RESPONSE_TO_GONADOTROPHINS_UP | 91 | http://www.broadinstitute.org/gsea/msigdb/cards/SASSON_RESPONSE_TO_GONADOTROPHINS_UP |
| TURASHVILI_BREAST_LOBULAR_CARCINOMA_VS_DUCTAL_NORMAL_DN | 91 | http://www.broadinstitute.org/gsea/msigdb/cards/TURASHVILI_BREAST_LOBULAR_CARCINOMA_VS_DUCTAL_NORMAL_DN |
| ZHU_CMV_24_HR_DN | 91 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHU_CMV_24_HR_DN |
| BILANGES_SERUM_SENSITIVE_GENES | 90 | http://www.broadinstitute.org/gsea/msigdb/cards/BILANGES_SERUM_SENSITIVE_GENES |
| GAUSSMANN_MLL_AF4_FUSION_TARGETS_A_DN | 90 | http://www.broadinstitute.org/gsea/msigdb/cards/GAUSSMANN_MLL_AF4_FUSION_TARGETS_A_DN |
| GAVIN_FOXP3_TARGETS_CLUSTER_P7 | 90 | http://www.broadinstitute.org/gsea/msigdb/cards/GAVIN_FOXP3_TARGETS_CLUSTER_P7 |
| HILLION_HMGA1_TARGETS | 90 | http://www.broadinstitute.org/gsea/msigdb/cards/HILLION_HMGA1_TARGETS |
| IGARASHI_ATF4_TARGETS_DN | 90 | http://www.broadinstitute.org/gsea/msigdb/cards/IGARASHI_ATF4_TARGETS_DN |
| IVANOVSKA_MIR106B_TARGETS | 90 | http://www.broadinstitute.org/gsea/msigdb/cards/IVANOVSKA_MIR106B_TARGETS |
| KEGG_GAP_JUNCTION | 90 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GAP_JUNCTION |
| MORI_IMMATURE_B_LYMPHOCYTE_DN | 90 | http://www.broadinstitute.org/gsea/msigdb/cards/MORI_IMMATURE_B_LYMPHOCYTE_DN |
| MORI_MATURE_B_LYMPHOCYTE_UP | 90 | http://www.broadinstitute.org/gsea/msigdb/cards/MORI_MATURE_B_LYMPHOCYTE_UP |
| REACTOME_SIGNALING_BY_ERBB4 | 90 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_ERBB4 |
| SANA_TNF_SIGNALING_DN | 90 | http://www.broadinstitute.org/gsea/msigdb/cards/SANA_TNF_SIGNALING_DN |
| SASSON_RESPONSE_TO_FORSKOLIN_UP | 90 | http://www.broadinstitute.org/gsea/msigdb/cards/SASSON_RESPONSE_TO_FORSKOLIN_UP |
| BHATTACHARYA_EMBRYONIC_STEM_CELL | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/BHATTACHARYA_EMBRYONIC_STEM_CELL |
| BOYLAN_MULTIPLE_MYELOMA_D_UP | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYLAN_MULTIPLE_MYELOMA_D_UP |
| DELASERNA_MYOD_TARGETS_UP | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/DELASERNA_MYOD_TARGETS_UP |
| KANG_IMMORTALIZED_BY_TERT_UP | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/KANG_IMMORTALIZED_BY_TERT_UP |
| KEGG_ANTIGEN_PROCESSING_AND_PRESENTATION | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ANTIGEN_PROCESSING_AND_PRESENTATION |
| KEGG_PROSTATE_CANCER | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PROSTATE_CANCER |
| LEE_AGING_NEOCORTEX_UP | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_AGING_NEOCORTEX_UP |
| LEIN_PONS_MARKERS | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/LEIN_PONS_MARKERS |
| LIU_VAV3_PROSTATE_CARCINOGENESIS_UP | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_VAV3_PROSTATE_CARCINOGENESIS_UP |
| LI_DCP2_BOUND_MRNA | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_DCP2_BOUND_MRNA |
| REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ |
| REACTOME_RNA_POL_I_TRANSCRIPTION | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RNA_POL_I_TRANSCRIPTION |
| REACTOME_TRANSPORT_OF_GLUCOSE_AND_OTHER_SUGARS_BILE_SALTS_AND_ORGANIC_ACIDS_METAL_IONS_AND_AMINE_COMPOUNDS | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANSPORT_OF_GLUCOSE_AND_OTHER_SUGARS_BILE_SALTS_AND_ORGANIC_ACIDS_METAL_IONS_AND_AMINE_COMPOUNDS |
| RICKMAN_HEAD_AND_NECK_CANCER_E | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/RICKMAN_HEAD_AND_NECK_CANCER_E |
| SWEET_KRAS_ONCOGENIC_SIGNATURE | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/SWEET_KRAS_ONCOGENIC_SIGNATURE |
| VANDESLUIS_COMMD1_TARGETS_GROUP_3_UP | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/VANDESLUIS_COMMD1_TARGETS_GROUP_3_UP |
| VANLOO_SP3_TARGETS_DN | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/VANLOO_SP3_TARGETS_DN |
| WNT_SIGNALING | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/WNT_SIGNALING |
| ZHANG_TLX_TARGETS_UP | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_TLX_TARGETS_UP |
| ZHAN_MULTIPLE_MYELOMA_CD1_AND_CD2_UP | 89 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_CD1_AND_CD2_UP |
| ALCALA_APOPTOSIS | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/ALCALA_APOPTOSIS |
| BHAT_ESR1_TARGETS_NOT_VIA_AKT1_DN | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/BHAT_ESR1_TARGETS_NOT_VIA_AKT1_DN |
| DACOSTA_UV_RESPONSE_VIA_ERCC3_XPCS_DN | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/DACOSTA_UV_RESPONSE_VIA_ERCC3_XPCS_DN |
| DURCHDEWALD_SKIN_CARCINOGENESIS_UP | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/DURCHDEWALD_SKIN_CARCINOGENESIS_UP |
| HUMMERICH_SKIN_CANCER_PROGRESSION_UP | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/HUMMERICH_SKIN_CANCER_PROGRESSION_UP |
| KEGG_APOPTOSIS | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_APOPTOSIS |
| KEGG_HEMATOPOIETIC_CELL_LINEAGE | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_HEMATOPOIETIC_CELL_LINEAGE |
| KEGG_RIBOSOME | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_RIBOSOME |
| KESHELAVA_MULTIPLE_DRUG_RESISTANCE | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/KESHELAVA_MULTIPLE_DRUG_RESISTANCE |
| KIM_BIPOLAR_DISORDER_OLIGODENDROCYTE_DENSITY_CORR_DN | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_BIPOLAR_DISORDER_OLIGODENDROCYTE_DENSITY_CORR_DN |
| KYNG_DNA_DAMAGE_BY_GAMMA_AND_UV_RADIATION | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_DNA_DAMAGE_BY_GAMMA_AND_UV_RADIATION |
| LEE_CALORIE_RESTRICTION_NEOCORTEX_DN | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_CALORIE_RESTRICTION_NEOCORTEX_DN |
| NEMETH_INFLAMMATORY_RESPONSE_LPS_UP | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/NEMETH_INFLAMMATORY_RESPONSE_LPS_UP |
| REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS |
| SASSON_RESPONSE_TO_FORSKOLIN_DN | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/SASSON_RESPONSE_TO_FORSKOLIN_DN |
| SCHLESINGER_METHYLATED_DE_NOVO_IN_CANCER | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHLESINGER_METHYLATED_DE_NOVO_IN_CANCER |
| SEIDEN_ONCOGENESIS_BY_MET | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/SEIDEN_ONCOGENESIS_BY_MET |
| TIEN_INTESTINE_PROBIOTICS_2HR_DN | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/TIEN_INTESTINE_PROBIOTICS_2HR_DN |
| UEDA_CENTRAL_CLOCK | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/UEDA_CENTRAL_CLOCK |
| WALLACE_PROSTATE_CANCER_RACE_DN | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/WALLACE_PROSTATE_CANCER_RACE_DN |
| ZHANG_TARGETS_OF_EWSR1_FLI1_FUSION | 88 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_TARGETS_OF_EWSR1_FLI1_FUSION |
| BIOCARTA_MAPK_PATHWAY | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_MAPK_PATHWAY |
| FERRANDO_T_ALL_WITH_MLL_ENL_FUSION_DN | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/FERRANDO_T_ALL_WITH_MLL_ENL_FUSION_DN |
| FERRANDO_T_ALL_WITH_MLL_ENL_FUSION_UP | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/FERRANDO_T_ALL_WITH_MLL_ENL_FUSION_UP |
| FLECHNER_BIOPSY_KIDNEY_TRANSPLANT_REJECTED_VS_OK_UP | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/FLECHNER_BIOPSY_KIDNEY_TRANSPLANT_REJECTED_VS_OK_UP |
| GRAHAM_CML_QUIESCENT_VS_NORMAL_QUIESCENT_UP | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAHAM_CML_QUIESCENT_VS_NORMAL_QUIESCENT_UP |
| GRAHAM_NORMAL_QUIESCENT_VS_NORMAL_DIVIDING_DN | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAHAM_NORMAL_QUIESCENT_VS_NORMAL_DIVIDING_DN |
| KAYO_CALORIE_RESTRICTION_MUSCLE_DN | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/KAYO_CALORIE_RESTRICTION_MUSCLE_DN |
| KEGG_ERBB_SIGNALING_PATHWAY | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ERBB_SIGNALING_PATHWAY |
| MOOTHA_VOXPHOS | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/MOOTHA_VOXPHOS |
| NAKAYAMA_SOFT_TISSUE_TUMORS_PCA2_UP | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAYAMA_SOFT_TISSUE_TUMORS_PCA2_UP |
| PURBEY_TARGETS_OF_CTBP1_AND_SATB1_UP | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/PURBEY_TARGETS_OF_CTBP1_AND_SATB1_UP |
| REACTOME_EXTRACELLULAR_MATRIX_ORGANIZATION | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_EXTRACELLULAR_MATRIX_ORGANIZATION |
| REACTOME_INSULIN_RECEPTOR_SIGNALLING_CASCADE | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INSULIN_RECEPTOR_SIGNALLING_CASCADE |
| REACTOME_MITOTIC_PROMETAPHASE | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MITOTIC_PROMETAPHASE |
| SAFFORD_T_LYMPHOCYTE_ANERGY | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/SAFFORD_T_LYMPHOCYTE_ANERGY |
| SASSON_RESPONSE_TO_GONADOTROPHINS_DN | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/SASSON_RESPONSE_TO_GONADOTROPHINS_DN |
| SMITH_TERT_TARGETS_DN | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/SMITH_TERT_TARGETS_DN |
| YORDY_RECIPROCAL_REGULATION_BY_ETS1_AND_SP100_DN | 87 | http://www.broadinstitute.org/gsea/msigdb/cards/YORDY_RECIPROCAL_REGULATION_BY_ETS1_AND_SP100_DN |
| BAELDE_DIABETIC_NEPHROPATHY_UP | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/BAELDE_DIABETIC_NEPHROPATHY_UP |
| BROWNE_HCMV_INFECTION_16HR_DN | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_16HR_DN |
| BURTON_ADIPOGENESIS_8 | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/BURTON_ADIPOGENESIS_8 |
| GHANDHI_BYSTANDER_IRRADIATION_UP | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/GHANDHI_BYSTANDER_IRRADIATION_UP |
| HOEBEKE_LYMPHOID_STEM_CELL_DN | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/HOEBEKE_LYMPHOID_STEM_CELL_DN |
| KEGG_PROGESTERONE_MEDIATED_OOCYTE_MATURATION | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PROGESTERONE_MEDIATED_OOCYTE_MATURATION |
| KEGG_TGF_BETA_SIGNALING_PATHWAY | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_TGF_BETA_SIGNALING_PATHWAY |
| LAIHO_COLORECTAL_CANCER_SERRATED_DN | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/LAIHO_COLORECTAL_CANCER_SERRATED_DN |
| LEE_AGING_CEREBELLUM_DN | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_AGING_CEREBELLUM_DN |
| MANTOVANI_VIRAL_GPCR_SIGNALING_UP | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/MANTOVANI_VIRAL_GPCR_SIGNALING_UP |
| MORI_LARGE_PRE_BII_LYMPHOCYTE_UP | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/MORI_LARGE_PRE_BII_LYMPHOCYTE_UP |
| MORI_SMALL_PRE_BII_LYMPHOCYTE_UP | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/MORI_SMALL_PRE_BII_LYMPHOCYTE_UP |
| NELSON_RESPONSE_TO_ANDROGEN_UP | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/NELSON_RESPONSE_TO_ANDROGEN_UP |
| NGUYEN_NOTCH1_TARGETS_DN | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/NGUYEN_NOTCH1_TARGETS_DN |
| REACTOME_L1CAM_INTERACTIONS | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_L1CAM_INTERACTIONS |
| REACTOME_MEIOTIC_RECOMBINATION | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MEIOTIC_RECOMBINATION |
| TAKAO_RESPONSE_TO_UVB_RADIATION_UP | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/TAKAO_RESPONSE_TO_UVB_RADIATION_UP |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_2 | 86 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_2 |
| CHIANG_LIVER_CANCER_SUBCLASS_UNANNOTATED_UP | 85 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIANG_LIVER_CANCER_SUBCLASS_UNANNOTATED_UP |
| COLDREN_GEFITINIB_RESISTANCE_UP | 85 | http://www.broadinstitute.org/gsea/msigdb/cards/COLDREN_GEFITINIB_RESISTANCE_UP |
| GALINDO_IMMUNE_RESPONSE_TO_ENTEROTOXIN | 85 | http://www.broadinstitute.org/gsea/msigdb/cards/GALINDO_IMMUNE_RESPONSE_TO_ENTEROTOXIN |
| KEGG_HYPERTROPHIC_CARDIOMYOPATHY_HCM | 85 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_HYPERTROPHIC_CARDIOMYOPATHY_HCM |
| LEIN_CEREBELLUM_MARKERS | 85 | http://www.broadinstitute.org/gsea/msigdb/cards/LEIN_CEREBELLUM_MARKERS |
| REACTOME_REGULATION_OF_MITOTIC_CELL_CYCLE | 85 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_MITOTIC_CELL_CYCLE |
| SANA_RESPONSE_TO_IFNG_DN | 85 | http://www.broadinstitute.org/gsea/msigdb/cards/SANA_RESPONSE_TO_IFNG_DN |
| SMID_BREAST_CANCER_RELAPSE_IN_BRAIN_DN | 85 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_RELAPSE_IN_BRAIN_DN |
| STEIN_ESR1_TARGETS | 85 | http://www.broadinstitute.org/gsea/msigdb/cards/STEIN_ESR1_TARGETS |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_6HR_UP | 85 | http://www.broadinstitute.org/gsea/msigdb/cards/TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_6HR_UP |
| XU_GH1_EXOGENOUS_TARGETS_UP | 85 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_GH1_EXOGENOUS_TARGETS_UP |
| ZHOU_CELL_CYCLE_GENES_IN_IR_RESPONSE_6HR | 85 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHOU_CELL_CYCLE_GENES_IN_IR_RESPONSE_6HR |
| ACOSTA_PROLIFERATION_INDEPENDENT_MYC_TARGETS_UP | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/ACOSTA_PROLIFERATION_INDEPENDENT_MYC_TARGETS_UP |
| CHANG_IMMORTALIZED_BY_HPV31_UP | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/CHANG_IMMORTALIZED_BY_HPV31_UP |
| DACOSTA_UV_RESPONSE_VIA_ERCC3_TTD_DN | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/DACOSTA_UV_RESPONSE_VIA_ERCC3_TTD_DN |
| GRAESSMANN_RESPONSE_TO_MC_AND_SERUM_DEPRIVATION_DN | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAESSMANN_RESPONSE_TO_MC_AND_SERUM_DEPRIVATION_DN |
| HINATA_NFKB_TARGETS_FIBROBLAST_UP | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/HINATA_NFKB_TARGETS_FIBROBLAST_UP |
| KEGG_ECM_RECEPTOR_INTERACTION | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ECM_RECEPTOR_INTERACTION |
| KEGG_SMALL_CELL_LUNG_CANCER | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_SMALL_CELL_LUNG_CANCER |
| KIM_GLIS2_TARGETS_UP | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_GLIS2_TARGETS_UP |
| LEE_AGING_CEREBELLUM_UP | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_AGING_CEREBELLUM_UP |
| PID_CMYB_PATHWAY | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_CMYB_PATHWAY |
| RASHI_RESPONSE_TO_IONIZING_RADIATION_6 | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/RASHI_RESPONSE_TO_IONIZING_RADIATION_6 |
| REACTOME_ACTIVATION_OF_THE_MRNA_UPON_BINDING_OF_THE_CAP_BINDING_COMPLEX_AND_EIFS_AND_SUBSEQUENT_BINDING_TO_43S | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATION_OF_THE_MRNA_UPON_BINDING_OF_THE_CAP_BINDING_COMPLEX_AND_EIFS_AND_SUBSEQUENT_BINDING_TO_43S |
| REACTOME_REGULATION_OF_MRNA_STABILITY_BY_PROTEINS_THAT_BIND_AU_RICH_ELEMENTS | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_MRNA_STABILITY_BY_PROTEINS_THAT_BIND_AU_RICH_ELEMENTS |
| RUAN_RESPONSE_TO_TNF_DN | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/RUAN_RESPONSE_TO_TNF_DN |
| SCHUETZ_BREAST_CANCER_DUCTAL_INVASIVE_DN | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHUETZ_BREAST_CANCER_DUCTAL_INVASIVE_DN |
| SLEBOS_HEAD_AND_NECK_CANCER_WITH_HPV_UP | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/SLEBOS_HEAD_AND_NECK_CANCER_WITH_HPV_UP |
| SMID_BREAST_CANCER_LUMINAL_A_UP | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_LUMINAL_A_UP |
| STAMBOLSKY_RESPONSE_TO_VITAMIN_D3_UP | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/STAMBOLSKY_RESPONSE_TO_VITAMIN_D3_UP |
| SWEET_KRAS_TARGETS_UP | 84 | http://www.broadinstitute.org/gsea/msigdb/cards/SWEET_KRAS_TARGETS_UP |
| JIANG_HYPOXIA_CANCER | 83 | http://www.broadinstitute.org/gsea/msigdb/cards/JIANG_HYPOXIA_CANCER |
| LEE_CALORIE_RESTRICTION_NEOCORTEX_UP | 83 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_CALORIE_RESTRICTION_NEOCORTEX_UP |
| LEE_TARGETS_OF_PTCH1_AND_SUFU_DN | 83 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_TARGETS_OF_PTCH1_AND_SUFU_DN |
| LIEN_BREAST_CARCINOMA_METAPLASTIC_VS_DUCTAL_UP | 83 | http://www.broadinstitute.org/gsea/msigdb/cards/LIEN_BREAST_CARCINOMA_METAPLASTIC_VS_DUCTAL_UP |
| MARIADASON_REGULATED_BY_HISTONE_ACETYLATION_UP | 83 | http://www.broadinstitute.org/gsea/msigdb/cards/MARIADASON_REGULATED_BY_HISTONE_ACETYLATION_UP |
| ODONNELL_TARGETS_OF_MYC_AND_TFRC_UP | 83 | http://www.broadinstitute.org/gsea/msigdb/cards/ODONNELL_TARGETS_OF_MYC_AND_TFRC_UP |
| ONDER_CDH1_SIGNALING_VIA_CTNNB1 | 83 | http://www.broadinstitute.org/gsea/msigdb/cards/ONDER_CDH1_SIGNALING_VIA_CTNNB1 |
| REACTOME_AMYLOIDS | 83 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_AMYLOIDS |
| REACTOME_MYD88_MAL_CASCADE_INITIATED_ON_PLASMA_MEMBRANE | 83 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MYD88_MAL_CASCADE_INITIATED_ON_PLASMA_MEMBRANE |
| SAKAI_CHRONIC_HEPATITIS_VS_LIVER_CANCER_UP | 83 | http://www.broadinstitute.org/gsea/msigdb/cards/SAKAI_CHRONIC_HEPATITIS_VS_LIVER_CANCER_UP |
| SANA_TNF_SIGNALING_UP | 83 | http://www.broadinstitute.org/gsea/msigdb/cards/SANA_TNF_SIGNALING_UP |
| WARTERS_RESPONSE_TO_IR_SKIN | 83 | http://www.broadinstitute.org/gsea/msigdb/cards/WARTERS_RESPONSE_TO_IR_SKIN |
| BHAT_ESR1_TARGETS_VIA_AKT1_DN | 82 | http://www.broadinstitute.org/gsea/msigdb/cards/BHAT_ESR1_TARGETS_VIA_AKT1_DN |
| BILD_CTNNB1_ONCOGENIC_SIGNATURE | 82 | http://www.broadinstitute.org/gsea/msigdb/cards/BILD_CTNNB1_ONCOGENIC_SIGNATURE |
| FRASOR_RESPONSE_TO_ESTRADIOL_DN | 82 | http://www.broadinstitute.org/gsea/msigdb/cards/FRASOR_RESPONSE_TO_ESTRADIOL_DN |
| KUNINGER_IGF1_VS_PDGFB_TARGETS_UP | 82 | http://www.broadinstitute.org/gsea/msigdb/cards/KUNINGER_IGF1_VS_PDGFB_TARGETS_UP |
| LEIN_MIDBRAIN_MARKERS | 82 | http://www.broadinstitute.org/gsea/msigdb/cards/LEIN_MIDBRAIN_MARKERS |
| ODONNELL_METASTASIS_UP | 82 | http://www.broadinstitute.org/gsea/msigdb/cards/ODONNELL_METASTASIS_UP |
| PID_REG_GR_PATHWAY | 82 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_REG_GR_PATHWAY |
| PID_SMAD2_3NUCLEARPATHWAY | 82 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_SMAD2_3NUCLEARPATHWAY |
| PUIFFE_INVASION_INHIBITED_BY_ASCITES_UP | 82 | http://www.broadinstitute.org/gsea/msigdb/cards/PUIFFE_INVASION_INHIBITED_BY_ASCITES_UP |
| REACTOME_GLYCEROPHOSPHOLIPID_BIOSYNTHESIS | 82 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GLYCEROPHOSPHOLIPID_BIOSYNTHESIS |
| ROSS_ACUTE_MYELOID_LEUKEMIA_CBF | 82 | http://www.broadinstitute.org/gsea/msigdb/cards/ROSS_ACUTE_MYELOID_LEUKEMIA_CBF |
| ST_INTEGRIN_SIGNALING_PATHWAY | 82 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_INTEGRIN_SIGNALING_PATHWAY |
| WONG_ENDMETRIUM_CANCER_DN | 82 | http://www.broadinstitute.org/gsea/msigdb/cards/WONG_ENDMETRIUM_CANCER_DN |
| COATES_MACROPHAGE_M1_VS_M2_UP | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/COATES_MACROPHAGE_M1_VS_M2_UP |
| CROMER_METASTASIS_DN | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/CROMER_METASTASIS_DN |
| GRABARCZYK_BCL11B_TARGETS_UP | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/GRABARCZYK_BCL11B_TARGETS_UP |
| GUO_HEX_TARGETS_UP | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/GUO_HEX_TARGETS_UP |
| HARRIS_HYPOXIA | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/HARRIS_HYPOXIA |
| HERNANDEZ_ABERRANT_MITOSIS_BY_DOCETACEL_2NM_UP | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/HERNANDEZ_ABERRANT_MITOSIS_BY_DOCETACEL_2NM_UP |
| HUANG_DASATINIB_RESISTANCE_UP | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/HUANG_DASATINIB_RESISTANCE_UP |
| KYNG_DNA_DAMAGE_BY_GAMMA_RADIATION | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_DNA_DAMAGE_BY_GAMMA_RADIATION |
| LEIN_MEDULLA_MARKERS | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/LEIN_MEDULLA_MARKERS |
| MAHAJAN_RESPONSE_TO_IL1A_UP | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/MAHAJAN_RESPONSE_TO_IL1A_UP |
| MUNSHI_MULTIPLE_MYELOMA_UP | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/MUNSHI_MULTIPLE_MYELOMA_UP |
| REACTOME_ASPARAGINE_N_LINKED_GLYCOSYLATION | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ASPARAGINE_N_LINKED_GLYCOSYLATION |
| REACTOME_MITOTIC_G2_G2_M_PHASES | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MITOTIC_G2_G2_M_PHASES |
| REACTOME_M_G1_TRANSITION | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_M_G1_TRANSITION |
| REACTOME_P75_NTR_RECEPTOR_MEDIATED_SIGNALLING | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_P75_NTR_RECEPTOR_MEDIATED_SIGNALLING |
| SAKAI_TUMOR_INFILTRATING_MONOCYTES_DN | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/SAKAI_TUMOR_INFILTRATING_MONOCYTES_DN |
| SCIBETTA_KDM5B_TARGETS_DN | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/SCIBETTA_KDM5B_TARGETS_DN |
| SHAFFER_IRF4_TARGETS_IN_ACTIVATED_B_LYMPHOCYTE | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/SHAFFER_IRF4_TARGETS_IN_ACTIVATED_B_LYMPHOCYTE |
| WATANABE_COLON_CANCER_MSI_VS_MSS_DN | 81 | http://www.broadinstitute.org/gsea/msigdb/cards/WATANABE_COLON_CANCER_MSI_VS_MSS_DN |
| BASSO_HAIRY_CELL_LEUKEMIA_DN | 80 | http://www.broadinstitute.org/gsea/msigdb/cards/BASSO_HAIRY_CELL_LEUKEMIA_DN |
| BOYLAN_MULTIPLE_MYELOMA_PCA3_UP | 80 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYLAN_MULTIPLE_MYELOMA_PCA3_UP |
| FALVELLA_SMOKERS_WITH_LUNG_CANCER | 80 | http://www.broadinstitute.org/gsea/msigdb/cards/FALVELLA_SMOKERS_WITH_LUNG_CANCER |
| FONTAINE_PAPILLARY_THYROID_CARCINOMA_DN | 80 | http://www.broadinstitute.org/gsea/msigdb/cards/FONTAINE_PAPILLARY_THYROID_CARCINOMA_DN |
| HELLEBREKERS_SILENCED_DURING_TUMOR_ANGIOGENESIS | 80 | http://www.broadinstitute.org/gsea/msigdb/cards/HELLEBREKERS_SILENCED_DURING_TUMOR_ANGIOGENESIS |
| KEGG_CARDIAC_MUSCLE_CONTRACTION | 80 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_CARDIAC_MUSCLE_CONTRACTION |
| LEE_AGING_NEOCORTEX_DN | 80 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_AGING_NEOCORTEX_DN |
| LINDVALL_IMMORTALIZED_BY_TERT_DN | 80 | http://www.broadinstitute.org/gsea/msigdb/cards/LINDVALL_IMMORTALIZED_BY_TERT_DN |
| MIKKELSEN_IPS_HCP_WITH_H3_UNMETHYLATED | 80 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_IPS_HCP_WITH_H3_UNMETHYLATED |
| MORI_PRE_BI_LYMPHOCYTE_UP | 80 | http://www.broadinstitute.org/gsea/msigdb/cards/MORI_PRE_BI_LYMPHOCYTE_UP |
| NAKAYAMA_SOFT_TISSUE_TUMORS_PCA2_DN | 80 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAYAMA_SOFT_TISSUE_TUMORS_PCA2_DN |
| PID_BETACATENIN_NUC_PATHWAY | 80 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_BETACATENIN_NUC_PATHWAY |
| PID_MET_PATHWAY | 80 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_MET_PATHWAY |
| REACTOME_UNFOLDED_PROTEIN_RESPONSE | 80 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_UNFOLDED_PROTEIN_RESPONSE |
| SCHUHMACHER_MYC_TARGETS_UP | 80 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHUHMACHER_MYC_TARGETS_UP |
| WOO_LIVER_CANCER_RECURRENCE_DN | 80 | http://www.broadinstitute.org/gsea/msigdb/cards/WOO_LIVER_CANCER_RECURRENCE_DN |
| CHIANG_LIVER_CANCER_SUBCLASS_POLYSOMY7_UP | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIANG_LIVER_CANCER_SUBCLASS_POLYSOMY7_UP |
| CROMER_METASTASIS_UP | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/CROMER_METASTASIS_UP |
| DUAN_PRDM5_TARGETS | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/DUAN_PRDM5_TARGETS |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_TURQUOISE_UP | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_TURQUOISE_UP |
| GAVIN_FOXP3_TARGETS_CLUSTER_P2 | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/GAVIN_FOXP3_TARGETS_CLUSTER_P2 |
| KEGG_FC_EPSILON_RI_SIGNALING_PATHWAY | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_FC_EPSILON_RI_SIGNALING_PATHWAY |
| LIU_COMMON_CANCER_GENES | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_COMMON_CANCER_GENES |
| MALONEY_RESPONSE_TO_17AAG_DN | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/MALONEY_RESPONSE_TO_17AAG_DN |
| MCBRYAN_PUBERTAL_BREAST_6_7WK_DN | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/MCBRYAN_PUBERTAL_BREAST_6_7WK_DN |
| MCCLUNG_COCAINE_REWARD_5D | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/MCCLUNG_COCAINE_REWARD_5D |
| MEISSNER_NPC_HCP_WITH_H3K27ME3 | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/MEISSNER_NPC_HCP_WITH_H3K27ME3 |
| PID_MYC_ACTIVPATHWAY | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_MYC_ACTIVPATHWAY |
| PID_P73PATHWAY | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_P73PATHWAY |
| REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS |
| REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RESPIRATORY_ELECTRON_TRANSPORT |
| TOOKER_GEMCITABINE_RESISTANCE_UP | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/TOOKER_GEMCITABINE_RESISTANCE_UP |
| YAO_HOXA10_TARGETS_VIA_PROGESTERONE_UP | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_HOXA10_TARGETS_VIA_PROGESTERONE_UP |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_12 | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_12 |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_16 | 79 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_16 |
| BOSCO_INTERFERON_INDUCED_ANTIVIRAL_MODULE | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/BOSCO_INTERFERON_INDUCED_ANTIVIRAL_MODULE |
| BOYLAN_MULTIPLE_MYELOMA_D_DN | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYLAN_MULTIPLE_MYELOMA_D_DN |
| BRUECKNER_TARGETS_OF_MIRLET7A3_DN | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/BRUECKNER_TARGETS_OF_MIRLET7A3_DN |
| COATES_MACROPHAGE_M1_VS_M2_DN | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/COATES_MACROPHAGE_M1_VS_M2_DN |
| GINESTIER_BREAST_CANCER_ZNF217_AMPLIFIED_UP | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/GINESTIER_BREAST_CANCER_ZNF217_AMPLIFIED_UP |
| HADDAD_T_LYMPHOCYTE_AND_NK_PROGENITOR_UP | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/HADDAD_T_LYMPHOCYTE_AND_NK_PROGENITOR_UP |
| KEGG_PEROXISOME | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PEROXISOME |
| LINDVALL_IMMORTALIZED_BY_TERT_UP | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/LINDVALL_IMMORTALIZED_BY_TERT_UP |
| RADMACHER_AML_PROGNOSIS | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/RADMACHER_AML_PROGNOSIS |
| REACTOME_CELL_JUNCTION_ORGANIZATION | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CELL_JUNCTION_ORGANIZATION |
| REACTOME_OPIOID_SIGNALLING | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_OPIOID_SIGNALLING |
| REACTOME_PLATELET_HOMEOSTASIS | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PLATELET_HOMEOSTASIS |
| REACTOME_SIGNALING_BY_SCF_KIT | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_SCF_KIT |
| RHEIN_ALL_GLUCOCORTICOID_THERAPY_UP | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/RHEIN_ALL_GLUCOCORTICOID_THERAPY_UP |
| ROSS_AML_WITH_MLL_FUSIONS | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/ROSS_AML_WITH_MLL_FUSIONS |
| ROSS_LEUKEMIA_WITH_MLL_FUSIONS | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/ROSS_LEUKEMIA_WITH_MLL_FUSIONS |
| SANA_RESPONSE_TO_IFNG_UP | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/SANA_RESPONSE_TO_IFNG_UP |
| TARTE_PLASMA_CELL_VS_B_LYMPHOCYTE_UP | 78 | http://www.broadinstitute.org/gsea/msigdb/cards/TARTE_PLASMA_CELL_VS_B_LYMPHOCYTE_UP |
| CARD_MIR302A_TARGETS | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/CARD_MIR302A_TARGETS |
| CHEBOTAEV_GR_TARGETS_UP | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEBOTAEV_GR_TARGETS_UP |
| DACOSTA_UV_RESPONSE_VIA_ERCC3_COMMON_UP | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/DACOSTA_UV_RESPONSE_VIA_ERCC3_COMMON_UP |
| FRIDMAN_SENESCENCE_UP | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/FRIDMAN_SENESCENCE_UP |
| GROSS_HYPOXIA_VIA_HIF1A_UP | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/GROSS_HYPOXIA_VIA_HIF1A_UP |
| HESS_TARGETS_OF_HOXA9_AND_MEIS1_DN | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/HESS_TARGETS_OF_HOXA9_AND_MEIS1_DN |
| JACKSON_DNMT1_TARGETS_UP | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/JACKSON_DNMT1_TARGETS_UP |
| KEGG_GLYCEROPHOSPHOLIPID_METABOLISM | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GLYCEROPHOSPHOLIPID_METABOLISM |
| LIN_APC_TARGETS | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/LIN_APC_TARGETS |
| MORI_PRE_BI_LYMPHOCYTE_DN | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/MORI_PRE_BI_LYMPHOCYTE_DN |
| MULLIGHAN_NPM1_MUTATED_SIGNATURE_2_DN | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/MULLIGHAN_NPM1_MUTATED_SIGNATURE_2_DN |
| REACTOME_TRAF6_MEDIATED_INDUCTION_OF_NFKB_AND_MAP_KINASES_UPON_TLR7_8_OR_9_ACTIVATION | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRAF6_MEDIATED_INDUCTION_OF_NFKB_AND_MAP_KINASES_UPON_TLR7_8_OR_9_ACTIVATION |
| RIZ_ERYTHROID_DIFFERENTIATION | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/RIZ_ERYTHROID_DIFFERENTIATION |
| RODRIGUES_THYROID_CARCINOMA_DN | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/RODRIGUES_THYROID_CARCINOMA_DN |
| ROSS_AML_WITH_PML_RARA_FUSION | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/ROSS_AML_WITH_PML_RARA_FUSION |
| SEKI_INFLAMMATORY_RESPONSE_LPS_UP | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/SEKI_INFLAMMATORY_RESPONSE_LPS_UP |
| TAYLOR_METHYLATED_IN_ACUTE_LYMPHOBLASTIC_LEUKEMIA | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/TAYLOR_METHYLATED_IN_ACUTE_LYMPHOBLASTIC_LEUKEMIA |
| THEILGAARD_NEUTROPHIL_AT_SKIN_WOUND_UP | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/THEILGAARD_NEUTROPHIL_AT_SKIN_WOUND_UP |
| ZHONG_SECRETOME_OF_LUNG_CANCER_AND_MACROPHAGE | 77 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHONG_SECRETOME_OF_LUNG_CANCER_AND_MACROPHAGE |
| KEGG_ARRHYTHMOGENIC_RIGHT_VENTRICULAR_CARDIOMYOPATHY_ARVC | 76 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ARRHYTHMOGENIC_RIGHT_VENTRICULAR_CARDIOMYOPATHY_ARVC |
| KEGG_PHOSPHATIDYLINOSITOL_SIGNALING_SYSTEM | 76 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PHOSPHATIDYLINOSITOL_SIGNALING_SYSTEM |
| KEGG_VEGF_SIGNALING_PATHWAY | 76 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_VEGF_SIGNALING_PATHWAY |
| MAHAJAN_RESPONSE_TO_IL1A_DN | 76 | http://www.broadinstitute.org/gsea/msigdb/cards/MAHAJAN_RESPONSE_TO_IL1A_DN |
| MORI_SMALL_PRE_BII_LYMPHOCYTE_DN | 76 | http://www.broadinstitute.org/gsea/msigdb/cards/MORI_SMALL_PRE_BII_LYMPHOCYTE_DN |
| NAKAYAMA_SOFT_TISSUE_TUMORS_PCA1_UP | 76 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAYAMA_SOFT_TISSUE_TUMORS_PCA1_UP |
| NIKOLSKY_BREAST_CANCER_7Q21_Q22_AMPLICON | 76 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_7Q21_Q22_AMPLICON |
| PEART_HDAC_PROLIFERATION_CLUSTER_DN | 76 | http://www.broadinstitute.org/gsea/msigdb/cards/PEART_HDAC_PROLIFERATION_CLUSTER_DN |
| PEDERSEN_TARGETS_OF_611CTF_ISOFORM_OF_ERBB2 | 76 | http://www.broadinstitute.org/gsea/msigdb/cards/PEDERSEN_TARGETS_OF_611CTF_ISOFORM_OF_ERBB2 |
| REACTOME_ANTIGEN_PROCESSING_CROSS_PRESENTATION | 76 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ANTIGEN_PROCESSING_CROSS_PRESENTATION |
| ROSS_AML_WITH_AML1_ETO_FUSION | 76 | http://www.broadinstitute.org/gsea/msigdb/cards/ROSS_AML_WITH_AML1_ETO_FUSION |
| SU_TESTIS | 76 | http://www.broadinstitute.org/gsea/msigdb/cards/SU_TESTIS |
| YAMASHITA_LIVER_CANCER_STEM_CELL_DN | 76 | http://www.broadinstitute.org/gsea/msigdb/cards/YAMASHITA_LIVER_CANCER_STEM_CELL_DN |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_0 | 76 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_0 |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_7 | 76 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_7 |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_9 | 76 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_9 |
| BAE_BRCA1_TARGETS_UP | 75 | http://www.broadinstitute.org/gsea/msigdb/cards/BAE_BRCA1_TARGETS_UP |
| CHESLER_BRAIN_QTL_CIS | 75 | http://www.broadinstitute.org/gsea/msigdb/cards/CHESLER_BRAIN_QTL_CIS |
| FONTAINE_FOLLICULAR_THYROID_ADENOMA_UP | 75 | http://www.broadinstitute.org/gsea/msigdb/cards/FONTAINE_FOLLICULAR_THYROID_ADENOMA_UP |
| HOFFMANN_PRE_BI_TO_LARGE_PRE_BII_LYMPHOCYTE_DN | 75 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFFMANN_PRE_BI_TO_LARGE_PRE_BII_LYMPHOCYTE_DN |
| JAIN_NFKB_SIGNALING | 75 | http://www.broadinstitute.org/gsea/msigdb/cards/JAIN_NFKB_SIGNALING |
| KEGG_ADHERENS_JUNCTION | 75 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ADHERENS_JUNCTION |
| KEGG_B_CELL_RECEPTOR_SIGNALING_PATHWAY | 75 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_B_CELL_RECEPTOR_SIGNALING_PATHWAY |
| KOKKINAKIS_METHIONINE_DEPRIVATION_96HR_DN | 75 | http://www.broadinstitute.org/gsea/msigdb/cards/KOKKINAKIS_METHIONINE_DEPRIVATION_96HR_DN |
| MCCLUNG_COCAIN_REWARD_4WK | 75 | http://www.broadinstitute.org/gsea/msigdb/cards/MCCLUNG_COCAIN_REWARD_4WK |
| MORI_MATURE_B_LYMPHOCYTE_DN | 75 | http://www.broadinstitute.org/gsea/msigdb/cards/MORI_MATURE_B_LYMPHOCYTE_DN |
| PID_AVB3_INTEGRIN_PATHWAY | 75 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_AVB3_INTEGRIN_PATHWAY |
| REACTOME_TELOMERE_MAINTENANCE | 75 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TELOMERE_MAINTENANCE |
| TCGA_GLIOBLASTOMA_COPY_NUMBER_UP | 75 | http://www.broadinstitute.org/gsea/msigdb/cards/TCGA_GLIOBLASTOMA_COPY_NUMBER_UP |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_6 | 75 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_6 |
| CAMPS_COLON_CANCER_COPY_NUMBER_DN | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/CAMPS_COLON_CANCER_COPY_NUMBER_DN |
| CORRE_MULTIPLE_MYELOMA_UP | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/CORRE_MULTIPLE_MYELOMA_UP |
| DARWICHE_PAPILLOMA_PROGRESSION_RISK | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/DARWICHE_PAPILLOMA_PROGRESSION_RISK |
| DER_IFN_ALPHA_RESPONSE_UP | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/DER_IFN_ALPHA_RESPONSE_UP |
| FURUKAWA_DUSP6_TARGETS_PCI35_DN | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/FURUKAWA_DUSP6_TARGETS_PCI35_DN |
| FURUKAWA_DUSP6_TARGETS_PCI35_UP | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/FURUKAWA_DUSP6_TARGETS_PCI35_UP |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_GREY_DN | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_GREY_DN |
| JI_RESPONSE_TO_FSH_UP | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/JI_RESPONSE_TO_FSH_UP |
| LEE_LIVER_CANCER_DENA_DN | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_DENA_DN |
| LEE_METASTASIS_AND_ALTERNATIVE_SPLICING_UP | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_METASTASIS_AND_ALTERNATIVE_SPLICING_UP |
| LEIN_OLIGODENDROCYTE_MARKERS | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/LEIN_OLIGODENDROCYTE_MARKERS |
| LIAN_LIPA_TARGETS_6M | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/LIAN_LIPA_TARGETS_6M |
| MIKKELSEN_MCV6_ICP_WITH_H3K27ME3 | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_MCV6_ICP_WITH_H3K27ME3 |
| MILI_PSEUDOPODIA_CHEMOTAXIS_UP | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/MILI_PSEUDOPODIA_CHEMOTAXIS_UP |
| NAKAYAMA_SOFT_TISSUE_TUMORS_PCA1_DN | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAYAMA_SOFT_TISSUE_TUMORS_PCA1_DN |
| OUILLETTE_CLL_13Q14_DELETION_UP | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/OUILLETTE_CLL_13Q14_DELETION_UP |
| PID_E2F_PATHWAY | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_E2F_PATHWAY |
| POTTI_ADRIAMYCIN_SENSITIVITY | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/POTTI_ADRIAMYCIN_SENSITIVITY |
| RAMALHO_STEMNESS_DN | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/RAMALHO_STEMNESS_DN |
| REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX |
| REACTOME_G_ALPHA1213_SIGNALLING_EVENTS | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_G_ALPHA1213_SIGNALLING_EVENTS |
| REACTOME_TRIF_MEDIATED_TLR3_SIGNALING | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRIF_MEDIATED_TLR3_SIGNALING |
| SHEPARD_BMYB_TARGETS | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/SHEPARD_BMYB_TARGETS |
| TURASHVILI_BREAST_LOBULAR_CARCINOMA_VS_LOBULAR_NORMAL_DN | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/TURASHVILI_BREAST_LOBULAR_CARCINOMA_VS_LOBULAR_NORMAL_DN |
| URS_ADIPOCYTE_DIFFERENTIATION_UP | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/URS_ADIPOCYTE_DIFFERENTIATION_UP |
| VANASSE_BCL2_TARGETS_DN | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/VANASSE_BCL2_TARGETS_DN |
| WESTON_VEGFA_TARGETS_3HR | 74 | http://www.broadinstitute.org/gsea/msigdb/cards/WESTON_VEGFA_TARGETS_3HR |
| ADDYA_ERYTHROID_DIFFERENTIATION_BY_HEMIN | 73 | http://www.broadinstitute.org/gsea/msigdb/cards/ADDYA_ERYTHROID_DIFFERENTIATION_BY_HEMIN |
| BILANGES_RAPAMYCIN_SENSITIVE_VIA_TSC1_AND_TSC2 | 73 | http://www.broadinstitute.org/gsea/msigdb/cards/BILANGES_RAPAMYCIN_SENSITIVE_VIA_TSC1_AND_TSC2 |
| DITTMER_PTHLH_TARGETS_DN | 73 | http://www.broadinstitute.org/gsea/msigdb/cards/DITTMER_PTHLH_TARGETS_DN |
| GAURNIER_PSMD4_TARGETS | 73 | http://www.broadinstitute.org/gsea/msigdb/cards/GAURNIER_PSMD4_TARGETS |
| HOSHIDA_LIVER_CANCER_SURVIVAL_UP | 73 | http://www.broadinstitute.org/gsea/msigdb/cards/HOSHIDA_LIVER_CANCER_SURVIVAL_UP |
| KEGG_CHRONIC_MYELOID_LEUKEMIA | 73 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_CHRONIC_MYELOID_LEUKEMIA |
| KEGG_VIRAL_MYOCARDITIS | 73 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_VIRAL_MYOCARDITIS |
| LIN_MELANOMA_COPY_NUMBER_UP | 73 | http://www.broadinstitute.org/gsea/msigdb/cards/LIN_MELANOMA_COPY_NUMBER_UP |
| MOREAUX_B_LYMPHOCYTE_MATURATION_BY_TACI_DN | 73 | http://www.broadinstitute.org/gsea/msigdb/cards/MOREAUX_B_LYMPHOCYTE_MATURATION_BY_TACI_DN |
| REACTOME_APC_C_CDC20_MEDIATED_DEGRADATION_OF_MITOTIC_PROTEINS | 73 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_APC_C_CDC20_MEDIATED_DEGRADATION_OF_MITOTIC_PROTEINS |
| REACTOME_MEIOTIC_SYNAPSIS | 73 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MEIOTIC_SYNAPSIS |
| REACTOME_RIG_I_MDA5_MEDIATED_INDUCTION_OF_IFN_ALPHA_BETA_PATHWAYS | 73 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RIG_I_MDA5_MEDIATED_INDUCTION_OF_IFN_ALPHA_BETA_PATHWAYS |
| SEITZ_NEOPLASTIC_TRANSFORMATION_BY_8P_DELETION_UP | 73 | http://www.broadinstitute.org/gsea/msigdb/cards/SEITZ_NEOPLASTIC_TRANSFORMATION_BY_8P_DELETION_UP |
| TURASHVILI_BREAST_DUCTAL_CARCINOMA_VS_LOBULAR_NORMAL_UP | 73 | http://www.broadinstitute.org/gsea/msigdb/cards/TURASHVILI_BREAST_DUCTAL_CARCINOMA_VS_LOBULAR_NORMAL_UP |
| WATTEL_AUTONOMOUS_THYROID_ADENOMA_UP | 73 | http://www.broadinstitute.org/gsea/msigdb/cards/WATTEL_AUTONOMOUS_THYROID_ADENOMA_UP |
| BURTON_ADIPOGENESIS_2 | 72 | http://www.broadinstitute.org/gsea/msigdb/cards/BURTON_ADIPOGENESIS_2 |
| CROONQUIST_NRAS_SIGNALING_DN | 72 | http://www.broadinstitute.org/gsea/msigdb/cards/CROONQUIST_NRAS_SIGNALING_DN |
| DANG_REGULATED_BY_MYC_UP | 72 | http://www.broadinstitute.org/gsea/msigdb/cards/DANG_REGULATED_BY_MYC_UP |
| DOANE_BREAST_CANCER_CLASSES_UP | 72 | http://www.broadinstitute.org/gsea/msigdb/cards/DOANE_BREAST_CANCER_CLASSES_UP |
| HOFFMANN_LARGE_TO_SMALL_PRE_BII_LYMPHOCYTE_DN | 72 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFFMANN_LARGE_TO_SMALL_PRE_BII_LYMPHOCYTE_DN |
| HUANG_GATA2_TARGETS_DN | 72 | http://www.broadinstitute.org/gsea/msigdb/cards/HUANG_GATA2_TARGETS_DN |
| KEGG_DRUG_METABOLISM_CYTOCHROME_P450 | 72 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_DRUG_METABOLISM_CYTOCHROME_P450 |
| KEGG_LEISHMANIA_INFECTION | 72 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_LEISHMANIA_INFECTION |
| MATZUK_SPERMATOCYTE | 72 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_SPERMATOCYTE |
| REACTOME_APC_C_CDH1_MEDIATED_DEGRADATION_OF_CDC20_AND_OTHER_APC_C_CDH1_TARGETED_PROTEINS_IN_LATE_MITOSIS_EARLY_G1 | 72 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_APC_C_CDH1_MEDIATED_DEGRADATION_OF_CDC20_AND_OTHER_APC_C_CDH1_TARGETED_PROTEINS_IN_LATE_MITOSIS_EARLY_G1 |
| REACTOME_METABOLISM_OF_NUCLEOTIDES | 72 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_METABOLISM_OF_NUCLEOTIDES |
| REACTOME_NFKB_AND_MAP_KINASES_ACTIVATION_MEDIATED_BY_TLR4_SIGNALING_REPERTOIRE | 72 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NFKB_AND_MAP_KINASES_ACTIVATION_MEDIATED_BY_TLR4_SIGNALING_REPERTOIRE |
| REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | 72 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION |
| SESTO_RESPONSE_TO_UV_C1 | 72 | http://www.broadinstitute.org/gsea/msigdb/cards/SESTO_RESPONSE_TO_UV_C1 |
| SESTO_RESPONSE_TO_UV_C8 | 72 | http://www.broadinstitute.org/gsea/msigdb/cards/SESTO_RESPONSE_TO_UV_C8 |
| BROWNE_HCMV_INFECTION_6HR_UP | 71 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_6HR_UP |
| DER_IFN_GAMMA_RESPONSE_UP | 71 | http://www.broadinstitute.org/gsea/msigdb/cards/DER_IFN_GAMMA_RESPONSE_UP |
| GEORGANTAS_HSC_MARKERS | 71 | http://www.broadinstitute.org/gsea/msigdb/cards/GEORGANTAS_HSC_MARKERS |
| GU_PDEF_TARGETS_UP | 71 | http://www.broadinstitute.org/gsea/msigdb/cards/GU_PDEF_TARGETS_UP |
| JECHLINGER_EPITHELIAL_TO_MESENCHYMAL_TRANSITION_UP | 71 | http://www.broadinstitute.org/gsea/msigdb/cards/JECHLINGER_EPITHELIAL_TO_MESENCHYMAL_TRANSITION_UP |
| KEGG_MELANOMA | 71 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_MELANOMA |
| KEGG_RIG_I_LIKE_RECEPTOR_SIGNALING_PATHWAY | 71 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_RIG_I_LIKE_RECEPTOR_SIGNALING_PATHWAY |
| KUMAR_AUTOPHAGY_NETWORK | 71 | http://www.broadinstitute.org/gsea/msigdb/cards/KUMAR_AUTOPHAGY_NETWORK |
| KYNG_RESPONSE_TO_H2O2 | 71 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_RESPONSE_TO_H2O2 |
| MARTINEZ_RESPONSE_TO_TRABECTEDIN_UP | 71 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTINEZ_RESPONSE_TO_TRABECTEDIN_UP |
| NATSUME_RESPONSE_TO_INTERFERON_BETA_UP | 71 | http://www.broadinstitute.org/gsea/msigdb/cards/NATSUME_RESPONSE_TO_INTERFERON_BETA_UP |
| REACTOME_PI3K_CASCADE | 71 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PI3K_CASCADE |
| CADWELL_ATG16L1_TARGETS_DN | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/CADWELL_ATG16L1_TARGETS_DN |
| ENGELMANN_CANCER_PROGENITORS_DN | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/ENGELMANN_CANCER_PROGENITORS_DN |
| HOFFMANN_SMALL_PRE_BII_TO_IMMATURE_B_LYMPHOCYTE_UP | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFFMANN_SMALL_PRE_BII_TO_IMMATURE_B_LYMPHOCYTE_UP |
| KEGG_LONG_TERM_DEPRESSION | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_LONG_TERM_DEPRESSION |
| KEGG_LONG_TERM_POTENTIATION | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_LONG_TERM_POTENTIATION |
| KEGG_METABOLISM_OF_XENOBIOTICS_BY_CYTOCHROME_P450 | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_METABOLISM_OF_XENOBIOTICS_BY_CYTOCHROME_P450 |
| KEGG_PANCREATIC_CANCER | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PANCREATIC_CANCER |
| KEGG_RENAL_CELL_CARCINOMA | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_RENAL_CELL_CARCINOMA |
| LANDIS_BREAST_CANCER_PROGRESSION_DN | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/LANDIS_BREAST_CANCER_PROGRESSION_DN |
| LIU_NASOPHARYNGEAL_CARCINOMA | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_NASOPHARYNGEAL_CARCINOMA |
| MIKKELSEN_MEF_LCP_WITH_H3K27ME3 | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_MEF_LCP_WITH_H3K27ME3 |
| PID_AP1_PATHWAY | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_AP1_PATHWAY |
| PID_CDC42_PATHWAY | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_CDC42_PATHWAY |
| RAGHAVACHARI_PLATELET_SPECIFIC_GENES | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/RAGHAVACHARI_PLATELET_SPECIFIC_GENES |
| REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL |
| REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS |
| REACTOME_PHASE_II_CONJUGATION | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PHASE_II_CONJUGATION |
| REACTOME_SIGNALING_BY_NOTCH1 | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_NOTCH1 |
| SHEDDEN_LUNG_CANCER_GOOD_SURVIVAL_A5 | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/SHEDDEN_LUNG_CANCER_GOOD_SURVIVAL_A5 |
| SHETH_LIVER_CANCER_VS_TXNIP_LOSS_PAM3 | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/SHETH_LIVER_CANCER_VS_TXNIP_LOSS_PAM3 |
| VERNELL_RETINOBLASTOMA_PATHWAY_UP | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/VERNELL_RETINOBLASTOMA_PATHWAY_UP |
| ZHONG_RESPONSE_TO_AZACITIDINE_AND_TSA_DN | 70 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHONG_RESPONSE_TO_AZACITIDINE_AND_TSA_DN |
| AMIT_EGF_RESPONSE_120_HELA | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_EGF_RESPONSE_120_HELA |
| BOSCO_EPITHELIAL_DIFFERENTIATION_MODULE | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/BOSCO_EPITHELIAL_DIFFERENTIATION_MODULE |
| BOYLAN_MULTIPLE_MYELOMA_PCA3_DN | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYLAN_MULTIPLE_MYELOMA_PCA3_DN |
| GOTZMANN_EPITHELIAL_TO_MESENCHYMAL_TRANSITION_UP | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/GOTZMANN_EPITHELIAL_TO_MESENCHYMAL_TRANSITION_UP |
| HOSHIDA_LIVER_CANCER_LATE_RECURRENCE_DN | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/HOSHIDA_LIVER_CANCER_LATE_RECURRENCE_DN |
| HOWLIN_PUBERTAL_MAMMARY_GLAND | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/HOWLIN_PUBERTAL_MAMMARY_GLAND |
| HUANG_DASATINIB_RESISTANCE_DN | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/HUANG_DASATINIB_RESISTANCE_DN |
| KEGG_COMPLEMENT_AND_COAGULATION_CASCADES | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_COMPLEMENT_AND_COAGULATION_CASCADES |
| KEGG_P53_SIGNALING_PATHWAY | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_P53_SIGNALING_PATHWAY |
| KEGG_PPAR_SIGNALING_PATHWAY | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PPAR_SIGNALING_PATHWAY |
| LEIN_NEURON_MARKERS | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/LEIN_NEURON_MARKERS |
| LINDSTEDT_DENDRITIC_CELL_MATURATION_C | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/LINDSTEDT_DENDRITIC_CELL_MATURATION_C |
| OUELLET_CULTURED_OVARIAN_CANCER_INVASIVE_VS_LMP_UP | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/OUELLET_CULTURED_OVARIAN_CANCER_INVASIVE_VS_LMP_UP |
| PID_MTOR_4PATHWAY | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_MTOR_4PATHWAY |
| PID_P75NTRPATHWAY | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_P75NTRPATHWAY |
| PID_VEGFR1_2_PATHWAY | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_VEGFR1_2_PATHWAY |
| REACTOME_GLUCOSE_METABOLISM | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GLUCOSE_METABOLISM |
| REACTOME_SPHINGOLIPID_METABOLISM | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SPHINGOLIPID_METABOLISM |
| RHODES_UNDIFFERENTIATED_CANCER | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/RHODES_UNDIFFERENTIATED_CANCER |
| RIZKI_TUMOR_INVASIVENESS_2D_UP | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/RIZKI_TUMOR_INVASIVENESS_2D_UP |
| TURASHVILI_BREAST_DUCTAL_CARCINOMA_VS_LOBULAR_NORMAL_DN | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/TURASHVILI_BREAST_DUCTAL_CARCINOMA_VS_LOBULAR_NORMAL_DN |
| TURASHVILI_BREAST_LOBULAR_CARCINOMA_VS_DUCTAL_NORMAL_UP | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/TURASHVILI_BREAST_LOBULAR_CARCINOMA_VS_DUCTAL_NORMAL_UP |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_10 | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_10 |
| ZAMORA_NOS2_TARGETS_UP | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/ZAMORA_NOS2_TARGETS_UP |
| ZHANG_RESPONSE_TO_CANTHARIDIN_DN | 69 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_RESPONSE_TO_CANTHARIDIN_DN |
| AKL_HTLV1_INFECTION_DN | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/AKL_HTLV1_INFECTION_DN |
| BASSO_CD40_SIGNALING_DN | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/BASSO_CD40_SIGNALING_DN |
| BILANGES_SERUM_AND_RAPAMYCIN_SENSITIVE_GENES | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/BILANGES_SERUM_AND_RAPAMYCIN_SENSITIVE_GENES |
| BROWNE_INTERFERON_RESPONSIVE_GENES | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_INTERFERON_RESPONSIVE_GENES |
| CASTELLANO_NRAS_TARGETS_UP | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/CASTELLANO_NRAS_TARGETS_UP |
| DAVICIONI_TARGETS_OF_PAX_FOXO1_FUSIONS_DN | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/DAVICIONI_TARGETS_OF_PAX_FOXO1_FUSIONS_DN |
| DELPUECH_FOXO3_TARGETS_UP | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/DELPUECH_FOXO3_TARGETS_UP |
| FONTAINE_FOLLICULAR_THYROID_ADENOMA_DN | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/FONTAINE_FOLLICULAR_THYROID_ADENOMA_DN |
| KEGG_EPITHELIAL_CELL_SIGNALING_IN_HELICOBACTER_PYLORI_INFECTION | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_EPITHELIAL_CELL_SIGNALING_IN_HELICOBACTER_PYLORI_INFECTION |
| LINDSTEDT_DENDRITIC_CELL_MATURATION_D | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/LINDSTEDT_DENDRITIC_CELL_MATURATION_D |
| LIN_NPAS4_TARGETS_DN | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/LIN_NPAS4_TARGETS_DN |
| NOUZOVA_METHYLATED_IN_APL | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/NOUZOVA_METHYLATED_IN_APL |
| PID_TELOMERASEPATHWAY | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_TELOMERASEPATHWAY |
| REACTOME_SEMAPHORIN_INTERACTIONS | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SEMAPHORIN_INTERACTIONS |
| ROSS_AML_OF_FAB_M7_TYPE | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/ROSS_AML_OF_FAB_M7_TYPE |
| SESTO_RESPONSE_TO_UV_C7 | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/SESTO_RESPONSE_TO_UV_C7 |
| TURASHVILI_BREAST_NORMAL_DUCTAL_VS_LOBULAR_UP | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/TURASHVILI_BREAST_NORMAL_DUCTAL_VS_LOBULAR_UP |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_1 | 68 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_1 |
| CHIARETTI_ACUTE_LYMPHOBLASTIC_LEUKEMIA_ZAP70 | 67 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIARETTI_ACUTE_LYMPHOBLASTIC_LEUKEMIA_ZAP70 |
| DAZARD_UV_RESPONSE_CLUSTER_G1 | 67 | http://www.broadinstitute.org/gsea/msigdb/cards/DAZARD_UV_RESPONSE_CLUSTER_G1 |
| ELVIDGE_HIF1A_TARGETS_UP | 67 | http://www.broadinstitute.org/gsea/msigdb/cards/ELVIDGE_HIF1A_TARGETS_UP |
| GAJATE_RESPONSE_TO_TRABECTEDIN_UP | 67 | http://www.broadinstitute.org/gsea/msigdb/cards/GAJATE_RESPONSE_TO_TRABECTEDIN_UP |
| GENTILE_UV_LOW_DOSE_DN | 67 | http://www.broadinstitute.org/gsea/msigdb/cards/GENTILE_UV_LOW_DOSE_DN |
| KEGG_ADIPOCYTOKINE_SIGNALING_PATHWAY | 67 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ADIPOCYTOKINE_SIGNALING_PATHWAY |
| LINDSTEDT_DENDRITIC_CELL_MATURATION_A | 67 | http://www.broadinstitute.org/gsea/msigdb/cards/LINDSTEDT_DENDRITIC_CELL_MATURATION_A |
| MCMURRAY_TP53_HRAS_COOPERATION_RESPONSE_DN | 67 | http://www.broadinstitute.org/gsea/msigdb/cards/MCMURRAY_TP53_HRAS_COOPERATION_RESPONSE_DN |
| MOLENAAR_TARGETS_OF_CCND1_AND_CDK4_UP | 67 | http://www.broadinstitute.org/gsea/msigdb/cards/MOLENAAR_TARGETS_OF_CCND1_AND_CDK4_UP |
| REACTOME_ORC1_REMOVAL_FROM_CHROMATIN | 67 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ORC1_REMOVAL_FROM_CHROMATIN |
| SETLUR_PROSTATE_CANCER_TMPRSS2_ERG_FUSION_UP | 67 | http://www.broadinstitute.org/gsea/msigdb/cards/SETLUR_PROSTATE_CANCER_TMPRSS2_ERG_FUSION_UP |
| SHAFFER_IRF4_TARGETS_IN_PLASMA_CELL_VS_MATURE_B_LYMPHOCYTE | 67 | http://www.broadinstitute.org/gsea/msigdb/cards/SHAFFER_IRF4_TARGETS_IN_PLASMA_CELL_VS_MATURE_B_LYMPHOCYTE |
| SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | 67 | http://www.broadinstitute.org/gsea/msigdb/cards/SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES |
| VANHARANTA_UTERINE_FIBROID_DN | 67 | http://www.broadinstitute.org/gsea/msigdb/cards/VANHARANTA_UTERINE_FIBROID_DN |
| VANHARANTA_UTERINE_FIBROID_WITH_7Q_DELETION_UP | 67 | http://www.broadinstitute.org/gsea/msigdb/cards/VANHARANTA_UTERINE_FIBROID_WITH_7Q_DELETION_UP |
| WAMUNYOKOLI_OVARIAN_CANCER_GRADES_1_2_DN | 67 | http://www.broadinstitute.org/gsea/msigdb/cards/WAMUNYOKOLI_OVARIAN_CANCER_GRADES_1_2_DN |
| BEIER_GLIOMA_STEM_CELL_DN | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/BEIER_GLIOMA_STEM_CELL_DN |
| BRUNO_HEMATOPOIESIS | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/BRUNO_HEMATOPOIESIS |
| DIRMEIER_LMP1_RESPONSE_EARLY | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/DIRMEIER_LMP1_RESPONSE_EARLY |
| FONTAINE_PAPILLARY_THYROID_CARCINOMA_UP | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/FONTAINE_PAPILLARY_THYROID_CARCINOMA_UP |
| GAZDA_DIAMOND_BLACKFAN_ANEMIA_PROGENITOR_DN | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/GAZDA_DIAMOND_BLACKFAN_ANEMIA_PROGENITOR_DN |
| GRAHAM_NORMAL_QUIESCENT_VS_NORMAL_DIVIDING_UP | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAHAM_NORMAL_QUIESCENT_VS_NORMAL_DIVIDING_UP |
| JAZAG_TGFB1_SIGNALING_VIA_SMAD4_DN | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/JAZAG_TGFB1_SIGNALING_VIA_SMAD4_DN |
| JECHLINGER_EPITHELIAL_TO_MESENCHYMAL_TRANSITION_DN | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/JECHLINGER_EPITHELIAL_TO_MESENCHYMAL_TRANSITION_DN |
| KIM_GERMINAL_CENTER_T_HELPER_UP | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_GERMINAL_CENTER_T_HELPER_UP |
| LEE_LIVER_CANCER_CIPROFIBRATE_DN | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_CIPROFIBRATE_DN |
| MARSON_FOXP3_TARGETS_UP | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/MARSON_FOXP3_TARGETS_UP |
| NAKAMURA_ADIPOGENESIS_EARLY_UP | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAMURA_ADIPOGENESIS_EARLY_UP |
| PID_HDAC_CLASSI_PATHWAY | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_HDAC_CLASSI_PATHWAY |
| PID_HIF1_TFPATHWAY | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_HIF1_TFPATHWAY |
| PID_INTEGRIN1_PATHWAY | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_INTEGRIN1_PATHWAY |
| PID_LYSOPHOSPHOLIPID_PATHWAY | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_LYSOPHOSPHOLIPID_PATHWAY |
| PID_TCR_PATHWAY | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_TCR_PATHWAY |
| RAMASWAMY_METASTASIS_UP | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/RAMASWAMY_METASTASIS_UP |
| REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES |
| REACTOME_RECRUITMENT_OF_MITOTIC_CENTROSOME_PROTEINS_AND_COMPLEXES | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RECRUITMENT_OF_MITOTIC_CENTROSOME_PROTEINS_AND_COMPLEXES |
| RIGGINS_TAMOXIFEN_RESISTANCE_UP | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/RIGGINS_TAMOXIFEN_RESISTANCE_UP |
| SWEET_KRAS_TARGETS_DN | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/SWEET_KRAS_TARGETS_DN |
| TENEDINI_MEGAKARYOCYTE_MARKERS | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/TENEDINI_MEGAKARYOCYTE_MARKERS |
| WANG_PROSTATE_CANCER_ANDROGEN_INDEPENDENT | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_PROSTATE_CANCER_ANDROGEN_INDEPENDENT |
| WILCOX_PRESPONSE_TO_ROGESTERONE_DN | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/WILCOX_PRESPONSE_TO_ROGESTERONE_DN |
| ZHAN_MULTIPLE_MYELOMA_CD1_VS_CD2_UP | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_CD1_VS_CD2_UP |
| ZHONG_SECRETOME_OF_LUNG_CANCER_AND_ENDOTHELIUM | 66 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHONG_SECRETOME_OF_LUNG_CANCER_AND_ENDOTHELIUM |
| AMIT_SERUM_RESPONSE_120_MCF10A | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_SERUM_RESPONSE_120_MCF10A |
| BOYAULT_LIVER_CANCER_SUBCLASS_G6_UP | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYAULT_LIVER_CANCER_SUBCLASS_G6_UP |
| BYSTRYKH_HEMATOPOIESIS_STEM_CELL_AND_BRAIN_QTL_CIS | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/BYSTRYKH_HEMATOPOIESIS_STEM_CELL_AND_BRAIN_QTL_CIS |
| CHANG_IMMORTALIZED_BY_HPV31_DN | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/CHANG_IMMORTALIZED_BY_HPV31_DN |
| GOLDRATH_IMMUNE_MEMORY | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/GOLDRATH_IMMUNE_MEMORY |
| GUO_HEX_TARGETS_DN | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/GUO_HEX_TARGETS_DN |
| HESS_TARGETS_OF_HOXA9_AND_MEIS1_UP | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/HESS_TARGETS_OF_HOXA9_AND_MEIS1_UP |
| KEGG_GLIOMA | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GLIOMA |
| LEE_LIVER_CANCER_ACOX1_DN | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_ACOX1_DN |
| LEE_LIVER_CANCER_MYC_TGFA_DN | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_MYC_TGFA_DN |
| PID_BCR_5PATHWAY | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_BCR_5PATHWAY |
| PID_CD8TCRDOWNSTREAMPATHWAY | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_CD8TCRDOWNSTREAMPATHWAY |
| PID_ERA_GENOMIC_PATHWAY | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ERA_GENOMIC_PATHWAY |
| PID_IL4_2PATHWAY | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_IL4_2PATHWAY |
| PID_RB_1PATHWAY | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_RB_1PATHWAY |
| REACTOME_ASSEMBLY_OF_THE_PRE_REPLICATIVE_COMPLEX | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ASSEMBLY_OF_THE_PRE_REPLICATIVE_COMPLEX |
| REACTOME_CYCLIN_E_ASSOCIATED_EVENTS_DURING_G1_S_TRANSITION_ | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CYCLIN_E_ASSOCIATED_EVENTS_DURING_G1_S_TRANSITION_ |
| REACTOME_SIGNALING_BY_WNT | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_WNT |
| SHAFFER_IRF4_TARGETS_IN_ACTIVATED_DENDRITIC_CELL | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/SHAFFER_IRF4_TARGETS_IN_ACTIVATED_DENDRITIC_CELL |
| ST_FAS_SIGNALING_PATHWAY | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_FAS_SIGNALING_PATHWAY |
| YOSHIOKA_LIVER_CANCER_EARLY_RECURRENCE_DN | 65 | http://www.broadinstitute.org/gsea/msigdb/cards/YOSHIOKA_LIVER_CANCER_EARLY_RECURRENCE_DN |
| ANASTASSIOU_CANCER_MESENCHYMAL_TRANSITION_SIGNATURE | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/ANASTASSIOU_CANCER_MESENCHYMAL_TRANSITION_SIGNATURE |
| BYSTROEM_CORRELATED_WITH_IL5_DN | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/BYSTROEM_CORRELATED_WITH_IL5_DN |
| DACOSTA_UV_RESPONSE_VIA_ERCC3_TTD_UP | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/DACOSTA_UV_RESPONSE_VIA_ERCC3_TTD_UP |
| DAVICIONI_RHABDOMYOSARCOMA_PAX_FOXO1_FUSION_UP | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/DAVICIONI_RHABDOMYOSARCOMA_PAX_FOXO1_FUSION_UP |
| GERHOLD_ADIPOGENESIS_DN | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/GERHOLD_ADIPOGENESIS_DN |
| HAHTOLA_MYCOSIS_FUNGOIDES_CD4_UP | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/HAHTOLA_MYCOSIS_FUNGOIDES_CD4_UP |
| HERNANDEZ_MITOTIC_ARREST_BY_DOCETAXEL_2_UP | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/HERNANDEZ_MITOTIC_ARREST_BY_DOCETAXEL_2_UP |
| INGRAM_SHH_TARGETS_DN | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/INGRAM_SHH_TARGETS_DN |
| JOSEPH_RESPONSE_TO_SODIUM_BUTYRATE_DN | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/JOSEPH_RESPONSE_TO_SODIUM_BUTYRATE_DN |
| KEGG_RETINOL_METABOLISM | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_RETINOL_METABOLISM |
| KOKKINAKIS_METHIONINE_DEPRIVATION_48HR_DN | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/KOKKINAKIS_METHIONINE_DEPRIVATION_48HR_DN |
| LEE_LIVER_CANCER_ACOX1_UP | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_ACOX1_UP |
| LEE_LIVER_CANCER_E2F1_DN | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_E2F1_DN |
| LEE_LIVER_CANCER_MYC_E2F1_DN | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_MYC_E2F1_DN |
| LIU_SMARCA4_TARGETS | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_SMARCA4_TARGETS |
| PELLICCIOTTA_HDAC_IN_ANTIGEN_PRESENTATION_UP | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/PELLICCIOTTA_HDAC_IN_ANTIGEN_PRESENTATION_UP |
| PETROVA_PROX1_TARGETS_DN | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/PETROVA_PROX1_TARGETS_DN |
| REACTOME_ACTIVATION_OF_NF_KAPPAB_IN_B_CELLS | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATION_OF_NF_KAPPAB_IN_B_CELLS |
| REACTOME_AUTODEGRADATION_OF_CDH1_BY_CDH1_APC_C | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_AUTODEGRADATION_OF_CDH1_BY_CDH1_APC_C |
| REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE |
| REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INTERFERON_ALPHA_BETA_SIGNALING |
| REACTOME_NCAM_SIGNALING_FOR_NEURITE_OUT_GROWTH | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NCAM_SIGNALING_FOR_NEURITE_OUT_GROWTH |
| RHODES_CANCER_META_SIGNATURE | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/RHODES_CANCER_META_SIGNATURE |
| RIZKI_TUMOR_INVASIVENESS_2D_DN | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/RIZKI_TUMOR_INVASIVENESS_2D_DN |
| ZHANG_GATA6_TARGETS_DN | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_GATA6_TARGETS_DN |
| ZHAN_MULTIPLE_MYELOMA_UP | 64 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_UP |
| BILBAN_B_CLL_LPL_UP | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/BILBAN_B_CLL_LPL_UP |
| BRIDEAU_IMPRINTED_GENES | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/BRIDEAU_IMPRINTED_GENES |
| BURTON_ADIPOGENESIS_PEAK_AT_0HR | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/BURTON_ADIPOGENESIS_PEAK_AT_0HR |
| CROMER_TUMORIGENESIS_UP | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/CROMER_TUMORIGENESIS_UP |
| FLECHNER_PBL_KIDNEY_TRANSPLANT_REJECTED_VS_OK_UP | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/FLECHNER_PBL_KIDNEY_TRANSPLANT_REJECTED_VS_OK_UP |
| GOTTWEIN_TARGETS_OF_KSHV_MIR_K12_11 | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/GOTTWEIN_TARGETS_OF_KSHV_MIR_K12_11 |
| HADDAD_T_LYMPHOCYTE_AND_NK_PROGENITOR_DN | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/HADDAD_T_LYMPHOCYTE_AND_NK_PROGENITOR_DN |
| JOHNSTONE_PARVB_TARGETS_1_DN | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/JOHNSTONE_PARVB_TARGETS_1_DN |
| KYNG_DNA_DAMAGE_BY_4NQO_OR_UV | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_DNA_DAMAGE_BY_4NQO_OR_UV |
| LEE_LIVER_CANCER_MYC_DN | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_MYC_DN |
| MIKKELSEN_ES_HCP_WITH_H3_UNMETHYLATED | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_ES_HCP_WITH_H3_UNMETHYLATED |
| MODY_HIPPOCAMPUS_POSTNATAL | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/MODY_HIPPOCAMPUS_POSTNATAL |
| PHONG_TNF_TARGETS_UP | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/PHONG_TNF_TARGETS_UP |
| PID_ENDOTHELINPATHWAY | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ENDOTHELINPATHWAY |
| PID_IL12_2PATHWAY | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_IL12_2PATHWAY |
| PID_MYC_REPRESSPATHWAY | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_MYC_REPRESSPATHWAY |
| REACTOME_COSTIMULATION_BY_THE_CD28_FAMILY | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_COSTIMULATION_BY_THE_CD28_FAMILY |
| REACTOME_INTERFERON_GAMMA_SIGNALING | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INTERFERON_GAMMA_SIGNALING |
| REACTOME_SIGNALING_BY_TGF_BETA_RECEPTOR_COMPLEX | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_TGF_BETA_RECEPTOR_COMPLEX |
| ROYLANCE_BREAST_CANCER_16Q_COPY_NUMBER_UP | 63 | http://www.broadinstitute.org/gsea/msigdb/cards/ROYLANCE_BREAST_CANCER_16Q_COPY_NUMBER_UP |
| BILD_SRC_ONCOGENIC_SIGNATURE | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/BILD_SRC_ONCOGENIC_SIGNATURE |
| BREDEMEYER_RAG_SIGNALING_NOT_VIA_ATM_UP | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/BREDEMEYER_RAG_SIGNALING_NOT_VIA_ATM_UP |
| CORRE_MULTIPLE_MYELOMA_DN | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/CORRE_MULTIPLE_MYELOMA_DN |
| GEORGES_CELL_CYCLE_MIR192_TARGETS | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/GEORGES_CELL_CYCLE_MIR192_TARGETS |
| HOSHIDA_LIVER_CANCER_LATE_RECURRENCE_UP | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/HOSHIDA_LIVER_CANCER_LATE_RECURRENCE_UP |
| ISSAEVA_MLL2_TARGETS | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/ISSAEVA_MLL2_TARGETS |
| KEGG_COLORECTAL_CANCER | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_COLORECTAL_CANCER |
| KEGG_GLYCOLYSIS_GLUCONEOGENESIS | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GLYCOLYSIS_GLUCONEOGENESIS |
| KEGG_NOD_LIKE_RECEPTOR_SIGNALING_PATHWAY | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_NOD_LIKE_RECEPTOR_SIGNALING_PATHWAY |
| KORKOLA_YOLK_SAC_TUMOR | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/KORKOLA_YOLK_SAC_TUMOR |
| KYNG_DNA_DAMAGE_BY_UV | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_DNA_DAMAGE_BY_UV |
| LEE_LIVER_CANCER_E2F1_UP | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_E2F1_UP |
| MCBRYAN_PUBERTAL_TGFB1_TARGETS_DN | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/MCBRYAN_PUBERTAL_TGFB1_TARGETS_DN |
| PID_FCER1PATHWAY | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_FCER1PATHWAY |
| PID_TRKRPATHWAY | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_TRKRPATHWAY |
| POMEROY_MEDULLOBLASTOMA_DESMOPLASIC_VS_CLASSIC_UP | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/POMEROY_MEDULLOBLASTOMA_DESMOPLASIC_VS_CLASSIC_UP |
| REACTOME_RNA_POL_I_PROMOTER_OPENING | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RNA_POL_I_PROMOTER_OPENING |
| SAMOLS_TARGETS_OF_KHSV_MIRNAS_DN | 62 | http://www.broadinstitute.org/gsea/msigdb/cards/SAMOLS_TARGETS_OF_KHSV_MIRNAS_DN |
| BAKKER_FOXO3_TARGETS_UP | 61 | http://www.broadinstitute.org/gsea/msigdb/cards/BAKKER_FOXO3_TARGETS_UP |
| BROWNE_HCMV_INFECTION_1HR_UP | 61 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_1HR_UP |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_BLUE_DN | 61 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_BLUE_DN |
| GENTILE_RESPONSE_CLUSTER_D3 | 61 | http://www.broadinstitute.org/gsea/msigdb/cards/GENTILE_RESPONSE_CLUSTER_D3 |
| KAUFFMANN_MELANOMA_RELAPSE_UP | 61 | http://www.broadinstitute.org/gsea/msigdb/cards/KAUFFMANN_MELANOMA_RELAPSE_UP |
| LEE_LIVER_CANCER_MYC_TGFA_UP | 61 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_MYC_TGFA_UP |
| MASSARWEH_RESPONSE_TO_ESTRADIOL | 61 | http://www.broadinstitute.org/gsea/msigdb/cards/MASSARWEH_RESPONSE_TO_ESTRADIOL |
| NADLER_OBESITY_UP | 61 | http://www.broadinstitute.org/gsea/msigdb/cards/NADLER_OBESITY_UP |
| PID_AR_PATHWAY | 61 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_AR_PATHWAY |
| RAMASWAMY_METASTASIS_DN | 61 | http://www.broadinstitute.org/gsea/msigdb/cards/RAMASWAMY_METASTASIS_DN |
| RASHI_RESPONSE_TO_IONIZING_RADIATION_4 | 61 | http://www.broadinstitute.org/gsea/msigdb/cards/RASHI_RESPONSE_TO_IONIZING_RADIATION_4 |
| REACTOME_ER_PHAGOSOME_PATHWAY | 61 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ER_PHAGOSOME_PATHWAY |
| REACTOME_RNA_POL_II_PRE_TRANSCRIPTION_EVENTS | 61 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RNA_POL_II_PRE_TRANSCRIPTION_EVENTS |
| REN_BOUND_BY_E2F | 61 | http://www.broadinstitute.org/gsea/msigdb/cards/REN_BOUND_BY_E2F |
| STEARMAN_LUNG_CANCER_EARLY_VS_LATE_DN | 61 | http://www.broadinstitute.org/gsea/msigdb/cards/STEARMAN_LUNG_CANCER_EARLY_VS_LATE_DN |
| ZWANG_EGF_PERSISTENTLY_DN | 61 | http://www.broadinstitute.org/gsea/msigdb/cards/ZWANG_EGF_PERSISTENTLY_DN |
| AMIT_EGF_RESPONSE_240_HELA | 60 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_EGF_RESPONSE_240_HELA |
| BACOLOD_RESISTANCE_TO_ALKYLATING_AGENTS_DN | 60 | http://www.broadinstitute.org/gsea/msigdb/cards/BACOLOD_RESISTANCE_TO_ALKYLATING_AGENTS_DN |
| BEGUM_TARGETS_OF_PAX3_FOXO1_FUSION_UP | 60 | http://www.broadinstitute.org/gsea/msigdb/cards/BEGUM_TARGETS_OF_PAX3_FOXO1_FUSION_UP |
| CROONQUIST_STROMAL_STIMULATION_UP | 60 | http://www.broadinstitute.org/gsea/msigdb/cards/CROONQUIST_STROMAL_STIMULATION_UP |
| CUI_GLUCOSE_DEPRIVATION | 60 | http://www.broadinstitute.org/gsea/msigdb/cards/CUI_GLUCOSE_DEPRIVATION |
| HUTTMANN_B_CLL_POOR_SURVIVAL_DN | 60 | http://www.broadinstitute.org/gsea/msigdb/cards/HUTTMANN_B_CLL_POOR_SURVIVAL_DN |
| KEGG_ACUTE_MYELOID_LEUKEMIA | 60 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ACUTE_MYELOID_LEUKEMIA |
| LEE_LIVER_CANCER_CIPROFIBRATE_UP | 60 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_CIPROFIBRATE_UP |
| LEE_LIVER_CANCER_DENA_UP | 60 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_DENA_UP |
| OUILLETTE_CLL_13Q14_DELETION_DN | 60 | http://www.broadinstitute.org/gsea/msigdb/cards/OUILLETTE_CLL_13Q14_DELETION_DN |
| REACTOME_CELL_DEATH_SIGNALLING_VIA_NRAGE_NRIF_AND_NADE | 60 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CELL_DEATH_SIGNALLING_VIA_NRAGE_NRIF_AND_NADE |
| REACTOME_TRANS_GOLGI_NETWORK_VESICLE_BUDDING | 60 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANS_GOLGI_NETWORK_VESICLE_BUDDING |
| SONG_TARGETS_OF_IE86_CMV_PROTEIN | 60 | http://www.broadinstitute.org/gsea/msigdb/cards/SONG_TARGETS_OF_IE86_CMV_PROTEIN |
| WEIGEL_OXIDATIVE_STRESS_BY_HNE_AND_TBH | 60 | http://www.broadinstitute.org/gsea/msigdb/cards/WEIGEL_OXIDATIVE_STRESS_BY_HNE_AND_TBH |
| WIERENGA_STAT5A_TARGETS_GROUP2 | 60 | http://www.broadinstitute.org/gsea/msigdb/cards/WIERENGA_STAT5A_TARGETS_GROUP2 |
| ASTIER_INTEGRIN_SIGNALING | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/ASTIER_INTEGRIN_SIGNALING |
| BARIS_THYROID_CANCER_DN | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/BARIS_THYROID_CANCER_DN |
| BOYLAN_MULTIPLE_MYELOMA_C_DN | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYLAN_MULTIPLE_MYELOMA_C_DN |
| CAFFAREL_RESPONSE_TO_THC_24HR_5_DN | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/CAFFAREL_RESPONSE_TO_THC_24HR_5_DN |
| DASU_IL6_SIGNALING_UP | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/DASU_IL6_SIGNALING_UP |
| DAVICIONI_PAX_FOXO1_SIGNATURE_IN_ARMS_UP | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/DAVICIONI_PAX_FOXO1_SIGNATURE_IN_ARMS_UP |
| DAWSON_METHYLATED_IN_LYMPHOMA_TCL1 | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/DAWSON_METHYLATED_IN_LYMPHOMA_TCL1 |
| ELVIDGE_HYPOXIA_BY_DMOG_DN | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/ELVIDGE_HYPOXIA_BY_DMOG_DN |
| HUPER_BREAST_BASAL_VS_LUMINAL_DN | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/HUPER_BREAST_BASAL_VS_LUMINAL_DN |
| KEGG_PATHOGENIC_ESCHERICHIA_COLI_INFECTION | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PATHOGENIC_ESCHERICHIA_COLI_INFECTION |
| KEGG_RNA_DEGRADATION | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_RNA_DEGRADATION |
| LIAN_LIPA_TARGETS_3M | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/LIAN_LIPA_TARGETS_3M |
| MEISSNER_BRAIN_HCP_WITH_H3K4ME2_AND_H3K27ME3 | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/MEISSNER_BRAIN_HCP_WITH_H3K4ME2_AND_H3K27ME3 |
| ONDER_CDH1_TARGETS_3_DN | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/ONDER_CDH1_TARGETS_3_DN |
| PID_ATF2_PATHWAY | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ATF2_PATHWAY |
| PID_FAK_PATHWAY | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_FAK_PATHWAY |
| PID_NOTCH_PATHWAY | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_NOTCH_PATHWAY |
| PID_P53REGULATIONPATHWAY | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_P53REGULATIONPATHWAY |
| POMEROY_MEDULLOBLASTOMA_DESMOPLASIC_VS_CLASSIC_DN | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/POMEROY_MEDULLOBLASTOMA_DESMOPLASIC_VS_CLASSIC_DN |
| REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES |
| REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS |
| WESTON_VEGFA_TARGETS_6HR | 59 | http://www.broadinstitute.org/gsea/msigdb/cards/WESTON_VEGFA_TARGETS_6HR |
| BERNARD_PPAPDC1B_TARGETS_DN | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/BERNARD_PPAPDC1B_TARGETS_DN |
| BIOCARTA_HIVNEF_PATHWAY | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_HIVNEF_PATHWAY |
| BIOCARTA_PPARA_PATHWAY | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_PPARA_PATHWAY |
| CHIARADONNA_NEOPLASTIC_TRANSFORMATION_KRAS_CDC25_UP | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIARADONNA_NEOPLASTIC_TRANSFORMATION_KRAS_CDC25_UP |
| JI_RESPONSE_TO_FSH_DN | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/JI_RESPONSE_TO_FSH_DN |
| JOHANSSON_GLIOMAGENESIS_BY_PDGFB_UP | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/JOHANSSON_GLIOMAGENESIS_BY_PDGFB_UP |
| KANNAN_TP53_TARGETS_UP | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/KANNAN_TP53_TARGETS_UP |
| KEGG_ARACHIDONIC_ACID_METABOLISM | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ARACHIDONIC_ACID_METABOLISM |
| KLEIN_PRIMARY_EFFUSION_LYMPHOMA_DN | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/KLEIN_PRIMARY_EFFUSION_LYMPHOMA_DN |
| KOHOUTEK_CCNT2_TARGETS | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/KOHOUTEK_CCNT2_TARGETS |
| LIM_MAMMARY_LUMINAL_PROGENITOR_UP | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/LIM_MAMMARY_LUMINAL_PROGENITOR_UP |
| LUCAS_HNF4A_TARGETS_UP | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/LUCAS_HNF4A_TARGETS_UP |
| MIKKELSEN_NPC_LCP_WITH_H3K4ME3 | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_NPC_LCP_WITH_H3K4ME3 |
| MOLENAAR_TARGETS_OF_CCND1_AND_CDK4_DN | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/MOLENAAR_TARGETS_OF_CCND1_AND_CDK4_DN |
| MORI_LARGE_PRE_BII_LYMPHOCYTE_DN | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/MORI_LARGE_PRE_BII_LYMPHOCYTE_DN |
| NADLER_HYPERGLYCEMIA_AT_OBESITY | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/NADLER_HYPERGLYCEMIA_AT_OBESITY |
| NAGASHIMA_EGF_SIGNALING_UP | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/NAGASHIMA_EGF_SIGNALING_UP |
| NAGASHIMA_NRG1_SIGNALING_DN | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/NAGASHIMA_NRG1_SIGNALING_DN |
| PID_SHP2_PATHWAY | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_SHP2_PATHWAY |
| REACTOME_COLLAGEN_FORMATION | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_COLLAGEN_FORMATION |
| REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT |
| REACTOME_REGULATION_OF_APOPTOSIS | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_APOPTOSIS |
| SANSOM_WNT_PATHWAY_REQUIRE_MYC | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/SANSOM_WNT_PATHWAY_REQUIRE_MYC |
| SCHEIDEREIT_IKK_INTERACTING_PROTEINS | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHEIDEREIT_IKK_INTERACTING_PROTEINS |
| VERRECCHIA_EARLY_RESPONSE_TO_TGFB1 | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/VERRECCHIA_EARLY_RESPONSE_TO_TGFB1 |
| WAKASUGI_HAVE_ZNF143_BINDING_SITES | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/WAKASUGI_HAVE_ZNF143_BINDING_SITES |
| WUNDER_INFLAMMATORY_RESPONSE_AND_CHOLESTEROL_UP | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/WUNDER_INFLAMMATORY_RESPONSE_AND_CHOLESTEROL_UP |
| ZHENG_GLIOBLASTOMA_PLASTICITY_DN | 58 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHENG_GLIOBLASTOMA_PLASTICITY_DN |
| AMIT_SERUM_RESPONSE_240_MCF10A | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_SERUM_RESPONSE_240_MCF10A |
| AMIT_SERUM_RESPONSE_60_MCF10A | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_SERUM_RESPONSE_60_MCF10A |
| BORLAK_LIVER_CANCER_EGF_UP | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/BORLAK_LIVER_CANCER_EGF_UP |
| BREDEMEYER_RAG_SIGNALING_NOT_VIA_ATM_DN | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/BREDEMEYER_RAG_SIGNALING_NOT_VIA_ATM_DN |
| BURTON_ADIPOGENESIS_11 | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/BURTON_ADIPOGENESIS_11 |
| DELASERNA_MYOD_TARGETS_DN | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/DELASERNA_MYOD_TARGETS_DN |
| DIRMEIER_LMP1_RESPONSE_LATE_UP | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/DIRMEIER_LMP1_RESPONSE_LATE_UP |
| GARCIA_TARGETS_OF_FLI1_AND_DAX1_UP | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/GARCIA_TARGETS_OF_FLI1_AND_DAX1_UP |
| GRABARCZYK_BCL11B_TARGETS_DN | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/GRABARCZYK_BCL11B_TARGETS_DN |
| GRAHAM_CML_QUIESCENT_VS_NORMAL_DIVIDING_UP | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAHAM_CML_QUIESCENT_VS_NORMAL_DIVIDING_UP |
| GYORFFY_MITOXANTRONE_RESISTANCE | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/GYORFFY_MITOXANTRONE_RESISTANCE |
| LEE_EARLY_T_LYMPHOCYTE_DN | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_EARLY_T_LYMPHOCYTE_DN |
| MCCLUNG_CREB1_TARGETS_DN | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/MCCLUNG_CREB1_TARGETS_DN |
| NIKOLSKY_BREAST_CANCER_8P12_P11_AMPLICON | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_8P12_P11_AMPLICON |
| PEART_HDAC_PROLIFERATION_CLUSTER_UP | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/PEART_HDAC_PROLIFERATION_CLUSTER_UP |
| PID_TXA2PATHWAY | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_TXA2PATHWAY |
| REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES |
| REACTOME_P53_DEPENDENT_G1_DNA_DAMAGE_RESPONSE | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_P53_DEPENDENT_G1_DNA_DAMAGE_RESPONSE |
| SCHAEFFER_PROSTATE_DEVELOPMENT_12HR_DN | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_PROSTATE_DEVELOPMENT_12HR_DN |
| TANG_SENESCENCE_TP53_TARGETS_DN | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/TANG_SENESCENCE_TP53_TARGETS_DN |
| TONG_INTERACT_WITH_PTTG1 | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/TONG_INTERACT_WITH_PTTG1 |
| WIEDERSCHAIN_TARGETS_OF_BMI1_AND_PCGF2 | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/WIEDERSCHAIN_TARGETS_OF_BMI1_AND_PCGF2 |
| YAMASHITA_METHYLATED_IN_PROSTATE_CANCER | 57 | http://www.broadinstitute.org/gsea/msigdb/cards/YAMASHITA_METHYLATED_IN_PROSTATE_CANCER |
| BIOCARTA_NFAT_PATHWAY | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_NFAT_PATHWAY |
| BROWNE_HCMV_INFECTION_10HR_DN | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_10HR_DN |
| BROWNE_HCMV_INFECTION_30MIN_UP | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_30MIN_UP |
| GALE_APL_WITH_FLT3_MUTATED_UP | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/GALE_APL_WITH_FLT3_MUTATED_UP |
| GRADE_COLON_VS_RECTAL_CANCER_DN | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/GRADE_COLON_VS_RECTAL_CANCER_DN |
| GUTIERREZ_CHRONIC_LYMPHOCYTIC_LEUKEMIA_DN | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/GUTIERREZ_CHRONIC_LYMPHOCYTIC_LEUKEMIA_DN |
| GYORFFY_DOXORUBICIN_RESISTANCE | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/GYORFFY_DOXORUBICIN_RESISTANCE |
| HENDRICKS_SMARCA4_TARGETS_UP | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/HENDRICKS_SMARCA4_TARGETS_UP |
| HOQUE_METHYLATED_IN_CANCER | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/HOQUE_METHYLATED_IN_CANCER |
| KEGG_CYTOSOLIC_DNA_SENSING_PATHWAY | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_CYTOSOLIC_DNA_SENSING_PATHWAY |
| KEGG_HEDGEHOG_SIGNALING_PATHWAY | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_HEDGEHOG_SIGNALING_PATHWAY |
| KEGG_VIBRIO_CHOLERAE_INFECTION | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_VIBRIO_CHOLERAE_INFECTION |
| KYNG_ENVIRONMENTAL_STRESS_RESPONSE_UP | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_ENVIRONMENTAL_STRESS_RESPONSE_UP |
| LEE_LIVER_CANCER_MYC_E2F1_UP | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_MYC_E2F1_UP |
| LY_AGING_OLD_DN | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/LY_AGING_OLD_DN |
| MAYBURD_RESPONSE_TO_L663536_DN | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/MAYBURD_RESPONSE_TO_L663536_DN |
| PUJANA_BREAST_CANCER_WITH_BRCA1_MUTATED_UP | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/PUJANA_BREAST_CANCER_WITH_BRCA1_MUTATED_UP |
| REACTOME_CDT1_ASSOCIATION_WITH_THE_CDC6_ORC_ORIGIN_COMPLEX | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CDT1_ASSOCIATION_WITH_THE_CDC6_ORC_ORIGIN_COMPLEX |
| REACTOME_CELL_CELL_JUNCTION_ORGANIZATION | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CELL_CELL_JUNCTION_ORGANIZATION |
| REACTOME_PI_3K_CASCADE | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PI_3K_CASCADE |
| REACTOME_SCFSKP2_MEDIATED_DEGRADATION_OF_P27_P21 | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SCFSKP2_MEDIATED_DEGRADATION_OF_P27_P21 |
| VANTVEER_BREAST_CANCER_METASTASIS_UP | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/VANTVEER_BREAST_CANCER_METASTASIS_UP |
| WORSCHECH_TUMOR_REJECTION_UP | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/WORSCHECH_TUMOR_REJECTION_UP |
| ZHENG_IL22_SIGNALING_UP | 56 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHENG_IL22_SIGNALING_UP |
| BROWNE_HCMV_INFECTION_4HR_UP | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_4HR_UP |
| DEMAGALHAES_AGING_UP | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/DEMAGALHAES_AGING_UP |
| GENTILE_UV_RESPONSE_CLUSTER_D4 | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/GENTILE_UV_RESPONSE_CLUSTER_D4 |
| HU_GENOTOXIN_ACTION_DIRECT_VS_INDIRECT_24HR | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/HU_GENOTOXIN_ACTION_DIRECT_VS_INDIRECT_24HR |
| KEGG_BASAL_CELL_CARCINOMA | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_BASAL_CELL_CARCINOMA |
| KEGG_STEROID_HORMONE_BIOSYNTHESIS | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_STEROID_HORMONE_BIOSYNTHESIS |
| LANDIS_ERBB2_BREAST_PRENEOPLASTIC_DN | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/LANDIS_ERBB2_BREAST_PRENEOPLASTIC_DN |
| LAU_APOPTOSIS_CDKN2A_UP | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/LAU_APOPTOSIS_CDKN2A_UP |
| MIDORIKAWA_AMPLIFIED_IN_LIVER_CANCER | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/MIDORIKAWA_AMPLIFIED_IN_LIVER_CANCER |
| PID_FGF_PATHWAY | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_FGF_PATHWAY |
| PID_IL2_1PATHWAY | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_IL2_1PATHWAY |
| PID_TGFBRPATHWAY | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_TGFBRPATHWAY |
| REACTOME_ION_CHANNEL_TRANSPORT | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ION_CHANNEL_TRANSPORT |
| SMIRNOV_RESPONSE_TO_IR_2HR_DN | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/SMIRNOV_RESPONSE_TO_IR_2HR_DN |
| SU_LIVER | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/SU_LIVER |
| TIEN_INTESTINE_PROBIOTICS_6HR_UP | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/TIEN_INTESTINE_PROBIOTICS_6HR_UP |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_GRANULOCYTE_UP | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_GRANULOCYTE_UP |
| VANTVEER_BREAST_CANCER_POOR_PROGNOSIS | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/VANTVEER_BREAST_CANCER_POOR_PROGNOSIS |
| WATTEL_AUTONOMOUS_THYROID_ADENOMA_DN | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/WATTEL_AUTONOMOUS_THYROID_ADENOMA_DN |
| WU_APOPTOSIS_BY_CDKN1A_VIA_TP53 | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/WU_APOPTOSIS_BY_CDKN1A_VIA_TP53 |
| WU_SILENCED_BY_METHYLATION_IN_BLADDER_CANCER | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/WU_SILENCED_BY_METHYLATION_IN_BLADDER_CANCER |
| YU_MYC_TARGETS_DN | 55 | http://www.broadinstitute.org/gsea/msigdb/cards/YU_MYC_TARGETS_DN |
| AUNG_GASTRIC_CANCER | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/AUNG_GASTRIC_CANCER |
| FIGUEROA_AML_METHYLATION_CLUSTER_2_UP | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/FIGUEROA_AML_METHYLATION_CLUSTER_2_UP |
| HUPER_BREAST_BASAL_VS_LUMINAL_UP | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/HUPER_BREAST_BASAL_VS_LUMINAL_UP |
| JIANG_AGING_CEREBRAL_CORTEX_DN | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/JIANG_AGING_CEREBRAL_CORTEX_DN |
| KANG_DOXORUBICIN_RESISTANCE_UP | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/KANG_DOXORUBICIN_RESISTANCE_UP |
| KEGG_ARGININE_AND_PROLINE_METABOLISM | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ARGININE_AND_PROLINE_METABOLISM |
| KEGG_INOSITOL_PHOSPHATE_METABOLISM | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_INOSITOL_PHOSPHATE_METABOLISM |
| KEGG_NON_SMALL_CELL_LUNG_CANCER | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_NON_SMALL_CELL_LUNG_CANCER |
| LEE_LIVER_CANCER_MYC_UP | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_MYC_UP |
| MARIADASON_REGULATED_BY_HISTONE_ACETYLATION_DN | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/MARIADASON_REGULATED_BY_HISTONE_ACETYLATION_DN |
| MARSON_FOXP3_TARGETS_DN | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/MARSON_FOXP3_TARGETS_DN |
| MIKKELSEN_IPS_ICP_WITH_H3K27ME3 | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_IPS_ICP_WITH_H3K27ME3 |
| PID_NFAT_3PATHWAY | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_NFAT_3PATHWAY |
| PID_RAC1_PATHWAY | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_RAC1_PATHWAY |
| PID_TAP63PATHWAY | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_TAP63PATHWAY |
| PLASARI_TGFB1_SIGNALING_VIA_NFIC_10HR_UP | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/PLASARI_TGFB1_SIGNALING_VIA_NFIC_10HR_UP |
| REACTOME_APOPTOTIC_EXECUTION_PHASE | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_APOPTOTIC_EXECUTION_PHASE |
| REACTOME_PHOSPHOLIPASE_C_MEDIATED_CASCADE | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PHOSPHOLIPASE_C_MEDIATED_CASCADE |
| REACTOME_TCR_SIGNALING | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TCR_SIGNALING |
| REACTOME_TRANSPORT_OF_MATURE_TRANSCRIPT_TO_CYTOPLASM | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANSPORT_OF_MATURE_TRANSCRIPT_TO_CYTOPLASM |
| RICKMAN_HEAD_AND_NECK_CANCER_F | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/RICKMAN_HEAD_AND_NECK_CANCER_F |
| ROVERSI_GLIOMA_COPY_NUMBER_DN | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/ROVERSI_GLIOMA_COPY_NUMBER_DN |
| SESTO_RESPONSE_TO_UV_C2 | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/SESTO_RESPONSE_TO_UV_C2 |
| SUNG_METASTASIS_STROMA_DN | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/SUNG_METASTASIS_STROMA_DN |
| SU_PANCREAS | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/SU_PANCREAS |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_MONOCYTE_DN | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_MONOCYTE_DN |
| ZHOU_TNF_SIGNALING_30MIN | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHOU_TNF_SIGNALING_30MIN |
| ZHOU_TNF_SIGNALING_4HR | 54 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHOU_TNF_SIGNALING_4HR |
| FLOTHO_PEDIATRIC_ALL_THERAPY_RESPONSE_UP | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/FLOTHO_PEDIATRIC_ALL_THERAPY_RESPONSE_UP |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_TURQUOISE_DN | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_TURQUOISE_DN |
| HENDRICKS_SMARCA4_TARGETS_DN | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/HENDRICKS_SMARCA4_TARGETS_DN |
| IKEDA_MIR1_TARGETS_UP | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/IKEDA_MIR1_TARGETS_UP |
| ISHIDA_E2F_TARGETS | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/ISHIDA_E2F_TARGETS |
| KEGG_AMYOTROPHIC_LATERAL_SCLEROSIS_ALS | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_AMYOTROPHIC_LATERAL_SCLEROSIS_ALS |
| KEGG_AUTOIMMUNE_THYROID_DISEASE | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_AUTOIMMUNE_THYROID_DISEASE |
| KRIEG_HYPOXIA_VIA_KDM3A | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/KRIEG_HYPOXIA_VIA_KDM3A |
| LEE_TARGETS_OF_PTCH1_AND_SUFU_UP | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_TARGETS_OF_PTCH1_AND_SUFU_UP |
| LIANG_SILENCED_BY_METHYLATION_2 | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/LIANG_SILENCED_BY_METHYLATION_2 |
| LINDSTEDT_DENDRITIC_CELL_MATURATION_B | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/LINDSTEDT_DENDRITIC_CELL_MATURATION_B |
| MENSSEN_MYC_TARGETS | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/MENSSEN_MYC_TARGETS |
| MORI_IMMATURE_B_LYMPHOCYTE_UP | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/MORI_IMMATURE_B_LYMPHOCYTE_UP |
| NIKOLSKY_BREAST_CANCER_16Q24_AMPLICON | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_16Q24_AMPLICON |
| PID_AR_TF_PATHWAY | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_AR_TF_PATHWAY |
| PID_CD8TCRPATHWAY | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_CD8TCRPATHWAY |
| REACTOME_CIRCADIAN_CLOCK | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CIRCADIAN_CLOCK |
| REACTOME_DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 |
| REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS |
| REACTOME_PROTEIN_FOLDING | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PROTEIN_FOLDING |
| ROESSLER_LIVER_CANCER_METASTASIS_DN | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/ROESSLER_LIVER_CANCER_METASTASIS_DN |
| SMIRNOV_RESPONSE_TO_IR_2HR_UP | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/SMIRNOV_RESPONSE_TO_IR_2HR_UP |
| STARK_HYPPOCAMPUS_22Q11_DELETION_UP | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/STARK_HYPPOCAMPUS_22Q11_DELETION_UP |
| YAMASHITA_LIVER_CANCER_WITH_EPCAM_UP | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/YAMASHITA_LIVER_CANCER_WITH_EPCAM_UP |
| ZHU_CMV_8_HR_DN | 53 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHU_CMV_8_HR_DN |
| BECKER_TAMOXIFEN_RESISTANCE_DN | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/BECKER_TAMOXIFEN_RESISTANCE_DN |
| BOYAULT_LIVER_CANCER_SUBCLASS_G23_UP | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYAULT_LIVER_CANCER_SUBCLASS_G23_UP |
| CHIANG_LIVER_CANCER_SUBCLASS_INTERFERON_DN | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIANG_LIVER_CANCER_SUBCLASS_INTERFERON_DN |
| CHIBA_RESPONSE_TO_TSA_UP | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIBA_RESPONSE_TO_TSA_UP |
| CHNG_MULTIPLE_MYELOMA_HYPERPLOID_UP | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/CHNG_MULTIPLE_MYELOMA_HYPERPLOID_UP |
| DUNNE_TARGETS_OF_AML1_MTG8_FUSION_UP | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/DUNNE_TARGETS_OF_AML1_MTG8_FUSION_UP |
| HALMOS_CEBPA_TARGETS_UP | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/HALMOS_CEBPA_TARGETS_UP |
| KEGG_ENDOMETRIAL_CANCER | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ENDOMETRIAL_CANCER |
| KEGG_MTOR_SIGNALING_PATHWAY | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_MTOR_SIGNALING_PATHWAY |
| KEGG_STARCH_AND_SUCROSE_METABOLISM | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_STARCH_AND_SUCROSE_METABOLISM |
| KEGG_TASTE_TRANSDUCTION | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_TASTE_TRANSDUCTION |
| KENNY_CTNNB1_TARGETS_DN | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/KENNY_CTNNB1_TARGETS_DN |
| MARIADASON_RESPONSE_TO_BUTYRATE_SULINDAC_6 | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/MARIADASON_RESPONSE_TO_BUTYRATE_SULINDAC_6 |
| NATSUME_RESPONSE_TO_INTERFERON_BETA_DN | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/NATSUME_RESPONSE_TO_INTERFERON_BETA_DN |
| PAPASPYRIDONOS_UNSTABLE_ATEROSCLEROTIC_PLAQUE_UP | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/PAPASPYRIDONOS_UNSTABLE_ATEROSCLEROTIC_PLAQUE_UP |
| PID_CASPASE_PATHWAY | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_CASPASE_PATHWAY |
| PID_KITPATHWAY | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_KITPATHWAY |
| PID_PTP1BPATHWAY | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_PTP1BPATHWAY |
| PRAMOONJAGO_SOX4_TARGETS_UP | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/PRAMOONJAGO_SOX4_TARGETS_UP |
| RADAEVA_RESPONSE_TO_IFNA1_UP | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/RADAEVA_RESPONSE_TO_IFNA1_UP |
| REACTOME_GABA_RECEPTOR_ACTIVATION | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GABA_RECEPTOR_ACTIVATION |
| REACTOME_HEPARAN_SULFATE_HEPARIN_HS_GAG_METABOLISM | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_HEPARAN_SULFATE_HEPARIN_HS_GAG_METABOLISM |
| REACTOME_VIF_MEDIATED_DEGRADATION_OF_APOBEC3G | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_VIF_MEDIATED_DEGRADATION_OF_APOBEC3G |
| ROSS_AML_WITH_CBFB_MYH11_FUSION | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/ROSS_AML_WITH_CBFB_MYH11_FUSION |
| SOTIRIOU_BREAST_CANCER_GRADE_1_VS_3_DN | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/SOTIRIOU_BREAST_CANCER_GRADE_1_VS_3_DN |
| VILIMAS_NOTCH1_TARGETS_UP | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/VILIMAS_NOTCH1_TARGETS_UP |
| WINTER_HYPOXIA_DN | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/WINTER_HYPOXIA_DN |
| ZHAN_MULTIPLE_MYELOMA_CD1_VS_CD2_DN | 52 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_CD1_VS_CD2_DN |
| APPIERTO_RESPONSE_TO_FENRETINIDE_DN | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/APPIERTO_RESPONSE_TO_FENRETINIDE_DN |
| BOYAULT_LIVER_CANCER_SUBCLASS_G123_DN | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYAULT_LIVER_CANCER_SUBCLASS_G123_DN |
| BOYAULT_LIVER_CANCER_SUBCLASS_G3_DN | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYAULT_LIVER_CANCER_SUBCLASS_G3_DN |
| BURTON_ADIPOGENESIS_7 | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/BURTON_ADIPOGENESIS_7 |
| BURTON_ADIPOGENESIS_PEAK_AT_2HR | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/BURTON_ADIPOGENESIS_PEAK_AT_2HR |
| BYSTROEM_CORRELATED_WITH_IL5_UP | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/BYSTROEM_CORRELATED_WITH_IL5_UP |
| CHAUHAN_RESPONSE_TO_METHOXYESTRADIOL_UP | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/CHAUHAN_RESPONSE_TO_METHOXYESTRADIOL_UP |
| CHIARADONNA_NEOPLASTIC_TRANSFORMATION_KRAS_CDC25_DN | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIARADONNA_NEOPLASTIC_TRANSFORMATION_KRAS_CDC25_DN |
| CROMER_TUMORIGENESIS_DN | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/CROMER_TUMORIGENESIS_DN |
| DAIRKEE_CANCER_PRONE_RESPONSE_BPA | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/DAIRKEE_CANCER_PRONE_RESPONSE_BPA |
| FLECHNER_PBL_KIDNEY_TRANSPLANT_REJECTED_VS_OK_DN | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/FLECHNER_PBL_KIDNEY_TRANSPLANT_REJECTED_VS_OK_DN |
| FRASOR_TAMOXIFEN_RESPONSE_UP | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/FRASOR_TAMOXIFEN_RESPONSE_UP |
| GUILLAUMOND_KLF10_TARGETS_UP | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/GUILLAUMOND_KLF10_TARGETS_UP |
| KEGG_DRUG_METABOLISM_OTHER_ENZYMES | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_DRUG_METABOLISM_OTHER_ENZYMES |
| KLEIN_PRIMARY_EFFUSION_LYMPHOMA_UP | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/KLEIN_PRIMARY_EFFUSION_LYMPHOMA_UP |
| LEE_CALORIE_RESTRICTION_MUSCLE_DN | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_CALORIE_RESTRICTION_MUSCLE_DN |
| LI_WILMS_TUMOR_VS_FETAL_KIDNEY_2_DN | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_WILMS_TUMOR_VS_FETAL_KIDNEY_2_DN |
| LUI_THYROID_CANCER_CLUSTER_1 | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/LUI_THYROID_CANCER_CLUSTER_1 |
| MORI_PLASMA_CELL_UP | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/MORI_PLASMA_CELL_UP |
| PRAMOONJAGO_SOX4_TARGETS_DN | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/PRAMOONJAGO_SOX4_TARGETS_DN |
| REACTOME_AQUAPORIN_MEDIATED_TRANSPORT | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_AQUAPORIN_MEDIATED_TRANSPORT |
| REACTOME_AUTODEGRADATION_OF_THE_E3_UBIQUITIN_LIGASE_COP1 | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_AUTODEGRADATION_OF_THE_E3_UBIQUITIN_LIGASE_COP1 |
| REACTOME_CYTOCHROME_P450_ARRANGED_BY_SUBSTRATE_TYPE | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CYTOCHROME_P450_ARRANGED_BY_SUBSTRATE_TYPE |
| REACTOME_DEFENSINS | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DEFENSINS |
| REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS |
| REACTOME_NUCLEOTIDE_EXCISION_REPAIR | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NUCLEOTIDE_EXCISION_REPAIR |
| REACTOME_P53_INDEPENDENT_G1_S_DNA_DAMAGE_CHECKPOINT | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_P53_INDEPENDENT_G1_S_DNA_DAMAGE_CHECKPOINT |
| REACTOME_SCF_BETA_TRCP_MEDIATED_DEGRADATION_OF_EMI1 | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SCF_BETA_TRCP_MEDIATED_DEGRADATION_OF_EMI1 |
| SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES |
| TSUNODA_CISPLATIN_RESISTANCE_DN | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/TSUNODA_CISPLATIN_RESISTANCE_DN |
| WANG_BARRETTS_ESOPHAGUS_UP | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_BARRETTS_ESOPHAGUS_UP |
| ZHANG_ANTIVIRAL_RESPONSE_TO_RIBAVIRIN_DN | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_ANTIVIRAL_RESPONSE_TO_RIBAVIRIN_DN |
| ZHANG_PROLIFERATING_VS_QUIESCENT | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_PROLIFERATING_VS_QUIESCENT |
| ZHAN_MULTIPLE_MYELOMA_CD1_AND_CD2_DN | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_CD1_AND_CD2_DN |
| ZWANG_CLASS_2_TRANSIENTLY_INDUCED_BY_EGF | 51 | http://www.broadinstitute.org/gsea/msigdb/cards/ZWANG_CLASS_2_TRANSIENTLY_INDUCED_BY_EGF |
| BECKER_TAMOXIFEN_RESISTANCE_UP | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/BECKER_TAMOXIFEN_RESISTANCE_UP |
| CHARAFE_BREAST_CANCER_BASAL_VS_MESENCHYMAL_DN | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/CHARAFE_BREAST_CANCER_BASAL_VS_MESENCHYMAL_DN |
| CHEN_NEUROBLASTOMA_COPY_NUMBER_GAINS | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEN_NEUROBLASTOMA_COPY_NUMBER_GAINS |
| CHIBA_RESPONSE_TO_TSA | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIBA_RESPONSE_TO_TSA |
| DAUER_STAT3_TARGETS_DN | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/DAUER_STAT3_TARGETS_DN |
| EBAUER_MYOGENIC_TARGETS_OF_PAX3_FOXO1_FUSION | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/EBAUER_MYOGENIC_TARGETS_OF_PAX3_FOXO1_FUSION |
| FIGUEROA_AML_METHYLATION_CLUSTER_5_DN | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/FIGUEROA_AML_METHYLATION_CLUSTER_5_DN |
| FRASOR_RESPONSE_TO_SERM_OR_FULVESTRANT_DN | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/FRASOR_RESPONSE_TO_SERM_OR_FULVESTRANT_DN |
| HEDENFALK_BREAST_CANCER_HEREDITARY_VS_SPORADIC | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/HEDENFALK_BREAST_CANCER_HEREDITARY_VS_SPORADIC |
| HOFFMANN_IMMATURE_TO_MATURE_B_LYMPHOCYTE_DN | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFFMANN_IMMATURE_TO_MATURE_B_LYMPHOCYTE_DN |
| HOFFMANN_SMALL_PRE_BII_TO_IMMATURE_B_LYMPHOCYTE_DN | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFFMANN_SMALL_PRE_BII_TO_IMMATURE_B_LYMPHOCYTE_DN |
| HOFMANN_CELL_LYMPHOMA_UP | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFMANN_CELL_LYMPHOMA_UP |
| KANG_GIST_WITH_PDGFRA_UP | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/KANG_GIST_WITH_PDGFRA_UP |
| KEGG_GLUTATHIONE_METABOLISM | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GLUTATHIONE_METABOLISM |
| KENNY_CTNNB1_TARGETS_UP | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/KENNY_CTNNB1_TARGETS_UP |
| KOHOUTEK_CCNT1_TARGETS | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/KOHOUTEK_CCNT1_TARGETS |
| MARTINEZ_RESPONSE_TO_TRABECTEDIN | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTINEZ_RESPONSE_TO_TRABECTEDIN |
| NADERI_BREAST_CANCER_PROGNOSIS_UP | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/NADERI_BREAST_CANCER_PROGNOSIS_UP |
| NOUSHMEHR_GBM_SILENCED_BY_METHYLATION | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/NOUSHMEHR_GBM_SILENCED_BY_METHYLATION |
| PARK_APL_PATHOGENESIS_DN | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/PARK_APL_PATHOGENESIS_DN |
| PARK_HSC_AND_MULTIPOTENT_PROGENITORS | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/PARK_HSC_AND_MULTIPOTENT_PROGENITORS |
| PID_ANGIOPOIETINRECEPTOR_PATHWAY | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ANGIOPOIETINRECEPTOR_PATHWAY |
| REACTOME_MAP_KINASE_ACTIVATION_IN_TLR_CASCADE | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MAP_KINASE_ACTIVATION_IN_TLR_CASCADE |
| ROY_WOUND_BLOOD_VESSEL_UP | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/ROY_WOUND_BLOOD_VESSEL_UP |
| SASAI_RESISTANCE_TO_NEOPLASTIC_TRANSFROMATION | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/SASAI_RESISTANCE_TO_NEOPLASTIC_TRANSFROMATION |
| SATO_SILENCED_BY_DEACETYLATION_IN_PANCREATIC_CANCER | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/SATO_SILENCED_BY_DEACETYLATION_IN_PANCREATIC_CANCER |
| SATO_SILENCED_BY_METHYLATION_IN_PANCREATIC_CANCER_2 | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/SATO_SILENCED_BY_METHYLATION_IN_PANCREATIC_CANCER_2 |
| STAMBOLSKY_TARGETS_OF_MUTATED_TP53_DN | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/STAMBOLSKY_TARGETS_OF_MUTATED_TP53_DN |
| STOSSI_RESPONSE_TO_ESTRADIOL | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/STOSSI_RESPONSE_TO_ESTRADIOL |
| YANG_BREAST_CANCER_ESR1_LASER_DN | 50 | http://www.broadinstitute.org/gsea/msigdb/cards/YANG_BREAST_CANCER_ESR1_LASER_DN |
| APRELIKOVA_BRCA1_TARGETS | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/APRELIKOVA_BRCA1_TARGETS |
| ASTON_MAJOR_DEPRESSIVE_DISORDER_UP | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/ASTON_MAJOR_DEPRESSIVE_DISORDER_UP |
| BIOCARTA_TCR_PATHWAY | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_TCR_PATHWAY |
| BREDEMEYER_RAG_SIGNALING_VIA_ATM_NOT_VIA_NFKB_UP | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/BREDEMEYER_RAG_SIGNALING_VIA_ATM_NOT_VIA_NFKB_UP |
| BROWNE_HCMV_INFECTION_2HR_DN | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_2HR_DN |
| DAUER_STAT3_TARGETS_UP | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/DAUER_STAT3_TARGETS_UP |
| FAELT_B_CLL_WITH_VH3_21_DN | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/FAELT_B_CLL_WITH_VH3_21_DN |
| GERHOLD_ADIPOGENESIS_UP | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/GERHOLD_ADIPOGENESIS_UP |
| ICHIBA_GRAFT_VERSUS_HOST_DISEASE_35D_DN | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/ICHIBA_GRAFT_VERSUS_HOST_DISEASE_35D_DN |
| JAZAERI_BREAST_CANCER_BRCA1_VS_BRCA2_UP | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/JAZAERI_BREAST_CANCER_BRCA1_VS_BRCA2_UP |
| KEGG_GLYCEROLIPID_METABOLISM | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GLYCEROLIPID_METABOLISM |
| LEE_LIVER_CANCER | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER |
| LINDGREN_BLADDER_CANCER_HIGH_RECURRENCE | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/LINDGREN_BLADDER_CANCER_HIGH_RECURRENCE |
| MANTOVANI_VIRAL_GPCR_SIGNALING_DN | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/MANTOVANI_VIRAL_GPCR_SIGNALING_DN |
| MARCHINI_TRABECTEDIN_RESISTANCE_DN | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/MARCHINI_TRABECTEDIN_RESISTANCE_DN |
| OLSSON_E2F3_TARGETS_DN | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/OLSSON_E2F3_TARGETS_DN |
| PELLICCIOTTA_HDAC_IN_ANTIGEN_PRESENTATION_DN | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/PELLICCIOTTA_HDAC_IN_ANTIGEN_PRESENTATION_DN |
| PID_ARF6_TRAFFICKINGPATHWAY | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ARF6_TRAFFICKINGPATHWAY |
| PID_FOXOPATHWAY | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_FOXOPATHWAY |
| PID_PI3KCIPATHWAY | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_PI3KCIPATHWAY |
| REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS |
| REACTOME_CHONDROITIN_SULFATE_DERMATAN_SULFATE_METABOLISM | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CHONDROITIN_SULFATE_DERMATAN_SULFATE_METABOLISM |
| REACTOME_METABOLISM_OF_NON_CODING_RNA | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_METABOLISM_OF_NON_CODING_RNA |
| REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY |
| REACTOME_REGULATION_OF_ORNITHINE_DECARBOXYLASE_ODC | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_ORNITHINE_DECARBOXYLASE_ODC |
| SATO_SILENCED_EPIGENETICALLY_IN_PANCREATIC_CANCER | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/SATO_SILENCED_EPIGENETICALLY_IN_PANCREATIC_CANCER |
| STAMBOLSKY_TARGETS_OF_MUTATED_TP53_UP | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/STAMBOLSKY_TARGETS_OF_MUTATED_TP53_UP |
| WONG_PROTEASOME_GENE_MODULE | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/WONG_PROTEASOME_GENE_MODULE |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_8 | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_8 |
| ZHAN_MULTIPLE_MYELOMA_HP_UP | 49 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_HP_UP |
| ABE_INNER_EAR | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/ABE_INNER_EAR |
| BERENJENO_ROCK_SIGNALING_NOT_VIA_RHOA_DN | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/BERENJENO_ROCK_SIGNALING_NOT_VIA_RHOA_DN |
| BURTON_ADIPOGENESIS_4 | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/BURTON_ADIPOGENESIS_4 |
| CHANGOLKAR_H2AFY_TARGETS_UP | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/CHANGOLKAR_H2AFY_TARGETS_UP |
| DELACROIX_RAR_TARGETS_UP | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/DELACROIX_RAR_TARGETS_UP |
| DOANE_BREAST_CANCER_ESR1_DN | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/DOANE_BREAST_CANCER_ESR1_DN |
| EBAUER_TARGETS_OF_PAX3_FOXO1_FUSION_DN | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/EBAUER_TARGETS_OF_PAX3_FOXO1_FUSION_DN |
| ENGELMANN_CANCER_PROGENITORS_UP | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/ENGELMANN_CANCER_PROGENITORS_UP |
| FAELT_B_CLL_WITH_VH_REARRANGEMENTS_DN | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/FAELT_B_CLL_WITH_VH_REARRANGEMENTS_DN |
| FAELT_B_CLL_WITH_VH_REARRANGEMENTS_UP | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/FAELT_B_CLL_WITH_VH_REARRANGEMENTS_UP |
| FIGUEROA_AML_METHYLATION_CLUSTER_1_DN | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/FIGUEROA_AML_METHYLATION_CLUSTER_1_DN |
| GNATENKO_PLATELET_SIGNATURE | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/GNATENKO_PLATELET_SIGNATURE |
| HANN_RESISTANCE_TO_BCL2_INHIBITOR_DN | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/HANN_RESISTANCE_TO_BCL2_INHIBITOR_DN |
| KEGG_INTESTINAL_IMMUNE_NETWORK_FOR_IGA_PRODUCTION | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_INTESTINAL_IMMUNE_NETWORK_FOR_IGA_PRODUCTION |
| KEGG_PROTEASOME | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PROTEASOME |
| MCCLUNG_DELTA_FOSB_TARGETS_2WK | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/MCCLUNG_DELTA_FOSB_TARGETS_2WK |
| MCDOWELL_ACUTE_LUNG_INJURY_DN | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/MCDOWELL_ACUTE_LUNG_INJURY_DN |
| NADLER_OBESITY_DN | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/NADLER_OBESITY_DN |
| OXFORD_RALA_OR_RALB_TARGETS_UP | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/OXFORD_RALA_OR_RALB_TARGETS_UP |
| PENG_GLUCOSE_DEPRIVATION_UP | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/PENG_GLUCOSE_DEPRIVATION_UP |
| PID_AJDISS_2PATHWAY | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_AJDISS_2PATHWAY |
| PID_CERAMIDE_PATHWAY | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_CERAMIDE_PATHWAY |
| PID_HEDGEHOG_GLIPATHWAY | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_HEDGEHOG_GLIPATHWAY |
| PID_HES_HEYPATHWAY | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_HES_HEYPATHWAY |
| RASHI_RESPONSE_TO_IONIZING_RADIATION_3 | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/RASHI_RESPONSE_TO_IONIZING_RADIATION_3 |
| REACTOME_CDK_MEDIATED_PHOSPHORYLATION_AND_REMOVAL_OF_CDC6 | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CDK_MEDIATED_PHOSPHORYLATION_AND_REMOVAL_OF_CDC6 |
| REACTOME_CROSS_PRESENTATION_OF_SOLUBLE_EXOGENOUS_ANTIGENS_ENDOSOMES | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CROSS_PRESENTATION_OF_SOLUBLE_EXOGENOUS_ANTIGENS_ENDOSOMES |
| REACTOME_DEADENYLATION_DEPENDENT_MRNA_DECAY | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DEADENYLATION_DEPENDENT_MRNA_DECAY |
| REACTOME_MUSCLE_CONTRACTION | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MUSCLE_CONTRACTION |
| REACTOME_PACKAGING_OF_TELOMERE_ENDS | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PACKAGING_OF_TELOMERE_ENDS |
| REACTOME_PI_METABOLISM | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PI_METABOLISM |
| REACTOME_PYRUVATE_METABOLISM_AND_CITRIC_ACID_TCA_CYCLE | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PYRUVATE_METABOLISM_AND_CITRIC_ACID_TCA_CYCLE |
| RICKMAN_HEAD_AND_NECK_CANCER_B | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/RICKMAN_HEAD_AND_NECK_CANCER_B |
| TAVOR_CEBPA_TARGETS_UP | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/TAVOR_CEBPA_TARGETS_UP |
| ZHAN_MULTIPLE_MYELOMA_LB_UP | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_LB_UP |
| ZHAN_MULTIPLE_MYELOMA_MS_UP | 48 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_MS_UP |
| BOHN_PRIMARY_IMMUNODEFICIENCY_SYNDROM_UP | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/BOHN_PRIMARY_IMMUNODEFICIENCY_SYNDROM_UP |
| BOYLAN_MULTIPLE_MYELOMA_C_UP | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYLAN_MULTIPLE_MYELOMA_C_UP |
| BROWNE_HCMV_INFECTION_8HR_DN | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_8HR_DN |
| GRAHAM_CML_QUIESCENT_VS_NORMAL_QUIESCENT_DN | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAHAM_CML_QUIESCENT_VS_NORMAL_QUIESCENT_DN |
| IRITANI_MAD1_TARGETS_DN | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/IRITANI_MAD1_TARGETS_DN |
| JIANG_AGING_HYPOTHALAMUS_UP | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/JIANG_AGING_HYPOTHALAMUS_UP |
| KEGG_NOTCH_SIGNALING_PATHWAY | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_NOTCH_SIGNALING_PATHWAY |
| KEGG_TYPE_II_DIABETES_MELLITUS | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_TYPE_II_DIABETES_MELLITUS |
| KHETCHOUMIAN_TRIM24_TARGETS_UP | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/KHETCHOUMIAN_TRIM24_TARGETS_UP |
| LEONARD_HYPOXIA | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/LEONARD_HYPOXIA |
| MCCLUNG_DELTA_FOSB_TARGETS_8WK | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/MCCLUNG_DELTA_FOSB_TARGETS_8WK |
| MELLMAN_TUT1_TARGETS_DN | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/MELLMAN_TUT1_TARGETS_DN |
| NAKAMURA_METASTASIS | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAMURA_METASTASIS |
| PID_DELTANP63PATHWAY | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_DELTANP63PATHWAY |
| PID_FANCONI_PATHWAY | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_FANCONI_PATHWAY |
| PID_IL6_7PATHWAY | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_IL6_7PATHWAY |
| PID_LKB1_PATHWAY | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_LKB1_PATHWAY |
| PID_NFAT_TFPATHWAY | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_NFAT_TFPATHWAY |
| POMEROY_MEDULLOBLASTOMA_PROGNOSIS_UP | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/POMEROY_MEDULLOBLASTOMA_PROGNOSIS_UP |
| POTTI_DOCETAXEL_SENSITIVITY | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/POTTI_DOCETAXEL_SENSITIVITY |
| SCHLOSSER_MYC_TARGETS_AND_SERUM_RESPONSE_DN | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHLOSSER_MYC_TARGETS_AND_SERUM_RESPONSE_DN |
| SCHLOSSER_MYC_TARGETS_AND_SERUM_RESPONSE_UP | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHLOSSER_MYC_TARGETS_AND_SERUM_RESPONSE_UP |
| SILIGAN_BOUND_BY_EWS_FLT1_FUSION | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/SILIGAN_BOUND_BY_EWS_FLT1_FUSION |
| WARTERS_IR_RESPONSE_5GY | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/WARTERS_IR_RESPONSE_5GY |
| WOOD_EBV_EBNA1_TARGETS_DN | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/WOOD_EBV_EBNA1_TARGETS_DN |
| YAMASHITA_LIVER_CANCER_STEM_CELL_UP | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/YAMASHITA_LIVER_CANCER_STEM_CELL_UP |
| ZHAN_MULTIPLE_MYELOMA_HP_DN | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_HP_DN |
| ZHAN_MULTIPLE_MYELOMA_MF_UP | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_MF_UP |
| ZHU_CMV_8_HR_UP | 47 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHU_CMV_8_HR_UP |
| ABBUD_LIF_SIGNALING_1_UP | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/ABBUD_LIF_SIGNALING_1_UP |
| AMIT_EGF_RESPONSE_60_HELA | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_EGF_RESPONSE_60_HELA |
| ASGHARZADEH_NEUROBLASTOMA_POOR_SURVIVAL_DN | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/ASGHARZADEH_NEUROBLASTOMA_POOR_SURVIVAL_DN |
| BERTUCCI_INVASIVE_CARCINOMA_DUCTAL_VS_LOBULAR_DN | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/BERTUCCI_INVASIVE_CARCINOMA_DUCTAL_VS_LOBULAR_DN |
| BIOCARTA_KERATINOCYTE_PATHWAY | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_KERATINOCYTE_PATHWAY |
| BRACHAT_RESPONSE_TO_CAMPTOTHECIN_DN | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/BRACHAT_RESPONSE_TO_CAMPTOTHECIN_DN |
| HALMOS_CEBPA_TARGETS_DN | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/HALMOS_CEBPA_TARGETS_DN |
| HEIDENBLAD_AMPLICON_8Q24_DN | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/HEIDENBLAD_AMPLICON_8Q24_DN |
| JIANG_TIP30_TARGETS_UP | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/JIANG_TIP30_TARGETS_UP |
| KEGG_N_GLYCAN_BIOSYNTHESIS | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_N_GLYCAN_BIOSYNTHESIS |
| KRASNOSELSKAYA_ILF3_TARGETS_DN | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/KRASNOSELSKAYA_ILF3_TARGETS_DN |
| KUNINGER_IGF1_VS_PDGFB_TARGETS_DN | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/KUNINGER_IGF1_VS_PDGFB_TARGETS_DN |
| KYNG_RESPONSE_TO_H2O2_VIA_ERCC6_DN | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_RESPONSE_TO_H2O2_VIA_ERCC6_DN |
| LEE_AGING_MUSCLE_DN | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_AGING_MUSCLE_DN |
| MADAN_DPPA4_TARGETS | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/MADAN_DPPA4_TARGETS |
| NAKAJIMA_MAST_CELL | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAJIMA_MAST_CELL |
| NAKAMURA_CANCER_MICROENVIRONMENT_DN | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAMURA_CANCER_MICROENVIRONMENT_DN |
| NIKOLSKY_BREAST_CANCER_12Q13_Q21_AMPLICON | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_12Q13_Q21_AMPLICON |
| NUTT_GBM_VS_AO_GLIOMA_UP | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/NUTT_GBM_VS_AO_GLIOMA_UP |
| OSADA_ASCL1_TARGETS_UP | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/OSADA_ASCL1_TARGETS_UP |
| PARENT_MTOR_SIGNALING_DN | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/PARENT_MTOR_SIGNALING_DN |
| PEDERSEN_METASTASIS_BY_ERBB2_ISOFORM_1 | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/PEDERSEN_METASTASIS_BY_ERBB2_ISOFORM_1 |
| PID_A6B1_A6B4_INTEGRIN_PATHWAY | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_A6B1_A6B4_INTEGRIN_PATHWAY |
| PID_PLK1_PATHWAY | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_PLK1_PATHWAY |
| PID_PS1PATHWAY | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_PS1PATHWAY |
| PID_RHOA_REG_PATHWAY | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_RHOA_REG_PATHWAY |
| PID_SYNDECAN_1_PATHWAY | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_SYNDECAN_1_PATHWAY |
| PID_TNFPATHWAY | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_TNFPATHWAY |
| REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S |
| REACTOME_LIPID_DIGESTION_MOBILIZATION_AND_TRANSPORT | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_LIPID_DIGESTION_MOBILIZATION_AND_TRANSPORT |
| REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION |
| REACTOME_NUCLEOTIDE_BINDING_DOMAIN_LEUCINE_RICH_REPEAT_CONTAINING_RECEPTOR_NLR_SIGNALING_PATHWAYS | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NUCLEOTIDE_BINDING_DOMAIN_LEUCINE_RICH_REPEAT_CONTAINING_RECEPTOR_NLR_SIGNALING_PATHWAYS |
| SAGIV_CD24_TARGETS_DN | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/SAGIV_CD24_TARGETS_DN |
| SESTO_RESPONSE_TO_UV_C5 | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/SESTO_RESPONSE_TO_UV_C5 |
| SHETH_LIVER_CANCER_VS_TXNIP_LOSS_PAM6 | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/SHETH_LIVER_CANCER_VS_TXNIP_LOSS_PAM6 |
| SIG_BCR_SIGNALING_PATHWAY | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/SIG_BCR_SIGNALING_PATHWAY |
| TSENG_ADIPOGENIC_POTENTIAL_DN | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/TSENG_ADIPOGENIC_POTENTIAL_DN |
| WINNEPENNINCKX_MELANOMA_METASTASIS_DN | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/WINNEPENNINCKX_MELANOMA_METASTASIS_DN |
| ZHAN_MULTIPLE_MYELOMA_CD2_DN | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_CD2_DN |
| ZHAN_MULTIPLE_MYELOMA_MS_DN | 46 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_MS_DN |
| ABRAMSON_INTERACT_WITH_AIRE | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/ABRAMSON_INTERACT_WITH_AIRE |
| BEGUM_TARGETS_OF_PAX3_FOXO1_FUSION_DN | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/BEGUM_TARGETS_OF_PAX3_FOXO1_FUSION_DN |
| BIOCARTA_BIOPEPTIDES_PATHWAY | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_BIOPEPTIDES_PATHWAY |
| BOYAULT_LIVER_CANCER_SUBCLASS_G123_UP | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYAULT_LIVER_CANCER_SUBCLASS_G123_UP |
| CERIBELLI_GENES_INACTIVE_AND_BOUND_BY_NFY | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/CERIBELLI_GENES_INACTIVE_AND_BOUND_BY_NFY |
| CHANDRAN_METASTASIS_TOP50_DN | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/CHANDRAN_METASTASIS_TOP50_DN |
| CHENG_RESPONSE_TO_NICKEL_ACETATE | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/CHENG_RESPONSE_TO_NICKEL_ACETATE |
| GRADE_METASTASIS_DN | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/GRADE_METASTASIS_DN |
| HUANG_FOXA2_TARGETS_UP | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/HUANG_FOXA2_TARGETS_UP |
| JOHANSSON_BRAIN_CANCER_EARLY_VS_LATE_DN | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/JOHANSSON_BRAIN_CANCER_EARLY_VS_LATE_DN |
| KLEIN_TARGETS_OF_BCR_ABL1_FUSION | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/KLEIN_TARGETS_OF_BCR_ABL1_FUSION |
| LEE_AGING_MUSCLE_UP | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_AGING_MUSCLE_UP |
| LEE_METASTASIS_AND_ALTERNATIVE_SPLICING_DN | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_METASTASIS_AND_ALTERNATIVE_SPLICING_DN |
| LIANG_HEMATOPOIESIS_STEM_CELL_NUMBER_LARGE_VS_TINY_DN | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/LIANG_HEMATOPOIESIS_STEM_CELL_NUMBER_LARGE_VS_TINY_DN |
| LUI_THYROID_CANCER_PAX8_PPARG_DN | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/LUI_THYROID_CANCER_PAX8_PPARG_DN |
| MCDOWELL_ACUTE_LUNG_INJURY_UP | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/MCDOWELL_ACUTE_LUNG_INJURY_UP |
| NAKAMURA_METASTASIS_MODEL_UP | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAMURA_METASTASIS_MODEL_UP |
| NUTT_GBM_VS_AO_GLIOMA_DN | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/NUTT_GBM_VS_AO_GLIOMA_DN |
| ODONNELL_TARGETS_OF_MYC_AND_TFRC_DN | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/ODONNELL_TARGETS_OF_MYC_AND_TFRC_DN |
| PID_HNF3BPATHWAY | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_HNF3BPATHWAY |
| PID_ILK_PATHWAY | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ILK_PATHWAY |
| PID_INSULIN_PATHWAY | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_INSULIN_PATHWAY |
| PID_RHOA_PATHWAY | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_RHOA_PATHWAY |
| RASHI_RESPONSE_TO_IONIZING_RADIATION_1 | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/RASHI_RESPONSE_TO_IONIZING_RADIATION_1 |
| REACTOME_FORMATION_OF_RNA_POL_II_ELONGATION_COMPLEX_ | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FORMATION_OF_RNA_POL_II_ELONGATION_COMPLEX_ |
| REACTOME_G2_M_CHECKPOINTS | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_G2_M_CHECKPOINTS |
| REACTOME_MRNA_SPLICING_MINOR_PATHWAY | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MRNA_SPLICING_MINOR_PATHWAY |
| REACTOME_TRANSCRIPTION_COUPLED_NER_TC_NER | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANSCRIPTION_COUPLED_NER_TC_NER |
| SCHAEFFER_SOX9_TARGETS_IN_PROSTATE_DEVELOPMENT_DN | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_SOX9_TARGETS_IN_PROSTATE_DEVELOPMENT_DN |
| SHIPP_DLBCL_CURED_VS_FATAL_DN | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/SHIPP_DLBCL_CURED_VS_FATAL_DN |
| SHIPP_DLBCL_VS_FOLLICULAR_LYMPHOMA_DN | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/SHIPP_DLBCL_VS_FOLLICULAR_LYMPHOMA_DN |
| SHIPP_DLBCL_VS_FOLLICULAR_LYMPHOMA_UP | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/SHIPP_DLBCL_VS_FOLLICULAR_LYMPHOMA_UP |
| SIG_CHEMOTAXIS | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/SIG_CHEMOTAXIS |
| SMITH_LIVER_CANCER | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/SMITH_LIVER_CANCER |
| ST_DIFFERENTIATION_PATHWAY_IN_PC12_CELLS | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_DIFFERENTIATION_PATHWAY_IN_PC12_CELLS |
| ST_T_CELL_SIGNAL_TRANSDUCTION | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_T_CELL_SIGNAL_TRANSDUCTION |
| TRAYNOR_RETT_SYNDROM_UP | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/TRAYNOR_RETT_SYNDROM_UP |
| VANHARANTA_UTERINE_FIBROID_UP | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/VANHARANTA_UTERINE_FIBROID_UP |
| WANG_TARGETS_OF_MLL_CBP_FUSION_DN | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_TARGETS_OF_MLL_CBP_FUSION_DN |
| ZHAN_MULTIPLE_MYELOMA_CD1_DN | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_CD1_DN |
| ZHAN_MULTIPLE_MYELOMA_CD1_UP | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_CD1_UP |
| ZHAN_MULTIPLE_MYELOMA_CD2_UP | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_CD2_UP |
| ZHAN_MULTIPLE_MYELOMA_PR_UP | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_PR_UP |
| ZHAN_V2_LATE_DIFFERENTIATION_GENES | 45 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_V2_LATE_DIFFERENTIATION_GENES |
| CERIBELLI_PROMOTERS_INACTIVE_AND_BOUND_BY_NFY | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/CERIBELLI_PROMOTERS_INACTIVE_AND_BOUND_BY_NFY |
| CHOI_ATL_STAGE_PREDICTOR | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/CHOI_ATL_STAGE_PREDICTOR |
| CHUNG_BLISTER_CYTOTOXICITY_DN | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/CHUNG_BLISTER_CYTOTOXICITY_DN |
| FAELT_B_CLL_WITH_VH3_21_UP | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/FAELT_B_CLL_WITH_VH3_21_UP |
| FARMER_BREAST_CANCER_CLUSTER_1 | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/FARMER_BREAST_CANCER_CLUSTER_1 |
| GROSS_HYPOXIA_VIA_ELK3_ONLY_DN | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/GROSS_HYPOXIA_VIA_ELK3_ONLY_DN |
| HARRIS_BRAIN_CANCER_PROGENITORS | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/HARRIS_BRAIN_CANCER_PROGENITORS |
| JAEGER_METASTASIS_UP | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/JAEGER_METASTASIS_UP |
| KEGG_ABC_TRANSPORTERS | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ABC_TRANSPORTERS |
| KEGG_AMINO_SUGAR_AND_NUCLEOTIDE_SUGAR_METABOLISM | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_AMINO_SUGAR_AND_NUCLEOTIDE_SUGAR_METABOLISM |
| KEGG_LYSINE_DEGRADATION | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_LYSINE_DEGRADATION |
| KEGG_NUCLEOTIDE_EXCISION_REPAIR | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_NUCLEOTIDE_EXCISION_REPAIR |
| KEGG_TYPE_I_DIABETES_MELLITUS | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_TYPE_I_DIABETES_MELLITUS |
| KEGG_VALINE_LEUCINE_AND_ISOLEUCINE_DEGRADATION | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_VALINE_LEUCINE_AND_ISOLEUCINE_DEGRADATION |
| KEGG_VASOPRESSIN_REGULATED_WATER_REABSORPTION | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_VASOPRESSIN_REGULATED_WATER_REABSORPTION |
| KERLEY_RESPONSE_TO_CISPLATIN_UP | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/KERLEY_RESPONSE_TO_CISPLATIN_UP |
| KORKOLA_SEMINOMA_UP | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/KORKOLA_SEMINOMA_UP |
| KUROZUMI_RESPONSE_TO_ONCOCYTIC_VIRUS | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/KUROZUMI_RESPONSE_TO_ONCOCYTIC_VIRUS |
| LANDIS_BREAST_CANCER_PROGRESSION_UP | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/LANDIS_BREAST_CANCER_PROGRESSION_UP |
| LIANG_HEMATOPOIESIS_STEM_CELL_NUMBER_LARGE_VS_TINY_UP | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/LIANG_HEMATOPOIESIS_STEM_CELL_NUMBER_LARGE_VS_TINY_UP |
| LUI_THYROID_CANCER_PAX8_PPARG_UP | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/LUI_THYROID_CANCER_PAX8_PPARG_UP |
| MARTIN_INTERACT_WITH_HDAC | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTIN_INTERACT_WITH_HDAC |
| MA_MYELOID_DIFFERENTIATION_DN | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/MA_MYELOID_DIFFERENTIATION_DN |
| MEDINA_SMARCA4_TARGETS | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/MEDINA_SMARCA4_TARGETS |
| OHGUCHI_LIVER_HNF4A_TARGETS_UP | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/OHGUCHI_LIVER_HNF4A_TARGETS_UP |
| PARK_HSC_MARKERS | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/PARK_HSC_MARKERS |
| PID_ERBB2ERBB3PATHWAY | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ERBB2ERBB3PATHWAY |
| PID_HNF3APATHWAY | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_HNF3APATHWAY |
| REACTOME_CLEAVAGE_OF_GROWING_TRANSCRIPT_IN_THE_TERMINATION_REGION_ | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CLEAVAGE_OF_GROWING_TRANSCRIPT_IN_THE_TERMINATION_REGION_ |
| REACTOME_G_ALPHA_Z_SIGNALLING_EVENTS | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_G_ALPHA_Z_SIGNALLING_EVENTS |
| REACTOME_PI3K_EVENTS_IN_ERBB2_SIGNALING | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PI3K_EVENTS_IN_ERBB2_SIGNALING |
| REACTOME_PRE_NOTCH_EXPRESSION_AND_PROCESSING | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PRE_NOTCH_EXPRESSION_AND_PROCESSING |
| REACTOME_REGULATION_OF_WATER_BALANCE_BY_RENAL_AQUAPORINS | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_WATER_BALANCE_BY_RENAL_AQUAPORINS |
| REACTOME_SIGNALING_BY_FGFR_MUTANTS | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_FGFR_MUTANTS |
| ROVERSI_GLIOMA_LOH_REGIONS | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/ROVERSI_GLIOMA_LOH_REGIONS |
| STREICHER_LSM1_TARGETS_UP | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/STREICHER_LSM1_TARGETS_UP |
| THILLAINADESAN_ZNF217_TARGETS_UP | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/THILLAINADESAN_ZNF217_TARGETS_UP |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_SUSTAINDED_IN_ERYTHROCYTE_UP | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_SUSTAINDED_IN_ERYTHROCYTE_UP |
| TURASHVILI_BREAST_DUCTAL_CARCINOMA_VS_DUCTAL_NORMAL_UP | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/TURASHVILI_BREAST_DUCTAL_CARCINOMA_VS_DUCTAL_NORMAL_UP |
| VANTVEER_BREAST_CANCER_BRCA1_DN | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/VANTVEER_BREAST_CANCER_BRCA1_DN |
| WANG_TARGETS_OF_MLL_CBP_FUSION_UP | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_TARGETS_OF_MLL_CBP_FUSION_UP |
| WHITFIELD_CELL_CYCLE_LITERATURE | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/WHITFIELD_CELL_CYCLE_LITERATURE |
| XU_CREBBP_TARGETS_DN | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_CREBBP_TARGETS_DN |
| ZHAN_MULTIPLE_MYELOMA_LB_DN | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_LB_DN |
| ZHAN_MULTIPLE_MYELOMA_PR_DN | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_PR_DN |
| ZUCCHI_METASTASIS_DN | 44 | http://www.broadinstitute.org/gsea/msigdb/cards/ZUCCHI_METASTASIS_DN |
| AMIT_EGF_RESPONSE_120_MCF10A | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_EGF_RESPONSE_120_MCF10A |
| AMIT_EGF_RESPONSE_480_MCF10A | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_EGF_RESPONSE_480_MCF10A |
| BURTON_ADIPOGENESIS_PEAK_AT_24HR | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/BURTON_ADIPOGENESIS_PEAK_AT_24HR |
| COWLING_MYCN_TARGETS | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/COWLING_MYCN_TARGETS |
| DORN_ADENOVIRUS_INFECTION_24HR_DN | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/DORN_ADENOVIRUS_INFECTION_24HR_DN |
| FUJII_YBX1_TARGETS_UP | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/FUJII_YBX1_TARGETS_UP |
| GALLUZZI_PERMEABILIZE_MITOCHONDRIA | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/GALLUZZI_PERMEABILIZE_MITOCHONDRIA |
| HOFFMANN_IMMATURE_TO_MATURE_B_LYMPHOCYTE_UP | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFFMANN_IMMATURE_TO_MATURE_B_LYMPHOCYTE_UP |
| HUMMEL_BURKITTS_LYMPHOMA_UP | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/HUMMEL_BURKITTS_LYMPHOMA_UP |
| IKEDA_MIR133_TARGETS_UP | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/IKEDA_MIR133_TARGETS_UP |
| JAZAERI_BREAST_CANCER_BRCA1_VS_BRCA2_DN | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/JAZAERI_BREAST_CANCER_BRCA1_VS_BRCA2_DN |
| LASTOWSKA_COAMPLIFIED_WITH_MYCN | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/LASTOWSKA_COAMPLIFIED_WITH_MYCN |
| LEE_CALORIE_RESTRICTION_MUSCLE_UP | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_CALORIE_RESTRICTION_MUSCLE_UP |
| LIU_TARGETS_OF_VMYB_VS_CMYB_DN | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_TARGETS_OF_VMYB_VS_CMYB_DN |
| MANTOVANI_NFKB_TARGETS_UP | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/MANTOVANI_NFKB_TARGETS_UP |
| MILI_PSEUDOPODIA | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/MILI_PSEUDOPODIA |
| NAKAMURA_METASTASIS_MODEL_DN | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAMURA_METASTASIS_MODEL_DN |
| NAKAYAMA_FRA2_TARGETS | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAYAMA_FRA2_TARGETS |
| PAPASPYRIDONOS_UNSTABLE_ATEROSCLEROTIC_PLAQUE_DN | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/PAPASPYRIDONOS_UNSTABLE_ATEROSCLEROTIC_PLAQUE_DN |
| PID_CXCR3PATHWAY | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_CXCR3PATHWAY |
| PID_INTEGRIN3_PATHWAY | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_INTEGRIN3_PATHWAY |
| PID_TCPTP_PATHWAY | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_TCPTP_PATHWAY |
| PID_THROMBIN_PAR1_PATHWAY | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_THROMBIN_PAR1_PATHWAY |
| POMEROY_MEDULLOBLASTOMA_PROGNOSIS_DN | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/POMEROY_MEDULLOBLASTOMA_PROGNOSIS_DN |
| POTTI_5FU_SENSITIVITY | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/POTTI_5FU_SENSITIVITY |
| POTTI_ETOPOSIDE_SENSITIVITY | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/POTTI_ETOPOSIDE_SENSITIVITY |
| REACTOME_IL_3_5_AND_GM_CSF_SIGNALING | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_IL_3_5_AND_GM_CSF_SIGNALING |
| REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK |
| REACTOME_PLC_BETA_MEDIATED_EVENTS | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PLC_BETA_MEDIATED_EVENTS |
| REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_GLUCAGON_LIKE_PEPTIDE1 | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_GLUCAGON_LIKE_PEPTIDE1 |
| REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS |
| RIZ_ERYTHROID_DIFFERENTIATION_12HR | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/RIZ_ERYTHROID_DIFFERENTIATION_12HR |
| ULE_SPLICING_VIA_NOVA2 | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/ULE_SPLICING_VIA_NOVA2 |
| ZUCCHI_METASTASIS_UP | 43 | http://www.broadinstitute.org/gsea/msigdb/cards/ZUCCHI_METASTASIS_UP |
| AMIT_EGF_RESPONSE_40_HELA | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_EGF_RESPONSE_40_HELA |
| BARRIER_COLON_CANCER_RECURRENCE_UP | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/BARRIER_COLON_CANCER_RECURRENCE_UP |
| BILBAN_B_CLL_LPL_DN | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/BILBAN_B_CLL_LPL_DN |
| BIOCARTA_CHREBP2_PATHWAY | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CHREBP2_PATHWAY |
| CHEN_LVAD_SUPPORT_OF_FAILING_HEART_DN | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEN_LVAD_SUPPORT_OF_FAILING_HEART_DN |
| FIGUEROA_AML_METHYLATION_CLUSTER_3_DN | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/FIGUEROA_AML_METHYLATION_CLUSTER_3_DN |
| HAHTOLA_SEZARY_SYNDROM_DN | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/HAHTOLA_SEZARY_SYNDROM_DN |
| KEGG_ALDOSTERONE_REGULATED_SODIUM_REABSORPTION | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ALDOSTERONE_REGULATED_SODIUM_REABSORPTION |
| KEGG_BLADDER_CANCER | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_BLADDER_CANCER |
| KEGG_FATTY_ACID_METABOLISM | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_FATTY_ACID_METABOLISM |
| KEGG_GRAFT_VERSUS_HOST_DISEASE | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GRAFT_VERSUS_HOST_DISEASE |
| KEGG_TYROSINE_METABOLISM | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_TYROSINE_METABOLISM |
| LEIN_ASTROCYTE_MARKERS | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/LEIN_ASTROCYTE_MARKERS |
| LUI_THYROID_CANCER_CLUSTER_2 | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/LUI_THYROID_CANCER_CLUSTER_2 |
| MAGRANGEAS_MULTIPLE_MYELOMA_IGLL_VS_IGLK_UP | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/MAGRANGEAS_MULTIPLE_MYELOMA_IGLL_VS_IGLK_UP |
| MCGARVEY_SILENCED_BY_METHYLATION_IN_COLON_CANCER | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/MCGARVEY_SILENCED_BY_METHYLATION_IN_COLON_CANCER |
| MIKKELSEN_ES_ICP_WITH_H3K27ME3 | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_ES_ICP_WITH_H3K27ME3 |
| MODY_HIPPOCAMPUS_PRENATAL | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/MODY_HIPPOCAMPUS_PRENATAL |
| ONO_FOXP3_TARGETS_DN | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/ONO_FOXP3_TARGETS_DN |
| PID_BMPPATHWAY | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_BMPPATHWAY |
| PID_ECADHERIN_STABILIZATION_PATHWAY | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ECADHERIN_STABILIZATION_PATHWAY |
| PID_UPA_UPAR_PATHWAY | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_UPA_UPAR_PATHWAY |
| REACTOME_BETA_DEFENSINS | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_BETA_DEFENSINS |
| REACTOME_TRNA_AMINOACYLATION | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRNA_AMINOACYLATION |
| RUAN_RESPONSE_TO_TNF_TROGLITAZONE_DN | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/RUAN_RESPONSE_TO_TNF_TROGLITAZONE_DN |
| SCHLINGEMANN_SKIN_CARCINOGENESIS_TPA_UP | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHLINGEMANN_SKIN_CARCINOGENESIS_TPA_UP |
| WEBER_METHYLATED_HCP_IN_FIBROBLAST_DN | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/WEBER_METHYLATED_HCP_IN_FIBROBLAST_DN |
| WIERENGA_PML_INTERACTOME | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/WIERENGA_PML_INTERACTOME |
| YU_MYC_TARGETS_UP | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/YU_MYC_TARGETS_UP |
| ZHAN_EARLY_DIFFERENTIATION_GENES_DN | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_EARLY_DIFFERENTIATION_GENES_DN |
| ZHENG_IL22_SIGNALING_DN | 42 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHENG_IL22_SIGNALING_DN |
| BIOCARTA_FCER1_PATHWAY | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_FCER1_PATHWAY |
| CHEN_HOXA5_TARGETS_9HR_DN | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEN_HOXA5_TARGETS_9HR_DN |
| CROONQUIST_NRAS_SIGNALING_UP | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/CROONQUIST_NRAS_SIGNALING_UP |
| CROONQUIST_NRAS_VS_STROMAL_STIMULATION_UP | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/CROONQUIST_NRAS_VS_STROMAL_STIMULATION_UP |
| DELPUECH_FOXO3_TARGETS_DN | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/DELPUECH_FOXO3_TARGETS_DN |
| EHLERS_ANEUPLOIDY_UP | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/EHLERS_ANEUPLOIDY_UP |
| ELVIDGE_HIF1A_AND_HIF2A_TARGETS_UP | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/ELVIDGE_HIF1A_AND_HIF2A_TARGETS_UP |
| FLECHNER_PBL_KIDNEY_TRANSPLANT_OK_VS_DONOR_DN | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/FLECHNER_PBL_KIDNEY_TRANSPLANT_OK_VS_DONOR_DN |
| GENTILE_UV_RESPONSE_CLUSTER_D2 | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/GENTILE_UV_RESPONSE_CLUSTER_D2 |
| KAAB_FAILED_HEART_VENTRICLE_DN | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/KAAB_FAILED_HEART_VENTRICLE_DN |
| KAMMINGA_EZH2_TARGETS | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/KAMMINGA_EZH2_TARGETS |
| KAMMINGA_SENESCENCE | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/KAMMINGA_SENESCENCE |
| KEGG_AMINOACYL_TRNA_BIOSYNTHESIS | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_AMINOACYL_TRNA_BIOSYNTHESIS |
| KEGG_PORPHYRIN_AND_CHLOROPHYLL_METABOLISM | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PORPHYRIN_AND_CHLOROPHYLL_METABOLISM |
| KORKOLA_EMBRYONAL_CARCINOMA_UP | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/KORKOLA_EMBRYONAL_CARCINOMA_UP |
| KUROKAWA_LIVER_CANCER_CHEMOTHERAPY_DN | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/KUROKAWA_LIVER_CANCER_CHEMOTHERAPY_DN |
| LIN_MELANOMA_COPY_NUMBER_DN | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/LIN_MELANOMA_COPY_NUMBER_DN |
| LI_LUNG_CANCER | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_LUNG_CANCER |
| MALONEY_RESPONSE_TO_17AAG_UP | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/MALONEY_RESPONSE_TO_17AAG_UP |
| MIKKELSEN_ES_HCP_WITH_H3K27ME3 | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_ES_HCP_WITH_H3K27ME3 |
| ONO_AML1_TARGETS_DN | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/ONO_AML1_TARGETS_DN |
| PID_AMB2_NEUTROPHILS_PATHWAY | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_AMB2_NEUTROPHILS_PATHWAY |
| PID_ERBB1_INTERNALIZATION_PATHWAY | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ERBB1_INTERNALIZATION_PATHWAY |
| PID_ER_NONGENOMIC_PATHWAY | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ER_NONGENOMIC_PATHWAY |
| REACTOME_IL_2_SIGNALING | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_IL_2_SIGNALING |
| REACTOME_NETRIN1_SIGNALING | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NETRIN1_SIGNALING |
| REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING |
| RIZ_ERYTHROID_DIFFERENTIATION_HBZ | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/RIZ_ERYTHROID_DIFFERENTIATION_HBZ |
| STEIN_ESRRA_TARGETS_RESPONSIVE_TO_ESTROGEN_DN | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/STEIN_ESRRA_TARGETS_RESPONSIVE_TO_ESTROGEN_DN |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_6HR_DN | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_6HR_DN |
| WEINMANN_ADAPTATION_TO_HYPOXIA_DN | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/WEINMANN_ADAPTATION_TO_HYPOXIA_DN |
| WENG_POR_TARGETS_LIVER_UP | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/WENG_POR_TARGETS_LIVER_UP |
| ZHAN_MULTIPLE_MYELOMA_DN | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_DN |
| ZHAN_MULTIPLE_MYELOMA_MF_DN | 41 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_MF_DN |
| AMUNDSON_GAMMA_RADIATION_RESPONSE | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/AMUNDSON_GAMMA_RADIATION_RESPONSE |
| BENPORATH_ES_2 | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/BENPORATH_ES_2 |
| BERNARD_PPAPDC1B_TARGETS_UP | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/BERNARD_PPAPDC1B_TARGETS_UP |
| BIOCARTA_P38MAPK_PATHWAY | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_P38MAPK_PATHWAY |
| BOHN_PRIMARY_IMMUNODEFICIENCY_SYNDROM_DN | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/BOHN_PRIMARY_IMMUNODEFICIENCY_SYNDROM_DN |
| BOYAULT_LIVER_CANCER_SUBCLASS_G1_DN | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYAULT_LIVER_CANCER_SUBCLASS_G1_DN |
| BOYLAN_MULTIPLE_MYELOMA_D_CLUSTER_DN | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYLAN_MULTIPLE_MYELOMA_D_CLUSTER_DN |
| BURTON_ADIPOGENESIS_PEAK_AT_16HR | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/BURTON_ADIPOGENESIS_PEAK_AT_16HR |
| CHANGOLKAR_H2AFY_TARGETS_DN | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/CHANGOLKAR_H2AFY_TARGETS_DN |
| CHESLER_BRAIN_HIGHEST_EXPRESSION | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/CHESLER_BRAIN_HIGHEST_EXPRESSION |
| DORN_ADENOVIRUS_INFECTION_48HR_DN | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/DORN_ADENOVIRUS_INFECTION_48HR_DN |
| ELLWOOD_MYC_TARGETS_DN | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/ELLWOOD_MYC_TARGETS_DN |
| GENTILE_UV_RESPONSE_CLUSTER_D7 | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/GENTILE_UV_RESPONSE_CLUSTER_D7 |
| GENTILE_UV_RESPONSE_CLUSTER_D8 | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/GENTILE_UV_RESPONSE_CLUSTER_D8 |
| GOBERT_CORE_OLIGODENDROCYTE_DIFFERENTIATION | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/GOBERT_CORE_OLIGODENDROCYTE_DIFFERENTIATION |
| HEIDENBLAD_AMPLICON_8Q24_UP | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/HEIDENBLAD_AMPLICON_8Q24_UP |
| ICHIBA_GRAFT_VERSUS_HOST_DISEASE_D7_DN | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/ICHIBA_GRAFT_VERSUS_HOST_DISEASE_D7_DN |
| JAERVINEN_AMPLIFIED_IN_LARYNGEAL_CANCER | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/JAERVINEN_AMPLIFIED_IN_LARYNGEAL_CANCER |
| JIANG_AGING_HYPOTHALAMUS_DN | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/JIANG_AGING_HYPOTHALAMUS_DN |
| KEGG_PYRUVATE_METABOLISM | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PYRUVATE_METABOLISM |
| KEGG_SPHINGOLIPID_METABOLISM | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_SPHINGOLIPID_METABOLISM |
| KEGG_TRYPTOPHAN_METABOLISM | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_TRYPTOPHAN_METABOLISM |
| KYNG_ENVIRONMENTAL_STRESS_RESPONSE_NOT_BY_4NQO_IN_WS | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_ENVIRONMENTAL_STRESS_RESPONSE_NOT_BY_4NQO_IN_WS |
| KYNG_RESPONSE_TO_H2O2_VIA_ERCC6_UP | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_RESPONSE_TO_H2O2_VIA_ERCC6_UP |
| LIAO_HAVE_SOX4_BINDING_SITES | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/LIAO_HAVE_SOX4_BINDING_SITES |
| MUELLER_METHYLATED_IN_GLIOBLASTOMA | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/MUELLER_METHYLATED_IN_GLIOBLASTOMA |
| PID_EPHBFWDPATHWAY | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_EPHBFWDPATHWAY |
| PID_FOXM1PATHWAY | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_FOXM1PATHWAY |
| PID_IFNGPATHWAY | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_IFNGPATHWAY |
| REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS |
| RIZ_ERYTHROID_DIFFERENTIATION_6HR | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/RIZ_ERYTHROID_DIFFERENTIATION_6HR |
| RIZ_ERYTHROID_DIFFERENTIATION_CCNE1 | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/RIZ_ERYTHROID_DIFFERENTIATION_CCNE1 |
| ST_B_CELL_ANTIGEN_RECEPTOR | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_B_CELL_ANTIGEN_RECEPTOR |
| ST_JNK_MAPK_PATHWAY | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_JNK_MAPK_PATHWAY |
| TOMLINS_PROSTATE_CANCER_DN | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/TOMLINS_PROSTATE_CANCER_DN |
| TOMLINS_PROSTATE_CANCER_UP | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/TOMLINS_PROSTATE_CANCER_UP |
| VALK_AML_WITH_FLT3_ITD | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_WITH_FLT3_ITD |
| VANASSE_BCL2_TARGETS_UP | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/VANASSE_BCL2_TARGETS_UP |
| VARELA_ZMPSTE24_TARGETS_UP | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/VARELA_ZMPSTE24_TARGETS_UP |
| YOSHIOKA_LIVER_CANCER_EARLY_RECURRENCE_UP | 40 | http://www.broadinstitute.org/gsea/msigdb/cards/YOSHIOKA_LIVER_CANCER_EARLY_RECURRENCE_UP |
| AIYAR_COBRA1_TARGETS_UP | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/AIYAR_COBRA1_TARGETS_UP |
| AMIT_EGF_RESPONSE_60_MCF10A | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_EGF_RESPONSE_60_MCF10A |
| AMIT_SERUM_RESPONSE_480_MCF10A | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_SERUM_RESPONSE_480_MCF10A |
| BEIER_GLIOMA_STEM_CELL_UP | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/BEIER_GLIOMA_STEM_CELL_UP |
| BILANGES_RAPAMYCIN_SENSITIVE_GENES | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/BILANGES_RAPAMYCIN_SENSITIVE_GENES |
| BILANGES_SERUM_SENSITIVE_VIA_TSC2 | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/BILANGES_SERUM_SENSITIVE_VIA_TSC2 |
| BIOCARTA_FMLP_PATHWAY | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_FMLP_PATHWAY |
| BOYAULT_LIVER_CANCER_SUBCLASS_G12_UP | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYAULT_LIVER_CANCER_SUBCLASS_G12_UP |
| BROWNE_HCMV_INFECTION_2HR_UP | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/BROWNE_HCMV_INFECTION_2HR_UP |
| BURTON_ADIPOGENESIS_PEAK_AT_8HR | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/BURTON_ADIPOGENESIS_PEAK_AT_8HR |
| CASORELLI_APL_SECONDARY_VS_DE_NOVO_UP | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/CASORELLI_APL_SECONDARY_VS_DE_NOVO_UP |
| CONRAD_STEM_CELL | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/CONRAD_STEM_CELL |
| DORN_ADENOVIRUS_INFECTION_32HR_DN | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/DORN_ADENOVIRUS_INFECTION_32HR_DN |
| GAZDA_DIAMOND_BLACKFAN_ANEMIA_PROGENITOR_UP | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/GAZDA_DIAMOND_BLACKFAN_ANEMIA_PROGENITOR_UP |
| GENTILE_UV_RESPONSE_CLUSTER_D5 | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/GENTILE_UV_RESPONSE_CLUSTER_D5 |
| GU_PDEF_TARGETS_DN | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/GU_PDEF_TARGETS_DN |
| HAN_JNK_SINGALING_DN | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/HAN_JNK_SINGALING_DN |
| HOFMANN_CELL_LYMPHOMA_DN | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFMANN_CELL_LYMPHOMA_DN |
| KORKOLA_TERATOMA | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/KORKOLA_TERATOMA |
| MATZUK_MEIOTIC_AND_DNA_REPAIR | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_MEIOTIC_AND_DNA_REPAIR |
| MA_MYELOID_DIFFERENTIATION_UP | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/MA_MYELOID_DIFFERENTIATION_UP |
| MCBRYAN_PUBERTAL_BREAST_3_4WK_DN | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/MCBRYAN_PUBERTAL_BREAST_3_4WK_DN |
| NEWMAN_ERCC6_TARGETS_DN | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/NEWMAN_ERCC6_TARGETS_DN |
| PID_ATR_PATHWAY | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ATR_PATHWAY |
| PID_AURORA_B_PATHWAY | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_AURORA_B_PATHWAY |
| PID_ECADHERIN_NASCENTAJ_PATHWAY | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ECADHERIN_NASCENTAJ_PATHWAY |
| PID_RET_PATHWAY | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_RET_PATHWAY |
| POTTI_PACLITAXEL_SENSITIVITY | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/POTTI_PACLITAXEL_SENSITIVITY |
| REACTOME_IL1_SIGNALING | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_IL1_SIGNALING |
| REACTOME_NCAM1_INTERACTIONS | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NCAM1_INTERACTIONS |
| SESTO_RESPONSE_TO_UV_C6 | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/SESTO_RESPONSE_TO_UV_C6 |
| SHIPP_DLBCL_CURED_VS_FATAL_UP | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/SHIPP_DLBCL_CURED_VS_FATAL_UP |
| SMID_BREAST_CANCER_RELAPSE_IN_BRAIN_UP | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_RELAPSE_IN_BRAIN_UP |
| STONER_ESOPHAGEAL_CARCINOGENESIS_UP | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/STONER_ESOPHAGEAL_CARCINOGENESIS_UP |
| VANDESLUIS_COMMD1_TARGETS_GROUP_3_DN | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/VANDESLUIS_COMMD1_TARGETS_GROUP_3_DN |
| VERRECCHIA_DELAYED_RESPONSE_TO_TGFB1 | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/VERRECCHIA_DELAYED_RESPONSE_TO_TGFB1 |
| WANG_LSD1_TARGETS_DN | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_LSD1_TARGETS_DN |
| WEIGEL_OXIDATIVE_STRESS_BY_HNE_AND_H2O2 | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/WEIGEL_OXIDATIVE_STRESS_BY_HNE_AND_H2O2 |
| WHITEHURST_PACLITAXEL_SENSITIVITY | 39 | http://www.broadinstitute.org/gsea/msigdb/cards/WHITEHURST_PACLITAXEL_SENSITIVITY |
| APPIERTO_RESPONSE_TO_FENRETINIDE_UP | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/APPIERTO_RESPONSE_TO_FENRETINIDE_UP |
| BIOCARTA_IL2RB_PATHWAY | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_IL2RB_PATHWAY |
| BIOCARTA_INTEGRIN_PATHWAY | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_INTEGRIN_PATHWAY |
| BOYLAN_MULTIPLE_MYELOMA_C_CLUSTER_UP | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYLAN_MULTIPLE_MYELOMA_C_CLUSTER_UP |
| BREDEMEYER_RAG_SIGNALING_VIA_ATM_NOT_VIA_NFKB_DN | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/BREDEMEYER_RAG_SIGNALING_VIA_ATM_NOT_VIA_NFKB_DN |
| CEBALLOS_TARGETS_OF_TP53_AND_MYC_DN | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/CEBALLOS_TARGETS_OF_TP53_AND_MYC_DN |
| CERVERA_SDHB_TARGETS_1_DN | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/CERVERA_SDHB_TARGETS_1_DN |
| CHANDRAN_METASTASIS_TOP50_UP | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/CHANDRAN_METASTASIS_TOP50_UP |
| FIGUEROA_AML_METHYLATION_CLUSTER_6_DN | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/FIGUEROA_AML_METHYLATION_CLUSTER_6_DN |
| GAUSSMANN_MLL_AF4_FUSION_TARGETS_D_UP | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/GAUSSMANN_MLL_AF4_FUSION_TARGETS_D_UP |
| GAZDA_DIAMOND_BLACKFAN_ANEMIA_MYELOID_DN | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/GAZDA_DIAMOND_BLACKFAN_ANEMIA_MYELOID_DN |
| GEISS_RESPONSE_TO_DSRNA_UP | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/GEISS_RESPONSE_TO_DSRNA_UP |
| GRADE_COLON_VS_RECTAL_CANCER_UP | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/GRADE_COLON_VS_RECTAL_CANCER_UP |
| HATADA_METHYLATED_IN_LUNG_CANCER_DN | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/HATADA_METHYLATED_IN_LUNG_CANCER_DN |
| HERNANDEZ_MITOTIC_ARREST_BY_DOCETAXEL_1_DN | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/HERNANDEZ_MITOTIC_ARREST_BY_DOCETAXEL_1_DN |
| HOLLEMAN_VINCRISTINE_RESISTANCE_B_ALL_UP | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLEMAN_VINCRISTINE_RESISTANCE_B_ALL_UP |
| KAAB_FAILED_HEART_ATRIUM_UP | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/KAAB_FAILED_HEART_ATRIUM_UP |
| KEEN_RESPONSE_TO_ROSIGLITAZONE_UP | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/KEEN_RESPONSE_TO_ROSIGLITAZONE_UP |
| KEGG_ALLOGRAFT_REJECTION | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ALLOGRAFT_REJECTION |
| KEGG_SNARE_INTERACTIONS_IN_VESICULAR_TRANSPORT | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_SNARE_INTERACTIONS_IN_VESICULAR_TRANSPORT |
| KRASNOSELSKAYA_ILF3_TARGETS_UP | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/KRASNOSELSKAYA_ILF3_TARGETS_UP |
| KYNG_DNA_DAMAGE_BY_4NQO | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_DNA_DAMAGE_BY_4NQO |
| LIANG_HEMATOPOIESIS_STEM_CELL_NUMBER_SMALL_VS_HUGE_UP | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/LIANG_HEMATOPOIESIS_STEM_CELL_NUMBER_SMALL_VS_HUGE_UP |
| LIU_LIVER_CANCER | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_LIVER_CANCER |
| MIKKELSEN_MEF_ICP_WITH_H3K4ME3_AND_H3K27ME3 | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_MEF_ICP_WITH_H3K4ME3_AND_H3K27ME3 |
| NAKAMURA_ADIPOGENESIS_EARLY_DN | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAMURA_ADIPOGENESIS_EARLY_DN |
| NAKAMURA_ADIPOGENESIS_LATE_DN | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAMURA_ADIPOGENESIS_LATE_DN |
| NIKOLSKY_BREAST_CANCER_1Q21_AMPLICON | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_1Q21_AMPLICON |
| NIKOLSKY_BREAST_CANCER_7P22_AMPLICON | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_7P22_AMPLICON |
| ONGUSAHA_TP53_TARGETS | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/ONGUSAHA_TP53_TARGETS |
| PENG_GLUTAMINE_DEPRIVATION_UP | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/PENG_GLUTAMINE_DEPRIVATION_UP |
| PID_ERBB4_PATHWAY | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ERBB4_PATHWAY |
| PID_FASPATHWAY | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_FASPATHWAY |
| PID_P38ALPHABETADOWNSTREAMPATHWAY | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_P38ALPHABETADOWNSTREAMPATHWAY |
| PID_RAC1_REG_PATHWAY | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_RAC1_REG_PATHWAY |
| PIONTEK_PKD1_TARGETS_UP | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/PIONTEK_PKD1_TARGETS_UP |
| REACTOME_ACTIVATION_OF_ATR_IN_RESPONSE_TO_REPLICATION_STRESS | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATION_OF_ATR_IN_RESPONSE_TO_REPLICATION_STRESS |
| REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_AMINE_LIGAND_BINDING_RECEPTORS |
| REACTOME_G1_PHASE | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_G1_PHASE |
| REACTOME_GAB1_SIGNALOSOME | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GAB1_SIGNALOSOME |
| REACTOME_GABA_B_RECEPTOR_ACTIVATION | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GABA_B_RECEPTOR_ACTIVATION |
| REACTOME_GLUCOSE_TRANSPORT | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GLUCOSE_TRANSPORT |
| REACTOME_GLYCOSPHINGOLIPID_METABOLISM | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GLYCOSPHINGOLIPID_METABOLISM |
| REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 |
| REACTOME_PI3K_AKT_ACTIVATION | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PI3K_AKT_ACTIVATION |
| REACTOME_PI3K_EVENTS_IN_ERBB4_SIGNALING | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PI3K_EVENTS_IN_ERBB4_SIGNALING |
| REACTOME_TRANSCRIPTIONAL_ACTIVITY_OF_SMAD2_SMAD3_SMAD4_HETEROTRIMER | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANSCRIPTIONAL_ACTIVITY_OF_SMAD2_SMAD3_SMAD4_HETEROTRIMER |
| REACTOME_TRIGLYCERIDE_BIOSYNTHESIS | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRIGLYCERIDE_BIOSYNTHESIS |
| SPIRA_SMOKERS_LUNG_CANCER_UP | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/SPIRA_SMOKERS_LUNG_CANCER_UP |
| TARTE_PLASMA_CELL_VS_B_LYMPHOCYTE_DN | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/TARTE_PLASMA_CELL_VS_B_LYMPHOCYTE_DN |
| TERAO_AOX4_TARGETS_SKIN_UP | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/TERAO_AOX4_TARGETS_SKIN_UP |
| VARELA_ZMPSTE24_TARGETS_DN | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/VARELA_ZMPSTE24_TARGETS_DN |
| YOKOE_CANCER_TESTIS_ANTIGENS | 38 | http://www.broadinstitute.org/gsea/msigdb/cards/YOKOE_CANCER_TESTIS_ANTIGENS |
| BILANGES_SERUM_RESPONSE_TRANSLATION | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/BILANGES_SERUM_RESPONSE_TRANSLATION |
| BIOCARTA_ALK_PATHWAY | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_ALK_PATHWAY |
| BIOCARTA_BCR_PATHWAY | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_BCR_PATHWAY |
| BIOCARTA_GPCR_PATHWAY | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_GPCR_PATHWAY |
| BIOCARTA_MET_PATHWAY | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_MET_PATHWAY |
| BIOCARTA_PAR1_PATHWAY | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_PAR1_PATHWAY |
| BIOCARTA_TOLL_PATHWAY | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_TOLL_PATHWAY |
| CHESLER_BRAIN_HIGHEST_GENETIC_VARIANCE | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/CHESLER_BRAIN_HIGHEST_GENETIC_VARIANCE |
| CUI_TCF21_TARGETS_UP | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/CUI_TCF21_TARGETS_UP |
| FRASOR_RESPONSE_TO_ESTRADIOL_UP | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/FRASOR_RESPONSE_TO_ESTRADIOL_UP |
| GENTILE_UV_RESPONSE_CLUSTER_D6 | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/GENTILE_UV_RESPONSE_CLUSTER_D6 |
| HOWLIN_CITED1_TARGETS_1_DN | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/HOWLIN_CITED1_TARGETS_1_DN |
| HU_ANGIOGENESIS_DN | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/HU_ANGIOGENESIS_DN |
| HU_GENOTOXIN_ACTION_DIRECT_VS_INDIRECT_4HR | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/HU_GENOTOXIN_ACTION_DIRECT_VS_INDIRECT_4HR |
| KIM_ALL_DISORDERS_CALB1_CORR_DN | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_ALL_DISORDERS_CALB1_CORR_DN |
| LANDIS_ERBB2_BREAST_TUMORS_65_DN | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/LANDIS_ERBB2_BREAST_TUMORS_65_DN |
| LEIN_LOCALIZED_TO_PROXIMAL_DENDRITES | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/LEIN_LOCALIZED_TO_PROXIMAL_DENDRITES |
| MATZUK_SPERMATID_DIFFERENTIATION | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_SPERMATID_DIFFERENTIATION |
| MEISSNER_BRAIN_HCP_WITH_H3_UNMETHYLATED | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/MEISSNER_BRAIN_HCP_WITH_H3_UNMETHYLATED |
| MURAKAMI_UV_RESPONSE_6HR_UP | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/MURAKAMI_UV_RESPONSE_6HR_UP |
| PID_FRA_PATHWAY | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_FRA_PATHWAY |
| PID_GMCSF_PATHWAY | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_GMCSF_PATHWAY |
| PID_IL23PATHWAY | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_IL23PATHWAY |
| QI_HYPOXIA_TARGETS_OF_HIF1A_AND_FOXA2 | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/QI_HYPOXIA_TARGETS_OF_HIF1A_AND_FOXA2 |
| REACTOME_ACTIVATION_OF_NMDA_RECEPTOR_UPON_GLUTAMATE_BINDING_AND_POSTSYNAPTIC_EVENTS | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATION_OF_NMDA_RECEPTOR_UPON_GLUTAMATE_BINDING_AND_POSTSYNAPTIC_EVENTS |
| REACTOME_DOWNSTREAM_TCR_SIGNALING | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DOWNSTREAM_TCR_SIGNALING |
| REACTOME_NEGATIVE_REGULATION_OF_FGFR_SIGNALING | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NEGATIVE_REGULATION_OF_FGFR_SIGNALING |
| RICKMAN_HEAD_AND_NECK_CANCER_D | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/RICKMAN_HEAD_AND_NECK_CANCER_D |
| ROLEF_GLIS3_TARGETS | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/ROLEF_GLIS3_TARGETS |
| SMID_BREAST_CANCER_RELAPSE_IN_LUNG_DN | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_RELAPSE_IN_LUNG_DN |
| SNIJDERS_AMPLIFIED_IN_HEAD_AND_NECK_TUMORS | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/SNIJDERS_AMPLIFIED_IN_HEAD_AND_NECK_TUMORS |
| ST_GA13_PATHWAY | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_GA13_PATHWAY |
| ST_P38_MAPK_PATHWAY | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_P38_MAPK_PATHWAY |
| ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY |
| UZONYI_RESPONSE_TO_LEUKOTRIENE_AND_THROMBIN | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/UZONYI_RESPONSE_TO_LEUKOTRIENE_AND_THROMBIN |
| VALK_AML_WITH_CEBPA | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_WITH_CEBPA |
| WANG_BARRETTS_ESOPHAGUS_AND_ESOPHAGUS_CANCER_DN | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_BARRETTS_ESOPHAGUS_AND_ESOPHAGUS_CANCER_DN |
| ZHENG_FOXP3_TARGETS_IN_T_LYMPHOCYTE_DN | 37 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHENG_FOXP3_TARGETS_IN_T_LYMPHOCYTE_DN |
| ALONSO_METASTASIS_EMT_UP | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/ALONSO_METASTASIS_EMT_UP |
| BIOCARTA_AGR_PATHWAY | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_AGR_PATHWAY |
| BIOCARTA_AT1R_PATHWAY | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_AT1R_PATHWAY |
| BURTON_ADIPOGENESIS_12 | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/BURTON_ADIPOGENESIS_12 |
| CLASPER_LYMPHATIC_VESSELS_DURING_METASTASIS_DN | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/CLASPER_LYMPHATIC_VESSELS_DURING_METASTASIS_DN |
| DACOSTA_ERCC3_ALLELE_XPCS_VS_TTD_DN | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/DACOSTA_ERCC3_ALLELE_XPCS_VS_TTD_DN |
| FONTAINE_THYROID_TUMOR_UNCERTAIN_MALIGNANCY_UP | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/FONTAINE_THYROID_TUMOR_UNCERTAIN_MALIGNANCY_UP |
| HANN_RESISTANCE_TO_BCL2_INHIBITOR_UP | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/HANN_RESISTANCE_TO_BCL2_INHIBITOR_UP |
| HERNANDEZ_MITOTIC_ARREST_BY_DOCETAXEL_1_UP | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/HERNANDEZ_MITOTIC_ARREST_BY_DOCETAXEL_1_UP |
| HOFFMANN_PRE_BI_TO_LARGE_PRE_BII_LYMPHOCYTE_UP | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFFMANN_PRE_BI_TO_LARGE_PRE_BII_LYMPHOCYTE_UP |
| HOUSTIS_ROS | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/HOUSTIS_ROS |
| HUANG_FOXA2_TARGETS_DN | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/HUANG_FOXA2_TARGETS_DN |
| JIANG_AGING_CEREBRAL_CORTEX_UP | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/JIANG_AGING_CEREBRAL_CORTEX_UP |
| KEGG_BASAL_TRANSCRIPTION_FACTORS | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_BASAL_TRANSCRIPTION_FACTORS |
| KEGG_DNA_REPLICATION | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_DNA_REPLICATION |
| KIM_ALL_DISORDERS_OLIGODENDROCYTE_NUMBER_CORR_DN | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_ALL_DISORDERS_OLIGODENDROCYTE_NUMBER_CORR_DN |
| LIU_CDX2_TARGETS_UP | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_CDX2_TARGETS_UP |
| MARZEC_IL2_SIGNALING_DN | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/MARZEC_IL2_SIGNALING_DN |
| MATTIOLI_MULTIPLE_MYELOMA_WITH_14Q32_TRANSLOCATIONS | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/MATTIOLI_MULTIPLE_MYELOMA_WITH_14Q32_TRANSLOCATIONS |
| MMS_MOUSE_LYMPH_HIGH_4HRS_UP | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/MMS_MOUSE_LYMPH_HIGH_4HRS_UP |
| PID_PI3KPLCTRKPATHWAY | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_PI3KPLCTRKPATHWAY |
| REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION |
| REACTOME_FRS2_MEDIATED_CASCADE | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FRS2_MEDIATED_CASCADE |
| REACTOME_IRON_UPTAKE_AND_TRANSPORT | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_IRON_UPTAKE_AND_TRANSPORT |
| REACTOME_PLATELET_AGGREGATION_PLUG_FORMATION | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PLATELET_AGGREGATION_PLUG_FORMATION |
| REACTOME_SIGNALLING_TO_ERKS | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALLING_TO_ERKS |
| SAKAI_CHRONIC_HEPATITIS_VS_LIVER_CANCER_DN | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/SAKAI_CHRONIC_HEPATITIS_VS_LIVER_CANCER_DN |
| SEMENZA_HIF1_TARGETS | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/SEMENZA_HIF1_TARGETS |
| SHAFFER_IRF4_MULTIPLE_MYELOMA_PROGRAM | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/SHAFFER_IRF4_MULTIPLE_MYELOMA_PROGRAM |
| SHIN_B_CELL_LYMPHOMA_CLUSTER_8 | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/SHIN_B_CELL_LYMPHOMA_CLUSTER_8 |
| SIG_PIP3_SIGNALING_IN_B_LYMPHOCYTES | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/SIG_PIP3_SIGNALING_IN_B_LYMPHOCYTES |
| STEARMAN_TUMOR_FIELD_EFFECT_UP | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/STEARMAN_TUMOR_FIELD_EFFECT_UP |
| ST_ADRENERGIC | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_ADRENERGIC |
| SUBTIL_PROGESTIN_TARGETS | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/SUBTIL_PROGESTIN_TARGETS |
| VALK_AML_CLUSTER_11 | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_CLUSTER_11 |
| VANHARANTA_UTERINE_FIBROID_WITH_7Q_DELETION_DN | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/VANHARANTA_UTERINE_FIBROID_WITH_7Q_DELETION_DN |
| VANTVEER_BREAST_CANCER_BRCA1_UP | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/VANTVEER_BREAST_CANCER_BRCA1_UP |
| WEIGEL_OXIDATIVE_STRESS_BY_TBH_AND_H2O2 | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/WEIGEL_OXIDATIVE_STRESS_BY_TBH_AND_H2O2 |
| YANG_BREAST_CANCER_ESR1_UP | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/YANG_BREAST_CANCER_ESR1_UP |
| YIH_RESPONSE_TO_ARSENITE_C3 | 36 | http://www.broadinstitute.org/gsea/msigdb/cards/YIH_RESPONSE_TO_ARSENITE_C3 |
| AIGNER_ZEB1_TARGETS | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/AIGNER_ZEB1_TARGETS |
| BALDWIN_PRKCI_TARGETS_UP | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/BALDWIN_PRKCI_TARGETS_UP |
| BIOCARTA_CARM_ER_PATHWAY | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CARM_ER_PATHWAY |
| CHEMELLO_SOLEUS_VS_EDL_MYOFIBERS_UP | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEMELLO_SOLEUS_VS_EDL_MYOFIBERS_UP |
| COLIN_PILOCYTIC_ASTROCYTOMA_VS_GLIOBLASTOMA_UP | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/COLIN_PILOCYTIC_ASTROCYTOMA_VS_GLIOBLASTOMA_UP |
| DORSAM_HOXA9_TARGETS_UP | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/DORSAM_HOXA9_TARGETS_UP |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_BLACK_UP | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_BLACK_UP |
| GAUSSMANN_MLL_AF4_FUSION_TARGETS_G_DN | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/GAUSSMANN_MLL_AF4_FUSION_TARGETS_G_DN |
| GUTIERREZ_MULTIPLE_MYELOMA_DN | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/GUTIERREZ_MULTIPLE_MYELOMA_DN |
| HAN_JNK_SINGALING_UP | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/HAN_JNK_SINGALING_UP |
| HOWLIN_CITED1_TARGETS_1_UP | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/HOWLIN_CITED1_TARGETS_1_UP |
| HU_GENOTOXIC_DAMAGE_24HR | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/HU_GENOTOXIC_DAMAGE_24HR |
| HU_GENOTOXIC_DAMAGE_4HR | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/HU_GENOTOXIC_DAMAGE_4HR |
| JAZAG_TGFB1_SIGNALING_DN | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/JAZAG_TGFB1_SIGNALING_DN |
| KEGG_BASE_EXCISION_REPAIR | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_BASE_EXCISION_REPAIR |
| KEGG_PRIMARY_IMMUNODEFICIENCY | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PRIMARY_IMMUNODEFICIENCY |
| KEGG_PRION_DISEASES | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PRION_DISEASES |
| KEGG_REGULATION_OF_AUTOPHAGY | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_REGULATION_OF_AUTOPHAGY |
| KUUSELO_PANCREATIC_CANCER_19Q13_AMPLIFICATION | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/KUUSELO_PANCREATIC_CANCER_19Q13_AMPLIFICATION |
| LIEN_BREAST_CARCINOMA_METAPLASTIC | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/LIEN_BREAST_CARCINOMA_METAPLASTIC |
| LI_CISPLATIN_RESISTANCE_DN | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_CISPLATIN_RESISTANCE_DN |
| LOPEZ_TRANSLATION_VIA_FN1_SIGNALING | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/LOPEZ_TRANSLATION_VIA_FN1_SIGNALING |
| MODY_HIPPOCAMPUS_NEONATAL | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/MODY_HIPPOCAMPUS_NEONATAL |
| OUELLET_CULTURED_OVARIAN_CANCER_INVASIVE_VS_LMP_DN | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/OUELLET_CULTURED_OVARIAN_CANCER_INVASIVE_VS_LMP_DN |
| PACHER_TARGETS_OF_IGF1_AND_IGF2_UP | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/PACHER_TARGETS_OF_IGF1_AND_IGF2_UP |
| PID_ARF6_PATHWAY | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ARF6_PATHWAY |
| PID_ERBB1_RECEPTOR_PROXIMAL_PATHWAY | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ERBB1_RECEPTOR_PROXIMAL_PATHWAY |
| PID_HIVNEFPATHWAY | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_HIVNEFPATHWAY |
| PID_PI3KCIAKTPATHWAY | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_PI3KCIAKTPATHWAY |
| REACTOME_E2F_MEDIATED_REGULATION_OF_DNA_REPLICATION | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_E2F_MEDIATED_REGULATION_OF_DNA_REPLICATION |
| REACTOME_GLOBAL_GENOMIC_NER_GG_NER | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GLOBAL_GENOMIC_NER_GG_NER |
| REACTOME_METABOLISM_OF_STEROID_HORMONES_AND_VITAMINS_A_AND_D | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_METABOLISM_OF_STEROID_HORMONES_AND_VITAMINS_A_AND_D |
| REACTOME_MRNA_3_END_PROCESSING | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MRNA_3_END_PROCESSING |
| RODRIGUES_NTN1_AND_DCC_TARGETS | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/RODRIGUES_NTN1_AND_DCC_TARGETS |
| ROZANOV_MMP14_TARGETS_DN | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/ROZANOV_MMP14_TARGETS_DN |
| SCHRAETS_MLL_TARGETS_UP | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHRAETS_MLL_TARGETS_UP |
| SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES |
| ST_G_ALPHA_I_PATHWAY | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_G_ALPHA_I_PATHWAY |
| VALK_AML_CLUSTER_9 | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_CLUSTER_9 |
| WANG_METHYLATED_IN_BREAST_CANCER | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_METHYLATED_IN_BREAST_CANCER |
| WEIGEL_OXIDATIVE_STRESS_RESPONSE | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/WEIGEL_OXIDATIVE_STRESS_RESPONSE |
| WESTON_VEGFA_TARGETS_12HR | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/WESTON_VEGFA_TARGETS_12HR |
| XU_HGF_SIGNALING_NOT_VIA_AKT1_48HR_UP | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_HGF_SIGNALING_NOT_VIA_AKT1_48HR_UP |
| YAGI_AML_RELAPSE_PROGNOSIS | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/YAGI_AML_RELAPSE_PROGNOSIS |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_15 | 35 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_15 |
| ABE_VEGFA_TARGETS_2HR | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/ABE_VEGFA_TARGETS_2HR |
| BIOCARTA_MPR_PATHWAY | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_MPR_PATHWAY |
| CAFFAREL_RESPONSE_TO_THC_24HR_5_UP | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/CAFFAREL_RESPONSE_TO_THC_24HR_5_UP |
| CREIGHTON_AKT1_SIGNALING_VIA_MTOR_UP | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/CREIGHTON_AKT1_SIGNALING_VIA_MTOR_UP |
| DOANE_BREAST_CANCER_CLASSES_DN | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/DOANE_BREAST_CANCER_CLASSES_DN |
| FRIDMAN_IMMORTALIZATION_DN | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/FRIDMAN_IMMORTALIZATION_DN |
| HONMA_DOCETAXEL_RESISTANCE | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/HONMA_DOCETAXEL_RESISTANCE |
| JIANG_HYPOXIA_VIA_VHL | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/JIANG_HYPOXIA_VIA_VHL |
| KEGG_BUTANOATE_METABOLISM | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_BUTANOATE_METABOLISM |
| KEGG_CYSTEINE_AND_METHIONINE_METABOLISM | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_CYSTEINE_AND_METHIONINE_METABOLISM |
| KEGG_FRUCTOSE_AND_MANNOSE_METABOLISM | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_FRUCTOSE_AND_MANNOSE_METABOLISM |
| KORKOLA_CORRELATED_WITH_POU5F1 | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/KORKOLA_CORRELATED_WITH_POU5F1 |
| LUI_TARGETS_OF_PAX8_PPARG_FUSION | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/LUI_TARGETS_OF_PAX8_PPARG_FUSION |
| MEISSNER_ES_ICP_WITH_H3K4ME3 | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/MEISSNER_ES_ICP_WITH_H3K4ME3 |
| MIKKELSEN_MCV6_ICP_WITH_H3K4ME3_AND_H3K27ME3 | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_MCV6_ICP_WITH_H3K4ME3_AND_H3K27ME3 |
| PETRETTO_CARDIAC_HYPERTROPHY | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/PETRETTO_CARDIAC_HYPERTROPHY |
| PID_ATM_PATHWAY | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ATM_PATHWAY |
| PID_EPHA_FWDPATHWAY | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_EPHA_FWDPATHWAY |
| PID_EPOPATHWAY | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_EPOPATHWAY |
| PID_HDAC_CLASSII_PATHWAY | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_HDAC_CLASSII_PATHWAY |
| PID_HIF2PATHWAY | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_HIF2PATHWAY |
| PID_IL1PATHWAY | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_IL1PATHWAY |
| PID_IL2_PI3KPATHWAY | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_IL2_PI3KPATHWAY |
| PID_IL8CXCR2_PATHWAY | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_IL8CXCR2_PATHWAY |
| PID_MAPKTRKPATHWAY | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_MAPKTRKPATHWAY |
| PLASARI_TGFB1_TARGETS_1HR_UP | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/PLASARI_TGFB1_TARGETS_1HR_UP |
| POTTI_CYTOXAN_SENSITIVITY | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/POTTI_CYTOXAN_SENSITIVITY |
| REACTOME_ABC_FAMILY_PROTEINS_MEDIATED_TRANSPORT | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ABC_FAMILY_PROTEINS_MEDIATED_TRANSPORT |
| REACTOME_FORMATION_OF_THE_HIV1_EARLY_ELONGATION_COMPLEX | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FORMATION_OF_THE_HIV1_EARLY_ELONGATION_COMPLEX |
| REACTOME_GLUCAGON_SIGNALING_IN_METABOLIC_REGULATION | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GLUCAGON_SIGNALING_IN_METABOLIC_REGULATION |
| REACTOME_GLUCONEOGENESIS | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GLUCONEOGENESIS |
| REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES |
| REACTOME_NEUROTRANSMITTER_RELEASE_CYCLE | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NEUROTRANSMITTER_RELEASE_CYCLE |
| REACTOME_SIGNAL_TRANSDUCTION_BY_L1 | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNAL_TRANSDUCTION_BY_L1 |
| SARTIPY_NORMAL_AT_INSULIN_RESISTANCE_UP | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/SARTIPY_NORMAL_AT_INSULIN_RESISTANCE_UP |
| SCHOEN_NFKB_SIGNALING | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHOEN_NFKB_SIGNALING |
| SIG_CD40PATHWAYMAP | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/SIG_CD40PATHWAYMAP |
| ST_WNT_BETA_CATENIN_PATHWAY | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_WNT_BETA_CATENIN_PATHWAY |
| SUMI_HNF4A_TARGETS | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/SUMI_HNF4A_TARGETS |
| WANG_RESPONSE_TO_BEXAROTENE_UP | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_RESPONSE_TO_BEXAROTENE_UP |
| WONG_IFNA2_RESISTANCE_DN | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/WONG_IFNA2_RESISTANCE_DN |
| YANG_BREAST_CANCER_ESR1_LASER_UP | 34 | http://www.broadinstitute.org/gsea/msigdb/cards/YANG_BREAST_CANCER_ESR1_LASER_UP |
| APPEL_IMATINIB_RESPONSE | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/APPEL_IMATINIB_RESPONSE |
| BANDRES_RESPONSE_TO_CARMUSTIN_MGMT_24HR_DN | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/BANDRES_RESPONSE_TO_CARMUSTIN_MGMT_24HR_DN |
| BIOCARTA_DEATH_PATHWAY | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_DEATH_PATHWAY |
| BIOCARTA_IL1R_PATHWAY | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_IL1R_PATHWAY |
| BIOCARTA_NO1_PATHWAY | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_NO1_PATHWAY |
| BOGNI_TREATMENT_RELATED_MYELOID_LEUKEMIA_DN | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/BOGNI_TREATMENT_RELATED_MYELOID_LEUKEMIA_DN |
| BURTON_ADIPOGENESIS_1 | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/BURTON_ADIPOGENESIS_1 |
| CAFFAREL_RESPONSE_TO_THC_UP | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/CAFFAREL_RESPONSE_TO_THC_UP |
| CHO_NR4A1_TARGETS | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/CHO_NR4A1_TARGETS |
| DORN_ADENOVIRUS_INFECTION_12HR_DN | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/DORN_ADENOVIRUS_INFECTION_12HR_DN |
| DUTTA_APOPTOSIS_VIA_NFKB | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/DUTTA_APOPTOSIS_VIA_NFKB |
| FARMER_BREAST_CANCER_CLUSTER_2 | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/FARMER_BREAST_CANCER_CLUSTER_2 |
| FIRESTEIN_CTNNB1_PATHWAY | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/FIRESTEIN_CTNNB1_PATHWAY |
| GAUSSMANN_MLL_AF4_FUSION_TARGETS_F_DN | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/GAUSSMANN_MLL_AF4_FUSION_TARGETS_F_DN |
| GHANDHI_DIRECT_IRRADIATION_DN | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/GHANDHI_DIRECT_IRRADIATION_DN |
| GRADE_COLON_CANCER_DN | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/GRADE_COLON_CANCER_DN |
| GREENBAUM_E2A_TARGETS_UP | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/GREENBAUM_E2A_TARGETS_UP |
| GROSS_HYPOXIA_VIA_ELK3_ONLY_UP | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/GROSS_HYPOXIA_VIA_ELK3_ONLY_UP |
| HEIDENBLAD_AMPLICON_12P11_12_UP | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/HEIDENBLAD_AMPLICON_12P11_12_UP |
| JEPSEN_SMRT_TARGETS | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/JEPSEN_SMRT_TARGETS |
| KEGG_ETHER_LIPID_METABOLISM | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ETHER_LIPID_METABOLISM |
| KEGG_PROPANOATE_METABOLISM | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PROPANOATE_METABOLISM |
| KYNG_ENVIRONMENTAL_STRESS_RESPONSE_NOT_BY_GAMMA_IN_WS | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_ENVIRONMENTAL_STRESS_RESPONSE_NOT_BY_GAMMA_IN_WS |
| LIANG_HEMATOPOIESIS_STEM_CELL_NUMBER_SMALL_VS_HUGE_DN | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/LIANG_HEMATOPOIESIS_STEM_CELL_NUMBER_SMALL_VS_HUGE_DN |
| MORI_PLASMA_CELL_DN | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/MORI_PLASMA_CELL_DN |
| PEPPER_CHRONIC_LYMPHOCYTIC_LEUKEMIA_UP | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/PEPPER_CHRONIC_LYMPHOCYTIC_LEUKEMIA_UP |
| PID_ALPHASYNUCLEIN_PATHWAY | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ALPHASYNUCLEIN_PATHWAY |
| PID_IL12_STAT4PATHWAY | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_IL12_STAT4PATHWAY |
| PID_INTEGRIN_A4B1_PATHWAY | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_INTEGRIN_A4B1_PATHWAY |
| PID_NCADHERINPATHWAY | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_NCADHERINPATHWAY |
| PID_SYNDECAN_2_PATHWAY | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_SYNDECAN_2_PATHWAY |
| PLASARI_TGFB1_SIGNALING_VIA_NFIC_1HR_UP | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/PLASARI_TGFB1_SIGNALING_VIA_NFIC_1HR_UP |
| REACTOME_GLUCAGON_TYPE_LIGAND_RECEPTORS | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GLUCAGON_TYPE_LIGAND_RECEPTORS |
| REACTOME_INTERACTIONS_OF_VPR_WITH_HOST_CELLULAR_PROTEINS | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INTERACTIONS_OF_VPR_WITH_HOST_CELLULAR_PROTEINS |
| REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS |
| REACTOME_POST_NMDA_RECEPTOR_ACTIVATION_EVENTS | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_POST_NMDA_RECEPTOR_ACTIVATION_EVENTS |
| REACTOME_PURINE_METABOLISM | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PURINE_METABOLISM |
| REACTOME_RNA_POL_III_TRANSCRIPTION | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RNA_POL_III_TRANSCRIPTION |
| REACTOME_TRANSPORT_OF_MATURE_MRNA_DERIVED_FROM_AN_INTRONLESS_TRANSCRIPT | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANSPORT_OF_MATURE_MRNA_DERIVED_FROM_AN_INTRONLESS_TRANSCRIPT |
| REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION |
| ROZANOV_MMP14_TARGETS_SUBSET | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/ROZANOV_MMP14_TARGETS_SUBSET |
| SCHRAETS_MLL_TARGETS_DN | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHRAETS_MLL_TARGETS_DN |
| SIMBULAN_UV_RESPONSE_NORMAL_DN | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/SIMBULAN_UV_RESPONSE_NORMAL_DN |
| STAEGE_EWING_FAMILY_TUMOR | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/STAEGE_EWING_FAMILY_TUMOR |
| TANG_SENESCENCE_TP53_TARGETS_UP | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/TANG_SENESCENCE_TP53_TARGETS_UP |
| VALK_AML_CLUSTER_10 | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_CLUSTER_10 |
| VALK_AML_CLUSTER_3 | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_CLUSTER_3 |
| VALK_AML_CLUSTER_5 | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_CLUSTER_5 |
| VALK_AML_CLUSTER_6 | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_CLUSTER_6 |
| WENDT_COHESIN_TARGETS_UP | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/WENDT_COHESIN_TARGETS_UP |
| ZHAN_LATE_DIFFERENTIATION_GENES_UP | 33 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_LATE_DIFFERENTIATION_GENES_UP |
| AMIT_SERUM_RESPONSE_40_MCF10A | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_SERUM_RESPONSE_40_MCF10A |
| BAE_BRCA1_TARGETS_DN | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/BAE_BRCA1_TARGETS_DN |
| BANDRES_RESPONSE_TO_CARMUSTIN_WITHOUT_MGMT_48HR_DN | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/BANDRES_RESPONSE_TO_CARMUSTIN_WITHOUT_MGMT_48HR_DN |
| BARRIER_CANCER_RELAPSE_NORMAL_SAMPLE_UP | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/BARRIER_CANCER_RELAPSE_NORMAL_SAMPLE_UP |
| BAUS_TFF2_TARGETS_UP | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/BAUS_TFF2_TARGETS_UP |
| BIOCARTA_HDAC_PATHWAY | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_HDAC_PATHWAY |
| BIOCARTA_PDGF_PATHWAY | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_PDGF_PATHWAY |
| BIOCARTA_RHO_PATHWAY | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_RHO_PATHWAY |
| BOYLAN_MULTIPLE_MYELOMA_C_CLUSTER_DN | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYLAN_MULTIPLE_MYELOMA_C_CLUSTER_DN |
| CAVARD_LIVER_CANCER_MALIGNANT_VS_BENIGN | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/CAVARD_LIVER_CANCER_MALIGNANT_VS_BENIGN |
| DARWICHE_PAPILLOMA_RISK_HIGH_VS_LOW_DN | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/DARWICHE_PAPILLOMA_RISK_HIGH_VS_LOW_DN |
| DIRMEIER_LMP1_RESPONSE_LATE_DN | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/DIRMEIER_LMP1_RESPONSE_LATE_DN |
| DORSAM_HOXA9_TARGETS_DN | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/DORSAM_HOXA9_TARGETS_DN |
| FARDIN_HYPOXIA_11 | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/FARDIN_HYPOXIA_11 |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_YELLOW_UP | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_YELLOW_UP |
| GROSS_ELK3_TARGETS_DN | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/GROSS_ELK3_TARGETS_DN |
| IVANOVA_HEMATOPOIESIS_STEM_CELL_SHORT_TERM | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/IVANOVA_HEMATOPOIESIS_STEM_CELL_SHORT_TERM |
| KASLER_HDAC7_TARGETS_2_DN | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/KASLER_HDAC7_TARGETS_2_DN |
| KEGG_ALANINE_ASPARTATE_AND_GLUTAMATE_METABOLISM | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ALANINE_ASPARTATE_AND_GLUTAMATE_METABOLISM |
| KEGG_CITRATE_CYCLE_TCA_CYCLE | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_CITRATE_CYCLE_TCA_CYCLE |
| KIM_TIAL1_TARGETS | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_TIAL1_TARGETS |
| LIANG_SILENCED_BY_METHYLATION_UP | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/LIANG_SILENCED_BY_METHYLATION_UP |
| MEISSNER_BRAIN_ICP_WITH_H3K4ME3 | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/MEISSNER_BRAIN_ICP_WITH_H3K4ME3 |
| MOOTHA_GLUCONEOGENESIS | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/MOOTHA_GLUCONEOGENESIS |
| NEMETH_INFLAMMATORY_RESPONSE_LPS_DN | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/NEMETH_INFLAMMATORY_RESPONSE_LPS_DN |
| PID_NETRIN_PATHWAY | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_NETRIN_PATHWAY |
| PID_SYNDECAN_4_PATHWAY | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_SYNDECAN_4_PATHWAY |
| PID_WNT_NONCANONICAL_PATHWAY | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_WNT_NONCANONICAL_PATHWAY |
| POS_HISTAMINE_RESPONSE_NETWORK | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/POS_HISTAMINE_RESPONSE_NETWORK |
| REACTOME_CD28_CO_STIMULATION | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CD28_CO_STIMULATION |
| REACTOME_COMPLEMENT_CASCADE | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_COMPLEMENT_CASCADE |
| REACTOME_DAG_AND_IP3_SIGNALING | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DAG_AND_IP3_SIGNALING |
| REACTOME_ELONGATION_ARREST_AND_RECOVERY | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ELONGATION_ARREST_AND_RECOVERY |
| REACTOME_FORMATION_OF_FIBRIN_CLOT_CLOTTING_CASCADE | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FORMATION_OF_FIBRIN_CLOT_CLOTTING_CASCADE |
| REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING |
| REACTOME_THROMBIN_SIGNALLING_THROUGH_PROTEINASE_ACTIVATED_RECEPTORS_PARS | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_THROMBIN_SIGNALLING_THROUGH_PROTEINASE_ACTIVATED_RECEPTORS_PARS |
| SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX4_DN | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX4_DN |
| SCHLOSSER_MYC_AND_SERUM_RESPONSE_SYNERGY | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHLOSSER_MYC_AND_SERUM_RESPONSE_SYNERGY |
| ST_ERK1_ERK2_MAPK_PATHWAY | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_ERK1_ERK2_MAPK_PATHWAY |
| TRACEY_RESISTANCE_TO_IFNA2_DN | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/TRACEY_RESISTANCE_TO_IFNA2_DN |
| TSAI_RESPONSE_TO_RADIATION_THERAPY | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/TSAI_RESPONSE_TO_RADIATION_THERAPY |
| ZHAN_V1_LATE_DIFFERENTIATION_GENES_UP | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_V1_LATE_DIFFERENTIATION_GENES_UP |
| ZWANG_EGF_PERSISTENTLY_UP | 32 | http://www.broadinstitute.org/gsea/msigdb/cards/ZWANG_EGF_PERSISTENTLY_UP |
| BARRIER_CANCER_RELAPSE_NORMAL_SAMPLE_DN | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/BARRIER_CANCER_RELAPSE_NORMAL_SAMPLE_DN |
| BENNETT_SYSTEMIC_LUPUS_ERYTHEMATOSUS | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/BENNETT_SYSTEMIC_LUPUS_ERYTHEMATOSUS |
| BERENJENO_TRANSFORMED_BY_RHOA_FOREVER_DN | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/BERENJENO_TRANSFORMED_BY_RHOA_FOREVER_DN |
| BIOCARTA_EGF_PATHWAY | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_EGF_PATHWAY |
| BIOCARTA_MYOSIN_PATHWAY | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_MYOSIN_PATHWAY |
| BIOCARTA_PYK2_PATHWAY | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_PYK2_PATHWAY |
| BONOME_OVARIAN_CANCER_POOR_SURVIVAL_UP | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/BONOME_OVARIAN_CANCER_POOR_SURVIVAL_UP |
| BOQUEST_STEM_CELL_CULTURED_VS_FRESH_DN | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/BOQUEST_STEM_CELL_CULTURED_VS_FRESH_DN |
| CAFFAREL_RESPONSE_TO_THC_DN | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/CAFFAREL_RESPONSE_TO_THC_DN |
| CUI_TCF21_TARGETS_DN | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/CUI_TCF21_TARGETS_DN |
| DANG_MYC_TARGETS_DN | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/DANG_MYC_TARGETS_DN |
| DORSEY_GAB2_TARGETS | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/DORSEY_GAB2_TARGETS |
| HEIDENBLAD_AMPLIFIED_IN_PANCREATIC_CANCER | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/HEIDENBLAD_AMPLIFIED_IN_PANCREATIC_CANCER |
| HUNSBERGER_EXERCISE_REGULATED_GENES | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/HUNSBERGER_EXERCISE_REGULATED_GENES |
| IWANAGA_E2F1_TARGETS_INDUCED_BY_SERUM | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/IWANAGA_E2F1_TARGETS_INDUCED_BY_SERUM |
| JOSEPH_RESPONSE_TO_SODIUM_BUTYRATE_UP | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/JOSEPH_RESPONSE_TO_SODIUM_BUTYRATE_UP |
| KEGG_GLYCINE_SERINE_AND_THREONINE_METABOLISM | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GLYCINE_SERINE_AND_THREONINE_METABOLISM |
| KYNG_ENVIRONMENTAL_STRESS_RESPONSE_NOT_BY_GAMMA_IN_OLD | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_ENVIRONMENTAL_STRESS_RESPONSE_NOT_BY_GAMMA_IN_OLD |
| MCCABE_HOXC6_TARGETS_CANCER_UP | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/MCCABE_HOXC6_TARGETS_CANCER_UP |
| MOSERLE_IFNA_RESPONSE | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/MOSERLE_IFNA_RESPONSE |
| MUELLER_COMMON_TARGETS_OF_AML_FUSIONS_DN | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/MUELLER_COMMON_TARGETS_OF_AML_FUSIONS_DN |
| NIKOLSKY_BREAST_CANCER_20Q11_AMPLICON | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_20Q11_AMPLICON |
| NOJIMA_SFRP2_TARGETS_UP | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/NOJIMA_SFRP2_TARGETS_UP |
| PID_AR_NONGENOMIC_PATHWAY | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_AR_NONGENOMIC_PATHWAY |
| PID_AURORA_A_PATHWAY | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_AURORA_A_PATHWAY |
| PID_AVB3_OPN_PATHWAY | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_AVB3_OPN_PATHWAY |
| PID_CD40_PATHWAY | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_CD40_PATHWAY |
| PID_NEPHRIN_NEPH1_PATHWAY | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_NEPHRIN_NEPH1_PATHWAY |
| PID_P38ALPHABETAPATHWAY | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_P38ALPHABETAPATHWAY |
| REACTOME_ACTIVATION_OF_KAINATE_RECEPTORS_UPON_GLUTAMATE_BINDING | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATION_OF_KAINATE_RECEPTORS_UPON_GLUTAMATE_BINDING |
| REACTOME_ACTIVATION_OF_THE_PRE_REPLICATIVE_COMPLEX | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATION_OF_THE_PRE_REPLICATIVE_COMPLEX |
| REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE |
| REACTOME_GPVI_MEDIATED_ACTIVATION_CASCADE | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GPVI_MEDIATED_ACTIVATION_CASCADE |
| REACTOME_HS_GAG_BIOSYNTHESIS | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_HS_GAG_BIOSYNTHESIS |
| REACTOME_INWARDLY_RECTIFYING_K_CHANNELS | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INWARDLY_RECTIFYING_K_CHANNELS |
| REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING |
| REACTOME_SIGNAL_AMPLIFICATION | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNAL_AMPLIFICATION |
| REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS |
| REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE |
| REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES |
| RIZ_ERYTHROID_DIFFERENTIATION_HEMGN | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/RIZ_ERYTHROID_DIFFERENTIATION_HEMGN |
| SCIAN_INVERSED_TARGETS_OF_TP53_AND_TP73_DN | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/SCIAN_INVERSED_TARGETS_OF_TP53_AND_TP73_DN |
| SIMBULAN_PARP1_TARGETS_UP | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/SIMBULAN_PARP1_TARGETS_UP |
| SIMBULAN_UV_RESPONSE_IMMORTALIZED_DN | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/SIMBULAN_UV_RESPONSE_IMMORTALIZED_DN |
| STEIN_ESRRA_TARGETS_RESPONSIVE_TO_ESTROGEN_UP | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/STEIN_ESRRA_TARGETS_RESPONSIVE_TO_ESTROGEN_UP |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_3D_DN | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_3D_DN |
| TCGA_GLIOBLASTOMA_COPY_NUMBER_DN | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/TCGA_GLIOBLASTOMA_COPY_NUMBER_DN |
| THEODOROU_MAMMARY_TUMORIGENESIS | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/THEODOROU_MAMMARY_TUMORIGENESIS |
| TING_SILENCED_BY_DICER | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/TING_SILENCED_BY_DICER |
| VALK_AML_CLUSTER_15 | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_CLUSTER_15 |
| VALK_AML_CLUSTER_4 | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_CLUSTER_4 |
| WANG_IMMORTALIZED_BY_HOXA9_AND_MEIS1_UP | 31 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_IMMORTALIZED_BY_HOXA9_AND_MEIS1_UP |
| BIOCARTA_FAS_PATHWAY | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_FAS_PATHWAY |
| BOGNI_TREATMENT_RELATED_MYELOID_LEUKEMIA_UP | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/BOGNI_TREATMENT_RELATED_MYELOID_LEUKEMIA_UP |
| BURTON_ADIPOGENESIS_10 | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/BURTON_ADIPOGENESIS_10 |
| CHIARETTI_T_ALL_REFRACTORY_TO_THERAPY | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIARETTI_T_ALL_REFRACTORY_TO_THERAPY |
| DASU_IL6_SIGNALING_SCAR_UP | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/DASU_IL6_SIGNALING_SCAR_UP |
| DAZARD_UV_RESPONSE_CLUSTER_G2 | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/DAZARD_UV_RESPONSE_CLUSTER_G2 |
| GAZDA_DIAMOND_BLACKFAN_ANEMIA_MYELOID_UP | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/GAZDA_DIAMOND_BLACKFAN_ANEMIA_MYELOID_UP |
| GILDEA_METASTASIS | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/GILDEA_METASTASIS |
| GUILLAUMOND_KLF10_TARGETS_DN | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/GUILLAUMOND_KLF10_TARGETS_DN |
| HEIDENBLAD_AMPLICON_12P11_12_DN | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/HEIDENBLAD_AMPLICON_12P11_12_DN |
| HOFMANN_MYELODYSPLASTIC_SYNDROM_LOW_RISK_DN | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFMANN_MYELODYSPLASTIC_SYNDROM_LOW_RISK_DN |
| KEGG_ASTHMA | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ASTHMA |
| KEGG_O_GLYCAN_BIOSYNTHESIS | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_O_GLYCAN_BIOSYNTHESIS |
| KIM_LRRC3B_TARGETS | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_LRRC3B_TARGETS |
| KYNG_NORMAL_AGING_DN | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_NORMAL_AGING_DN |
| LIU_BREAST_CANCER | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_BREAST_CANCER |
| LI_PROSTATE_CANCER_EPIGENETIC | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_PROSTATE_CANCER_EPIGENETIC |
| LI_WILMS_TUMOR_VS_FETAL_KIDNEY_2_UP | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_WILMS_TUMOR_VS_FETAL_KIDNEY_2_UP |
| LY_AGING_PREMATURE_DN | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/LY_AGING_PREMATURE_DN |
| NAKAJIMA_EOSINOPHIL | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAJIMA_EOSINOPHIL |
| PARK_TRETINOIN_RESPONSE_AND_PML_RARA_FUSION | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/PARK_TRETINOIN_RESPONSE_AND_PML_RARA_FUSION |
| PID_CDC42_REG_PATHWAY | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_CDC42_REG_PATHWAY |
| PID_EPHRINBREVPATHWAY | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_EPHRINBREVPATHWAY |
| PID_IGF1_PATHWAY | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_IGF1_PATHWAY |
| PID_IL2_STAT5PATHWAY | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_IL2_STAT5PATHWAY |
| PID_NECTIN_PATHWAY | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_NECTIN_PATHWAY |
| PID_RAS_PATHWAY | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_RAS_PATHWAY |
| PID_RETINOIC_ACID_PATHWAY | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_RETINOIC_ACID_PATHWAY |
| PLASARI_TGFB1_SIGNALING_VIA_NFIC_10HR_DN | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/PLASARI_TGFB1_SIGNALING_VIA_NFIC_10HR_DN |
| REACTOME_BASIGIN_INTERACTIONS | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_BASIGIN_INTERACTIONS |
| REACTOME_CA_DEPENDENT_EVENTS | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CA_DEPENDENT_EVENTS |
| REACTOME_DNA_STRAND_ELONGATION | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DNA_STRAND_ELONGATION |
| REACTOME_FORMATION_OF_TRANSCRIPTION_COUPLED_NER_TC_NER_REPAIR_COMPLEX | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FORMATION_OF_TRANSCRIPTION_COUPLED_NER_TC_NER_REPAIR_COMPLEX |
| REACTOME_INTRINSIC_PATHWAY_FOR_APOPTOSIS | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INTRINSIC_PATHWAY_FOR_APOPTOSIS |
| REACTOME_KERATAN_SULFATE_KERATIN_METABOLISM | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_KERATAN_SULFATE_KERATIN_METABOLISM |
| REACTOME_MAPK_TARGETS_NUCLEAR_EVENTS_MEDIATED_BY_MAP_KINASES | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MAPK_TARGETS_NUCLEAR_EVENTS_MEDIATED_BY_MAP_KINASES |
| REACTOME_MRNA_CAPPING | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MRNA_CAPPING |
| REACTOME_NOD1_2_SIGNALING_PATHWAY | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NOD1_2_SIGNALING_PATHWAY |
| REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT |
| REACTOME_SIGNALING_BY_FGFR1_MUTANTS | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_FGFR1_MUTANTS |
| REACTOME_SIGNALING_BY_ROBO_RECEPTOR | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_ROBO_RECEPTOR |
| REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION |
| RORIE_TARGETS_OF_EWSR1_FLI1_FUSION_DN | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/RORIE_TARGETS_OF_EWSR1_FLI1_FUSION_DN |
| RORIE_TARGETS_OF_EWSR1_FLI1_FUSION_UP | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/RORIE_TARGETS_OF_EWSR1_FLI1_FUSION_UP |
| SEITZ_NEOPLASTIC_TRANSFORMATION_BY_8P_DELETION_DN | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/SEITZ_NEOPLASTIC_TRANSFORMATION_BY_8P_DELETION_DN |
| SHIN_B_CELL_LYMPHOMA_CLUSTER_2 | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/SHIN_B_CELL_LYMPHOMA_CLUSTER_2 |
| SU_PLACENTA | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/SU_PLACENTA |
| TAVOR_CEBPA_TARGETS_DN | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/TAVOR_CEBPA_TARGETS_DN |
| TSENG_ADIPOGENIC_POTENTIAL_UP | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/TSENG_ADIPOGENIC_POTENTIAL_UP |
| URS_ADIPOCYTE_DIFFERENTIATION_DN | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/URS_ADIPOCYTE_DIFFERENTIATION_DN |
| VALK_AML_CLUSTER_12 | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_CLUSTER_12 |
| VALK_AML_CLUSTER_13 | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_CLUSTER_13 |
| WANG_METASTASIS_OF_BREAST_CANCER_ESR1_DN | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_METASTASIS_OF_BREAST_CANCER_ESR1_DN |
| WORSCHECH_TUMOR_EVASION_AND_TOLEROGENICITY_UP | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/WORSCHECH_TUMOR_EVASION_AND_TOLEROGENICITY_UP |
| YANAGIHARA_ESX1_TARGETS | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/YANAGIHARA_ESX1_TARGETS |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_5 | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_5 |
| ZHANG_ANTIVIRAL_RESPONSE_TO_RIBAVIRIN_UP | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_ANTIVIRAL_RESPONSE_TO_RIBAVIRIN_UP |
| ZHAN_MULTIPLE_MYELOMA_SUBGROUPS | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_SUBGROUPS |
| ZHAN_VARIABLE_EARLY_DIFFERENTIATION_GENES_DN | 30 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_VARIABLE_EARLY_DIFFERENTIATION_GENES_DN |
| ABE_VEGFA_TARGETS_30MIN | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/ABE_VEGFA_TARGETS_30MIN |
| AIYAR_COBRA1_TARGETS_DN | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/AIYAR_COBRA1_TARGETS_DN |
| BERENJENO_ROCK_SIGNALING_NOT_VIA_RHOA_UP | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/BERENJENO_ROCK_SIGNALING_NOT_VIA_RHOA_UP |
| BERENJENO_TRANSFORMED_BY_RHOA_REVERSIBLY_DN | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/BERENJENO_TRANSFORMED_BY_RHOA_REVERSIBLY_DN |
| BIOCARTA_INFLAM_PATHWAY | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_INFLAM_PATHWAY |
| BIOCARTA_NKT_PATHWAY | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_NKT_PATHWAY |
| BIOCARTA_TNFR1_PATHWAY | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_TNFR1_PATHWAY |
| BIOCARTA_VEGF_PATHWAY | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_VEGF_PATHWAY |
| BIOCARTA_VIP_PATHWAY | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_VIP_PATHWAY |
| CHOW_RASSF1_TARGETS_DN | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/CHOW_RASSF1_TARGETS_DN |
| DALESSIO_TSA_RESPONSE | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/DALESSIO_TSA_RESPONSE |
| DORN_ADENOVIRUS_INFECTION_12HR_UP | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/DORN_ADENOVIRUS_INFECTION_12HR_UP |
| FLOTHO_PEDIATRIC_ALL_THERAPY_RESPONSE_DN | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/FLOTHO_PEDIATRIC_ALL_THERAPY_RESPONSE_DN |
| GENTLES_LEUKEMIC_STEM_CELL_UP | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/GENTLES_LEUKEMIC_STEM_CELL_UP |
| KEGG_HISTIDINE_METABOLISM | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_HISTIDINE_METABOLISM |
| KEGG_LINOLEIC_ACID_METABOLISM | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_LINOLEIC_ACID_METABOLISM |
| KEGG_RNA_POLYMERASE | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_RNA_POLYMERASE |
| KEGG_THYROID_CANCER | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_THYROID_CANCER |
| KRIGE_AMINO_ACID_DEPRIVATION | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/KRIGE_AMINO_ACID_DEPRIVATION |
| KYNG_WERNER_SYNDROM_DN | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_WERNER_SYNDROM_DN |
| LANG_MYB_FAMILY_TARGETS | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/LANG_MYB_FAMILY_TARGETS |
| LU_TUMOR_VASCULATURE_UP | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/LU_TUMOR_VASCULATURE_UP |
| MARSHALL_VIRAL_INFECTION_RESPONSE_DN | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/MARSHALL_VIRAL_INFECTION_RESPONSE_DN |
| MARSON_FOXP3_TARGETS_STIMULATED_UP | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/MARSON_FOXP3_TARGETS_STIMULATED_UP |
| MATZUK_CENTRAL_FOR_FEMALE_FERTILITY | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_CENTRAL_FOR_FEMALE_FERTILITY |
| MAYBURD_RESPONSE_TO_L663536_UP | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/MAYBURD_RESPONSE_TO_L663536_UP |
| MA_PITUITARY_FETAL_VS_ADULT_UP | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/MA_PITUITARY_FETAL_VS_ADULT_UP |
| MIKKELSEN_MCV6_LCP_WITH_H3K27ME3 | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_MCV6_LCP_WITH_H3K27ME3 |
| NAKAYAMA_FGF2_TARGETS | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAYAMA_FGF2_TARGETS |
| NGUYEN_NOTCH1_TARGETS_UP | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/NGUYEN_NOTCH1_TARGETS_UP |
| NUNODA_RESPONSE_TO_DASATINIB_IMATINIB_UP | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/NUNODA_RESPONSE_TO_DASATINIB_IMATINIB_UP |
| PAL_PRMT5_TARGETS_DN | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/PAL_PRMT5_TARGETS_DN |
| PATTERSON_DOCETAXEL_RESISTANCE | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/PATTERSON_DOCETAXEL_RESISTANCE |
| PEDERSEN_METASTASIS_BY_ERBB2_ISOFORM_6 | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/PEDERSEN_METASTASIS_BY_ERBB2_ISOFORM_6 |
| PID_BARD1PATHWAY | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_BARD1PATHWAY |
| PID_INTEGRIN2_PATHWAY | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_INTEGRIN2_PATHWAY |
| PID_REELINPATHWAY | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_REELINPATHWAY |
| PID_S1P_S1P3_PATHWAY | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_S1P_S1P3_PATHWAY |
| PID_TCRCALCIUMPATHWAY | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_TCRCALCIUMPATHWAY |
| PYEON_CANCER_HEAD_AND_NECK_VS_CERVICAL_DN | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/PYEON_CANCER_HEAD_AND_NECK_VS_CERVICAL_DN |
| REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS |
| REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN |
| REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX |
| REACTOME_GLYCOLYSIS | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GLYCOLYSIS |
| REACTOME_PERK_REGULATED_GENE_EXPRESSION | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PERK_REGULATED_GENE_EXPRESSION |
| REACTOME_PIP3_ACTIVATES_AKT_SIGNALING | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PIP3_ACTIVATES_AKT_SIGNALING |
| REACTOME_PKB_MEDIATED_EVENTS | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PKB_MEDIATED_EVENTS |
| REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION |
| REACTOME_STEROID_HORMONES | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_STEROID_HORMONES |
| REACTOME_TIGHT_JUNCTION_INTERACTIONS | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TIGHT_JUNCTION_INTERACTIONS |
| SCHLINGEMANN_SKIN_CARCINOGENESIS_TPA_DN | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHLINGEMANN_SKIN_CARCINOGENESIS_TPA_DN |
| STANELLE_E2F1_TARGETS | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/STANELLE_E2F1_TARGETS |
| ST_TUMOR_NECROSIS_FACTOR_PATHWAY | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_TUMOR_NECROSIS_FACTOR_PATHWAY |
| TAKADA_GASTRIC_CANCER_COPY_NUMBER_DN | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/TAKADA_GASTRIC_CANCER_COPY_NUMBER_DN |
| VALK_AML_CLUSTER_2 | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_CLUSTER_2 |
| WAKABAYASHI_ADIPOGENESIS_PPARG_BOUND_36HR | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/WAKABAYASHI_ADIPOGENESIS_PPARG_BOUND_36HR |
| WANG_RESPONSE_TO_ANDROGEN_UP | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_RESPONSE_TO_ANDROGEN_UP |
| WANG_RESPONSE_TO_BEXAROTENE_DN | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_RESPONSE_TO_BEXAROTENE_DN |
| WATANABE_COLON_CANCER_MSI_VS_MSS_UP | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/WATANABE_COLON_CANCER_MSI_VS_MSS_UP |
| WEINMANN_ADAPTATION_TO_HYPOXIA_UP | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/WEINMANN_ADAPTATION_TO_HYPOXIA_UP |
| WILENSKY_RESPONSE_TO_DARAPLADIB | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/WILENSKY_RESPONSE_TO_DARAPLADIB |
| WOTTON_RUNX_TARGETS_DN | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/WOTTON_RUNX_TARGETS_DN |
| YU_BAP1_TARGETS | 29 | http://www.broadinstitute.org/gsea/msigdb/cards/YU_BAP1_TARGETS |
| BIOCARTA_ERK_PATHWAY | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_ERK_PATHWAY |
| BIOCARTA_G1_PATHWAY | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_G1_PATHWAY |
| BIOCARTA_GH_PATHWAY | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_GH_PATHWAY |
| BIOCARTA_PROTEASOME_PATHWAY | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_PROTEASOME_PATHWAY |
| BRACHAT_RESPONSE_TO_CAMPTOTHECIN_UP | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/BRACHAT_RESPONSE_TO_CAMPTOTHECIN_UP |
| CHEN_LUNG_CANCER_SURVIVAL | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEN_LUNG_CANCER_SURVIVAL |
| CHNG_MULTIPLE_MYELOMA_HYPERPLOID_DN | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/CHNG_MULTIPLE_MYELOMA_HYPERPLOID_DN |
| CHUANG_OXIDATIVE_STRESS_RESPONSE_UP | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/CHUANG_OXIDATIVE_STRESS_RESPONSE_UP |
| COLIN_PILOCYTIC_ASTROCYTOMA_VS_GLIOBLASTOMA_DN | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/COLIN_PILOCYTIC_ASTROCYTOMA_VS_GLIOBLASTOMA_DN |
| DACOSTA_ERCC3_ALLELE_XPCS_VS_TTD_UP | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/DACOSTA_ERCC3_ALLELE_XPCS_VS_TTD_UP |
| DACOSTA_UV_RESPONSE_VIA_ERCC3_XPCS_UP | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/DACOSTA_UV_RESPONSE_VIA_ERCC3_XPCS_UP |
| DAIRKEE_CANCER_PRONE_RESPONSE_E2 | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/DAIRKEE_CANCER_PRONE_RESPONSE_E2 |
| DAVIES_MULTIPLE_MYELOMA_VS_MGUS_DN | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/DAVIES_MULTIPLE_MYELOMA_VS_MGUS_DN |
| DAZARD_UV_RESPONSE_CLUSTER_G24 | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/DAZARD_UV_RESPONSE_CLUSTER_G24 |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_MAGENTA_UP | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_MAGENTA_UP |
| GENTILE_UV_RESPONSE_CLUSTER_D9 | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/GENTILE_UV_RESPONSE_CLUSTER_D9 |
| HWANG_PROSTATE_CANCER_MARKERS | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/HWANG_PROSTATE_CANCER_MARKERS |
| IIZUKA_LIVER_CANCER_PROGRESSION_G2_G3_UP | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/IIZUKA_LIVER_CANCER_PROGRESSION_G2_G3_UP |
| KAMIKUBO_MYELOID_CEBPA_NETWORK | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/KAMIKUBO_MYELOID_CEBPA_NETWORK |
| KEGG_HOMOLOGOUS_RECOMBINATION | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_HOMOLOGOUS_RECOMBINATION |
| KEGG_PENTOSE_AND_GLUCURONATE_INTERCONVERSIONS | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PENTOSE_AND_GLUCURONATE_INTERCONVERSIONS |
| LI_CISPLATIN_RESISTANCE_UP | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_CISPLATIN_RESISTANCE_UP |
| LOPES_METHYLATED_IN_COLON_CANCER_DN | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/LOPES_METHYLATED_IN_COLON_CANCER_DN |
| LUI_THYROID_CANCER_CLUSTER_3 | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/LUI_THYROID_CANCER_CLUSTER_3 |
| MATZUK_MALE_REPRODUCTION_SERTOLI | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_MALE_REPRODUCTION_SERTOLI |
| MOREIRA_RESPONSE_TO_TSA_UP | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/MOREIRA_RESPONSE_TO_TSA_UP |
| OLSSON_E2F3_TARGETS_UP | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/OLSSON_E2F3_TARGETS_UP |
| PETRETTO_HEART_MASS_QTL_CIS_UP | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/PETRETTO_HEART_MASS_QTL_CIS_UP |
| PETROVA_PROX1_TARGETS_UP | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/PETROVA_PROX1_TARGETS_UP |
| PID_IL8CXCR1_PATHWAY | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_IL8CXCR1_PATHWAY |
| PID_LIS1PATHWAY | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_LIS1PATHWAY |
| PID_TRAIL_PATHWAY | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_TRAIL_PATHWAY |
| PID_WNT_SIGNALING_PATHWAY | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_WNT_SIGNALING_PATHWAY |
| REACTOME_APC_CDC20_MEDIATED_DEGRADATION_OF_NEK2A | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_APC_CDC20_MEDIATED_DEGRADATION_OF_NEK2A |
| REACTOME_G_PROTEIN_BETA_GAMMA_SIGNALLING | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_G_PROTEIN_BETA_GAMMA_SIGNALLING |
| REACTOME_LIPOPROTEIN_METABOLISM | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_LIPOPROTEIN_METABOLISM |
| REACTOME_MYOGENESIS | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MYOGENESIS |
| REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC |
| REACTOME_SHC_MEDIATED_CASCADE | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SHC_MEDIATED_CASCADE |
| REACTOME_THE_ROLE_OF_NEF_IN_HIV1_REPLICATION_AND_DISEASE_PATHOGENESIS | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_THE_ROLE_OF_NEF_IN_HIV1_REPLICATION_AND_DISEASE_PATHOGENESIS |
| REACTOME_TRAFFICKING_OF_AMPA_RECEPTORS | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRAFFICKING_OF_AMPA_RECEPTORS |
| REICHERT_MITOSIS_LIN9_TARGETS | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/REICHERT_MITOSIS_LIN9_TARGETS |
| SCHLESINGER_H3K27ME3_IN_NORMAL_AND_METHYLATED_IN_CANCER | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHLESINGER_H3K27ME3_IN_NORMAL_AND_METHYLATED_IN_CANCER |
| SHIN_B_CELL_LYMPHOMA_CLUSTER_3 | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/SHIN_B_CELL_LYMPHOMA_CLUSTER_3 |
| ST_GAQ_PATHWAY | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_GAQ_PATHWAY |
| TERAO_AOX4_TARGETS_HG_UP | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/TERAO_AOX4_TARGETS_HG_UP |
| TIAN_TNF_SIGNALING_VIA_NFKB | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/TIAN_TNF_SIGNALING_VIA_NFKB |
| VALK_AML_CLUSTER_1 | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_CLUSTER_1 |
| VALK_AML_CLUSTER_7 | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_CLUSTER_7 |
| WANG_ADIPOGENIC_GENES_REPRESSED_BY_SIRT1 | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_ADIPOGENIC_GENES_REPRESSED_BY_SIRT1 |
| WEBER_METHYLATED_HCP_IN_SPERM_DN | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/WEBER_METHYLATED_HCP_IN_SPERM_DN |
| WIKMAN_ASBESTOS_LUNG_CANCER_DN | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/WIKMAN_ASBESTOS_LUNG_CANCER_DN |
| WILLIAMS_ESR2_TARGETS_UP | 28 | http://www.broadinstitute.org/gsea/msigdb/cards/WILLIAMS_ESR2_TARGETS_UP |
| AKL_HTLV1_INFECTION_UP | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/AKL_HTLV1_INFECTION_UP |
| BIOCARTA_CREB_PATHWAY | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CREB_PATHWAY |
| BIOCARTA_EDG1_PATHWAY | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_EDG1_PATHWAY |
| BIOCARTA_GSK3_PATHWAY | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_GSK3_PATHWAY |
| BOYAULT_LIVER_CANCER_SUBCLASS_G2 | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYAULT_LIVER_CANCER_SUBCLASS_G2 |
| BOYAULT_LIVER_CANCER_SUBCLASS_G5_DN | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYAULT_LIVER_CANCER_SUBCLASS_G5_DN |
| BOYLAN_MULTIPLE_MYELOMA_D_CLUSTER_UP | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYLAN_MULTIPLE_MYELOMA_D_CLUSTER_UP |
| BRACHAT_RESPONSE_TO_METHOTREXATE_DN | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/BRACHAT_RESPONSE_TO_METHOTREXATE_DN |
| CHIN_BREAST_CANCER_COPY_NUMBER_UP | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIN_BREAST_CANCER_COPY_NUMBER_UP |
| CHOW_RASSF1_TARGETS_UP | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/CHOW_RASSF1_TARGETS_UP |
| EINAV_INTERFERON_SIGNATURE_IN_CANCER | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/EINAV_INTERFERON_SIGNATURE_IN_CANCER |
| GENTILE_UV_LOW_DOSE_UP | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/GENTILE_UV_LOW_DOSE_UP |
| GROSS_ELK3_TARGETS_UP | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/GROSS_ELK3_TARGETS_UP |
| HAHTOLA_MYCOSIS_FUNGOIDES_SKIN_DN | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/HAHTOLA_MYCOSIS_FUNGOIDES_SKIN_DN |
| HOEGERKORP_CD44_TARGETS_DIRECT_UP | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/HOEGERKORP_CD44_TARGETS_DIRECT_UP |
| HOLLEMAN_VINCRISTINE_RESISTANCE_ALL_UP | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLEMAN_VINCRISTINE_RESISTANCE_ALL_UP |
| IKEDA_MIR30_TARGETS_DN | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/IKEDA_MIR30_TARGETS_DN |
| JI_METASTASIS_REPRESSED_BY_STK11 | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/JI_METASTASIS_REPRESSED_BY_STK11 |
| KEGG_PENTOSE_PHOSPHATE_PATHWAY | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PENTOSE_PHOSPHATE_PATHWAY |
| LEE_DOUBLE_POLAR_THYMOCYTE | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_DOUBLE_POLAR_THYMOCYTE |
| LI_WILMS_TUMOR | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_WILMS_TUMOR |
| LOPES_METHYLATED_IN_COLON_CANCER_UP | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/LOPES_METHYLATED_IN_COLON_CANCER_UP |
| NICK_RESPONSE_TO_PROC_TREATMENT_DN | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/NICK_RESPONSE_TO_PROC_TREATMENT_DN |
| OHM_METHYLATED_IN_ADULT_CANCERS | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/OHM_METHYLATED_IN_ADULT_CANCERS |
| PID_GLYPICAN_1PATHWAY | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_GLYPICAN_1PATHWAY |
| PID_IL3_PATHWAY | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_IL3_PATHWAY |
| PLASARI_NFIC_TARGETS_BASAL_UP | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/PLASARI_NFIC_TARGETS_BASAL_UP |
| REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS |
| REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS |
| REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS |
| REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS |
| REACTOME_BILE_ACID_AND_BILE_SALT_METABOLISM | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_BILE_ACID_AND_BILE_SALT_METABOLISM |
| REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_RAS | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_RAS |
| REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT |
| REACTOME_EXTENSION_OF_TELOMERES | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_EXTENSION_OF_TELOMERES |
| REACTOME_GAP_JUNCTION_TRAFFICKING | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GAP_JUNCTION_TRAFFICKING |
| REACTOME_GENERATION_OF_SECOND_MESSENGER_MOLECULES | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GENERATION_OF_SECOND_MESSENGER_MOLECULES |
| REACTOME_G_PROTEIN_ACTIVATION | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_G_PROTEIN_ACTIVATION |
| REACTOME_IL_RECEPTOR_SHC_SIGNALING | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_IL_RECEPTOR_SHC_SIGNALING |
| REACTOME_INTEGRIN_ALPHAIIB_BETA3_SIGNALING | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INTEGRIN_ALPHAIIB_BETA3_SIGNALING |
| REACTOME_NEP_NS2_INTERACTS_WITH_THE_CELLULAR_EXPORT_MACHINERY | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NEP_NS2_INTERACTS_WITH_THE_CELLULAR_EXPORT_MACHINERY |
| REACTOME_PTM_GAMMA_CARBOXYLATION_HYPUSINE_FORMATION_AND_ARYLSULFATASE_ACTIVATION | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PTM_GAMMA_CARBOXYLATION_HYPUSINE_FORMATION_AND_ARYLSULFATASE_ACTIVATION |
| REACTOME_RECYCLING_PATHWAY_OF_L1 | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RECYCLING_PATHWAY_OF_L1 |
| REACTOME_REGULATION_OF_GLUCOKINASE_BY_GLUCOKINASE_REGULATORY_PROTEIN | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_GLUCOKINASE_BY_GLUCOKINASE_REGULATORY_PROTEIN |
| REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE |
| REACTOME_SIGNALLING_TO_RAS | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALLING_TO_RAS |
| REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION |
| REACTOME_STRIATED_MUSCLE_CONTRACTION | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_STRIATED_MUSCLE_CONTRACTION |
| REACTOME_SYNTHESIS_OF_PA | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_OF_PA |
| REACTOME_TRANSPORT_OF_RIBONUCLEOPROTEINS_INTO_THE_HOST_NUCLEUS | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANSPORT_OF_RIBONUCLEOPROTEINS_INTO_THE_HOST_NUCLEUS |
| RIZ_ERYTHROID_DIFFERENTIATION_APOBEC2 | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/RIZ_ERYTHROID_DIFFERENTIATION_APOBEC2 |
| SAKAI_TUMOR_INFILTRATING_MONOCYTES_UP | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/SAKAI_TUMOR_INFILTRATING_MONOCYTES_UP |
| SHIN_B_CELL_LYMPHOMA_CLUSTER_7 | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/SHIN_B_CELL_LYMPHOMA_CLUSTER_7 |
| SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/SIG_IL4RECEPTOR_IN_B_LYPHOCYTES |
| SMID_BREAST_CANCER_RELAPSE_IN_PLEURA_DN | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_RELAPSE_IN_PLEURA_DN |
| ST_GRANULE_CELL_SURVIVAL_PATHWAY | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_GRANULE_CELL_SURVIVAL_PATHWAY |
| ST_MYOCYTE_AD_PATHWAY | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_MYOCYTE_AD_PATHWAY |
| TERAMOTO_OPN_TARGETS_CLUSTER_6 | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/TERAMOTO_OPN_TARGETS_CLUSTER_6 |
| TERAO_AOX4_TARGETS_SKIN_DN | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/TERAO_AOX4_TARGETS_SKIN_DN |
| TIEN_INTESTINE_PROBIOTICS_2HR_UP | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/TIEN_INTESTINE_PROBIOTICS_2HR_UP |
| XU_AKT1_TARGETS_6HR | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_AKT1_TARGETS_6HR |
| XU_HGF_TARGETS_INDUCED_BY_AKT1_48HR_DN | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_HGF_TARGETS_INDUCED_BY_AKT1_48HR_DN |
| YANG_BREAST_CANCER_ESR1_BULK_UP | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/YANG_BREAST_CANCER_ESR1_BULK_UP |
| YE_METASTATIC_LIVER_CANCER | 27 | http://www.broadinstitute.org/gsea/msigdb/cards/YE_METASTATIC_LIVER_CANCER |
| ABBUD_LIF_SIGNALING_1_DN | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/ABBUD_LIF_SIGNALING_1_DN |
| ABRAHAM_ALPC_VS_MULTIPLE_MYELOMA_UP | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/ABRAHAM_ALPC_VS_MULTIPLE_MYELOMA_UP |
| ALONSO_METASTASIS_DN | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/ALONSO_METASTASIS_DN |
| BACOLOD_RESISTANCE_TO_ALKYLATING_AGENTS_UP | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/BACOLOD_RESISTANCE_TO_ALKYLATING_AGENTS_UP |
| BIOCARTA_BAD_PATHWAY | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_BAD_PATHWAY |
| BIOCARTA_PGC1A_PATHWAY | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_PGC1A_PATHWAY |
| BIOCARTA_RACCYCD_PATHWAY | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_RACCYCD_PATHWAY |
| BIOCARTA_WNT_PATHWAY | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_WNT_PATHWAY |
| BRACHAT_RESPONSE_TO_METHOTREXATE_UP | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/BRACHAT_RESPONSE_TO_METHOTREXATE_UP |
| CHIANG_LIVER_CANCER_SUBCLASS_INTERFERON_UP | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIANG_LIVER_CANCER_SUBCLASS_INTERFERON_UP |
| DEN_INTERACT_WITH_LCA5 | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/DEN_INTERACT_WITH_LCA5 |
| DING_LUNG_CANCER_MUTATED_SIGNIFICANTLY | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/DING_LUNG_CANCER_MUTATED_SIGNIFICANTLY |
| FINAK_BREAST_CANCER_SDPP_SIGNATURE | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/FINAK_BREAST_CANCER_SDPP_SIGNATURE |
| FONTAINE_THYROID_TUMOR_UNCERTAIN_MALIGNANCY_DN | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/FONTAINE_THYROID_TUMOR_UNCERTAIN_MALIGNANCY_DN |
| GAUSSMANN_MLL_AF4_FUSION_TARGETS_B_UP | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/GAUSSMANN_MLL_AF4_FUSION_TARGETS_B_UP |
| GAZIN_EPIGENETIC_SILENCING_BY_KRAS | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/GAZIN_EPIGENETIC_SILENCING_BY_KRAS |
| GUENTHER_GROWTH_SPHERICAL_VS_ADHERENT_DN | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/GUENTHER_GROWTH_SPHERICAL_VS_ADHERENT_DN |
| HAHTOLA_CTCL_CUTANEOUS | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/HAHTOLA_CTCL_CUTANEOUS |
| HOLLEMAN_ASPARAGINASE_RESISTANCE_B_ALL_UP | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLEMAN_ASPARAGINASE_RESISTANCE_B_ALL_UP |
| KEGG_GALACTOSE_METABOLISM | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GALACTOSE_METABOLISM |
| KEGG_GLYCOSAMINOGLYCAN_BIOSYNTHESIS_HEPARAN_SULFATE | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GLYCOSAMINOGLYCAN_BIOSYNTHESIS_HEPARAN_SULFATE |
| KEGG_GLYCOSPHINGOLIPID_BIOSYNTHESIS_LACTO_AND_NEOLACTO_SERIES | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GLYCOSPHINGOLIPID_BIOSYNTHESIS_LACTO_AND_NEOLACTO_SERIES |
| KEGG_SELENOAMINO_ACID_METABOLISM | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_SELENOAMINO_ACID_METABOLISM |
| KONDO_COLON_CANCER_HCP_WITH_H3K27ME1 | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/KONDO_COLON_CANCER_HCP_WITH_H3K27ME1 |
| MAGRANGEAS_MULTIPLE_MYELOMA_IGG_VS_IGA_DN | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/MAGRANGEAS_MULTIPLE_MYELOMA_IGG_VS_IGA_DN |
| MCMURRAY_TP53_HRAS_COOPERATION_RESPONSE_UP | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/MCMURRAY_TP53_HRAS_COOPERATION_RESPONSE_UP |
| NAKAMURA_CANCER_MICROENVIRONMENT_UP | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAMURA_CANCER_MICROENVIRONMENT_UP |
| NEWMAN_ERCC6_TARGETS_UP | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/NEWMAN_ERCC6_TARGETS_UP |
| NIKOLSKY_BREAST_CANCER_5P15_AMPLICON | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_5P15_AMPLICON |
| PID_ALK1PATHWAY | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ALK1PATHWAY |
| PID_IL27PATHWAY | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_IL27PATHWAY |
| PID_INSULIN_GLUCOSE_PATHWAY | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_INSULIN_GLUCOSE_PATHWAY |
| PID_INTEGRIN_CS_PATHWAY | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_INTEGRIN_CS_PATHWAY |
| PID_P38_MKK3_6PATHWAY | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_P38_MKK3_6PATHWAY |
| PID_RXR_VDR_PATHWAY | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_RXR_VDR_PATHWAY |
| PID_VEGFR1_PATHWAY | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_VEGFR1_PATHWAY |
| REACTOME_ACTIVATION_OF_GENES_BY_ATF4 | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATION_OF_GENES_BY_ATF4 |
| REACTOME_APC_C_CDC20_MEDIATED_DEGRADATION_OF_CYCLIN_B | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_APC_C_CDC20_MEDIATED_DEGRADATION_OF_CYCLIN_B |
| REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE |
| REACTOME_KERATAN_SULFATE_BIOSYNTHESIS | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_KERATAN_SULFATE_BIOSYNTHESIS |
| REACTOME_POST_TRANSLATIONAL_MODIFICATION_SYNTHESIS_OF_GPI_ANCHORED_PROTEINS | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_POST_TRANSLATIONAL_MODIFICATION_SYNTHESIS_OF_GPI_ANCHORED_PROTEINS |
| REACTOME_REGULATORY_RNA_PATHWAYS | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATORY_RNA_PATHWAYS |
| REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER |
| REACTOME_TGF_BETA_RECEPTOR_SIGNALING_ACTIVATES_SMADS | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TGF_BETA_RECEPTOR_SIGNALING_ACTIVATES_SMADS |
| ROYLANCE_BREAST_CANCER_16Q_COPY_NUMBER_DN | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/ROYLANCE_BREAST_CANCER_16Q_COPY_NUMBER_DN |
| SCHMIDT_POR_TARGETS_IN_LIMB_BUD_UP | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHMIDT_POR_TARGETS_IN_LIMB_BUD_UP |
| STAMBOLSKY_RESPONSE_TO_VITAMIN_D3_DN | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/STAMBOLSKY_RESPONSE_TO_VITAMIN_D3_DN |
| ST_INTERLEUKIN_4_PATHWAY | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_INTERLEUKIN_4_PATHWAY |
| TOMIDA_METASTASIS_UP | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/TOMIDA_METASTASIS_UP |
| VALK_AML_CLUSTER_16 | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_CLUSTER_16 |
| VALK_AML_CLUSTER_8 | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_CLUSTER_8 |
| WALLACE_JAK2_TARGETS_UP | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/WALLACE_JAK2_TARGETS_UP |
| WANG_BARRETTS_ESOPHAGUS_AND_ESOPHAGUS_CANCER_UP | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_BARRETTS_ESOPHAGUS_AND_ESOPHAGUS_CANCER_UP |
| WANG_CLASSIC_ADIPOGENIC_TARGETS_OF_PPARG | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_CLASSIC_ADIPOGENIC_TARGETS_OF_PPARG |
| WILLIAMS_ESR1_TARGETS_UP | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/WILLIAMS_ESR1_TARGETS_UP |
| XU_CREBBP_TARGETS_UP | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_CREBBP_TARGETS_UP |
| XU_HGF_SIGNALING_NOT_VIA_AKT1_6HR | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_HGF_SIGNALING_NOT_VIA_AKT1_6HR |
| ZHENG_FOXP3_TARGETS_UP | 26 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHENG_FOXP3_TARGETS_UP |
| BERTUCCI_INVASIVE_CARCINOMA_DUCTAL_VS_LOBULAR_UP | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/BERTUCCI_INVASIVE_CARCINOMA_DUCTAL_VS_LOBULAR_UP |
| BIOCARTA_MCALPAIN_PATHWAY | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_MCALPAIN_PATHWAY |
| BIOCARTA_STRESS_PATHWAY | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_STRESS_PATHWAY |
| BYSTRYKH_HEMATOPOIESIS_STEM_CELL_SCP2_QTL_TRANS | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/BYSTRYKH_HEMATOPOIESIS_STEM_CELL_SCP2_QTL_TRANS |
| CHEOK_RESPONSE_TO_MERCAPTOPURINE_AND_HD_MTX_DN | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEOK_RESPONSE_TO_MERCAPTOPURINE_AND_HD_MTX_DN |
| COLLER_MYC_TARGETS_UP | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/COLLER_MYC_TARGETS_UP |
| FLECHNER_BIOPSY_KIDNEY_TRANSPLANT_OK_VS_DONOR_DN | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/FLECHNER_BIOPSY_KIDNEY_TRANSPLANT_OK_VS_DONOR_DN |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_GREEN_DN | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_GREEN_DN |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_RED_DN | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_RED_DN |
| GAZDA_DIAMOND_BLACKFAN_ANEMIA_ERYTHROID_UP | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/GAZDA_DIAMOND_BLACKFAN_ANEMIA_ERYTHROID_UP |
| GENTILE_UV_HIGH_DOSE_UP | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/GENTILE_UV_HIGH_DOSE_UP |
| GROSS_HIF1A_TARGETS_DN | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/GROSS_HIF1A_TARGETS_DN |
| HERNANDEZ_ABERRANT_MITOSIS_BY_DOCETACEL_2NM_DN | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/HERNANDEZ_ABERRANT_MITOSIS_BY_DOCETACEL_2NM_DN |
| HOEGERKORP_CD44_TARGETS_TEMPORAL_DN | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/HOEGERKORP_CD44_TARGETS_TEMPORAL_DN |
| HOLLEMAN_ASPARAGINASE_RESISTANCE_ALL_DN | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLEMAN_ASPARAGINASE_RESISTANCE_ALL_DN |
| HORTON_SREBF_TARGETS | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/HORTON_SREBF_TARGETS |
| IIZUKA_LIVER_CANCER_PROGRESSION_G1_G2_DN | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/IIZUKA_LIVER_CANCER_PROGRESSION_G1_G2_DN |
| IIZUKA_LIVER_CANCER_PROGRESSION_L1_G1_UP | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/IIZUKA_LIVER_CANCER_PROGRESSION_L1_G1_UP |
| JACKSON_DNMT1_TARGETS_DN | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/JACKSON_DNMT1_TARGETS_DN |
| KEGG_ASCORBATE_AND_ALDARATE_METABOLISM | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ASCORBATE_AND_ALDARATE_METABOLISM |
| KEGG_DORSO_VENTRAL_AXIS_FORMATION | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_DORSO_VENTRAL_AXIS_FORMATION |
| KEGG_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI_ANCHOR_BIOSYNTHESIS | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI_ANCHOR_BIOSYNTHESIS |
| KEGG_MATURITY_ONSET_DIABETES_OF_THE_YOUNG | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_MATURITY_ONSET_DIABETES_OF_THE_YOUNG |
| KIM_HYPOXIA | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_HYPOXIA |
| KORKOLA_EMBRYONIC_CARCINOMA_VS_SEMINOMA_DN | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/KORKOLA_EMBRYONIC_CARCINOMA_VS_SEMINOMA_DN |
| KYNG_ENVIRONMENTAL_STRESS_RESPONSE_NOT_BY_UV_IN_OLD | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_ENVIRONMENTAL_STRESS_RESPONSE_NOT_BY_UV_IN_OLD |
| LIAN_NEUTROPHIL_GRANULE_CONSTITUENTS | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/LIAN_NEUTROPHIL_GRANULE_CONSTITUENTS |
| LU_TUMOR_ANGIOGENESIS_UP | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/LU_TUMOR_ANGIOGENESIS_UP |
| MARCINIAK_ER_STRESS_RESPONSE_VIA_CHOP | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/MARCINIAK_ER_STRESS_RESPONSE_VIA_CHOP |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_17 | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_17 |
| NOJIMA_SFRP2_TARGETS_DN | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/NOJIMA_SFRP2_TARGETS_DN |
| OKAMOTO_LIVER_CANCER_MULTICENTRIC_OCCURRENCE_UP | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/OKAMOTO_LIVER_CANCER_MULTICENTRIC_OCCURRENCE_UP |
| OZEN_MIR125B1_TARGETS | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/OZEN_MIR125B1_TARGETS |
| PID_HDAC_CLASSIII_PATHWAY | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_HDAC_CLASSIII_PATHWAY |
| PID_INTEGRIN_A9B1_PATHWAY | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_INTEGRIN_A9B1_PATHWAY |
| PID_LYMPHANGIOGENESIS_PATHWAY | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_LYMPHANGIOGENESIS_PATHWAY |
| PID_MYC_PATHWAY | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_MYC_PATHWAY |
| PID_TOLL_ENDOGENOUS_PATHWAY | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_TOLL_ENDOGENOUS_PATHWAY |
| REACTOME_ADP_SIGNALLING_THROUGH_P2RY1 | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ADP_SIGNALLING_THROUGH_P2RY1 |
| REACTOME_A_TETRASACCHARIDE_LINKER_SEQUENCE_IS_REQUIRED_FOR_GAG_SYNTHESIS | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_A_TETRASACCHARIDE_LINKER_SEQUENCE_IS_REQUIRED_FOR_GAG_SYNTHESIS |
| REACTOME_DARPP_32_EVENTS | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DARPP_32_EVENTS |
| REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS |
| REACTOME_EGFR_DOWNREGULATION | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_EGFR_DOWNREGULATION |
| REACTOME_FANCONI_ANEMIA_PATHWAY | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FANCONI_ANEMIA_PATHWAY |
| REACTOME_G0_AND_EARLY_G1 | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_G0_AND_EARLY_G1 |
| REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PI3KGAMMA | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PI3KGAMMA |
| REACTOME_INHIBITION_OF_INSULIN_SECRETION_BY_ADRENALINE_NORADRENALINE | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INHIBITION_OF_INSULIN_SECRETION_BY_ADRENALINE_NORADRENALINE |
| REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS |
| REACTOME_NITRIC_OXIDE_STIMULATES_GUANYLATE_CYCLASE | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NITRIC_OXIDE_STIMULATES_GUANYLATE_CYCLASE |
| REACTOME_REGULATION_OF_HYPOXIA_INDUCIBLE_FACTOR_HIF_BY_OXYGEN | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_HYPOXIA_INDUCIBLE_FACTOR_HIF_BY_OXYGEN |
| REACTOME_RNA_POL_I_TRANSCRIPTION_INITIATION | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RNA_POL_I_TRANSCRIPTION_INITIATION |
| REACTOME_SMOOTH_MUSCLE_CONTRACTION | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SMOOTH_MUSCLE_CONTRACTION |
| REACTOME_TRANSFERRIN_ENDOCYTOSIS_AND_RECYCLING | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANSFERRIN_ENDOCYTOSIS_AND_RECYCLING |
| RUAN_RESPONSE_TO_TROGLITAZONE_UP | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/RUAN_RESPONSE_TO_TROGLITAZONE_UP |
| SHARMA_PILOCYTIC_ASTROCYTOMA_LOCATION_UP | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/SHARMA_PILOCYTIC_ASTROCYTOMA_LOCATION_UP |
| STEGER_ADIPOGENESIS_DN | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/STEGER_ADIPOGENESIS_DN |
| VALK_AML_WITH_EVI1 | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_WITH_EVI1 |
| VANDESLUIS_NORMAL_EMBRYOS_DN | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/VANDESLUIS_NORMAL_EMBRYOS_DN |
| VERRECCHIA_RESPONSE_TO_TGFB1_C2 | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/VERRECCHIA_RESPONSE_TO_TGFB1_C2 |
| WAGNER_APO2_SENSITIVITY | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/WAGNER_APO2_SENSITIVITY |
| WANG_BARRETTS_ESOPHAGUS_DN | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_BARRETTS_ESOPHAGUS_DN |
| WANG_NFKB_TARGETS | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_NFKB_TARGETS |
| WONG_ENDMETRIUM_CANCER_UP | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/WONG_ENDMETRIUM_CANCER_UP |
| YANG_BREAST_CANCER_ESR1_DN | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/YANG_BREAST_CANCER_ESR1_DN |
| YORDY_RECIPROCAL_REGULATION_BY_ETS1_AND_SP100_UP | 25 | http://www.broadinstitute.org/gsea/msigdb/cards/YORDY_RECIPROCAL_REGULATION_BY_ETS1_AND_SP100_UP |
| AZARE_STAT3_TARGETS | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/AZARE_STAT3_TARGETS |
| BIOCARTA_CSK_PATHWAY | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CSK_PATHWAY |
| BIOCARTA_CXCR4_PATHWAY | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CXCR4_PATHWAY |
| BIOCARTA_ECM_PATHWAY | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_ECM_PATHWAY |
| BIOCARTA_EIF4_PATHWAY | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_EIF4_PATHWAY |
| BIOCARTA_G2_PATHWAY | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_G2_PATHWAY |
| BIOCARTA_NOS1_PATHWAY | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_NOS1_PATHWAY |
| BIOCARTA_NTHI_PATHWAY | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_NTHI_PATHWAY |
| BIOCARTA_TPO_PATHWAY | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_TPO_PATHWAY |
| BUCKANOVICH_T_LYMPHOCYTE_HOMING_ON_TUMOR_DN | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/BUCKANOVICH_T_LYMPHOCYTE_HOMING_ON_TUMOR_DN |
| BUCKANOVICH_T_LYMPHOCYTE_HOMING_ON_TUMOR_UP | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/BUCKANOVICH_T_LYMPHOCYTE_HOMING_ON_TUMOR_UP |
| CHEOK_RESPONSE_TO_HD_MTX_DN | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEOK_RESPONSE_TO_HD_MTX_DN |
| CHIANG_LIVER_CANCER_SUBCLASS_POLYSOMY7_DN | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIANG_LIVER_CANCER_SUBCLASS_POLYSOMY7_DN |
| DEBOSSCHER_NFKB_TARGETS_REPRESSED_BY_GLUCOCORTICOIDS | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/DEBOSSCHER_NFKB_TARGETS_REPRESSED_BY_GLUCOCORTICOIDS |
| DELACROIX_RAR_TARGETS_DN | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/DELACROIX_RAR_TARGETS_DN |
| FRASOR_RESPONSE_TO_SERM_OR_FULVESTRANT_UP | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/FRASOR_RESPONSE_TO_SERM_OR_FULVESTRANT_UP |
| GOLUB_ALL_VS_AML_DN | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/GOLUB_ALL_VS_AML_DN |
| GOLUB_ALL_VS_AML_UP | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/GOLUB_ALL_VS_AML_UP |
| HASLINGER_B_CLL_WITH_13Q14_DELETION | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/HASLINGER_B_CLL_WITH_13Q14_DELETION |
| HASLINGER_B_CLL_WITH_CHROMOSOME_12_TRISOMY | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/HASLINGER_B_CLL_WITH_CHROMOSOME_12_TRISOMY |
| HOFMANN_MYELODYSPLASTIC_SYNDROM_RISK_UP | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFMANN_MYELODYSPLASTIC_SYNDROM_RISK_UP |
| JEON_SMAD6_TARGETS_UP | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/JEON_SMAD6_TARGETS_UP |
| JIANG_TIP30_TARGETS_DN | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/JIANG_TIP30_TARGETS_DN |
| KEGG_NICOTINATE_AND_NICOTINAMIDE_METABOLISM | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_NICOTINATE_AND_NICOTINAMIDE_METABOLISM |
| KEGG_PROTEIN_EXPORT | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PROTEIN_EXPORT |
| KIM_GERMINAL_CENTER_T_HELPER_DN | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_GERMINAL_CENTER_T_HELPER_DN |
| LA_MEN1_TARGETS | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/LA_MEN1_TARGETS |
| MAEKAWA_ATF2_TARGETS | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/MAEKAWA_ATF2_TARGETS |
| MAGRANGEAS_MULTIPLE_MYELOMA_IGLL_VS_IGLK_DN | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/MAGRANGEAS_MULTIPLE_MYELOMA_IGLL_VS_IGLK_DN |
| MATZUK_SPERMATOGONIA | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_SPERMATOGONIA |
| MEISSNER_NPC_ICP_WITH_H3_UNMETHYLATED | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/MEISSNER_NPC_ICP_WITH_H3_UNMETHYLATED |
| MISHRA_CARCINOMA_ASSOCIATED_FIBROBLAST_DN | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/MISHRA_CARCINOMA_ASSOCIATED_FIBROBLAST_DN |
| MISHRA_CARCINOMA_ASSOCIATED_FIBROBLAST_UP | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/MISHRA_CARCINOMA_ASSOCIATED_FIBROBLAST_UP |
| MURATA_VIRULENCE_OF_H_PILORI | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/MURATA_VIRULENCE_OF_H_PILORI |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_23 | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_23 |
| ODONNELL_METASTASIS_DN | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/ODONNELL_METASTASIS_DN |
| ONO_AML1_TARGETS_UP | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/ONO_AML1_TARGETS_UP |
| OSADA_ASCL1_TARGETS_DN | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/OSADA_ASCL1_TARGETS_DN |
| OXFORD_RALA_OR_RALB_TARGETS_DN | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/OXFORD_RALA_OR_RALB_TARGETS_DN |
| PETRETTO_HEART_MASS_QTL_CIS_DN | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/PETRETTO_HEART_MASS_QTL_CIS_DN |
| PID_RHODOPSIN_PATHWAY | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_RHODOPSIN_PATHWAY |
| PID_S1P_S1P2_PATHWAY | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_S1P_S1P2_PATHWAY |
| REACTOME_CHOLESTEROL_BIOSYNTHESIS | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CHOLESTEROL_BIOSYNTHESIS |
| REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION |
| REACTOME_DOUBLE_STRAND_BREAK_REPAIR | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DOUBLE_STRAND_BREAK_REPAIR |
| REACTOME_GROWTH_HORMONE_RECEPTOR_SIGNALING | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GROWTH_HORMONE_RECEPTOR_SIGNALING |
| REACTOME_INHIBITION_OF_THE_PROTEOLYTIC_ACTIVITY_OF_APC_C_REQUIRED_FOR_THE_ONSET_OF_ANAPHASE_BY_MITOTIC_SPINDLE_CHECKPOINT_COMPONENTS | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INHIBITION_OF_THE_PROTEOLYTIC_ACTIVITY_OF_APC_C_REQUIRED_FOR_THE_ONSET_OF_ANAPHASE_BY_MITOTIC_SPINDLE_CHECKPOINT_COMPONENTS |
| REACTOME_KINESINS | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_KINESINS |
| REACTOME_NUCLEAR_EVENTS_KINASE_AND_TRANSCRIPTION_FACTOR_ACTIVATION | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NUCLEAR_EVENTS_KINASE_AND_TRANSCRIPTION_FACTOR_ACTIVATION |
| REACTOME_PYRIMIDINE_METABOLISM | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PYRIMIDINE_METABOLISM |
| REACTOME_REGULATION_OF_IFNA_SIGNALING | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_IFNA_SIGNALING |
| REACTOME_RORA_ACTIVATES_CIRCADIAN_EXPRESSION | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RORA_ACTIVATES_CIRCADIAN_EXPRESSION |
| REACTOME_SULFUR_AMINO_ACID_METABOLISM | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SULFUR_AMINO_ACID_METABOLISM |
| REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS |
| REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION |
| SA_B_CELL_RECEPTOR_COMPLEXES | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/SA_B_CELL_RECEPTOR_COMPLEXES |
| SCHRAMM_INHBA_TARGETS_DN | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHRAMM_INHBA_TARGETS_DN |
| SHI_SPARC_TARGETS_UP | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/SHI_SPARC_TARGETS_UP |
| WANG_IMMORTALIZED_BY_HOXA9_AND_MEIS1_DN | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_IMMORTALIZED_BY_HOXA9_AND_MEIS1_DN |
| WANG_LSD1_TARGETS_UP | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_LSD1_TARGETS_UP |
| WANG_TNF_TARGETS | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_TNF_TARGETS |
| WEST_ADRENOCORTICAL_CARCINOMA_VS_ADENOMA_DN | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/WEST_ADRENOCORTICAL_CARCINOMA_VS_ADENOMA_DN |
| WILLERT_WNT_SIGNALING | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/WILLERT_WNT_SIGNALING |
| YAN_ESCAPE_FROM_ANOIKIS | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/YAN_ESCAPE_FROM_ANOIKIS |
| YIH_RESPONSE_TO_ARSENITE_C1 | 24 | http://www.broadinstitute.org/gsea/msigdb/cards/YIH_RESPONSE_TO_ARSENITE_C1 |
| BARIS_THYROID_CANCER_UP | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/BARIS_THYROID_CANCER_UP |
| BILANGES_SERUM_SENSITIVE_VIA_TSC1 | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/BILANGES_SERUM_SENSITIVE_VIA_TSC1 |
| BIOCARTA_CASPASE_PATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CASPASE_PATHWAY |
| BIOCARTA_CCR3_PATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CCR3_PATHWAY |
| BIOCARTA_CELLCYCLE_PATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CELLCYCLE_PATHWAY |
| BIOCARTA_CTCF_PATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CTCF_PATHWAY |
| BIOCARTA_GLEEVEC_PATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_GLEEVEC_PATHWAY |
| BIOCARTA_IGF1R_PATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_IGF1R_PATHWAY |
| BIOCARTA_IL12_PATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_IL12_PATHWAY |
| BIOCARTA_INTRINSIC_PATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_INTRINSIC_PATHWAY |
| BIOCARTA_MEF2D_PATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_MEF2D_PATHWAY |
| BIOCARTA_MTOR_PATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_MTOR_PATHWAY |
| BIOCARTA_NFKB_PATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_NFKB_PATHWAY |
| BIOCARTA_P53HYPOXIA_PATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_P53HYPOXIA_PATHWAY |
| BIOCARTA_PTDINS_PATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_PTDINS_PATHWAY |
| BIOCARTA_RAC1_PATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_RAC1_PATHWAY |
| BIOCARTA_RAS_PATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_RAS_PATHWAY |
| CAIRO_PML_TARGETS_BOUND_BY_MYC_UP | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/CAIRO_PML_TARGETS_BOUND_BY_MYC_UP |
| CHEN_ETV5_TARGETS_TESTIS | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEN_ETV5_TARGETS_TESTIS |
| CHEOK_RESPONSE_TO_HD_MTX_UP | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEOK_RESPONSE_TO_HD_MTX_UP |
| CHIBA_RESPONSE_TO_TSA_DN | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIBA_RESPONSE_TO_TSA_DN |
| CLIMENT_BREAST_CANCER_COPY_NUMBER_UP | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/CLIMENT_BREAST_CANCER_COPY_NUMBER_UP |
| CREIGHTON_AKT1_SIGNALING_VIA_MTOR_DN | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/CREIGHTON_AKT1_SIGNALING_VIA_MTOR_DN |
| EGUCHI_CELL_CYCLE_RB1_TARGETS | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/EGUCHI_CELL_CYCLE_RB1_TARGETS |
| FERRANDO_HOX11_NEIGHBORS | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/FERRANDO_HOX11_NEIGHBORS |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_GREEN_UP | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_GREEN_UP |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_YELLOW_DN | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_YELLOW_DN |
| GRAHAM_CML_QUIESCENT_VS_CML_DIVIDING_UP | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAHAM_CML_QUIESCENT_VS_CML_DIVIDING_UP |
| HASLINGER_B_CLL_WITH_11Q23_DELETION | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/HASLINGER_B_CLL_WITH_11Q23_DELETION |
| HERNANDEZ_ABERRANT_MITOSIS_BY_DOCETACEL_4NM_UP | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/HERNANDEZ_ABERRANT_MITOSIS_BY_DOCETACEL_4NM_UP |
| HINATA_NFKB_TARGETS_KERATINOCYTE_DN | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/HINATA_NFKB_TARGETS_KERATINOCYTE_DN |
| HOFMANN_MYELODYSPLASTIC_SYNDROM_RISK_DN | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFMANN_MYELODYSPLASTIC_SYNDROM_RISK_DN |
| KEGG_MISMATCH_REPAIR | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_MISMATCH_REPAIR |
| KEGG_NITROGEN_METABOLISM | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_NITROGEN_METABOLISM |
| KEGG_PROXIMAL_TUBULE_BICARBONATE_RECLAMATION | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PROXIMAL_TUBULE_BICARBONATE_RECLAMATION |
| MAHADEVAN_IMATINIB_RESISTANCE_UP | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/MAHADEVAN_IMATINIB_RESISTANCE_UP |
| MARIADASON_RESPONSE_TO_CURCUMIN_SULINDAC_5 | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/MARIADASON_RESPONSE_TO_CURCUMIN_SULINDAC_5 |
| MARKS_HDAC_TARGETS_UP | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/MARKS_HDAC_TARGETS_UP |
| MULLIGAN_NTF3_SIGNALING_VIA_INSR_AND_IGF1R_UP | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/MULLIGAN_NTF3_SIGNALING_VIA_INSR_AND_IGF1R_UP |
| ONO_FOXP3_TARGETS_UP | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/ONO_FOXP3_TARGETS_UP |
| PID_CONE_PATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_CONE_PATHWAY |
| PID_NFKAPPABCANONICALPATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_NFKAPPABCANONICALPATHWAY |
| PID_PRLSIGNALINGEVENTSPATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_PRLSIGNALINGEVENTSPATHWAY |
| REACTOME_ABORTIVE_ELONGATION_OF_HIV1_TRANSCRIPT_IN_THE_ABSENCE_OF_TAT | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ABORTIVE_ELONGATION_OF_HIV1_TRANSCRIPT_IN_THE_ABSENCE_OF_TAT |
| REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA |
| REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING |
| REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER |
| REACTOME_GLUTATHIONE_CONJUGATION | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GLUTATHIONE_CONJUGATION |
| REACTOME_INSULIN_RECEPTOR_RECYCLING | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INSULIN_RECEPTOR_RECYCLING |
| REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS |
| REACTOME_LYSOSOME_VESICLE_BIOGENESIS | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_LYSOSOME_VESICLE_BIOGENESIS |
| REACTOME_MICRORNA_MIRNA_BIOGENESIS | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MICRORNA_MIRNA_BIOGENESIS |
| REACTOME_PHOSPHORYLATION_OF_THE_APC_C | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PHOSPHORYLATION_OF_THE_APC_C |
| REACTOME_PROCESSING_OF_CAPPED_INTRONLESS_PRE_MRNA | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PROCESSING_OF_CAPPED_INTRONLESS_PRE_MRNA |
| REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_2_PROMOTER | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_2_PROMOTER |
| REACTOME_SIGNALING_BY_BMP | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_BMP |
| REACTOME_TAK1_ACTIVATES_NFKB_BY_PHOSPHORYLATION_AND_ACTIVATION_OF_IKKS_COMPLEX | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TAK1_ACTIVATES_NFKB_BY_PHOSPHORYLATION_AND_ACTIVATION_OF_IKKS_COMPLEX |
| REACTOME_THROMBOXANE_SIGNALLING_THROUGH_TP_RECEPTOR | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_THROMBOXANE_SIGNALLING_THROUGH_TP_RECEPTOR |
| SABATES_COLORECTAL_ADENOMA_SIZE_UP | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/SABATES_COLORECTAL_ADENOMA_SIZE_UP |
| SAGIV_CD24_TARGETS_UP | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/SAGIV_CD24_TARGETS_UP |
| SEKI_INFLAMMATORY_RESPONSE_LPS_DN | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/SEKI_INFLAMMATORY_RESPONSE_LPS_DN |
| ST_GA12_PATHWAY | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_GA12_PATHWAY |
| SUZUKI_RESPONSE_TO_TSA_AND_DECITABINE_1A | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/SUZUKI_RESPONSE_TO_TSA_AND_DECITABINE_1A |
| SUZUKI_RESPONSE_TO_TSA_AND_DECITABINE_1B | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/SUZUKI_RESPONSE_TO_TSA_AND_DECITABINE_1B |
| UROSEVIC_RESPONSE_TO_IMIQUIMOD | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/UROSEVIC_RESPONSE_TO_IMIQUIMOD |
| VIETOR_IFRD1_TARGETS | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/VIETOR_IFRD1_TARGETS |
| WANG_RESPONSE_TO_FORSKOLIN_UP | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_RESPONSE_TO_FORSKOLIN_UP |
| WENG_POR_TARGETS_GLOBAL_DN | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/WENG_POR_TARGETS_GLOBAL_DN |
| WU_HBX_TARGETS_1_DN | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/WU_HBX_TARGETS_1_DN |
| WU_HBX_TARGETS_2_UP | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/WU_HBX_TARGETS_2_UP |
| YANG_BREAST_CANCER_ESR1_BULK_DN | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/YANG_BREAST_CANCER_ESR1_BULK_DN |
| YANG_MUC2_TARGETS_DUODENUM_3MO_DN | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/YANG_MUC2_TARGETS_DUODENUM_3MO_DN |
| ZHANG_INTERFERON_RESPONSE | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_INTERFERON_RESPONSE |
| ZIRN_TRETINOIN_RESPONSE_WT1_UP | 23 | http://www.broadinstitute.org/gsea/msigdb/cards/ZIRN_TRETINOIN_RESPONSE_WT1_UP |
| BIOCARTA_AKT_PATHWAY | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_AKT_PATHWAY |
| BIOCARTA_CERAMIDE_PATHWAY | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CERAMIDE_PATHWAY |
| BIOCARTA_CHEMICAL_PATHWAY | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CHEMICAL_PATHWAY |
| BIOCARTA_CYTOKINE_PATHWAY | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CYTOKINE_PATHWAY |
| BIOCARTA_DC_PATHWAY | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_DC_PATHWAY |
| BIOCARTA_GCR_PATHWAY | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_GCR_PATHWAY |
| BIOCARTA_HER2_PATHWAY | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_HER2_PATHWAY |
| BIOCARTA_IL2_PATHWAY | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_IL2_PATHWAY |
| BIOCARTA_IL6_PATHWAY | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_IL6_PATHWAY |
| BIOCARTA_INSULIN_PATHWAY | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_INSULIN_PATHWAY |
| BIOCARTA_SPPA_PATHWAY | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_SPPA_PATHWAY |
| BONOME_OVARIAN_CANCER_POOR_SURVIVAL_DN | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/BONOME_OVARIAN_CANCER_POOR_SURVIVAL_DN |
| BRACHAT_RESPONSE_TO_CISPLATIN | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/BRACHAT_RESPONSE_TO_CISPLATIN |
| BREUHAHN_GROWTH_FACTOR_SIGNALING_IN_LIVER_CANCER | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/BREUHAHN_GROWTH_FACTOR_SIGNALING_IN_LIVER_CANCER |
| CHEOK_RESPONSE_TO_MERCAPTOPURINE_DN | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEOK_RESPONSE_TO_MERCAPTOPURINE_DN |
| FERRARI_RESPONSE_TO_FENRETINIDE_UP | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/FERRARI_RESPONSE_TO_FENRETINIDE_UP |
| GALLUZZI_PREVENT_MITOCHONDIAL_PERMEABILIZATION | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/GALLUZZI_PREVENT_MITOCHONDIAL_PERMEABILIZATION |
| GAUSSMANN_MLL_AF4_FUSION_TARGETS_E_DN | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/GAUSSMANN_MLL_AF4_FUSION_TARGETS_E_DN |
| GAVIN_PDE3B_TARGETS | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/GAVIN_PDE3B_TARGETS |
| HANSON_HRAS_SIGNALING_VIA_NFKB | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/HANSON_HRAS_SIGNALING_VIA_NFKB |
| HOFMANN_MYELODYSPLASTIC_SYNDROM_LOW_RISK_UP | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFMANN_MYELODYSPLASTIC_SYNDROM_LOW_RISK_UP |
| HOLLEMAN_ASPARAGINASE_RESISTANCE_ALL_UP | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLEMAN_ASPARAGINASE_RESISTANCE_ALL_UP |
| HOLLEMAN_PREDNISOLONE_RESISTANCE_B_ALL_UP | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLEMAN_PREDNISOLONE_RESISTANCE_B_ALL_UP |
| KANG_FLUOROURACIL_RESISTANCE_UP | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/KANG_FLUOROURACIL_RESISTANCE_UP |
| KEGG_BETA_ALANINE_METABOLISM | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_BETA_ALANINE_METABOLISM |
| KEGG_BIOSYNTHESIS_OF_UNSATURATED_FATTY_ACIDS | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_BIOSYNTHESIS_OF_UNSATURATED_FATTY_ACIDS |
| KEGG_GLYCOSAMINOGLYCAN_BIOSYNTHESIS_CHONDROITIN_SULFATE | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GLYCOSAMINOGLYCAN_BIOSYNTHESIS_CHONDROITIN_SULFATE |
| KORKOLA_EMBRYONIC_CARCINOMA_VS_SEMINOMA_UP | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/KORKOLA_EMBRYONIC_CARCINOMA_VS_SEMINOMA_UP |
| LANDIS_ERBB2_BREAST_TUMORS_65_UP | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/LANDIS_ERBB2_BREAST_TUMORS_65_UP |
| LU_TUMOR_ENDOTHELIAL_MARKERS_UP | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/LU_TUMOR_ENDOTHELIAL_MARKERS_UP |
| MAGRANGEAS_MULTIPLE_MYELOMA_IGG_VS_IGA_UP | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/MAGRANGEAS_MULTIPLE_MYELOMA_IGG_VS_IGA_UP |
| MATZUK_IMPLANTATION_AND_UTERINE | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_IMPLANTATION_AND_UTERINE |
| MOOTHA_FFA_OXYDATION | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/MOOTHA_FFA_OXYDATION |
| NIKOLSKY_BREAST_CANCER_15Q26_AMPLICON | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_15Q26_AMPLICON |
| NIKOLSKY_BREAST_CANCER_19Q13.1_AMPLICON | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_19Q13.1_AMPLICON |
| NIKOLSKY_OVERCONNECTED_IN_BREAST_CANCER | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_OVERCONNECTED_IN_BREAST_CANCER |
| NUMATA_CSF3_SIGNALING_VIA_STAT3 | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/NUMATA_CSF3_SIGNALING_VIA_STAT3 |
| OKAWA_NEUROBLASTOMA_1P36_31_DELETION | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/OKAWA_NEUROBLASTOMA_1P36_31_DELETION |
| PARK_TRETINOIN_RESPONSE_AND_RARA_PLZF_FUSION | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/PARK_TRETINOIN_RESPONSE_AND_RARA_PLZF_FUSION |
| PID_HEDGEHOG_2PATHWAY | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_HEDGEHOG_2PATHWAY |
| PID_PDGFRAPATHWAY | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_PDGFRAPATHWAY |
| RAMPON_ENRICHED_LEARNING_ENVIRONMENT_LATE_UP | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/RAMPON_ENRICHED_LEARNING_ENVIRONMENT_LATE_UP |
| REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACYL_CHAIN_REMODELLING_OF_PC |
| REACTOME_CD28_DEPENDENT_PI3K_AKT_SIGNALING | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CD28_DEPENDENT_PI3K_AKT_SIGNALING |
| REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE |
| REACTOME_DEADENYLATION_OF_MRNA | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DEADENYLATION_OF_MRNA |
| REACTOME_FGFR_LIGAND_BINDING_AND_ACTIVATION | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FGFR_LIGAND_BINDING_AND_ACTIVATION |
| REACTOME_FORMATION_OF_TUBULIN_FOLDING_INTERMEDIATES_BY_CCT_TRIC | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FORMATION_OF_TUBULIN_FOLDING_INTERMEDIATES_BY_CCT_TRIC |
| REACTOME_GRB2_EVENTS_IN_ERBB2_SIGNALING | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GRB2_EVENTS_IN_ERBB2_SIGNALING |
| REACTOME_INCRETIN_SYNTHESIS_SECRETION_AND_INACTIVATION | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INCRETIN_SYNTHESIS_SECRETION_AND_INACTIVATION |
| REACTOME_METAL_ION_SLC_TRANSPORTERS | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_METAL_ION_SLC_TRANSPORTERS |
| REACTOME_RNA_POL_I_TRANSCRIPTION_TERMINATION | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RNA_POL_I_TRANSCRIPTION_TERMINATION |
| REACTOME_SIGNALING_BY_HIPPO | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_HIPPO |
| ROY_WOUND_BLOOD_VESSEL_DN | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/ROY_WOUND_BLOOD_VESSEL_DN |
| SCIAN_CELL_CYCLE_TARGETS_OF_TP53_AND_TP73_DN | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/SCIAN_CELL_CYCLE_TARGETS_OF_TP53_AND_TP73_DN |
| SPIRA_SMOKERS_LUNG_CANCER_DN | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/SPIRA_SMOKERS_LUNG_CANCER_DN |
| SUZUKI_RESPONSE_TO_TSA | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/SUZUKI_RESPONSE_TO_TSA |
| TIAN_TNF_SIGNALING_NOT_VIA_NFKB | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/TIAN_TNF_SIGNALING_NOT_VIA_NFKB |
| VALK_AML_WITH_11Q23_REARRANGED | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_WITH_11Q23_REARRANGED |
| VERNELL_RETINOBLASTOMA_PATHWAY_DN | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/VERNELL_RETINOBLASTOMA_PATHWAY_DN |
| WANG_METASTASIS_OF_BREAST_CANCER_ESR1_UP | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_METASTASIS_OF_BREAST_CANCER_ESR1_UP |
| WEBER_METHYLATED_ICP_IN_FIBROBLAST | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/WEBER_METHYLATED_ICP_IN_FIBROBLAST |
| WELCH_GATA1_TARGETS | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/WELCH_GATA1_TARGETS |
| YUAN_ZNF143_PARTNERS | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/YUAN_ZNF143_PARTNERS |
| ZHAN_MULTIPLE_MYELOMA_SPIKED | 22 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_MULTIPLE_MYELOMA_SPIKED |
| AMIT_SERUM_RESPONSE_20_MCF10A | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_SERUM_RESPONSE_20_MCF10A |
| BIOCARTA_ATRBRCA_PATHWAY | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_ATRBRCA_PATHWAY |
| BIOCARTA_CALCINEURIN_PATHWAY | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CALCINEURIN_PATHWAY |
| BIOCARTA_CTLA4_PATHWAY | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CTLA4_PATHWAY |
| BIOCARTA_IGF1_PATHWAY | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_IGF1_PATHWAY |
| BIOCARTA_MITOCHONDRIA_PATHWAY | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_MITOCHONDRIA_PATHWAY |
| BIOCARTA_NDKDYNAMIN_PATHWAY | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_NDKDYNAMIN_PATHWAY |
| BIOCARTA_TFF_PATHWAY | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_TFF_PATHWAY |
| BIOCARTA_TOB1_PATHWAY | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_TOB1_PATHWAY |
| CEBALLOS_TARGETS_OF_TP53_AND_MYC_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/CEBALLOS_TARGETS_OF_TP53_AND_MYC_UP |
| CHEOK_RESPONSE_TO_MERCAPTOPURINE_AND_LD_MTX_DN | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEOK_RESPONSE_TO_MERCAPTOPURINE_AND_LD_MTX_DN |
| DAZARD_UV_RESPONSE_CLUSTER_G4 | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/DAZARD_UV_RESPONSE_CLUSTER_G4 |
| DELLA_RESPONSE_TO_TSA_AND_BUTYRATE | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/DELLA_RESPONSE_TO_TSA_AND_BUTYRATE |
| FERRANDO_TAL1_NEIGHBORS | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/FERRANDO_TAL1_NEIGHBORS |
| FINETTI_BREAST_CANCERS_KINOME_BLUE | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/FINETTI_BREAST_CANCERS_KINOME_BLUE |
| FOURNIER_ACINAR_DEVELOPMENT_EARLY_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/FOURNIER_ACINAR_DEVELOPMENT_EARLY_UP |
| FOURNIER_ACINAR_DEVELOPMENT_LATE_DN | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/FOURNIER_ACINAR_DEVELOPMENT_LATE_DN |
| GESERICK_TERT_TARGETS_DN | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/GESERICK_TERT_TARGETS_DN |
| GREENBAUM_E2A_TARGETS_DN | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/GREENBAUM_E2A_TARGETS_DN |
| GUENTHER_GROWTH_SPHERICAL_VS_ADHERENT_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/GUENTHER_GROWTH_SPHERICAL_VS_ADHERENT_UP |
| HASLINGER_B_CLL_WITH_17P13_DELETION | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/HASLINGER_B_CLL_WITH_17P13_DELETION |
| HUI_MAPK14_TARGETS_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/HUI_MAPK14_TARGETS_UP |
| HU_ANGIOGENESIS_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/HU_ANGIOGENESIS_UP |
| IIZUKA_LIVER_CANCER_PROGRESSION_L0_L1_DN | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/IIZUKA_LIVER_CANCER_PROGRESSION_L0_L1_DN |
| JOHANSSON_GLIOMAGENESIS_BY_PDGFB_DN | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/JOHANSSON_GLIOMAGENESIS_BY_PDGFB_DN |
| KANNAN_TP53_TARGETS_DN | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/KANNAN_TP53_TARGETS_DN |
| KEGG_GLYCOSAMINOGLYCAN_DEGRADATION | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GLYCOSAMINOGLYCAN_DEGRADATION |
| KUROZUMI_RESPONSE_TO_ONCOCYTIC_VIRUS_AND_CYCLIC_RGD | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/KUROZUMI_RESPONSE_TO_ONCOCYTIC_VIRUS_AND_CYCLIC_RGD |
| KYNG_ENVIRONMENTAL_STRESS_RESPONSE_DN | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_ENVIRONMENTAL_STRESS_RESPONSE_DN |
| LANDEMAINE_LUNG_METASTASIS | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/LANDEMAINE_LUNG_METASTASIS |
| LEE_INTRATHYMIC_T_PROGENITOR | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_INTRATHYMIC_T_PROGENITOR |
| MACLACHLAN_BRCA1_TARGETS_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/MACLACHLAN_BRCA1_TARGETS_UP |
| MARCHINI_TRABECTEDIN_RESISTANCE_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/MARCHINI_TRABECTEDIN_RESISTANCE_UP |
| MARIADASON_RESPONSE_TO_BUTYRATE_SULINDAC_4 | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/MARIADASON_RESPONSE_TO_BUTYRATE_SULINDAC_4 |
| MCCABE_HOXC6_TARGETS_DN | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/MCCABE_HOXC6_TARGETS_DN |
| MOOTHA_GLYCOGEN_METABOLISM | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/MOOTHA_GLYCOGEN_METABOLISM |
| MOOTHA_GLYCOLYSIS | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/MOOTHA_GLYCOLYSIS |
| MURAKAMI_UV_RESPONSE_6HR_DN | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/MURAKAMI_UV_RESPONSE_6HR_DN |
| NIKOLSKY_BREAST_CANCER_6P24_P22_AMPLICON | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_6P24_P22_AMPLICON |
| OUYANG_PROSTATE_CANCER_PROGRESSION_DN | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/OUYANG_PROSTATE_CANCER_PROGRESSION_DN |
| PEPPER_CHRONIC_LYMPHOCYTIC_LEUKEMIA_DN | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/PEPPER_CHRONIC_LYMPHOCYTIC_LEUKEMIA_DN |
| PID_ECADHERIN_KERATINOCYTE_PATHWAY | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ECADHERIN_KERATINOCYTE_PATHWAY |
| PID_P38_MK2PATHWAY | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_P38_MK2PATHWAY |
| PID_S1P_META_PATHWAY | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_S1P_META_PATHWAY |
| PID_S1P_S1P1_PATHWAY | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_S1P_S1P1_PATHWAY |
| RAHMAN_TP53_TARGETS_PHOSPHORYLATED | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/RAHMAN_TP53_TARGETS_PHOSPHORYLATED |
| REACTOME_ACYL_CHAIN_REMODELLING_OF_PE | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACYL_CHAIN_REMODELLING_OF_PE |
| REACTOME_ADP_SIGNALLING_THROUGH_P2RY12 | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ADP_SIGNALLING_THROUGH_P2RY12 |
| REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC |
| REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS |
| REACTOME_CTLA4_INHIBITORY_SIGNALING | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CTLA4_INHIBITORY_SIGNALING |
| REACTOME_EARLY_PHASE_OF_HIV_LIFE_CYCLE | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_EARLY_PHASE_OF_HIV_LIFE_CYCLE |
| REACTOME_ERK_MAPK_TARGETS | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ERK_MAPK_TARGETS |
| REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING |
| REACTOME_LIGAND_GATED_ION_CHANNEL_TRANSPORT | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_LIGAND_GATED_ION_CHANNEL_TRANSPORT |
| REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION |
| REACTOME_NEF_MEDIATES_DOWN_MODULATION_OF_CELL_SURFACE_RECEPTORS_BY_RECRUITING_THEM_TO_CLATHRIN_ADAPTERS | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NEF_MEDIATES_DOWN_MODULATION_OF_CELL_SURFACE_RECEPTORS_BY_RECRUITING_THEM_TO_CLATHRIN_ADAPTERS |
| REACTOME_PEROXISOMAL_LIPID_METABOLISM | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PEROXISOMAL_LIPID_METABOLISM |
| REACTOME_TRAF6_MEDIATED_NFKB_ACTIVATION | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRAF6_MEDIATED_NFKB_ACTIVATION |
| SARTIPY_NORMAL_AT_INSULIN_RESISTANCE_DN | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/SARTIPY_NORMAL_AT_INSULIN_RESISTANCE_DN |
| SCHAEFFER_SOX9_TARGETS_IN_PROSTATE_DEVELOPMENT_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_SOX9_TARGETS_IN_PROSTATE_DEVELOPMENT_UP |
| SCHURINGA_STAT5A_TARGETS_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHURINGA_STAT5A_TARGETS_UP |
| SMID_BREAST_CANCER_RELAPSE_IN_LUNG_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_RELAPSE_IN_LUNG_UP |
| SPIELMAN_LYMPHOBLAST_EUROPEAN_VS_ASIAN_2FC_DN | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/SPIELMAN_LYMPHOBLAST_EUROPEAN_VS_ASIAN_2FC_DN |
| STEGER_ADIPOGENESIS_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/STEGER_ADIPOGENESIS_UP |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_SUSTAINED_IN_MONOCYTE_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_SUSTAINED_IN_MONOCYTE_UP |
| TURASHVILI_BREAST_CARCINOMA_DUCTAL_VS_LOBULAR_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/TURASHVILI_BREAST_CARCINOMA_DUCTAL_VS_LOBULAR_UP |
| VERRECCHIA_RESPONSE_TO_TGFB1_C5 | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/VERRECCHIA_RESPONSE_TO_TGFB1_C5 |
| WANG_NEOPLASTIC_TRANSFORMATION_BY_CCND1_MYC | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_NEOPLASTIC_TRANSFORMATION_BY_CCND1_MYC |
| WENG_POR_DOSAGE | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/WENG_POR_DOSAGE |
| WEST_ADRENOCORTICAL_CARCINOMA_VS_ADENOMA_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/WEST_ADRENOCORTICAL_CARCINOMA_VS_ADENOMA_UP |
| WEST_ADRENOCORTICAL_TUMOR_MARKERS_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/WEST_ADRENOCORTICAL_TUMOR_MARKERS_UP |
| WHITE_NEUROBLASTOMA_WITH_1P36.3_DELETION | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/WHITE_NEUROBLASTOMA_WITH_1P36.3_DELETION |
| WILSON_PROTEASES_AT_TUMOR_BONE_INTERFACE_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/WILSON_PROTEASES_AT_TUMOR_BONE_INTERFACE_UP |
| WOTTON_RUNX_TARGETS_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/WOTTON_RUNX_TARGETS_UP |
| YANG_MUC2_TARGETS_DUODENUM_6MO_DN | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/YANG_MUC2_TARGETS_DUODENUM_6MO_DN |
| ZIRN_TRETINOIN_RESPONSE_UP | 21 | http://www.broadinstitute.org/gsea/msigdb/cards/ZIRN_TRETINOIN_RESPONSE_UP |
| ABE_VEGFA_TARGETS | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/ABE_VEGFA_TARGETS |
| AMIT_EGF_RESPONSE_240_MCF10A | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_EGF_RESPONSE_240_MCF10A |
| AMUNDSON_GAMMA_RADIATION_RESISTANCE | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/AMUNDSON_GAMMA_RADIATION_RESISTANCE |
| BARRIER_COLON_CANCER_RECURRENCE_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/BARRIER_COLON_CANCER_RECURRENCE_DN |
| BIOCARTA_ACTINY_PATHWAY | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_ACTINY_PATHWAY |
| BIOCARTA_AMI_PATHWAY | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_AMI_PATHWAY |
| BIOCARTA_ATM_PATHWAY | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_ATM_PATHWAY |
| BIOCARTA_CCR5_PATHWAY | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CCR5_PATHWAY |
| BIOCARTA_IGF1MTOR_PATHWAY | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_IGF1MTOR_PATHWAY |
| BIOCARTA_NKCELLS_PATHWAY | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_NKCELLS_PATHWAY |
| CHEN_ETV5_TARGETS_SERTOLI | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEN_ETV5_TARGETS_SERTOLI |
| CLASPER_LYMPHATIC_VESSELS_DURING_METASTASIS_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/CLASPER_LYMPHATIC_VESSELS_DURING_METASTASIS_UP |
| COLLIS_PRKDC_SUBSTRATES | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/COLLIS_PRKDC_SUBSTRATES |
| CROONQUIST_IL6_DEPRIVATION_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/CROONQUIST_IL6_DEPRIVATION_UP |
| DAVICIONI_PAX_FOXO1_SIGNATURE_IN_ARMS_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/DAVICIONI_PAX_FOXO1_SIGNATURE_IN_ARMS_DN |
| DAZARD_UV_RESPONSE_CLUSTER_G28 | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/DAZARD_UV_RESPONSE_CLUSTER_G28 |
| DE_YY1_TARGETS_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/DE_YY1_TARGETS_UP |
| DING_LUNG_CANCER_BY_MUTATION_RATE | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/DING_LUNG_CANCER_BY_MUTATION_RATE |
| FARMER_BREAST_CANCER_CLUSTER_7 | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/FARMER_BREAST_CANCER_CLUSTER_7 |
| HASLINGER_B_CLL_WITH_6Q21_DELETION | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/HASLINGER_B_CLL_WITH_6Q21_DELETION |
| HEDENFALK_BREAST_CANCER_BRACX_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/HEDENFALK_BREAST_CANCER_BRACX_DN |
| HEDENFALK_BREAST_CANCER_BRACX_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/HEDENFALK_BREAST_CANCER_BRACX_UP |
| HOFMANN_MYELODYSPLASTIC_SYNDROM_HIGH_RISK_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFMANN_MYELODYSPLASTIC_SYNDROM_HIGH_RISK_DN |
| KIM_MYCL1_AMPLIFICATION_TARGETS_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_MYCL1_AMPLIFICATION_TARGETS_DN |
| KORKOLA_YOLK_SAC_TUMOR_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/KORKOLA_YOLK_SAC_TUMOR_UP |
| KYNG_WERNER_SYNDROM_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_WERNER_SYNDROM_UP |
| LANDIS_ERBB2_BREAST_PRENEOPLASTIC_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/LANDIS_ERBB2_BREAST_PRENEOPLASTIC_UP |
| LIU_TARGETS_OF_VMYB_VS_CMYB_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_TARGETS_OF_VMYB_VS_CMYB_UP |
| MAHADEVAN_IMATINIB_RESISTANCE_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/MAHADEVAN_IMATINIB_RESISTANCE_DN |
| MAHADEVAN_RESPONSE_TO_MP470_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/MAHADEVAN_RESPONSE_TO_MP470_UP |
| MANN_RESPONSE_TO_AMIFOSTINE_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/MANN_RESPONSE_TO_AMIFOSTINE_UP |
| MASRI_RESISTANCE_TO_TAMOXIFEN_AND_AROMATASE_INHIBITORS_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/MASRI_RESISTANCE_TO_TAMOXIFEN_AND_AROMATASE_INHIBITORS_DN |
| MASRI_RESISTANCE_TO_TAMOXIFEN_AND_AROMATASE_INHIBITORS_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/MASRI_RESISTANCE_TO_TAMOXIFEN_AND_AROMATASE_INHIBITORS_UP |
| MCCABE_HOXC6_TARGETS_CANCER_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/MCCABE_HOXC6_TARGETS_CANCER_DN |
| MEINHOLD_OVARIAN_CANCER_LOW_GRADE_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/MEINHOLD_OVARIAN_CANCER_LOW_GRADE_DN |
| MEISSNER_NPC_ICP_WITH_H3K4ME3 | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/MEISSNER_NPC_ICP_WITH_H3K4ME3 |
| MOROSETTI_FACIOSCAPULOHUMERAL_MUSCULAR_DISTROPHY_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/MOROSETTI_FACIOSCAPULOHUMERAL_MUSCULAR_DISTROPHY_UP |
| MURAKAMI_UV_RESPONSE_24HR | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/MURAKAMI_UV_RESPONSE_24HR |
| NIELSEN_GIST_AND_SYNOVIAL_SARCOMA_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/NIELSEN_GIST_AND_SYNOVIAL_SARCOMA_DN |
| NIELSEN_GIST_AND_SYNOVIAL_SARCOMA_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/NIELSEN_GIST_AND_SYNOVIAL_SARCOMA_UP |
| NIELSEN_GIST_VS_SYNOVIAL_SARCOMA_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/NIELSEN_GIST_VS_SYNOVIAL_SARCOMA_DN |
| NIELSEN_LEIOMYOSARCOMA_CNN1_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/NIELSEN_LEIOMYOSARCOMA_CNN1_DN |
| NIELSEN_SYNOVIAL_SARCOMA_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/NIELSEN_SYNOVIAL_SARCOMA_DN |
| OUYANG_PROSTATE_CANCER_PROGRESSION_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/OUYANG_PROSTATE_CANCER_PROGRESSION_UP |
| PEDERSEN_METASTASIS_BY_ERBB2_ISOFORM_3 | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/PEDERSEN_METASTASIS_BY_ERBB2_ISOFORM_3 |
| PHESSE_TARGETS_OF_APC_AND_MBD2_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/PHESSE_TARGETS_OF_APC_AND_MBD2_UP |
| PID_WNT_CANONICAL_PATHWAY | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_WNT_CANONICAL_PATHWAY |
| REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY |
| REACTOME_ENOS_ACTIVATION_AND_REGULATION | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ENOS_ACTIVATION_AND_REGULATION |
| REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PLC_BETA | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PLC_BETA |
| REACTOME_HS_GAG_DEGRADATION | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_HS_GAG_DEGRADATION |
| REACTOME_NEPHRIN_INTERACTIONS | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NEPHRIN_INTERACTIONS |
| REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS |
| REACTOME_SHC1_EVENTS_IN_ERBB4_SIGNALING | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SHC1_EVENTS_IN_ERBB4_SIGNALING |
| SESTO_RESPONSE_TO_UV_C3 | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/SESTO_RESPONSE_TO_UV_C3 |
| SESTO_RESPONSE_TO_UV_C4 | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/SESTO_RESPONSE_TO_UV_C4 |
| SETLUR_PROSTATE_CANCER_TMPRSS2_ERG_FUSION_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/SETLUR_PROSTATE_CANCER_TMPRSS2_ERG_FUSION_DN |
| STARK_HYPPOCAMPUS_22Q11_DELETION_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/STARK_HYPPOCAMPUS_22Q11_DELETION_DN |
| ST_WNT_CA2_CYCLIC_GMP_PATHWAY | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_WNT_CA2_CYCLIC_GMP_PATHWAY |
| SU_THYMUS | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/SU_THYMUS |
| TOMLINS_METASTASIS_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/TOMLINS_METASTASIS_DN |
| TORCHIA_TARGETS_OF_EWSR1_FLI1_FUSION_TOP20_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/TORCHIA_TARGETS_OF_EWSR1_FLI1_FUSION_TOP20_UP |
| VILIMAS_NOTCH1_TARGETS_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/VILIMAS_NOTCH1_TARGETS_DN |
| WALLACE_PROSTATE_CANCER_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/WALLACE_PROSTATE_CANCER_UP |
| WANG_RECURRENT_LIVER_CANCER_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_RECURRENT_LIVER_CANCER_UP |
| WANG_THOC1_TARGETS_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_THOC1_TARGETS_DN |
| WEBER_METHYLATED_HCP_IN_SPERM_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/WEBER_METHYLATED_HCP_IN_SPERM_UP |
| WENG_POR_TARGETS_GLOBAL_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/WENG_POR_TARGETS_GLOBAL_UP |
| WENG_POR_TARGETS_LIVER_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/WENG_POR_TARGETS_LIVER_DN |
| WEST_ADRENOCORTICAL_TUMOR_MARKERS_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/WEST_ADRENOCORTICAL_TUMOR_MARKERS_DN |
| XU_HGF_SIGNALING_NOT_VIA_AKT1_48HR_DN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_HGF_SIGNALING_NOT_VIA_AKT1_48HR_DN |
| ZEMBUTSU_SENSITIVITY_TO_CISPLATIN | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/ZEMBUTSU_SENSITIVITY_TO_CISPLATIN |
| ZHU_SKIL_TARGETS_UP | 20 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHU_SKIL_TARGETS_UP |
| ABRAHAM_ALPC_VS_MULTIPLE_MYELOMA_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/ABRAHAM_ALPC_VS_MULTIPLE_MYELOMA_DN |
| AMIT_EGF_RESPONSE_40_MCF10A | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_EGF_RESPONSE_40_MCF10A |
| BERENJENO_TRANSFORMED_BY_RHOA_FOREVER_UP | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/BERENJENO_TRANSFORMED_BY_RHOA_FOREVER_UP |
| BIOCARTA_COMP_PATHWAY | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_COMP_PATHWAY |
| BIOCARTA_EPO_PATHWAY | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_EPO_PATHWAY |
| BIOCARTA_MAL_PATHWAY | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_MAL_PATHWAY |
| BIOCARTA_MTA3_PATHWAY | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_MTA3_PATHWAY |
| BIOCARTA_STATHMIN_PATHWAY | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_STATHMIN_PATHWAY |
| BIOCARTA_TGFB_PATHWAY | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_TGFB_PATHWAY |
| BIOCARTA_TH1TH2_PATHWAY | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_TH1TH2_PATHWAY |
| BIOCARTA_TID_PATHWAY | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_TID_PATHWAY |
| BOYAULT_LIVER_CANCER_SUBCLASS_G6_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYAULT_LIVER_CANCER_SUBCLASS_G6_DN |
| CHEMELLO_SOLEUS_VS_EDL_MYOFIBERS_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEMELLO_SOLEUS_VS_EDL_MYOFIBERS_DN |
| CHEN_PDGF_TARGETS | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEN_PDGF_TARGETS |
| CHIARETTI_T_ALL_RELAPSE_PROGNOSIS | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIARETTI_T_ALL_RELAPSE_PROGNOSIS |
| DUNNE_TARGETS_OF_AML1_MTG8_FUSION_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/DUNNE_TARGETS_OF_AML1_MTG8_FUSION_DN |
| FARMER_BREAST_CANCER_CLUSTER_5 | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/FARMER_BREAST_CANCER_CLUSTER_5 |
| GAJATE_RESPONSE_TO_TRABECTEDIN_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/GAJATE_RESPONSE_TO_TRABECTEDIN_DN |
| GAUSSMANN_MLL_AF4_FUSION_TARGETS_C_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/GAUSSMANN_MLL_AF4_FUSION_TARGETS_C_DN |
| GAVIN_IL2_RESPONSIVE_FOXP3_TARGETS_UP | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/GAVIN_IL2_RESPONSIVE_FOXP3_TARGETS_UP |
| GENTLES_LEUKEMIC_STEM_CELL_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/GENTLES_LEUKEMIC_STEM_CELL_DN |
| GRANDVAUX_IRF3_TARGETS_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/GRANDVAUX_IRF3_TARGETS_DN |
| HAHTOLA_MYCOSIS_FUNGOIDES_UP | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/HAHTOLA_MYCOSIS_FUNGOIDES_UP |
| HERNANDEZ_MITOTIC_ARREST_BY_DOCETAXEL_2_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/HERNANDEZ_MITOTIC_ARREST_BY_DOCETAXEL_2_DN |
| HOLLEMAN_PREDNISOLONE_RESISTANCE_ALL_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLEMAN_PREDNISOLONE_RESISTANCE_ALL_DN |
| HOLLEMAN_VINCRISTINE_RESISTANCE_ALL_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLEMAN_VINCRISTINE_RESISTANCE_ALL_DN |
| JEON_SMAD6_TARGETS_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/JEON_SMAD6_TARGETS_DN |
| KAMIKUBO_MYELOID_MN1_NETWORK | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/KAMIKUBO_MYELOID_MN1_NETWORK |
| KANG_AR_TARGETS_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/KANG_AR_TARGETS_DN |
| KANG_CISPLATIN_RESISTANCE_UP | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/KANG_CISPLATIN_RESISTANCE_UP |
| KANG_DOXORUBICIN_RESISTANCE_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/KANG_DOXORUBICIN_RESISTANCE_DN |
| KEGG_ALPHA_LINOLENIC_ACID_METABOLISM | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ALPHA_LINOLENIC_ACID_METABOLISM |
| KOBAYASHI_RESPONSE_TO_ROMIDEPSIN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/KOBAYASHI_RESPONSE_TO_ROMIDEPSIN |
| KYNG_NORMAL_AGING_UP | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_NORMAL_AGING_UP |
| LAMB_CCND1_TARGETS | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/LAMB_CCND1_TARGETS |
| LEE_NAIVE_T_LYMPHOCYTE | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_NAIVE_T_LYMPHOCYTE |
| LE_NEURONAL_DIFFERENTIATION_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/LE_NEURONAL_DIFFERENTIATION_DN |
| LI_WILMS_TUMOR_ANAPLASTIC_UP | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_WILMS_TUMOR_ANAPLASTIC_UP |
| MAHADEVAN_RESPONSE_TO_MP470_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/MAHADEVAN_RESPONSE_TO_MP470_DN |
| MARIADASON_RESPONSE_TO_CURCUMIN_SULINDAC_7 | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/MARIADASON_RESPONSE_TO_CURCUMIN_SULINDAC_7 |
| MARSON_FOXP3_CORE_DIRECT_TARGETS | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/MARSON_FOXP3_CORE_DIRECT_TARGETS |
| MATZUK_EMBRYONIC_GERM_CELL | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_EMBRYONIC_GERM_CELL |
| MA_PITUITARY_FETAL_VS_ADULT_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/MA_PITUITARY_FETAL_VS_ADULT_DN |
| MEINHOLD_OVARIAN_CANCER_LOW_GRADE_UP | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/MEINHOLD_OVARIAN_CANCER_LOW_GRADE_UP |
| MELLMAN_TUT1_TARGETS_UP | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/MELLMAN_TUT1_TARGETS_UP |
| MIZUSHIMA_AUTOPHAGOSOME_FORMATION | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/MIZUSHIMA_AUTOPHAGOSOME_FORMATION |
| NAGY_TFTC_COMPONENTS_HUMAN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/NAGY_TFTC_COMPONENTS_HUMAN |
| NELSON_RESPONSE_TO_ANDROGEN_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/NELSON_RESPONSE_TO_ANDROGEN_DN |
| NIELSEN_GIST_VS_SYNOVIAL_SARCOMA_UP | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/NIELSEN_GIST_VS_SYNOVIAL_SARCOMA_UP |
| NIELSEN_LEIOMYOSARCOMA_CNN1_UP | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/NIELSEN_LEIOMYOSARCOMA_CNN1_UP |
| NIELSEN_LIPOSARCOMA_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/NIELSEN_LIPOSARCOMA_DN |
| OUYANG_PROSTATE_CANCER_MARKERS | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/OUYANG_PROSTATE_CANCER_MARKERS |
| PARK_HSC_VS_MULTIPOTENT_PROGENITORS_UP | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/PARK_HSC_VS_MULTIPOTENT_PROGENITORS_UP |
| PID_ARF_3PATHWAY | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ARF_3PATHWAY |
| PID_EPHA2_FWDPATHWAY | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_EPHA2_FWDPATHWAY |
| PID_HIF1APATHWAY | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_HIF1APATHWAY |
| RASHI_NFKB1_TARGETS | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/RASHI_NFKB1_TARGETS |
| REACTOME_ACTIVATED_AMPK_STIMULATES_FATTY_ACID_OXIDATION_IN_MUSCLE | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATED_AMPK_STIMULATES_FATTY_ACID_OXIDATION_IN_MUSCLE |
| REACTOME_BASE_EXCISION_REPAIR | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_BASE_EXCISION_REPAIR |
| REACTOME_BOTULINUM_NEUROTOXICITY | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_BOTULINUM_NEUROTOXICITY |
| REACTOME_CGMP_EFFECTS | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CGMP_EFFECTS |
| REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_G1_S_SPECIFIC_TRANSCRIPTION |
| REACTOME_LAGGING_STRAND_SYNTHESIS | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_LAGGING_STRAND_SYNTHESIS |
| REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY |
| REACTOME_PROLONGED_ERK_ACTIVATION_EVENTS | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PROLONGED_ERK_ACTIVATION_EVENTS |
| REACTOME_PROSTACYCLIN_SIGNALLING_THROUGH_PROSTACYCLIN_RECEPTOR | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PROSTACYCLIN_SIGNALLING_THROUGH_PROSTACYCLIN_RECEPTOR |
| REACTOME_PYRUVATE_METABOLISM | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PYRUVATE_METABOLISM |
| REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION |
| REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS |
| REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES |
| REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS |
| REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GLP1 | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GLP1 |
| RUAN_RESPONSE_TO_TROGLITAZONE_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/RUAN_RESPONSE_TO_TROGLITAZONE_DN |
| SARTIPY_BLUNTED_BY_INSULIN_RESISTANCE_UP | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/SARTIPY_BLUNTED_BY_INSULIN_RESISTANCE_UP |
| SA_CASPASE_CASCADE | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/SA_CASPASE_CASCADE |
| SCIBETTA_KDM5B_TARGETS_UP | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/SCIBETTA_KDM5B_TARGETS_UP |
| SEIDEN_MET_SIGNALING | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/SEIDEN_MET_SIGNALING |
| SHIN_B_CELL_LYMPHOMA_CLUSTER_9 | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/SHIN_B_CELL_LYMPHOMA_CLUSTER_9 |
| STREICHER_LSM1_TARGETS_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/STREICHER_LSM1_TARGETS_DN |
| SUH_COEXPRESSED_WITH_ID1_AND_ID2_UP | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/SUH_COEXPRESSED_WITH_ID1_AND_ID2_UP |
| SYED_ESTRADIOL_RESPONSE | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/SYED_ESTRADIOL_RESPONSE |
| TERAMOTO_OPN_TARGETS_CLUSTER_7 | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/TERAMOTO_OPN_TARGETS_CLUSTER_7 |
| TRAYNOR_RETT_SYNDROM_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/TRAYNOR_RETT_SYNDROM_DN |
| VANDESLUIS_COMMD1_TARGETS_GROUP_4_UP | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/VANDESLUIS_COMMD1_TARGETS_GROUP_4_UP |
| VERNOCHET_ADIPOGENESIS | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/VERNOCHET_ADIPOGENESIS |
| VERRECCHIA_RESPONSE_TO_TGFB1_C1 | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/VERRECCHIA_RESPONSE_TO_TGFB1_C1 |
| VISALA_AGING_LYMPHOCYTE_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/VISALA_AGING_LYMPHOCYTE_DN |
| WU_ALZHEIMER_DISEASE_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/WU_ALZHEIMER_DISEASE_DN |
| XU_HGF_TARGETS_INDUCED_BY_AKT1_6HR | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_HGF_TARGETS_INDUCED_BY_AKT1_6HR |
| YAO_HOXA10_TARGETS_VIA_PROGESTERONE_DN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_HOXA10_TARGETS_VIA_PROGESTERONE_DN |
| ZEMBUTSU_SENSITIVITY_TO_MITOMYCIN | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/ZEMBUTSU_SENSITIVITY_TO_MITOMYCIN |
| ZEMBUTSU_SENSITIVITY_TO_VINCRISTINE | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/ZEMBUTSU_SENSITIVITY_TO_VINCRISTINE |
| ZHANG_RESPONSE_TO_CANTHARIDIN_UP | 19 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_RESPONSE_TO_CANTHARIDIN_UP |
| ALONSO_METASTASIS_NEURAL_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/ALONSO_METASTASIS_NEURAL_UP |
| AMIT_DELAYED_EARLY_GENES | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_DELAYED_EARLY_GENES |
| BANDRES_RESPONSE_TO_CARMUSTIN_MGMT_48HR_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/BANDRES_RESPONSE_TO_CARMUSTIN_MGMT_48HR_UP |
| BANDRES_RESPONSE_TO_CARMUSTIN_WITHOUT_MGMT_48HR_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/BANDRES_RESPONSE_TO_CARMUSTIN_WITHOUT_MGMT_48HR_UP |
| BIOCARTA_ARAP_PATHWAY | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_ARAP_PATHWAY |
| BIOCARTA_CARDIACEGF_PATHWAY | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CARDIACEGF_PATHWAY |
| BIOCARTA_ERK5_PATHWAY | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_ERK5_PATHWAY |
| BIOCARTA_ETS_PATHWAY | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_ETS_PATHWAY |
| BIOCARTA_MCM_PATHWAY | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_MCM_PATHWAY |
| BIOCARTA_NGF_PATHWAY | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_NGF_PATHWAY |
| BIOCARTA_PTEN_PATHWAY | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_PTEN_PATHWAY |
| BIOCARTA_SPRY_PATHWAY | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_SPRY_PATHWAY |
| BIOCARTA_TEL_PATHWAY | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_TEL_PATHWAY |
| BIOCARTA_TNFR2_PATHWAY | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_TNFR2_PATHWAY |
| BIOCARTA_UCALPAIN_PATHWAY | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_UCALPAIN_PATHWAY |
| BOWIE_RESPONSE_TO_TAMOXIFEN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/BOWIE_RESPONSE_TO_TAMOXIFEN |
| CARDOSO_RESPONSE_TO_GAMMA_RADIATION_AND_3AB | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/CARDOSO_RESPONSE_TO_GAMMA_RADIATION_AND_3AB |
| CHOI_ATL_CHRONIC_VS_ACUTE_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/CHOI_ATL_CHRONIC_VS_ACUTE_DN |
| DACOSTA_LOW_DOSE_UV_RESPONSE_VIA_ERCC3_XPCS_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/DACOSTA_LOW_DOSE_UV_RESPONSE_VIA_ERCC3_XPCS_UP |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_GREY_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_GREY_UP |
| GENTILE_UV_RESPONSE_CLUSTER_D1 | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/GENTILE_UV_RESPONSE_CLUSTER_D1 |
| HASLINGER_B_CLL_WITH_MUTATED_VH_GENES | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/HASLINGER_B_CLL_WITH_MUTATED_VH_GENES |
| HONRADO_BREAST_CANCER_BRCA1_VS_BRCA2 | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/HONRADO_BREAST_CANCER_BRCA1_VS_BRCA2 |
| HUMMERICH_BENIGN_SKIN_TUMOR_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/HUMMERICH_BENIGN_SKIN_TUMOR_DN |
| HUMMERICH_BENIGN_SKIN_TUMOR_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/HUMMERICH_BENIGN_SKIN_TUMOR_UP |
| HUMMERICH_MALIGNANT_SKIN_TUMOR_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/HUMMERICH_MALIGNANT_SKIN_TUMOR_DN |
| KAPOSI_LIVER_CANCER_MET_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/KAPOSI_LIVER_CANCER_MET_UP |
| KARAKAS_TGFB1_SIGNALING | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/KARAKAS_TGFB1_SIGNALING |
| KEGG_PHENYLALANINE_METABOLISM | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PHENYLALANINE_METABOLISM |
| KIM_PTEN_TARGETS_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_PTEN_TARGETS_UP |
| KOBAYASHI_EGFR_SIGNALING_6HR_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/KOBAYASHI_EGFR_SIGNALING_6HR_DN |
| LE_NEURONAL_DIFFERENTIATION_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/LE_NEURONAL_DIFFERENTIATION_UP |
| LIN_TUMOR_ESCAPE_FROM_IMMUNE_ATTACK | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/LIN_TUMOR_ESCAPE_FROM_IMMUNE_ATTACK |
| MAINA_VHL_TARGETS_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/MAINA_VHL_TARGETS_DN |
| MCGOWAN_RSP6_TARGETS_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/MCGOWAN_RSP6_TARGETS_UP |
| MOREIRA_RESPONSE_TO_TSA_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/MOREIRA_RESPONSE_TO_TSA_DN |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_8 | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_8 |
| NADERI_BREAST_CANCER_PROGNOSIS_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/NADERI_BREAST_CANCER_PROGNOSIS_DN |
| NAM_FXYD5_TARGETS_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/NAM_FXYD5_TARGETS_DN |
| NIELSEN_LEIOMYOSARCOMA_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/NIELSEN_LEIOMYOSARCOMA_DN |
| NIELSEN_LEIOMYOSARCOMA_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/NIELSEN_LEIOMYOSARCOMA_UP |
| NIELSEN_LIPOSARCOMA_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/NIELSEN_LIPOSARCOMA_UP |
| NIELSEN_MALIGNAT_FIBROUS_HISTIOCYTOMA_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/NIELSEN_MALIGNAT_FIBROUS_HISTIOCYTOMA_DN |
| NIELSEN_MALIGNAT_FIBROUS_HISTIOCYTOMA_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/NIELSEN_MALIGNAT_FIBROUS_HISTIOCYTOMA_UP |
| NIELSEN_SCHWANNOMA_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/NIELSEN_SCHWANNOMA_DN |
| NIELSEN_SCHWANNOMA_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/NIELSEN_SCHWANNOMA_UP |
| NIELSEN_SYNOVIAL_SARCOMA_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/NIELSEN_SYNOVIAL_SARCOMA_UP |
| OZANNE_AP1_TARGETS_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/OZANNE_AP1_TARGETS_UP |
| PARK_HSC_VS_MULTIPOTENT_PROGENITORS_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/PARK_HSC_VS_MULTIPOTENT_PROGENITORS_DN |
| PID_BETACATENIN_DEG_PATHWAY | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_BETACATENIN_DEG_PATHWAY |
| PIONTEK_PKD1_TARGETS_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/PIONTEK_PKD1_TARGETS_DN |
| PLASARI_NFIC_TARGETS_BASAL_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/PLASARI_NFIC_TARGETS_BASAL_DN |
| PODAR_RESPONSE_TO_ADAPHOSTIN_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/PODAR_RESPONSE_TO_ADAPHOSTIN_DN |
| RAY_TARGETS_OF_P210_BCR_ABL_FUSION_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/RAY_TARGETS_OF_P210_BCR_ABL_FUSION_UP |
| REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS |
| REACTOME_ACTIVATED_TAK1_MEDIATES_P38_MAPK_ACTIVATION | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATED_TAK1_MEDIATES_P38_MAPK_ACTIVATION |
| REACTOME_ENERGY_DEPENDENT_REGULATION_OF_MTOR_BY_LKB1_AMPK | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ENERGY_DEPENDENT_REGULATION_OF_MTOR_BY_LKB1_AMPK |
| REACTOME_FATTY_ACYL_COA_BIOSYNTHESIS | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FATTY_ACYL_COA_BIOSYNTHESIS |
| REACTOME_GAP_JUNCTION_ASSEMBLY | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GAP_JUNCTION_ASSEMBLY |
| REACTOME_GLUCURONIDATION | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GLUCURONIDATION |
| REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS |
| REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI |
| REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA |
| REACTOME_PD1_SIGNALING | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PD1_SIGNALING |
| REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB |
| REACTOME_PLATELET_CALCIUM_HOMEOSTASIS | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PLATELET_CALCIUM_HOMEOSTASIS |
| REACTOME_REGULATION_OF_SIGNALING_BY_CBL | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_SIGNALING_BY_CBL |
| REACTOME_RIP_MEDIATED_NFKB_ACTIVATION_VIA_DAI | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RIP_MEDIATED_NFKB_ACTIVATION_VIA_DAI |
| REACTOME_SIGNALING_BY_CONSTITUTIVELY_ACTIVE_EGFR | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_CONSTITUTIVELY_ACTIVE_EGFR |
| REACTOME_SIGNALING_BY_NODAL | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_NODAL |
| REACTOME_SYNTHESIS_OF_PC | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_OF_PC |
| SARTIPY_BLUNTED_BY_INSULIN_RESISTANCE_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/SARTIPY_BLUNTED_BY_INSULIN_RESISTANCE_DN |
| SCHEIDEREIT_IKK_TARGETS | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHEIDEREIT_IKK_TARGETS |
| SCHURINGA_STAT5A_TARGETS_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHURINGA_STAT5A_TARGETS_DN |
| SHIN_B_CELL_LYMPHOMA_CLUSTER_5 | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/SHIN_B_CELL_LYMPHOMA_CLUSTER_5 |
| SILIGAN_TARGETS_OF_EWS_FLI1_FUSION_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/SILIGAN_TARGETS_OF_EWS_FLI1_FUSION_DN |
| SMID_BREAST_CANCER_LUMINAL_A_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_LUMINAL_A_DN |
| STAMBOLSKY_BOUND_BY_MUTATED_TP53 | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/STAMBOLSKY_BOUND_BY_MUTATED_TP53 |
| STEIN_ESTROGEN_RESPONSE_NOT_VIA_ESRRA | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/STEIN_ESTROGEN_RESPONSE_NOT_VIA_ESRRA |
| TOMIDA_METASTASIS_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/TOMIDA_METASTASIS_DN |
| TORCHIA_TARGETS_OF_EWSR1_FLI1_FUSION_TOP20_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/TORCHIA_TARGETS_OF_EWSR1_FLI1_FUSION_TOP20_DN |
| TUOMISTO_TUMOR_SUPPRESSION_BY_COL13A1_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/TUOMISTO_TUMOR_SUPPRESSION_BY_COL13A1_UP |
| WATANABE_ULCERATIVE_COLITIS_WITH_CANCER_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/WATANABE_ULCERATIVE_COLITIS_WITH_CANCER_UP |
| WEBER_METHYLATED_IN_COLON_CANCER | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/WEBER_METHYLATED_IN_COLON_CANCER |
| WU_HBX_TARGETS_3_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/WU_HBX_TARGETS_3_UP |
| YIH_RESPONSE_TO_ARSENITE_C2 | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/YIH_RESPONSE_TO_ARSENITE_C2 |
| YIH_RESPONSE_TO_ARSENITE_C4 | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/YIH_RESPONSE_TO_ARSENITE_C4 |
| ZEMBUTSU_SENSITIVITY_TO_CYCLOPHOSPHAMIDE | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/ZEMBUTSU_SENSITIVITY_TO_CYCLOPHOSPHAMIDE |
| ZEMBUTSU_SENSITIVITY_TO_METHOTREXATE | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/ZEMBUTSU_SENSITIVITY_TO_METHOTREXATE |
| ZEMBUTSU_SENSITIVITY_TO_VINBLASTINE | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/ZEMBUTSU_SENSITIVITY_TO_VINBLASTINE |
| ZHENG_RESPONSE_TO_ARSENITE_DN | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHENG_RESPONSE_TO_ARSENITE_DN |
| ZHENG_RESPONSE_TO_ARSENITE_UP | 18 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHENG_RESPONSE_TO_ARSENITE_UP |
| AZARE_NEOPLASTIC_TRANSFORMATION_BY_STAT3_UP | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/AZARE_NEOPLASTIC_TRANSFORMATION_BY_STAT3_UP |
| BIOCARTA_41BB_PATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_41BB_PATHWAY |
| BIOCARTA_ARF_PATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_ARF_PATHWAY |
| BIOCARTA_CK1_PATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CK1_PATHWAY |
| BIOCARTA_HCMV_PATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_HCMV_PATHWAY |
| BIOCARTA_IL10_PATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_IL10_PATHWAY |
| BIOCARTA_IL17_PATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_IL17_PATHWAY |
| BIOCARTA_IL7_PATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_IL7_PATHWAY |
| BIOCARTA_LAIR_PATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_LAIR_PATHWAY |
| BIOCARTA_NO2IL12_PATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_NO2IL12_PATHWAY |
| BIOCARTA_PML_PATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_PML_PATHWAY |
| BOWIE_RESPONSE_TO_EXTRACELLULAR_MATRIX | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/BOWIE_RESPONSE_TO_EXTRACELLULAR_MATRIX |
| BOYAULT_LIVER_CANCER_SUBCLASS_G56_DN | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYAULT_LIVER_CANCER_SUBCLASS_G56_DN |
| DORMOY_ELAVL1_TARGETS | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/DORMOY_ELAVL1_TARGETS |
| GALE_APL_WITH_FLT3_MUTATED_DN | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/GALE_APL_WITH_FLT3_MUTATED_DN |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_RED_UP | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_RED_UP |
| GOUYER_TATI_TARGETS_DN | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/GOUYER_TATI_TARGETS_DN |
| GRATIAS_RETINOBLASTOMA_16Q24 | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/GRATIAS_RETINOBLASTOMA_16Q24 |
| HOWLIN_CITED1_TARGETS_2_DN | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/HOWLIN_CITED1_TARGETS_2_DN |
| HOWLIN_CITED1_TARGETS_2_UP | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/HOWLIN_CITED1_TARGETS_2_UP |
| IIZUKA_LIVER_CANCER_PROGRESSION_L0_L1_UP | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/IIZUKA_LIVER_CANCER_PROGRESSION_L0_L1_UP |
| INGA_TP53_TARGETS | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/INGA_TP53_TARGETS |
| JI_CARCINOGENESIS_BY_KRAS_AND_STK11_DN | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/JI_CARCINOGENESIS_BY_KRAS_AND_STK11_DN |
| KANG_AR_TARGETS_UP | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/KANG_AR_TARGETS_UP |
| KASLER_HDAC7_TARGETS_1_DN | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/KASLER_HDAC7_TARGETS_1_DN |
| KEGG_ONE_CARBON_POOL_BY_FOLATE | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_ONE_CARBON_POOL_BY_FOLATE |
| KEGG_RENIN_ANGIOTENSIN_SYSTEM | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_RENIN_ANGIOTENSIN_SYSTEM |
| KEGG_STEROID_BIOSYNTHESIS | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_STEROID_BIOSYNTHESIS |
| KYNG_RESPONSE_TO_H2O2_VIA_ERCC6 | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_RESPONSE_TO_H2O2_VIA_ERCC6 |
| LEE_METASTASIS_AND_RNA_PROCESSING_UP | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_METASTASIS_AND_RNA_PROCESSING_UP |
| LEIN_LOCALIZED_TO_DISTAL_AND_PROXIMAL_DENDRITES | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/LEIN_LOCALIZED_TO_DISTAL_AND_PROXIMAL_DENDRITES |
| LEI_HOXC8_TARGETS_DN | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/LEI_HOXC8_TARGETS_DN |
| LE_SKI_TARGETS_UP | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/LE_SKI_TARGETS_UP |
| LIANG_HEMATOPOIESIS_STEM_CELL_NUMBER_QTL | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/LIANG_HEMATOPOIESIS_STEM_CELL_NUMBER_QTL |
| LIU_IL13_MEMORY_MODEL_UP | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_IL13_MEMORY_MODEL_UP |
| LIU_VAV3_PROSTATE_CARCINOGENESIS_DN | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_VAV3_PROSTATE_CARCINOGENESIS_DN |
| LI_ADIPOGENESIS_BY_ACTIVATED_PPARG | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_ADIPOGENESIS_BY_ACTIVATED_PPARG |
| LI_CYTIDINE_ANALOG_PATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_CYTIDINE_ANALOG_PATHWAY |
| LOPEZ_EPITHELIOID_MESOTHELIOMA | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/LOPEZ_EPITHELIOID_MESOTHELIOMA |
| LOPEZ_MBD_TARGETS_IMPRINTED_AND_X_LINKED | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/LOPEZ_MBD_TARGETS_IMPRINTED_AND_X_LINKED |
| MATTHEWS_AP1_TARGETS | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/MATTHEWS_AP1_TARGETS |
| MATTHEWS_SKIN_CARCINOGENESIS_VIA_JUN | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/MATTHEWS_SKIN_CARCINOGENESIS_VIA_JUN |
| MEISSNER_BRAIN_HCP_WITH_H3K4ME2 | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/MEISSNER_BRAIN_HCP_WITH_H3K4ME2 |
| MORI_EMU_MYC_LYMPHOMA_BY_ONSET_TIME_DN | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/MORI_EMU_MYC_LYMPHOMA_BY_ONSET_TIME_DN |
| MURAKAMI_UV_RESPONSE_1HR_UP | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/MURAKAMI_UV_RESPONSE_1HR_UP |
| NGO_MALIGNANT_GLIOMA_1P_LOH | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/NGO_MALIGNANT_GLIOMA_1P_LOH |
| NIKOLSKY_BREAST_CANCER_22Q13_AMPLICON | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_22Q13_AMPLICON |
| ONDER_CDH1_TARGETS_3_UP | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/ONDER_CDH1_TARGETS_3_UP |
| PID_ANTHRAXPATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ANTHRAXPATHWAY |
| PID_INTEGRIN5_PATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_INTEGRIN5_PATHWAY |
| PID_NFKAPPABATYPICALPATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_NFKAPPABATYPICALPATHWAY |
| PID_SMAD2_3PATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_SMAD2_3PATHWAY |
| PID_SYNDECAN_3_PATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_SYNDECAN_3_PATHWAY |
| REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS |
| REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION |
| REACTOME_ARMS_MEDIATED_ACTIVATION | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ARMS_MEDIATED_ACTIVATION |
| REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM |
| REACTOME_DESTABILIZATION_OF_MRNA_BY_BRF1 | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DESTABILIZATION_OF_MRNA_BY_BRF1 |
| REACTOME_DESTABILIZATION_OF_MRNA_BY_KSRP | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DESTABILIZATION_OF_MRNA_BY_KSRP |
| REACTOME_DESTABILIZATION_OF_MRNA_BY_TRISTETRAPROLIN_TTP | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DESTABILIZATION_OF_MRNA_BY_TRISTETRAPROLIN_TTP |
| REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION |
| REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS |
| REACTOME_INFLAMMASOMES | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INFLAMMASOMES |
| REACTOME_INTRINSIC_PATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INTRINSIC_PATHWAY |
| REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS |
| REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS |
| REACTOME_RAP1_SIGNALLING | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RAP1_SIGNALLING |
| REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR |
| REACTOME_REGULATION_OF_KIT_SIGNALING | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_KIT_SIGNALING |
| REACTOME_RESOLUTION_OF_AP_SITES_VIA_THE_MULTIPLE_NUCLEOTIDE_PATCH_REPLACEMENT_PATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RESOLUTION_OF_AP_SITES_VIA_THE_MULTIPLE_NUCLEOTIDE_PATCH_REPLACEMENT_PATHWAY |
| REACTOME_RNA_POL_III_CHAIN_ELONGATION | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RNA_POL_III_CHAIN_ELONGATION |
| REACTOME_SHC_RELATED_EVENTS | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SHC_RELATED_EVENTS |
| REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI |
| REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE |
| REACTOME_TIE2_SIGNALING | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TIE2_SIGNALING |
| RODRIGUES_NTN1_TARGETS_UP | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/RODRIGUES_NTN1_TARGETS_UP |
| RUAN_RESPONSE_TO_TNF_TROGLITAZONE_UP | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/RUAN_RESPONSE_TO_TNF_TROGLITAZONE_UP |
| SA_PTEN_PATHWAY | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/SA_PTEN_PATHWAY |
| SA_TRKA_RECEPTOR | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/SA_TRKA_RECEPTOR |
| SCHWAB_TARGETS_OF_BMYB_POLYMORPHIC_VARIANTS_DN | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHWAB_TARGETS_OF_BMYB_POLYMORPHIC_VARIANTS_DN |
| SIMBULAN_PARP1_TARGETS_DN | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/SIMBULAN_PARP1_TARGETS_DN |
| THUM_MIR21_TARGETS_HEART_DISEASE_UP | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/THUM_MIR21_TARGETS_HEART_DISEASE_UP |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_GRANULOCYTE_DN | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_GRANULOCYTE_DN |
| TUOMISTO_TUMOR_SUPPRESSION_BY_COL13A1_DN | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/TUOMISTO_TUMOR_SUPPRESSION_BY_COL13A1_DN |
| WIKMAN_ASBESTOS_LUNG_CANCER_UP | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/WIKMAN_ASBESTOS_LUNG_CANCER_UP |
| XU_RESPONSE_TO_TRETINOIN_AND_NSC682994_UP | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_RESPONSE_TO_TRETINOIN_AND_NSC682994_UP |
| ZEMBUTSU_SENSITIVITY_TO_DOXORUBICIN | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/ZEMBUTSU_SENSITIVITY_TO_DOXORUBICIN |
| ZEMBUTSU_SENSITIVITY_TO_FLUOROURACIL | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/ZEMBUTSU_SENSITIVITY_TO_FLUOROURACIL |
| ZEMBUTSU_SENSITIVITY_TO_NIMUSTINE | 17 | http://www.broadinstitute.org/gsea/msigdb/cards/ZEMBUTSU_SENSITIVITY_TO_NIMUSTINE |
| ABDELMOHSEN_ELAVL4_TARGETS | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/ABDELMOHSEN_ELAVL4_TARGETS |
| AMUNDSON_DNA_DAMAGE_RESPONSE_TP53 | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/AMUNDSON_DNA_DAMAGE_RESPONSE_TP53 |
| BAKER_HEMATOPOIESIS_STAT3_TARGETS | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/BAKER_HEMATOPOIESIS_STAT3_TARGETS |
| BARRIER_CANCER_RELAPSE_TUMOR_SAMPLE_UP | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/BARRIER_CANCER_RELAPSE_TUMOR_SAMPLE_UP |
| BIOCARTA_ACH_PATHWAY | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_ACH_PATHWAY |
| BIOCARTA_BCELLSURVIVAL_PATHWAY | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_BCELLSURVIVAL_PATHWAY |
| BIOCARTA_CACAM_PATHWAY | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CACAM_PATHWAY |
| BIOCARTA_CDC42RAC_PATHWAY | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CDC42RAC_PATHWAY |
| BIOCARTA_CDMAC_PATHWAY | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CDMAC_PATHWAY |
| BIOCARTA_EIF_PATHWAY | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_EIF_PATHWAY |
| BIOCARTA_GATA3_PATHWAY | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_GATA3_PATHWAY |
| BIOCARTA_IL22BP_PATHWAY | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_IL22BP_PATHWAY |
| BIOCARTA_P53_PATHWAY | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_P53_PATHWAY |
| BIOCARTA_RELA_PATHWAY | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_RELA_PATHWAY |
| BIOCARTA_SHH_PATHWAY | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_SHH_PATHWAY |
| CAFFAREL_RESPONSE_TO_THC_8HR_5_UP | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/CAFFAREL_RESPONSE_TO_THC_8HR_5_UP |
| CAIRO_HEPATOBLASTOMA_POOR_SURVIVAL | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/CAIRO_HEPATOBLASTOMA_POOR_SURVIVAL |
| CHIN_BREAST_CANCER_COPY_NUMBER_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/CHIN_BREAST_CANCER_COPY_NUMBER_DN |
| DASU_IL6_SIGNALING_SCAR_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/DASU_IL6_SIGNALING_SCAR_DN |
| DEMAGALHAES_AGING_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/DEMAGALHAES_AGING_DN |
| FARMER_BREAST_CANCER_CLUSTER_4 | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/FARMER_BREAST_CANCER_CLUSTER_4 |
| FARMER_BREAST_CANCER_CLUSTER_6 | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/FARMER_BREAST_CANCER_CLUSTER_6 |
| FINETTI_BREAST_CANCER_KINOME_GREEN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/FINETTI_BREAST_CANCER_KINOME_GREEN |
| FINETTI_BREAST_CANCER_KINOME_RED | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/FINETTI_BREAST_CANCER_KINOME_RED |
| FUKUSHIMA_TNFSF11_TARGETS | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/FUKUSHIMA_TNFSF11_TARGETS |
| GEISS_RESPONSE_TO_DSRNA_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/GEISS_RESPONSE_TO_DSRNA_DN |
| GHO_ATF5_TARGETS_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/GHO_ATF5_TARGETS_DN |
| HAHTOLA_CTCL_PATHOGENESIS | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/HAHTOLA_CTCL_PATHOGENESIS |
| HAHTOLA_MYCOSIS_FUNGOIDES_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/HAHTOLA_MYCOSIS_FUNGOIDES_DN |
| HE_PTEN_TARGETS_UP | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/HE_PTEN_TARGETS_UP |
| HUMMERICH_MALIGNANT_SKIN_TUMOR_UP | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/HUMMERICH_MALIGNANT_SKIN_TUMOR_UP |
| IGLESIAS_E2F_TARGETS_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/IGLESIAS_E2F_TARGETS_DN |
| KANG_FLUOROURACIL_RESISTANCE_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/KANG_FLUOROURACIL_RESISTANCE_DN |
| KEGG_GLYOXYLATE_AND_DICARBOXYLATE_METABOLISM | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GLYOXYLATE_AND_DICARBOXYLATE_METABOLISM |
| KEGG_OTHER_GLYCAN_DEGRADATION | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_OTHER_GLYCAN_DEGRADATION |
| KEGG_PANTOTHENATE_AND_COA_BIOSYNTHESIS | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PANTOTHENATE_AND_COA_BIOSYNTHESIS |
| KEGG_PRIMARY_BILE_ACID_BIOSYNTHESIS | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_PRIMARY_BILE_ACID_BIOSYNTHESIS |
| KEGG_RIBOFLAVIN_METABOLISM | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_RIBOFLAVIN_METABOLISM |
| KOINUMA_COLON_CANCER_MSI_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/KOINUMA_COLON_CANCER_MSI_DN |
| KORKOLA_TERATOMA_UP | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/KORKOLA_TERATOMA_UP |
| LEE_LIVER_CANCER_HEPATOBLAST | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_LIVER_CANCER_HEPATOBLAST |
| LIU_TOPBP1_TARGETS | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_TOPBP1_TARGETS |
| LOPEZ_MESOTHELIOMA_SURVIVAL_OVERALL_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/LOPEZ_MESOTHELIOMA_SURVIVAL_OVERALL_DN |
| LUI_THYROID_CANCER_CLUSTER_4 | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/LUI_THYROID_CANCER_CLUSTER_4 |
| LUND_SILENCED_BY_METHYLATION | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/LUND_SILENCED_BY_METHYLATION |
| LY_AGING_MIDDLE_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/LY_AGING_MIDDLE_DN |
| MACLACHLAN_BRCA1_TARGETS_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/MACLACHLAN_BRCA1_TARGETS_DN |
| MARIADASON_RESPONSE_TO_BUTYRATE_CURCUMIN_SULINDAC_TSA_8 | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/MARIADASON_RESPONSE_TO_BUTYRATE_CURCUMIN_SULINDAC_TSA_8 |
| MATTIOLI_MGUS_VS_MULTIPLE_MYELOMA | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/MATTIOLI_MGUS_VS_MULTIPLE_MYELOMA |
| MATTIOLI_MULTIPLE_MYELOMA_SUBGROUPS | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/MATTIOLI_MULTIPLE_MYELOMA_SUBGROUPS |
| MOOTHA_TCA | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/MOOTHA_TCA |
| NIKOLSKY_BREAST_CANCER_21Q22_AMPLICON | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_21Q22_AMPLICON |
| PID_CIRCADIANPATHWAY | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_CIRCADIANPATHWAY |
| PID_DNAPK_PATHWAY | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_DNAPK_PATHWAY |
| RAY_TARGETS_OF_P210_BCR_ABL_FUSION_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/RAY_TARGETS_OF_P210_BCR_ABL_FUSION_DN |
| REACTOME_ACETYLCHOLINE_BINDING_AND_DOWNSTREAM_EVENTS | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACETYLCHOLINE_BINDING_AND_DOWNSTREAM_EVENTS |
| REACTOME_ACTIVATED_POINT_MUTANTS_OF_FGFR2 | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATED_POINT_MUTANTS_OF_FGFR2 |
| REACTOME_ACYL_CHAIN_REMODELLING_OF_PG | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACYL_CHAIN_REMODELLING_OF_PG |
| REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT |
| REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE |
| REACTOME_EICOSANOID_LIGAND_BINDING_RECEPTORS | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_EICOSANOID_LIGAND_BINDING_RECEPTORS |
| REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING |
| REACTOME_INITIAL_TRIGGERING_OF_COMPLEMENT | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INITIAL_TRIGGERING_OF_COMPLEMENT |
| REACTOME_INTEGRATION_OF_PROVIRUS | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INTEGRATION_OF_PROVIRUS |
| REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 |
| REACTOME_NUCLEOTIDE_LIKE_PURINERGIC_RECEPTORS | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NUCLEOTIDE_LIKE_PURINERGIC_RECEPTORS |
| REACTOME_PHOSPHORYLATION_OF_CD3_AND_TCR_ZETA_CHAINS | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PHOSPHORYLATION_OF_CD3_AND_TCR_ZETA_CHAINS |
| REACTOME_PLATELET_SENSITIZATION_BY_LDL | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PLATELET_SENSITIZATION_BY_LDL |
| REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI |
| REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS |
| REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN |
| REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION |
| REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS |
| REACTOME_XENOBIOTICS | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_XENOBIOTICS |
| RODRIGUES_THYROID_CARCINOMA_UP | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/RODRIGUES_THYROID_CARCINOMA_UP |
| SCHAVOLT_TARGETS_OF_TP53_AND_TP63 | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAVOLT_TARGETS_OF_TP53_AND_TP63 |
| ST_G_ALPHA_S_PATHWAY | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_G_ALPHA_S_PATHWAY |
| SUZUKI_AMPLIFIED_IN_ORAL_CANCER | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/SUZUKI_AMPLIFIED_IN_ORAL_CANCER |
| SU_SALIVARY_GLAND | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/SU_SALIVARY_GLAND |
| TIAN_BHLHA15_TARGETS | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/TIAN_BHLHA15_TARGETS |
| TIMOFEEVA_GROWTH_STRESS_VIA_STAT1_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/TIMOFEEVA_GROWTH_STRESS_VIA_STAT1_DN |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_ERYTHROCYTE_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_ERYTHROCYTE_DN |
| VANDESLUIS_COMMD1_TARGETS_GROUP_4_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/VANDESLUIS_COMMD1_TARGETS_GROUP_4_DN |
| VETTER_TARGETS_OF_PRKCA_AND_ETS1_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/VETTER_TARGETS_OF_PRKCA_AND_ETS1_DN |
| VETTER_TARGETS_OF_PRKCA_AND_ETS1_UP | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/VETTER_TARGETS_OF_PRKCA_AND_ETS1_UP |
| WANG_RECURRENT_LIVER_CANCER_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_RECURRENT_LIVER_CANCER_DN |
| WONG_IFNA2_RESISTANCE_UP | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/WONG_IFNA2_RESISTANCE_UP |
| WU_HBX_TARGETS_1_UP | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/WU_HBX_TARGETS_1_UP |
| WU_HBX_TARGETS_2_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/WU_HBX_TARGETS_2_DN |
| XU_RESPONSE_TO_TRETINOIN_UP | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_RESPONSE_TO_TRETINOIN_UP |
| YAMASHITA_LIVER_CANCER_WITH_EPCAM_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/YAMASHITA_LIVER_CANCER_WITH_EPCAM_DN |
| ZHAN_LATE_DIFFERENTIATION_GENES_DN | 16 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_LATE_DIFFERENTIATION_GENES_DN |
| BIOCARTA_AKAPCENTROSOME_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_AKAPCENTROSOME_PATHWAY |
| BIOCARTA_BARRESTIN_SRC_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_BARRESTIN_SRC_PATHWAY |
| BIOCARTA_CD40_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CD40_PATHWAY |
| BIOCARTA_CTL_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CTL_PATHWAY |
| BIOCARTA_ERYTH_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_ERYTH_PATHWAY |
| BIOCARTA_HIF_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_HIF_PATHWAY |
| BIOCARTA_HSP27_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_HSP27_PATHWAY |
| BIOCARTA_IL3_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_IL3_PATHWAY |
| BIOCARTA_LONGEVITY_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_LONGEVITY_PATHWAY |
| BIOCARTA_NUCLEARRS_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_NUCLEARRS_PATHWAY |
| BIOCARTA_PITX2_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_PITX2_PATHWAY |
| BIOCARTA_RARRXR_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_RARRXR_PATHWAY |
| BIOCARTA_STEM_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_STEM_PATHWAY |
| BIOCARTA_TALL1_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_TALL1_PATHWAY |
| BOYAULT_LIVER_CANCER_SUBCLASS_G12_DN | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYAULT_LIVER_CANCER_SUBCLASS_G12_DN |
| CHANG_POU5F1_TARGETS_UP | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/CHANG_POU5F1_TARGETS_UP |
| COLLIS_PRKDC_REGULATORS | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/COLLIS_PRKDC_REGULATORS |
| DAVICIONI_RHABDOMYOSARCOMA_PAX_FOXO1_FUSION_DN | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/DAVICIONI_RHABDOMYOSARCOMA_PAX_FOXO1_FUSION_DN |
| DAZARD_UV_RESPONSE_CLUSTER_G3 | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/DAZARD_UV_RESPONSE_CLUSTER_G3 |
| EHRLICH_ICF_SYNDROM_DN | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/EHRLICH_ICF_SYNDROM_DN |
| FERRANDO_LYL1_NEIGHBORS | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/FERRANDO_LYL1_NEIGHBORS |
| FIGUEROA_AML_METHYLATION_CLUSTER_4_DN | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/FIGUEROA_AML_METHYLATION_CLUSTER_4_DN |
| FINETTI_BREAST_CANCERS_KINOME_GRAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/FINETTI_BREAST_CANCERS_KINOME_GRAY |
| GRANDVAUX_IRF3_TARGETS_UP | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/GRANDVAUX_IRF3_TARGETS_UP |
| HOLLEMAN_ASPARAGINASE_RESISTANCE_B_ALL_DN | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLEMAN_ASPARAGINASE_RESISTANCE_B_ALL_DN |
| HOLLEMAN_VINCRISTINE_RESISTANCE_B_ALL_DN | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLEMAN_VINCRISTINE_RESISTANCE_B_ALL_DN |
| HUMMEL_BURKITTS_LYMPHOMA_DN | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/HUMMEL_BURKITTS_LYMPHOMA_DN |
| KEGG_GLYCOSAMINOGLYCAN_BIOSYNTHESIS_KERATAN_SULFATE | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GLYCOSAMINOGLYCAN_BIOSYNTHESIS_KERATAN_SULFATE |
| KEGG_GLYCOSPHINGOLIPID_BIOSYNTHESIS_GANGLIO_SERIES | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GLYCOSPHINGOLIPID_BIOSYNTHESIS_GANGLIO_SERIES |
| KEGG_TERPENOID_BACKBONE_BIOSYNTHESIS | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_TERPENOID_BACKBONE_BIOSYNTHESIS |
| KYNG_DNA_DAMAGE_BY_4NQO_OR_GAMMA_RADIATION | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_DNA_DAMAGE_BY_4NQO_OR_GAMMA_RADIATION |
| LIU_IL13_PRIMING_MODEL | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_IL13_PRIMING_MODEL |
| LI_CYTIDINE_ANALOGS_CYCTOTOXICITY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_CYTIDINE_ANALOGS_CYCTOTOXICITY |
| MAHADEVAN_GIST_MORPHOLOGICAL_SWITCH | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/MAHADEVAN_GIST_MORPHOLOGICAL_SWITCH |
| MARKS_ACETYLATED_NON_HISTONE_PROTEINS | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/MARKS_ACETYLATED_NON_HISTONE_PROTEINS |
| MARKS_HDAC_TARGETS_DN | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/MARKS_HDAC_TARGETS_DN |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_25 | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_25 |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_27 | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_27 |
| NAGY_STAGA_COMPONENTS_HUMAN | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/NAGY_STAGA_COMPONENTS_HUMAN |
| NIKOLSKY_BREAST_CANCER_12Q24_AMPLICON | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_12Q24_AMPLICON |
| PID_ARF6DOWNSTREAMPATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ARF6DOWNSTREAMPATHWAY |
| PID_ERBB_NETWORK_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ERBB_NETWORK_PATHWAY |
| PID_ERB_GENOMIC_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ERB_GENOMIC_PATHWAY |
| PID_LPA4_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_LPA4_PATHWAY |
| PID_THROMBIN_PAR4_PATHWAY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_THROMBIN_PAR4_PATHWAY |
| PIEPOLI_LGI1_TARGETS_UP | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/PIEPOLI_LGI1_TARGETS_UP |
| RAMPON_ENRICHED_LEARNING_ENVIRONMENT_EARLY_UP | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/RAMPON_ENRICHED_LEARNING_ENVIRONMENT_EARLY_UP |
| REACTOME_ACYL_CHAIN_REMODELLING_OF_PI | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACYL_CHAIN_REMODELLING_OF_PI |
| REACTOME_ACYL_CHAIN_REMODELLING_OF_PS | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACYL_CHAIN_REMODELLING_OF_PS |
| REACTOME_AMINE_DERIVED_HORMONES | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_AMINE_DERIVED_HORMONES |
| REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS |
| REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII |
| REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION |
| REACTOME_ENDOGENOUS_STEROLS | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ENDOGENOUS_STEROLS |
| REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE |
| REACTOME_GRB2_SOS_PROVIDES_LINKAGE_TO_MAPK_SIGNALING_FOR_INTERGRINS_ | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GRB2_SOS_PROVIDES_LINKAGE_TO_MAPK_SIGNALING_FOR_INTERGRINS_ |
| REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_HDL_MEDIATED_LIPID_TRANSPORT |
| REACTOME_METABOLISM_OF_POLYAMINES | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_METABOLISM_OF_POLYAMINES |
| REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE |
| REACTOME_NRIF_SIGNALS_CELL_DEATH_FROM_THE_NUCLEUS | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NRIF_SIGNALS_CELL_DEATH_FROM_THE_NUCLEUS |
| REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_OTHER_SEMAPHORIN_INTERACTIONS |
| REACTOME_P130CAS_LINKAGE_TO_MAPK_SIGNALING_FOR_INTEGRINS | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_P130CAS_LINKAGE_TO_MAPK_SIGNALING_FOR_INTEGRINS |
| REACTOME_PROCESSIVE_SYNTHESIS_ON_THE_LAGGING_STRAND | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PROCESSIVE_SYNTHESIS_ON_THE_LAGGING_STRAND |
| REACTOME_REGULATION_OF_AMPK_ACTIVITY_VIA_LKB1 | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_AMPK_ACTIVITY_VIA_LKB1 |
| REACTOME_SEMA3A_PAK_DEPENDENT_AXON_REPULSION | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SEMA3A_PAK_DEPENDENT_AXON_REPULSION |
| REACTOME_SHC1_EVENTS_IN_EGFR_SIGNALING | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SHC1_EVENTS_IN_EGFR_SIGNALING |
| REACTOME_SHC_MEDIATED_SIGNALLING | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SHC_MEDIATED_SIGNALLING |
| REACTOME_SIGNALLING_TO_P38_VIA_RIT_AND_RIN | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALLING_TO_P38_VIA_RIT_AND_RIN |
| REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_7ALPHA_HYDROXYCHOLESTEROL | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_7ALPHA_HYDROXYCHOLESTEROL |
| REACTOME_UNBLOCKING_OF_NMDA_RECEPTOR_GLUTAMATE_BINDING_AND_ACTIVATION | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_UNBLOCKING_OF_NMDA_RECEPTOR_GLUTAMATE_BINDING_AND_ACTIVATION |
| REACTOME_ZINC_TRANSPORTERS | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ZINC_TRANSPORTERS |
| RICKMAN_TUMOR_DIFFERENTIATED_MODERATELY_VS_POORLY_DN | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/RICKMAN_TUMOR_DIFFERENTIATED_MODERATELY_VS_POORLY_DN |
| SANCHEZ_MDM2_TARGETS | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/SANCHEZ_MDM2_TARGETS |
| SANDERSON_PPARA_TARGETS | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/SANDERSON_PPARA_TARGETS |
| SA_G1_AND_S_PHASES | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/SA_G1_AND_S_PHASES |
| SA_MMP_CYTOKINE_CONNECTION | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/SA_MMP_CYTOKINE_CONNECTION |
| SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX1_UP | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX1_UP |
| SILIGAN_TARGETS_OF_EWS_FLI1_FUSION_UP | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/SILIGAN_TARGETS_OF_EWS_FLI1_FUSION_UP |
| SINGH_NFE2L2_TARGETS | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/SINGH_NFE2L2_TARGETS |
| STEINER_ERYTHROCYTE_MEMBRANE_GENES | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/STEINER_ERYTHROCYTE_MEMBRANE_GENES |
| SU_KIDNEY | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/SU_KIDNEY |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_SUSTAINED_IN_GRANULOCYTE_UP | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_SUSTAINED_IN_GRANULOCYTE_UP |
| TSUNODA_CISPLATIN_RESISTANCE_UP | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/TSUNODA_CISPLATIN_RESISTANCE_UP |
| VANDESLUIS_COMMD1_TARGETS_GROUP_2_UP | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/VANDESLUIS_COMMD1_TARGETS_GROUP_2_UP |
| VISALA_RESPONSE_TO_HEAT_SHOCK_AND_AGING_UP | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/VISALA_RESPONSE_TO_HEAT_SHOCK_AND_AGING_UP |
| WANG_METASTASIS_OF_BREAST_CANCER | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_METASTASIS_OF_BREAST_CANCER |
| WEBER_METHYLATED_LCP_IN_SPERM_UP | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/WEBER_METHYLATED_LCP_IN_SPERM_UP |
| XU_RESPONSE_TO_TRETINOIN_AND_NSC682994_DN | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_RESPONSE_TO_TRETINOIN_AND_NSC682994_DN |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_3 | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_3 |
| YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_4 | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/YAO_TEMPORAL_RESPONSE_TO_PROGESTERONE_CLUSTER_4 |
| ZHANG_GATA6_TARGETS_UP | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_GATA6_TARGETS_UP |
| ZHAN_V1_LATE_DIFFERENTIATION_GENES_DN | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_V1_LATE_DIFFERENTIATION_GENES_DN |
| ZHAN_VARIABLE_EARLY_DIFFERENTIATION_GENES_UP | 15 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_VARIABLE_EARLY_DIFFERENTIATION_GENES_UP |
| ABBUD_LIF_SIGNALING_2_UP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/ABBUD_LIF_SIGNALING_2_UP |
| ABDULRAHMAN_KIDNEY_CANCER_VHL_DN | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/ABDULRAHMAN_KIDNEY_CANCER_VHL_DN |
| AMIT_EGF_RESPONSE_20_MCF10A | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_EGF_RESPONSE_20_MCF10A |
| BANDRES_RESPONSE_TO_CARMUSTIN_WITHOUT_MGMT_24HR_UP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/BANDRES_RESPONSE_TO_CARMUSTIN_WITHOUT_MGMT_24HR_UP |
| BIOCARTA_CELL2CELL_PATHWAY | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CELL2CELL_PATHWAY |
| BIOCARTA_CLASSIC_PATHWAY | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CLASSIC_PATHWAY |
| BIOCARTA_DREAM_PATHWAY | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_DREAM_PATHWAY |
| BIOCARTA_GRANULOCYTES_PATHWAY | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_GRANULOCYTES_PATHWAY |
| BIOCARTA_PLATELETAPP_PATHWAY | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_PLATELETAPP_PATHWAY |
| BIOCARTA_PS1_PATHWAY | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_PS1_PATHWAY |
| BIOCARTA_RANKL_PATHWAY | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_RANKL_PATHWAY |
| BIOCARTA_TCYTOTOXIC_PATHWAY | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_TCYTOTOXIC_PATHWAY |
| BIOCARTA_THELPER_PATHWAY | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_THELPER_PATHWAY |
| CAIRO_PML_TARGETS_BOUND_BY_MYC_DN | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/CAIRO_PML_TARGETS_BOUND_BY_MYC_DN |
| CASTELLANO_NRAS_TARGETS_DN | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/CASTELLANO_NRAS_TARGETS_DN |
| DORN_ADENOVIRUS_INFECTION_48HR_UP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/DORN_ADENOVIRUS_INFECTION_48HR_UP |
| GILMORE_CORE_NFKB_PATHWAY | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/GILMORE_CORE_NFKB_PATHWAY |
| GOERING_BLOOD_HDL_CHOLESTEROL_QTL_TRANS | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/GOERING_BLOOD_HDL_CHOLESTEROL_QTL_TRANS |
| GRANDVAUX_IFN_RESPONSE_NOT_VIA_IRF3 | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/GRANDVAUX_IFN_RESPONSE_NOT_VIA_IRF3 |
| GRASEMANN_RETINOBLASTOMA_WITH_6P_AMPLIFICATION | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/GRASEMANN_RETINOBLASTOMA_WITH_6P_AMPLIFICATION |
| HOEGERKORP_CD44_TARGETS_DIRECT_DN | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/HOEGERKORP_CD44_TARGETS_DIRECT_DN |
| INAMURA_LUNG_CANCER_SCC_DN | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/INAMURA_LUNG_CANCER_SCC_DN |
| INAMURA_LUNG_CANCER_SCC_SUBTYPES_UP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/INAMURA_LUNG_CANCER_SCC_SUBTYPES_UP |
| INAMURA_LUNG_CANCER_SCC_UP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/INAMURA_LUNG_CANCER_SCC_UP |
| KEGG_GLYCOSPHINGOLIPID_BIOSYNTHESIS_GLOBO_SERIES | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_GLYCOSPHINGOLIPID_BIOSYNTHESIS_GLOBO_SERIES |
| KEGG_NON_HOMOLOGOUS_END_JOINING | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_NON_HOMOLOGOUS_END_JOINING |
| KINNEY_DNMT1_METHYLATION_TARGETS | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/KINNEY_DNMT1_METHYLATION_TARGETS |
| LEE_SP4_THYMOCYTE | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/LEE_SP4_THYMOCYTE |
| LIM_MAMMARY_LUMINAL_PROGENITOR_DN | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/LIM_MAMMARY_LUMINAL_PROGENITOR_DN |
| LOPEZ_MESOTELIOMA_SURVIVAL_TIME_UP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/LOPEZ_MESOTELIOMA_SURVIVAL_TIME_UP |
| LOPEZ_MESOTHELIOMA_SURVIVAL_WORST_VS_BEST_UP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/LOPEZ_MESOTHELIOMA_SURVIVAL_WORST_VS_BEST_UP |
| LY_AGING_MIDDLE_UP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/LY_AGING_MIDDLE_UP |
| MATZUK_OVULATION | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_OVULATION |
| MATZUK_POST-IMPLANTATION_AND_POST-PARTUM | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_POST-IMPLANTATION_AND_POST-PARTUM |
| MEISSNER_ES_ICP_WITH_H3K4ME3_AND_H3K27ME3 | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/MEISSNER_ES_ICP_WITH_H3K4ME3_AND_H3K27ME3 |
| MIKKELSEN_ES_LCP_WITH_H3K27ME3 | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_ES_LCP_WITH_H3K27ME3 |
| MUELLER_COMMON_TARGETS_OF_AML_FUSIONS_UP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/MUELLER_COMMON_TARGETS_OF_AML_FUSIONS_UP |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_16 | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_16 |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_24 | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_24 |
| NAKAMURA_LUNG_CANCER_DIFFERENTIATION_MARKERS | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAMURA_LUNG_CANCER_DIFFERENTIATION_MARKERS |
| NIKOLSKY_BREAST_CANCER_14Q22_AMPLICON | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_14Q22_AMPLICON |
| ONGUSAHA_BRCA1_TARGETS_DN | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/ONGUSAHA_BRCA1_TARGETS_DN |
| PARK_APL_PATHOGENESIS_UP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/PARK_APL_PATHOGENESIS_UP |
| PID_IL5_PATHWAY | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_IL5_PATHWAY |
| PID_S1P_S1P4_PATHWAY | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_S1P_S1P4_PATHWAY |
| PID_TCRJNKPATHWAY | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_TCRJNKPATHWAY |
| PID_TCRRASPATHWAY | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_TCRRASPATHWAY |
| PIEPOLI_LGI1_TARGETS_DN | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/PIEPOLI_LGI1_TARGETS_DN |
| REACTOME_ACTIVATION_OF_IRF3_IRF7_MEDIATED_BY_TBK1_IKK_EPSILON | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATION_OF_IRF3_IRF7_MEDIATED_BY_TBK1_IKK_EPSILON |
| REACTOME_ACTIVATION_OF_RAC | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATION_OF_RAC |
| REACTOME_ASSOCIATION_OF_LICENSING_FACTORS_WITH_THE_PRE_REPLICATIVE_COMPLEX | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ASSOCIATION_OF_LICENSING_FACTORS_WITH_THE_PRE_REPLICATIVE_COMPLEX |
| REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS |
| REACTOME_COMMON_PATHWAY | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_COMMON_PATHWAY |
| REACTOME_CRMPS_IN_SEMA3A_SIGNALING | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CRMPS_IN_SEMA3A_SIGNALING |
| REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES |
| REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION |
| REACTOME_HYALURONAN_METABOLISM | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_HYALURONAN_METABOLISM |
| REACTOME_METABOLISM_OF_PORPHYRINS | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_METABOLISM_OF_PORPHYRINS |
| REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION |
| REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_N_GLYCAN_ANTENNAE_ELONGATION |
| REACTOME_P75NTR_SIGNALS_VIA_NFKB | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_P75NTR_SIGNALS_VIA_NFKB |
| REACTOME_PEPTIDE_HORMONE_BIOSYNTHESIS | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PEPTIDE_HORMONE_BIOSYNTHESIS |
| REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS |
| REACTOME_PROLACTIN_RECEPTOR_SIGNALING | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PROLACTIN_RECEPTOR_SIGNALING |
| REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_COMPLEMENT_CASCADE |
| REACTOME_REGULATION_OF_IFNG_SIGNALING | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_IFNG_SIGNALING |
| REACTOME_REPAIR_SYNTHESIS_FOR_GAP_FILLING_BY_DNA_POL_IN_TC_NER | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REPAIR_SYNTHESIS_FOR_GAP_FILLING_BY_DNA_POL_IN_TC_NER |
| REACTOME_SIGNAL_ATTENUATION | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNAL_ATTENUATION |
| REACTOME_SOS_MEDIATED_SIGNALLING | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SOS_MEDIATED_SIGNALLING |
| REACTOME_SPRY_REGULATION_OF_FGF_SIGNALING | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SPRY_REGULATION_OF_FGF_SIGNALING |
| REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS |
| REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS |
| REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP |
| REACTOME_TRAF3_DEPENDENT_IRF_ACTIVATION_PATHWAY | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRAF3_DEPENDENT_IRF_ACTIVATION_PATHWAY |
| REACTOME_TRAF6_MEDIATED_INDUCTION_OF_TAK1_COMPLEX | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRAF6_MEDIATED_INDUCTION_OF_TAK1_COMPLEX |
| REACTOME_TRAFFICKING_AND_PROCESSING_OF_ENDOSOMAL_TLR | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRAFFICKING_AND_PROCESSING_OF_ENDOSOMAL_TLR |
| REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE |
| REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS |
| ROETH_TERT_TARGETS_UP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/ROETH_TERT_TARGETS_UP |
| SABATES_COLORECTAL_ADENOMA_SIZE_DN | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/SABATES_COLORECTAL_ADENOMA_SIZE_DN |
| SPIELMAN_LYMPHOBLAST_EUROPEAN_VS_ASIAN_2FC_UP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/SPIELMAN_LYMPHOBLAST_EUROPEAN_VS_ASIAN_2FC_UP |
| TOMLINS_METASTASIS_UP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/TOMLINS_METASTASIS_UP |
| VERRECCHIA_RESPONSE_TO_TGFB1_C3 | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/VERRECCHIA_RESPONSE_TO_TGFB1_C3 |
| VICENT_METASTASIS_UP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/VICENT_METASTASIS_UP |
| VISALA_RESPONSE_TO_HEAT_SHOCK_AND_AGING_DN | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/VISALA_RESPONSE_TO_HEAT_SHOCK_AND_AGING_DN |
| WANG_RESPONSE_TO_PACLITAXEL_VIA_MAPK8_UP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_RESPONSE_TO_PACLITAXEL_VIA_MAPK8_UP |
| WATANABE_ULCERATIVE_COLITIS_WITH_CANCER_DN | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/WATANABE_ULCERATIVE_COLITIS_WITH_CANCER_DN |
| WEBER_METHYLATED_ICP_IN_SPERM_DN | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/WEBER_METHYLATED_ICP_IN_SPERM_DN |
| WORSCHECH_TUMOR_EVASION_AND_TOLEROGENICITY_DN | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/WORSCHECH_TUMOR_EVASION_AND_TOLEROGENICITY_DN |
| WU_ALZHEIMER_DISEASE_UP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/WU_ALZHEIMER_DISEASE_UP |
| XU_HGF_TARGETS_INDUCED_BY_AKT1_48HR_UP | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_HGF_TARGETS_INDUCED_BY_AKT1_48HR_UP |
| ZAIDI_OSTEOBLAST_TRANSCRIPTION_FACTORS | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/ZAIDI_OSTEOBLAST_TRANSCRIPTION_FACTORS |
| ZHANG_ADIPOGENESIS_BY_BMP7 | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHANG_ADIPOGENESIS_BY_BMP7 |
| ZHOU_PANCREATIC_ENDOCRINE_PROGENITOR | 14 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHOU_PANCREATIC_ENDOCRINE_PROGENITOR |
| BALLIF_DEVELOPMENTAL_DISABILITY_P16_P12_DELETION | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/BALLIF_DEVELOPMENTAL_DISABILITY_P16_P12_DELETION |
| BARRIER_CANCER_RELAPSE_TUMOR_SAMPLE_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/BARRIER_CANCER_RELAPSE_TUMOR_SAMPLE_DN |
| BIOCARTA_ACE2_PATHWAY | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_ACE2_PATHWAY |
| BIOCARTA_AGPCR_PATHWAY | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_AGPCR_PATHWAY |
| BIOCARTA_AHSP_PATHWAY | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_AHSP_PATHWAY |
| BIOCARTA_ARENRF2_PATHWAY | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_ARENRF2_PATHWAY |
| BIOCARTA_CARM1_PATHWAY | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CARM1_PATHWAY |
| BIOCARTA_CBL_PATHWAY | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CBL_PATHWAY |
| BIOCARTA_D4GDI_PATHWAY | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_D4GDI_PATHWAY |
| BIOCARTA_EXTRINSIC_PATHWAY | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_EXTRINSIC_PATHWAY |
| BIOCARTA_P27_PATHWAY | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_P27_PATHWAY |
| BIOCARTA_PARKIN_PATHWAY | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_PARKIN_PATHWAY |
| BIOCARTA_RB_PATHWAY | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_RB_PATHWAY |
| BIOCARTA_SALMONELLA_PATHWAY | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_SALMONELLA_PATHWAY |
| BIOCARTA_TCRA_PATHWAY | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_TCRA_PATHWAY |
| CAFFAREL_RESPONSE_TO_THC_24HR_3_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/CAFFAREL_RESPONSE_TO_THC_24HR_3_DN |
| CHEOK_RESPONSE_TO_MERCAPTOPURINE_UP | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEOK_RESPONSE_TO_MERCAPTOPURINE_UP |
| CONRAD_GERMLINE_STEM_CELL | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/CONRAD_GERMLINE_STEM_CELL |
| CROONQUIST_STROMAL_STIMULATION_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/CROONQUIST_STROMAL_STIMULATION_DN |
| DAVIES_MULTIPLE_MYELOMA_VS_MGUS_UP | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/DAVIES_MULTIPLE_MYELOMA_VS_MGUS_UP |
| DISTECHE_ESCAPED_FROM_X_INACTIVATION | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/DISTECHE_ESCAPED_FROM_X_INACTIVATION |
| EHRLICH_ICF_SYNDROM_UP | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/EHRLICH_ICF_SYNDROM_UP |
| ELLWOOD_MYC_TARGETS_UP | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/ELLWOOD_MYC_TARGETS_UP |
| FRIDMAN_SENESCENCE_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/FRIDMAN_SENESCENCE_DN |
| FU_INTERACT_WITH_ALKBH8 | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/FU_INTERACT_WITH_ALKBH8 |
| GERHOLD_RESPONSE_TO_TZD_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/GERHOLD_RESPONSE_TO_TZD_DN |
| GHO_ATF5_TARGETS_UP | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/GHO_ATF5_TARGETS_UP |
| GOERING_BLOOD_HDL_CHOLESTEROL_QTL_CIS | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/GOERING_BLOOD_HDL_CHOLESTEROL_QTL_CIS |
| GRAHAM_CML_DIVIDING_VS_NORMAL_DIVIDING_UP | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAHAM_CML_DIVIDING_VS_NORMAL_DIVIDING_UP |
| GUTIERREZ_WALDENSTROEMS_MACROGLOBULINEMIA_2 | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/GUTIERREZ_WALDENSTROEMS_MACROGLOBULINEMIA_2 |
| HASINA_NOL7_TARGETS_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/HASINA_NOL7_TARGETS_DN |
| HASINA_NOL7_TARGETS_UP | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/HASINA_NOL7_TARGETS_UP |
| HEIDENBLAD_AMPLIFIED_IN_SOFT_TISSUE_CANCER | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/HEIDENBLAD_AMPLIFIED_IN_SOFT_TISSUE_CANCER |
| HOEGERKORP_CD44_TARGETS_TEMPORAL_UP | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/HOEGERKORP_CD44_TARGETS_TEMPORAL_UP |
| HO_LIVER_CANCER_VASCULAR_INVASION | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/HO_LIVER_CANCER_VASCULAR_INVASION |
| IRITANI_MAD1_TARGETS_UP | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/IRITANI_MAD1_TARGETS_UP |
| IVANOV_MUTATED_IN_COLON_CANCER | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/IVANOV_MUTATED_IN_COLON_CANCER |
| JIANG_CORE_DUPLICON_GENES | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/JIANG_CORE_DUPLICON_GENES |
| KEGG_CIRCADIAN_RHYTHM_MAMMAL | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_CIRCADIAN_RHYTHM_MAMMAL |
| KEGG_SULFUR_METABOLISM | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_SULFUR_METABOLISM |
| KIM_MYCL1_AMPLIFICATION_TARGETS_UP | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_MYCL1_AMPLIFICATION_TARGETS_UP |
| KIM_RESPONSE_TO_TSA_AND_DECITABINE_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_RESPONSE_TO_TSA_AND_DECITABINE_DN |
| KORKOLA_EMBRYONAL_CARCINOMA | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/KORKOLA_EMBRYONAL_CARCINOMA |
| KORKOLA_EMBRYONAL_CARCINOMA_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/KORKOLA_EMBRYONAL_CARCINOMA_DN |
| KORKOLA_SEMINOMA_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/KORKOLA_SEMINOMA_DN |
| KRISHNAN_FURIN_TARGETS_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/KRISHNAN_FURIN_TARGETS_DN |
| KYNG_ENVIRONMENTAL_STRESS_RESPONSE_NOT_BY_4NQO_IN_OLD | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_ENVIRONMENTAL_STRESS_RESPONSE_NOT_BY_4NQO_IN_OLD |
| LUI_THYROID_CANCER_CLUSTER_5 | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/LUI_THYROID_CANCER_CLUSTER_5 |
| LU_TUMOR_VASCULATURE_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/LU_TUMOR_VASCULATURE_DN |
| MARTINELLI_IMMATURE_NEUTROPHIL_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTINELLI_IMMATURE_NEUTROPHIL_DN |
| MATZUK_EARLY_ANTRAL_FOLLICLE | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_EARLY_ANTRAL_FOLLICLE |
| MULLIGAN_NTF3_SIGNALING_VIA_INSR_AND_IGF1R_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/MULLIGAN_NTF3_SIGNALING_VIA_INSR_AND_IGF1R_DN |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_15 | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_15 |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_22 | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_22 |
| NIKOLSKY_BREAST_CANCER_1Q32_AMPLICON | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_1Q32_AMPLICON |
| NUNODA_RESPONSE_TO_DASATINIB_IMATINIB_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/NUNODA_RESPONSE_TO_DASATINIB_IMATINIB_DN |
| ONGUSAHA_BRCA1_TARGETS_UP | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/ONGUSAHA_BRCA1_TARGETS_UP |
| PARK_OSTEOBLAST_DIFFERENTIATION_BY_PHENYLAMIL_UP | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/PARK_OSTEOBLAST_DIFFERENTIATION_BY_PHENYLAMIL_UP |
| PHESSE_TARGETS_OF_APC_AND_MBD2_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/PHESSE_TARGETS_OF_APC_AND_MBD2_DN |
| POS_RESPONSE_TO_HISTAMINE_UP | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/POS_RESPONSE_TO_HISTAMINE_UP |
| RAY_ALZHEIMERS_DISEASE | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/RAY_ALZHEIMERS_DISEASE |
| REACTOME_ACTIVATION_OF_CHAPERONES_BY_ATF6_ALPHA | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATION_OF_CHAPERONES_BY_ATF6_ALPHA |
| REACTOME_ADENYLATE_CYCLASE_INHIBITORY_PATHWAY | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ADENYLATE_CYCLASE_INHIBITORY_PATHWAY |
| REACTOME_ADVANCED_GLYCOSYLATION_ENDPRODUCT_RECEPTOR_SIGNALING | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ADVANCED_GLYCOSYLATION_ENDPRODUCT_RECEPTOR_SIGNALING |
| REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION |
| REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS |
| REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING |
| REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS |
| REACTOME_HIGHLY_CALCIUM_PERMEABLE_POSTSYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_HIGHLY_CALCIUM_PERMEABLE_POSTSYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS |
| REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS |
| REACTOME_INHIBITION_OF_REPLICATION_INITIATION_OF_DAMAGED_DNA_BY_RB1_E2F1 | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INHIBITION_OF_REPLICATION_INITIATION_OF_DAMAGED_DNA_BY_RB1_E2F1 |
| REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY |
| REACTOME_N_GLYCAN_TRIMMING_IN_THE_ER_AND_CALNEXIN_CALRETICULIN_CYCLE | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_N_GLYCAN_TRIMMING_IN_THE_ER_AND_CALNEXIN_CALRETICULIN_CYCLE |
| REACTOME_ORGANIC_CATION_ANION_ZWITTERION_TRANSPORT | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ORGANIC_CATION_ANION_ZWITTERION_TRANSPORT |
| REACTOME_P38MAPK_EVENTS | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_P38MAPK_EVENTS |
| REACTOME_POL_SWITCHING | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_POL_SWITCHING |
| REACTOME_PURINE_SALVAGE | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PURINE_SALVAGE |
| REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX |
| REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING |
| REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION |
| REACTOME_TETRAHYDROBIOPTERIN_BH4_SYNTHESIS_RECYCLING_SALVAGE_AND_REGULATION | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TETRAHYDROBIOPTERIN_BH4_SYNTHESIS_RECYCLING_SALVAGE_AND_REGULATION |
| SAENZ_DETOX_PATHWAY_AND_CARCINOGENESIS_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/SAENZ_DETOX_PATHWAY_AND_CARCINOGENESIS_DN |
| SASAI_TARGETS_OF_CXCR6_AND_PTCH1_UP | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/SASAI_TARGETS_OF_CXCR6_AND_PTCH1_UP |
| SA_REG_CASCADE_OF_CYCLIN_EXPR | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/SA_REG_CASCADE_OF_CYCLIN_EXPR |
| SCHWAB_TARGETS_OF_BMYB_POLYMORPHIC_VARIANTS_UP | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHWAB_TARGETS_OF_BMYB_POLYMORPHIC_VARIANTS_UP |
| SHIN_B_CELL_LYMPHOMA_CLUSTER_1 | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/SHIN_B_CELL_LYMPHOMA_CLUSTER_1 |
| SHI_SPARC_TARGETS_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/SHI_SPARC_TARGETS_DN |
| STARK_BRAIN_22Q11_DELETION | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/STARK_BRAIN_22Q11_DELETION |
| TERAMOTO_OPN_TARGETS_CLUSTER_1 | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/TERAMOTO_OPN_TARGETS_CLUSTER_1 |
| TERAMOTO_OPN_TARGETS_CLUSTER_4 | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/TERAMOTO_OPN_TARGETS_CLUSTER_4 |
| TSAI_DNAJB4_TARGETS_UP | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/TSAI_DNAJB4_TARGETS_UP |
| VERRECCHIA_RESPONSE_TO_TGFB1_C4 | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/VERRECCHIA_RESPONSE_TO_TGFB1_C4 |
| WACKER_HYPOXIA_TARGETS_OF_VHL | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/WACKER_HYPOXIA_TARGETS_OF_VHL |
| WAGSCHAL_EHMT2_TARGETS_UP | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/WAGSCHAL_EHMT2_TARGETS_UP |
| WHITESIDE_CISPLATIN_RESISTANCE_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/WHITESIDE_CISPLATIN_RESISTANCE_DN |
| WU_HBX_TARGETS_3_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/WU_HBX_TARGETS_3_DN |
| XU_RESPONSE_TO_TRETINOIN_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_RESPONSE_TO_TRETINOIN_DN |
| YAGUE_PRETUMOR_DRUG_RESISTANCE_DN | 13 | http://www.broadinstitute.org/gsea/msigdb/cards/YAGUE_PRETUMOR_DRUG_RESISTANCE_DN |
| BIOCARTA_AKAP13_PATHWAY | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_AKAP13_PATHWAY |
| BIOCARTA_AKAP95_PATHWAY | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_AKAP95_PATHWAY |
| BIOCARTA_ASBCELL_PATHWAY | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_ASBCELL_PATHWAY |
| BIOCARTA_BARR_MAPK_PATHWAY | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_BARR_MAPK_PATHWAY |
| BIOCARTA_CFTR_PATHWAY | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CFTR_PATHWAY |
| BIOCARTA_ETC_PATHWAY | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_ETC_PATHWAY |
| BIOCARTA_FIBRINOLYSIS_PATHWAY | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_FIBRINOLYSIS_PATHWAY |
| BIOCARTA_LECTIN_PATHWAY | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_LECTIN_PATHWAY |
| BIOCARTA_PLCE_PATHWAY | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_PLCE_PATHWAY |
| BIOCARTA_RAB_PATHWAY | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_RAB_PATHWAY |
| BIOCARTA_TRKA_PATHWAY | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_TRKA_PATHWAY |
| BIOCARTA_VDR_PATHWAY | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_VDR_PATHWAY |
| BOYAULT_LIVER_CANCER_SUBCLASS_G56_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYAULT_LIVER_CANCER_SUBCLASS_G56_UP |
| BOYERINAS_ONCOFETAL_TARGETS_OF_LET7A1 | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYERINAS_ONCOFETAL_TARGETS_OF_LET7A1 |
| CHAN_INTERFERON_PRODUCING_DENDRITIC_CELL | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/CHAN_INTERFERON_PRODUCING_DENDRITIC_CELL |
| CLAUS_PGR_POSITIVE_MENINGIOMA_DN | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/CLAUS_PGR_POSITIVE_MENINGIOMA_DN |
| DAZARD_UV_RESPONSE_CLUSTER_G5 | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/DAZARD_UV_RESPONSE_CLUSTER_G5 |
| DING_LUNG_CANCER_MUTATED_FREQUENTLY | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/DING_LUNG_CANCER_MUTATED_FREQUENTLY |
| DORN_ADENOVIRUS_INFECTION_24HR_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/DORN_ADENOVIRUS_INFECTION_24HR_UP |
| EHLERS_ANEUPLOIDY_DN | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/EHLERS_ANEUPLOIDY_DN |
| FIGUEROA_AML_METHYLATION_CLUSTER_5_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/FIGUEROA_AML_METHYLATION_CLUSTER_5_UP |
| FUNG_IL2_SIGNALING_2 | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/FUNG_IL2_SIGNALING_2 |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_MAGENTA_DN | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_MAGENTA_DN |
| GHANDHI_BYSTANDER_IRRADIATION_DN | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/GHANDHI_BYSTANDER_IRRADIATION_DN |
| GRAHAM_CML_QUIESCENT_VS_NORMAL_DIVIDING_DN | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAHAM_CML_QUIESCENT_VS_NORMAL_DIVIDING_DN |
| GUTIERREZ_CHRONIC_LYMPHOCYTIC_LEUKEMIA_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/GUTIERREZ_CHRONIC_LYMPHOCYTIC_LEUKEMIA_UP |
| HEDVAT_ELF4_TARGETS_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/HEDVAT_ELF4_TARGETS_UP |
| HOLLEMAN_DAUNORUBICIN_B_ALL_DN | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLEMAN_DAUNORUBICIN_B_ALL_DN |
| HOLLEMAN_PREDNISOLONE_RESISTANCE_B_ALL_DN | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLEMAN_PREDNISOLONE_RESISTANCE_B_ALL_DN |
| IIZUKA_LIVER_CANCER_PROGRESSION_G1_G2_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/IIZUKA_LIVER_CANCER_PROGRESSION_G1_G2_UP |
| IIZUKA_LIVER_CANCER_PROGRESSION_L1_G1_DN | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/IIZUKA_LIVER_CANCER_PROGRESSION_L1_G1_DN |
| ITO_PTTG1_TARGETS_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/ITO_PTTG1_TARGETS_UP |
| JI_CARCINOGENESIS_BY_KRAS_AND_STK11_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/JI_CARCINOGENESIS_BY_KRAS_AND_STK11_UP |
| KRISHNAN_FURIN_TARGETS_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/KRISHNAN_FURIN_TARGETS_UP |
| KUROKAWA_LIVER_CANCER_CHEMOTHERAPY_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/KUROKAWA_LIVER_CANCER_CHEMOTHERAPY_UP |
| KUROKAWA_LIVER_CANCER_EARLY_RECURRENCE_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/KUROKAWA_LIVER_CANCER_EARLY_RECURRENCE_UP |
| KYNG_ENVIRONMENTAL_STRESS_RESPONSE_NOT_BY_UV_IN_WS | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/KYNG_ENVIRONMENTAL_STRESS_RESPONSE_NOT_BY_UV_IN_WS |
| LEI_HOXC8_TARGETS_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/LEI_HOXC8_TARGETS_UP |
| LOPEZ_MESOTHELIOMA_SURVIVAL_DN | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/LOPEZ_MESOTHELIOMA_SURVIVAL_DN |
| MALIK_REPRESSED_BY_ESTROGEN | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/MALIK_REPRESSED_BY_ESTROGEN |
| MCBRYAN_TERMINAL_END_BUD_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/MCBRYAN_TERMINAL_END_BUD_UP |
| MIKKELSEN_NPC_ICP_WITH_H3K27ME3 | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_NPC_ICP_WITH_H3K27ME3 |
| MIZUKAMI_HYPOXIA_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/MIZUKAMI_HYPOXIA_UP |
| MONTERO_THYROID_CANCER_POOR_SURVIVAL_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/MONTERO_THYROID_CANCER_POOR_SURVIVAL_UP |
| MOROSETTI_FACIOSCAPULOHUMERAL_MUSCULAR_DISTROPHY_DN | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/MOROSETTI_FACIOSCAPULOHUMERAL_MUSCULAR_DISTROPHY_DN |
| NEBEN_AML_WITH_FLT3_OR_NRAS_DN | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/NEBEN_AML_WITH_FLT3_OR_NRAS_DN |
| PARK_TRETINOIN_RESPONSE | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/PARK_TRETINOIN_RESPONSE |
| PETRETTO_BLOOD_PRESSURE_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/PETRETTO_BLOOD_PRESSURE_UP |
| REACTOME_AKT_PHOSPHORYLATES_TARGETS_IN_THE_CYTOSOL | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_AKT_PHOSPHORYLATES_TARGETS_IN_THE_CYTOSOL |
| REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM |
| REACTOME_APOBEC3G_MEDIATED_RESISTANCE_TO_HIV1_INFECTION | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_APOBEC3G_MEDIATED_RESISTANCE_TO_HIV1_INFECTION |
| REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS |
| REACTOME_CS_DS_DEGRADATION | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CS_DS_DEGRADATION |
| REACTOME_DIGESTION_OF_DIETARY_CARBOHYDRATE | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DIGESTION_OF_DIETARY_CARBOHYDRATE |
| REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING |
| REACTOME_ERKS_ARE_INACTIVATED | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ERKS_ARE_INACTIVATED |
| REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS |
| REACTOME_FGFR2C_LIGAND_BINDING_AND_ACTIVATION | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FGFR2C_LIGAND_BINDING_AND_ACTIVATION |
| REACTOME_FGFR4_LIGAND_BINDING_AND_ACTIVATION | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_FGFR4_LIGAND_BINDING_AND_ACTIVATION |
| REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT |
| REACTOME_GABA_A_RECEPTOR_ACTIVATION | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GABA_A_RECEPTOR_ACTIVATION |
| REACTOME_GLYCOPROTEIN_HORMONES | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GLYCOPROTEIN_HORMONES |
| REACTOME_NFKB_ACTIVATION_THROUGH_FADD_RIP1_PATHWAY_MEDIATED_BY_CASPASE_8_AND10 | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NFKB_ACTIVATION_THROUGH_FADD_RIP1_PATHWAY_MEDIATED_BY_CASPASE_8_AND10 |
| REACTOME_P2Y_RECEPTORS | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_P2Y_RECEPTORS |
| REACTOME_P75NTR_RECRUITS_SIGNALLING_COMPLEXES | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_P75NTR_RECRUITS_SIGNALLING_COMPLEXES |
| REACTOME_PLATELET_ADHESION_TO_EXPOSED_COLLAGEN | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PLATELET_ADHESION_TO_EXPOSED_COLLAGEN |
| REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS |
| REACTOME_PYRIMIDINE_CATABOLISM | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PYRIMIDINE_CATABOLISM |
| REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR |
| REACTOME_RESOLUTION_OF_AP_SITES_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RESOLUTION_OF_AP_SITES_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY |
| REACTOME_REVERSIBLE_HYDRATION_OF_CARBON_DIOXIDE | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REVERSIBLE_HYDRATION_OF_CARBON_DIOXIDE |
| REACTOME_SEROTONIN_RECEPTORS | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SEROTONIN_RECEPTORS |
| REACTOME_SIGNALING_BY_NOTCH2 | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_NOTCH2 |
| REACTOME_SIGNALING_BY_NOTCH3 | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_NOTCH3 |
| REACTOME_SIGNALING_BY_NOTCH4 | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_NOTCH4 |
| REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS |
| REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE |
| REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS |
| REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES |
| REACTOME_THE_NLRP3_INFLAMMASOME | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_THE_NLRP3_INFLAMMASOME |
| RUAN_RESPONSE_TO_TNF_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/RUAN_RESPONSE_TO_TNF_UP |
| SA_PROGRAMMED_CELL_DEATH | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/SA_PROGRAMMED_CELL_DEATH |
| SUZUKI_CTCFL_TARGETS_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/SUZUKI_CTCFL_TARGETS_UP |
| TAGHAVI_NEOPLASTIC_TRANSFORMATION | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/TAGHAVI_NEOPLASTIC_TRANSFORMATION |
| TURJANSKI_MAPK1_AND_MAPK2_TARGETS | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/TURJANSKI_MAPK1_AND_MAPK2_TARGETS |
| WAESCH_ANAPHASE_PROMOTING_COMPLEX | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/WAESCH_ANAPHASE_PROMOTING_COMPLEX |
| WEBER_METHYLATED_LCP_IN_FIBROBLAST_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/WEBER_METHYLATED_LCP_IN_FIBROBLAST_UP |
| WUNDER_INFLAMMATORY_RESPONSE_AND_CHOLESTEROL_DN | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/WUNDER_INFLAMMATORY_RESPONSE_AND_CHOLESTEROL_DN |
| WU_APOPTOSIS_BY_CDKN1A_NOT_VIA_TP53 | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/WU_APOPTOSIS_BY_CDKN1A_NOT_VIA_TP53 |
| ZHENG_FOXP3_TARGETS_IN_THYMUS_DN | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHENG_FOXP3_TARGETS_IN_THYMUS_DN |
| ZHENG_FOXP3_TARGETS_IN_T_LYMPHOCYTE_UP | 12 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHENG_FOXP3_TARGETS_IN_T_LYMPHOCYTE_UP |
| AMIT_EGF_RESPONSE_20_HELA | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/AMIT_EGF_RESPONSE_20_HELA |
| AUJLA_IL22_AND_IL17A_SIGNALING | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/AUJLA_IL22_AND_IL17A_SIGNALING |
| BAUS_TFF2_TARGETS_DN | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/BAUS_TFF2_TARGETS_DN |
| BIOCARTA_BLYMPHOCYTE_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_BLYMPHOCYTE_PATHWAY |
| BIOCARTA_CDK5_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_CDK5_PATHWAY |
| BIOCARTA_EGFR_SMRTE_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_EGFR_SMRTE_PATHWAY |
| BIOCARTA_EIF2_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_EIF2_PATHWAY |
| BIOCARTA_EPONFKB_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_EPONFKB_PATHWAY |
| BIOCARTA_IL4_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_IL4_PATHWAY |
| BIOCARTA_LEPTIN_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_LEPTIN_PATHWAY |
| BIOCARTA_LYM_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_LYM_PATHWAY |
| BIOCARTA_MONOCYTE_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_MONOCYTE_PATHWAY |
| BIOCARTA_P35ALZHEIMERS_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_P35ALZHEIMERS_PATHWAY |
| BIOCARTA_PTC1_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_PTC1_PATHWAY |
| BIOCARTA_SET_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_SET_PATHWAY |
| BIOCARTA_SRCRPTP_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_SRCRPTP_PATHWAY |
| BIOCARTA_TCAPOPTOSIS_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_TCAPOPTOSIS_PATHWAY |
| BIOCARTA_VITCB_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_VITCB_PATHWAY |
| CAFFAREL_RESPONSE_TO_THC_8HR_5_DN | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/CAFFAREL_RESPONSE_TO_THC_8HR_5_DN |
| CHUANG_OXIDATIVE_STRESS_RESPONSE_DN | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/CHUANG_OXIDATIVE_STRESS_RESPONSE_DN |
| DELASERNA_TARGETS_OF_MYOD_AND_SMARCA4 | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/DELASERNA_TARGETS_OF_MYOD_AND_SMARCA4 |
| DER_IFN_GAMMA_RESPONSE_DN | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/DER_IFN_GAMMA_RESPONSE_DN |
| DORN_ADENOVIRUS_INFECTION_32HR_UP | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/DORN_ADENOVIRUS_INFECTION_32HR_UP |
| FOURNIER_ACINAR_DEVELOPMENT_LATE_UP | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/FOURNIER_ACINAR_DEVELOPMENT_LATE_UP |
| FRASOR_TAMOXIFEN_RESPONSE_DN | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/FRASOR_TAMOXIFEN_RESPONSE_DN |
| FUNG_IL2_SIGNALING_1 | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/FUNG_IL2_SIGNALING_1 |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_BLACK_DN | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_BLACK_DN |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_LIGHTYELLOW_DN | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_LIGHTYELLOW_DN |
| GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_LIGHTYELLOW_UP | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/GARGALOVIC_RESPONSE_TO_OXIDIZED_PHOSPHOLIPIDS_LIGHTYELLOW_UP |
| GRAHAM_CML_QUIESCENT_VS_CML_DIVIDING_DN | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAHAM_CML_QUIESCENT_VS_CML_DIVIDING_DN |
| HASEGAWA_TUMORIGENESIS_BY_RET_C634R | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/HASEGAWA_TUMORIGENESIS_BY_RET_C634R |
| HOFFMAN_CLOCK_TARGETS_UP | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFFMAN_CLOCK_TARGETS_UP |
| HOLLEMAN_PREDNISOLONE_RESISTANCE_ALL_UP | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLEMAN_PREDNISOLONE_RESISTANCE_ALL_UP |
| IIZUKA_LIVER_CANCER_EARLY_RECURRENCE | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/IIZUKA_LIVER_CANCER_EARLY_RECURRENCE |
| KALMA_E2F1_TARGETS | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/KALMA_E2F1_TARGETS |
| KEGG_FOLATE_BIOSYNTHESIS | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_FOLATE_BIOSYNTHESIS |
| KEGG_VALINE_LEUCINE_AND_ISOLEUCINE_BIOSYNTHESIS | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_VALINE_LEUCINE_AND_ISOLEUCINE_BIOSYNTHESIS |
| KORKOLA_CHORIOCARCINOMA_DN | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/KORKOLA_CHORIOCARCINOMA_DN |
| LIANG_SILENCED_BY_METHYLATION_DN | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/LIANG_SILENCED_BY_METHYLATION_DN |
| LOPEZ_MESOTHELIOMA_SURVIVAL_UP | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/LOPEZ_MESOTHELIOMA_SURVIVAL_UP |
| MANTOVANI_NFKB_TARGETS_DN | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/MANTOVANI_NFKB_TARGETS_DN |
| MARSON_FOXP3_TARGETS_STIMULATED_DN | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/MARSON_FOXP3_TARGETS_STIMULATED_DN |
| MARTINELLI_IMMATURE_NEUTROPHIL_UP | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/MARTINELLI_IMMATURE_NEUTROPHIL_UP |
| MATHEW_FANCONI_ANEMIA_GENES | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/MATHEW_FANCONI_ANEMIA_GENES |
| MIKKELSEN_PLURIPOTENT_STATE_UP | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_PLURIPOTENT_STATE_UP |
| MONTERO_THYROID_CANCER_POOR_SURVIVAL_DN | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/MONTERO_THYROID_CANCER_POOR_SURVIVAL_DN |
| MUNSHI_MULTIPLE_MYELOMA_DN | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/MUNSHI_MULTIPLE_MYELOMA_DN |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_12 | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_12 |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_29 | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_29 |
| NIKOLSKY_BREAST_CANCER_7P15_AMPLICON | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_7P15_AMPLICON |
| OUELLET_OVARIAN_CANCER_INVASIVE_VS_LMP_DN | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/OUELLET_OVARIAN_CANCER_INVASIVE_VS_LMP_DN |
| PASTURAL_RIZ1_TARGETS_UP | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/PASTURAL_RIZ1_TARGETS_UP |
| PEDERSEN_METASTASIS_BY_ERBB2_ISOFORM_5 | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/PEDERSEN_METASTASIS_BY_ERBB2_ISOFORM_5 |
| PID_ALK2PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_ALK2PATHWAY |
| PID_INTEGRIN4_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_INTEGRIN4_PATHWAY |
| PID_P38GAMMADELTAPATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_P38GAMMADELTAPATHWAY |
| PID_RANBP2PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_RANBP2PATHWAY |
| POS_RESPONSE_TO_HISTAMINE_DN | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/POS_RESPONSE_TO_HISTAMINE_DN |
| REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA |
| REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS |
| REACTOME_CALNEXIN_CALRETICULIN_CYCLE | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CALNEXIN_CALRETICULIN_CYCLE |
| REACTOME_CD28_DEPENDENT_VAV1_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CD28_DEPENDENT_VAV1_PATHWAY |
| REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX |
| REACTOME_DOPAMINE_NEUROTRANSMITTER_RELEASE_CYCLE | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DOPAMINE_NEUROTRANSMITTER_RELEASE_CYCLE |
| REACTOME_DSCAM_INTERACTIONS | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_DSCAM_INTERACTIONS |
| REACTOME_GAMMA_CARBOXYLATION_TRANSPORT_AND_AMINO_TERMINAL_CLEAVAGE_OF_PROTEINS | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GAMMA_CARBOXYLATION_TRANSPORT_AND_AMINO_TERMINAL_CLEAVAGE_OF_PROTEINS |
| REACTOME_IL_6_SIGNALING | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_IL_6_SIGNALING |
| REACTOME_IL_7_SIGNALING | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_IL_7_SIGNALING |
| REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS |
| REACTOME_KERATAN_SULFATE_DEGRADATION | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_KERATAN_SULFATE_DEGRADATION |
| REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE |
| REACTOME_MTORC1_MEDIATED_SIGNALLING | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MTORC1_MEDIATED_SIGNALLING |
| REACTOME_NFKB_IS_ACTIVATED_AND_SIGNALS_SURVIVAL | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NFKB_IS_ACTIVATED_AND_SIGNALS_SURVIVAL |
| REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS |
| REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS |
| REACTOME_RECYCLING_OF_BILE_ACIDS_AND_SALTS | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RECYCLING_OF_BILE_ACIDS_AND_SALTS |
| REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE |
| REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY |
| REACTOME_ROLE_OF_SECOND_MESSENGERS_IN_NETRIN1_SIGNALING | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ROLE_OF_SECOND_MESSENGERS_IN_NETRIN1_SIGNALING |
| REACTOME_SIGNALING_BY_ACTIVATED_POINT_MUTANTS_OF_FGFR1 | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_ACTIVATED_POINT_MUTANTS_OF_FGFR1 |
| REACTOME_SIGNALING_BY_FGFR3_MUTANTS | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SIGNALING_BY_FGFR3_MUTANTS |
| REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS |
| REACTOME_SYNTHESIS_OF_PE | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_OF_PE |
| REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRANSPORT_OF_ORGANIC_ANIONS |
| REACTOME_TRYPTOPHAN_CATABOLISM | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRYPTOPHAN_CATABOLISM |
| REACTOME_UNWINDING_OF_DNA | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_UNWINDING_OF_DNA |
| REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM |
| ROZANOV_MMP14_CORRELATED | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/ROZANOV_MMP14_CORRELATED |
| SASAKI_TARGETS_OF_TP73_AND_TP63 | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/SASAKI_TARGETS_OF_TP73_AND_TP63 |
| SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX4_UP | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX4_UP |
| SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX5_UP | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX5_UP |
| SCIAN_INVERSED_TARGETS_OF_TP53_AND_TP73_UP | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/SCIAN_INVERSED_TARGETS_OF_TP53_AND_TP73_UP |
| SEIKE_LUNG_CANCER_POOR_SURVIVAL | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/SEIKE_LUNG_CANCER_POOR_SURVIVAL |
| SEMBA_FHIT_TARGETS_UP | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/SEMBA_FHIT_TARGETS_UP |
| SHIN_B_CELL_LYMPHOMA_CLUSTER_6 | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/SHIN_B_CELL_LYMPHOMA_CLUSTER_6 |
| STEGMEIER_PRE-MITOTIC_CELL_CYCLE_REGULATORS | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/STEGMEIER_PRE-MITOTIC_CELL_CYCLE_REGULATORS |
| ST_STAT3_PATHWAY | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_STAT3_PATHWAY |
| WHITESIDE_CISPLATIN_RESISTANCE_UP | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/WHITESIDE_CISPLATIN_RESISTANCE_UP |
| WILLIAMS_ESR2_TARGETS_DN | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/WILLIAMS_ESR2_TARGETS_DN |
| YAMANAKA_GLIOBLASTOMA_SURVIVAL_UP | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/YAMANAKA_GLIOBLASTOMA_SURVIVAL_UP |
| ZHOU_PANCREATIC_BETA_CELL | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHOU_PANCREATIC_BETA_CELL |
| ZHOU_PANCREATIC_EXOCRINE_PROGENITOR | 11 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHOU_PANCREATIC_EXOCRINE_PROGENITOR |
| AGARWAL_AKT_PATHWAY_TARGETS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/AGARWAL_AKT_PATHWAY_TARGETS |
| BAKER_HEMATOPOESIS_STAT1_TARGETS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/BAKER_HEMATOPOESIS_STAT1_TARGETS |
| BIOCARTA_BARRESTIN_PATHWAY | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_BARRESTIN_PATHWAY |
| BIOCARTA_DNAFRAGMENT_PATHWAY | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_DNAFRAGMENT_PATHWAY |
| BIOCARTA_EPHA4_PATHWAY | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_EPHA4_PATHWAY |
| BIOCARTA_FREE_PATHWAY | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_FREE_PATHWAY |
| BIOCARTA_GABA_PATHWAY | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_GABA_PATHWAY |
| BIOCARTA_GLYCOLYSIS_PATHWAY | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_GLYCOLYSIS_PATHWAY |
| BIOCARTA_IL5_PATHWAY | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_IL5_PATHWAY |
| BIOCARTA_RANMS_PATHWAY | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_RANMS_PATHWAY |
| BIOCARTA_RNA_PATHWAY | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_RNA_PATHWAY |
| BIOCARTA_SARS_PATHWAY | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_SARS_PATHWAY |
| BIOCARTA_SKP2E2F_PATHWAY | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_SKP2E2F_PATHWAY |
| BIOCARTA_SODD_PATHWAY | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_SODD_PATHWAY |
| BOYAULT_LIVER_CANCER_SUBCLASS_G23_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYAULT_LIVER_CANCER_SUBCLASS_G23_DN |
| BRUNEAU_SEPTATION_VENTRICULAR | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/BRUNEAU_SEPTATION_VENTRICULAR |
| BUDHU_LIVER_CANCER_METASTASIS_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/BUDHU_LIVER_CANCER_METASTASIS_UP |
| CAFFAREL_RESPONSE_TO_THC_8HR_3_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/CAFFAREL_RESPONSE_TO_THC_8HR_3_DN |
| CHASSOT_SKIN_WOUND | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/CHASSOT_SKIN_WOUND |
| CHEN_HOXA5_TARGETS_6HR_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEN_HOXA5_TARGETS_6HR_UP |
| CHEOK_RESPONSE_TO_MERCAPTOPURINE_AND_LD_MTX_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEOK_RESPONSE_TO_MERCAPTOPURINE_AND_LD_MTX_UP |
| CLAUS_PGR_POSITIVE_MENINGIOMA_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/CLAUS_PGR_POSITIVE_MENINGIOMA_UP |
| FREDERICK_PRKCI_TARGETS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/FREDERICK_PRKCI_TARGETS |
| FUJIWARA_PARK2_HEPATOCYTE_PROLIFERATION_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/FUJIWARA_PARK2_HEPATOCYTE_PROLIFERATION_UP |
| GOUYER_TATI_TARGETS_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/GOUYER_TATI_TARGETS_UP |
| GRAHAM_CML_DIVIDING_VS_NORMAL_DIVIDING_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/GRAHAM_CML_DIVIDING_VS_NORMAL_DIVIDING_DN |
| HOFFMAN_CLOCK_TARGETS_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFFMAN_CLOCK_TARGETS_DN |
| HOFMANN_MYELODYSPLASTIC_SYNDROM_HIGH_RISK_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/HOFMANN_MYELODYSPLASTIC_SYNDROM_HIGH_RISK_UP |
| HOLLEMAN_DAUNORUBICIN_B_ALL_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLEMAN_DAUNORUBICIN_B_ALL_UP |
| IYENGAR_RESPONSE_TO_ADIPOCYTE_FACTORS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/IYENGAR_RESPONSE_TO_ADIPOCYTE_FACTORS |
| JU_AGING_TERC_TARGETS_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/JU_AGING_TERC_TARGETS_UP |
| KEGG_LIMONENE_AND_PINENE_DEGRADATION | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_LIMONENE_AND_PINENE_DEGRADATION |
| KEGG_TAURINE_AND_HYPOTAURINE_METABOLISM | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/KEGG_TAURINE_AND_HYPOTAURINE_METABOLISM |
| KONG_E2F1_TARGETS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/KONG_E2F1_TARGETS |
| KUMAMOTO_RESPONSE_TO_NUTLIN_3A_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/KUMAMOTO_RESPONSE_TO_NUTLIN_3A_DN |
| MAINA_VHL_TARGETS_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/MAINA_VHL_TARGETS_UP |
| MANN_RESPONSE_TO_AMIFOSTINE_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/MANN_RESPONSE_TO_AMIFOSTINE_DN |
| MARIADASON_RESPONSE_TO_BUTYRATE_CURCUMIN_SULINDAC_TSA_1 | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/MARIADASON_RESPONSE_TO_BUTYRATE_CURCUMIN_SULINDAC_TSA_1 |
| MATZUK_PREOVULATORY_FOLLICLE | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_PREOVULATORY_FOLLICLE |
| MCCABE_HOXC6_TARGETS_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/MCCABE_HOXC6_TARGETS_UP |
| MCCOLLUM_GELDANAMYCIN_RESISTANCE_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/MCCOLLUM_GELDANAMYCIN_RESISTANCE_UP |
| MIKKELSEN_IPS_LCP_WITH_H3K27ME3 | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_IPS_LCP_WITH_H3K27ME3 |
| MIKKELSEN_PARTIALLY_REPROGRAMMED_TO_PLURIPOTENCY | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_PARTIALLY_REPROGRAMMED_TO_PLURIPOTENCY |
| MILICIC_FAMILIAL_ADENOMATOUS_POLYPOSIS_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/MILICIC_FAMILIAL_ADENOMATOUS_POLYPOSIS_DN |
| MILICIC_FAMILIAL_ADENOMATOUS_POLYPOSIS_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/MILICIC_FAMILIAL_ADENOMATOUS_POLYPOSIS_UP |
| MURAKAMI_UV_RESPONSE_1HR_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/MURAKAMI_UV_RESPONSE_1HR_DN |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_13 | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_13 |
| NIKOLSKY_BREAST_CANCER_17P11_AMPLICON | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_17P11_AMPLICON |
| OHASHI_AURKB_TARGETS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/OHASHI_AURKB_TARGETS |
| OSAWA_TNF_TARGETS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/OSAWA_TNF_TARGETS |
| OXFORD_RALA_AND_RALB_TARGETS_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/OXFORD_RALA_AND_RALB_TARGETS_UP |
| OXFORD_RALA_TARGETS_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/OXFORD_RALA_TARGETS_DN |
| PID_VEGF_VEGFR_PATHWAY | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/PID_VEGF_VEGFR_PATHWAY |
| PYEON_HPV_POSITIVE_TUMORS_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/PYEON_HPV_POSITIVE_TUMORS_DN |
| RADAEVA_RESPONSE_TO_IFNA1_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/RADAEVA_RESPONSE_TO_IFNA1_DN |
| RAMPON_ENRICHED_LEARNING_ENVIRONMENT_EARLY_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/RAMPON_ENRICHED_LEARNING_ENVIRONMENT_EARLY_DN |
| REACTOME_ABACAVIR_TRANSPORT_AND_METABOLISM | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ABACAVIR_TRANSPORT_AND_METABOLISM |
| REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE |
| REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS |
| REACTOME_ADENYLATE_CYCLASE_ACTIVATING_PATHWAY | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ADENYLATE_CYCLASE_ACTIVATING_PATHWAY |
| REACTOME_ANDROGEN_BIOSYNTHESIS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ANDROGEN_BIOSYNTHESIS |
| REACTOME_BASE_FREE_SUGAR_PHOSPHATE_REMOVAL_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_BASE_FREE_SUGAR_PHOSPHATE_REMOVAL_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY |
| REACTOME_BINDING_AND_ENTRY_OF_HIV_VIRION | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_BINDING_AND_ENTRY_OF_HIV_VIRION |
| REACTOME_COPI_MEDIATED_TRANSPORT | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_COPI_MEDIATED_TRANSPORT |
| REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS |
| REACTOME_E2F_ENABLED_INHIBITION_OF_PRE_REPLICATION_COMPLEX_FORMATION | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_E2F_ENABLED_INHIBITION_OF_PRE_REPLICATION_COMPLEX_FORMATION |
| REACTOME_ELEVATION_OF_CYTOSOLIC_CA2_LEVELS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ELEVATION_OF_CYTOSOLIC_CA2_LEVELS |
| REACTOME_ETHANOL_OXIDATION | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ETHANOL_OXIDATION |
| REACTOME_GAP_JUNCTION_DEGRADATION | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_GAP_JUNCTION_DEGRADATION |
| REACTOME_HORMONE_LIGAND_BINDING_RECEPTORS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_HORMONE_LIGAND_BINDING_RECEPTORS |
| REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION |
| REACTOME_IKK_COMPLEX_RECRUITMENT_MEDIATED_BY_RIP1 | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_IKK_COMPLEX_RECRUITMENT_MEDIATED_BY_RIP1 |
| REACTOME_IRAK1_RECRUITS_IKK_COMPLEX | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_IRAK1_RECRUITS_IKK_COMPLEX |
| REACTOME_MEMBRANE_BINDING_AND_TARGETTING_OF_GAG_PROTEINS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_MEMBRANE_BINDING_AND_TARGETTING_OF_GAG_PROTEINS |
| REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION |
| REACTOME_NOREPINEPHRINE_NEUROTRANSMITTER_RELEASE_CYCLE | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NOREPINEPHRINE_NEUROTRANSMITTER_RELEASE_CYCLE |
| REACTOME_OPSINS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_OPSINS |
| REACTOME_PECAM1_INTERACTIONS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PECAM1_INTERACTIONS |
| REACTOME_PROSTANOID_LIGAND_RECEPTORS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PROSTANOID_LIGAND_RECEPTORS |
| REACTOME_PURINE_CATABOLISM | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_PURINE_CATABOLISM |
| REACTOME_RAF_MAP_KINASE_CASCADE | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RAF_MAP_KINASE_CASCADE |
| REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES |
| REACTOME_REGULATED_PROTEOLYSIS_OF_P75NTR | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATED_PROTEOLYSIS_OF_P75NTR |
| REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK |
| REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND |
| REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS |
| REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_24_HYDROXYCHOLESTEROL | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_24_HYDROXYCHOLESTEROL |
| REACTOME_SYNTHESIS_OF_PIPS_AT_THE_LATE_ENDOSOME_MEMBRANE | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_SYNTHESIS_OF_PIPS_AT_THE_LATE_ENDOSOME_MEMBRANE |
| REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION_IN_TLR7_8_OR_9_SIGNALING | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION_IN_TLR7_8_OR_9_SIGNALING |
| REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS |
| SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX1_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX1_DN |
| SCHLESINGER_METHYLATED_IN_COLON_CANCER | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHLESINGER_METHYLATED_IN_COLON_CANCER |
| SEMBA_FHIT_TARGETS_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/SEMBA_FHIT_TARGETS_DN |
| SHANK_TAL1_TARGETS_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/SHANK_TAL1_TARGETS_DN |
| SHIRAISHI_PLZF_TARGETS_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/SHIRAISHI_PLZF_TARGETS_UP |
| SMID_BREAST_CANCER_RELAPSE_IN_LIVER_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_RELAPSE_IN_LIVER_DN |
| ST_INTERFERON_GAMMA_PATHWAY | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_INTERFERON_GAMMA_PATHWAY |
| TAKAYAMA_BOUND_BY_AR | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/TAKAYAMA_BOUND_BY_AR |
| TSUDA_ALVEOLAR_SOFT_PART_SARCOMA | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/TSUDA_ALVEOLAR_SOFT_PART_SARCOMA |
| TURJANSKI_MAPK14_TARGETS | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/TURJANSKI_MAPK14_TARGETS |
| VISALA_AGING_LYMPHOCYTE_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/VISALA_AGING_LYMPHOCYTE_UP |
| WEBER_METHYLATED_LCP_IN_FIBROBLAST_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/WEBER_METHYLATED_LCP_IN_FIBROBLAST_DN |
| WONG_ENDOMETRIAL_CANCER_LATE | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/WONG_ENDOMETRIAL_CANCER_LATE |
| WORSCHECH_TUMOR_REJECTION_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/WORSCHECH_TUMOR_REJECTION_DN |
| XU_HGF_TARGETS_REPRESSED_BY_AKT1_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_HGF_TARGETS_REPRESSED_BY_AKT1_UP |
| YANG_MUC2_TARGETS_DUODENUM_6MO_UP | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/YANG_MUC2_TARGETS_DUODENUM_6MO_UP |
| YEMELYANOV_GR_TARGETS_DN | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/YEMELYANOV_GR_TARGETS_DN |
| YIH_RESPONSE_TO_ARSENITE_C5 | 10 | http://www.broadinstitute.org/gsea/msigdb/cards/YIH_RESPONSE_TO_ARSENITE_C5 |
| BANDRES_RESPONSE_TO_CARMUSTIN_MGMT_24HR_UP | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/BANDRES_RESPONSE_TO_CARMUSTIN_MGMT_24HR_UP |
| BANDRES_RESPONSE_TO_CARMUSTIN_WITHOUT_MGMT_24HR_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/BANDRES_RESPONSE_TO_CARMUSTIN_WITHOUT_MGMT_24HR_DN |
| BENPORATH_ES_CORE_NINE | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/BENPORATH_ES_CORE_NINE |
| BIOCARTA_FEEDER_PATHWAY | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_FEEDER_PATHWAY |
| BYSTRYKH_HEMATOPOIESIS_STEM_CELL_FLI1 | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/BYSTRYKH_HEMATOPOIESIS_STEM_CELL_FLI1 |
| BYSTRYKH_HEMATOPOIESIS_STEM_CELL_IL3RA | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/BYSTRYKH_HEMATOPOIESIS_STEM_CELL_IL3RA |
| BYSTRYKH_HEMATOPOIESIS_STEM_CELL_RUNX1 | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/BYSTRYKH_HEMATOPOIESIS_STEM_CELL_RUNX1 |
| CASORELLI_APL_SECONDARY_VS_DE_NOVO_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/CASORELLI_APL_SECONDARY_VS_DE_NOVO_DN |
| CHESLER_BRAIN_D6MIT150_QTL_TRANS | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/CHESLER_BRAIN_D6MIT150_QTL_TRANS |
| COLLER_MYC_TARGETS_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/COLLER_MYC_TARGETS_DN |
| DACOSTA_LOW_DOSE_UV_RESPONSE_VIA_ERCC3_XPCS_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/DACOSTA_LOW_DOSE_UV_RESPONSE_VIA_ERCC3_XPCS_DN |
| FIGUEROA_AML_METHYLATION_CLUSTER_2_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/FIGUEROA_AML_METHYLATION_CLUSTER_2_DN |
| FIRESTEIN_CTNNB1_PATHWAY_AND_PROLIFERATION | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/FIRESTEIN_CTNNB1_PATHWAY_AND_PROLIFERATION |
| FUNG_IL2_TARGETS_WITH_STAT5_BINDING_SITES_T1 | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/FUNG_IL2_TARGETS_WITH_STAT5_BINDING_SITES_T1 |
| GAUSSMANN_MLL_AF4_FUSION_TARGETS_D_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/GAUSSMANN_MLL_AF4_FUSION_TARGETS_D_DN |
| GOUYER_TUMOR_INVASIVENESS | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/GOUYER_TUMOR_INVASIVENESS |
| GUTIERREZ_WALDENSTROEMS_MACROGLOBULINEMIA_1_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/GUTIERREZ_WALDENSTROEMS_MACROGLOBULINEMIA_1_DN |
| GUTIERREZ_WALDENSTROEMS_MACROGLOBULINEMIA_1_UP | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/GUTIERREZ_WALDENSTROEMS_MACROGLOBULINEMIA_1_UP |
| HAEGERSTRAND_RESPONSE_TO_IMATINIB | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/HAEGERSTRAND_RESPONSE_TO_IMATINIB |
| IIZUKA_LIVER_CANCER_PROGRESSION_G2_G3_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/IIZUKA_LIVER_CANCER_PROGRESSION_G2_G3_DN |
| ITO_PTTG1_TARGETS_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/ITO_PTTG1_TARGETS_DN |
| KIM_ALL_DISORDERS_DURATION_CORR_UP | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_ALL_DISORDERS_DURATION_CORR_UP |
| KOMMAGANI_TP63_GAMMA_TARGETS | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/KOMMAGANI_TP63_GAMMA_TARGETS |
| KUMAMOTO_RESPONSE_TO_NUTLIN_3A_UP | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/KUMAMOTO_RESPONSE_TO_NUTLIN_3A_UP |
| MATZUK_CUMULUS_EXPANSION | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_CUMULUS_EXPANSION |
| MATZUK_MATERNAL_EFFECT | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_MATERNAL_EFFECT |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_21 | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_21 |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_30 | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_30 |
| NADELLA_PRKAR1A_TARGETS_UP | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/NADELLA_PRKAR1A_TARGETS_UP |
| NAGY_PCAF_COMPONENTS_HUMAN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/NAGY_PCAF_COMPONENTS_HUMAN |
| NIKOLSKY_BREAST_CANCER_10Q22_AMPLICON | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_10Q22_AMPLICON |
| NOUSHMEHR_GBM_SOMATIC_MUTATED | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/NOUSHMEHR_GBM_SOMATIC_MUTATED |
| OXFORD_RALA_AND_RALB_TARGETS_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/OXFORD_RALA_AND_RALB_TARGETS_DN |
| OXFORD_RALA_TARGETS_UP | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/OXFORD_RALA_TARGETS_UP |
| OXFORD_RALB_TARGETS_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/OXFORD_RALB_TARGETS_DN |
| PUJANA_BREAST_CANCER_WITH_BRCA1_MUTATED_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/PUJANA_BREAST_CANCER_WITH_BRCA1_MUTATED_DN |
| RAFFEL_VEGFA_TARGETS_UP | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/RAFFEL_VEGFA_TARGETS_UP |
| REACTOME_ENDOSOMAL_VACUOLAR_PATHWAY | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_ENDOSOMAL_VACUOLAR_PATHWAY |
| REACTOME_IRAK2_MEDIATED_ACTIVATION_OF_TAK1_COMPLEX_UPON_TLR7_8_OR_9_STIMULATION | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_IRAK2_MEDIATED_ACTIVATION_OF_TAK1_COMPLEX_UPON_TLR7_8_OR_9_STIMULATION |
| REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK |
| RUNNE_GENDER_EFFECT_UP | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/RUNNE_GENDER_EFFECT_UP |
| SASSON_FSH_RESPONSE | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/SASSON_FSH_RESPONSE |
| SA_FAS_SIGNALING | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/SA_FAS_SIGNALING |
| SCHMAHL_PDGF_SIGNALING | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHMAHL_PDGF_SIGNALING |
| SCIAN_CELL_CYCLE_TARGETS_OF_TP53_AND_TP73_UP | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/SCIAN_CELL_CYCLE_TARGETS_OF_TP53_AND_TP73_UP |
| SHIRAISHI_PLZF_TARGETS_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/SHIRAISHI_PLZF_TARGETS_DN |
| SIMBULAN_UV_RESPONSE_NORMAL_UP | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/SIMBULAN_UV_RESPONSE_NORMAL_UP |
| ST_JAK_STAT_PATHWAY | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_JAK_STAT_PATHWAY |
| ST_TYPE_I_INTERFERON_PATHWAY | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_TYPE_I_INTERFERON_PATHWAY |
| TESAR_JAK_TARGETS_MOUSE_ES_D3_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/TESAR_JAK_TARGETS_MOUSE_ES_D3_DN |
| THILLAINADESAN_ZNF217_TARGETS_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/THILLAINADESAN_ZNF217_TARGETS_DN |
| TSUTSUMI_FBXW8_TARGETS | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/TSUTSUMI_FBXW8_TARGETS |
| VANLOO_SP3_TARGETS_UP | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/VANLOO_SP3_TARGETS_UP |
| WANG_ESOPHAGUS_CANCER_PROGRESSION_UP | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_ESOPHAGUS_CANCER_PROGRESSION_UP |
| WANG_RESPONSE_TO_FORSKOLIN_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_RESPONSE_TO_FORSKOLIN_DN |
| XU_AKT1_TARGETS_48HR | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/XU_AKT1_TARGETS_48HR |
| YAMANAKA_GLIOBLASTOMA_SURVIVAL_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/YAMANAKA_GLIOBLASTOMA_SURVIVAL_DN |
| YANAGISAWA_LUNG_CANCER_RECURRENCE | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/YANAGISAWA_LUNG_CANCER_RECURRENCE |
| ZERBINI_RESPONSE_TO_SULINDAC_UP | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/ZERBINI_RESPONSE_TO_SULINDAC_UP |
| ZHU_SKIL_TARGETS_DN | 9 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHU_SKIL_TARGETS_DN |
| BERENJENO_TRANSFORMED_BY_RHOA_REVERSIBLY_UP | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/BERENJENO_TRANSFORMED_BY_RHOA_REVERSIBLY_UP |
| BIOCARTA_KREB_PATHWAY | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_KREB_PATHWAY |
| BOYLAN_MULTIPLE_MYELOMA_PCA1_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/BOYLAN_MULTIPLE_MYELOMA_PCA1_DN |
| BRUNEAU_HEART_GREAT_VESSELS_AND_VALVULOGENESIS | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/BRUNEAU_HEART_GREAT_VESSELS_AND_VALVULOGENESIS |
| BYSTRYKH_HEMATOPOIESIS_STEM_CELL_FGF3 | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/BYSTRYKH_HEMATOPOIESIS_STEM_CELL_FGF3 |
| CAFFAREL_RESPONSE_TO_THC_24HR_3_UP | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/CAFFAREL_RESPONSE_TO_THC_24HR_3_UP |
| CASTELLANO_HRAS_AND_NRAS_TARGETS_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/CASTELLANO_HRAS_AND_NRAS_TARGETS_DN |
| CHANG_POU5F1_TARGETS_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/CHANG_POU5F1_TARGETS_DN |
| CLIMENT_BREAST_CANCER_COPY_NUMBER_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/CLIMENT_BREAST_CANCER_COPY_NUMBER_DN |
| DER_IFN_BETA_RESPONSE_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/DER_IFN_BETA_RESPONSE_DN |
| FUJIWARA_PARK2_HEPATOCYTE_PROLIFERATION_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/FUJIWARA_PARK2_HEPATOCYTE_PROLIFERATION_DN |
| FUJIWARA_PARK2_IN_LIVER_CANCER_UP | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/FUJIWARA_PARK2_IN_LIVER_CANCER_UP |
| GALIE_TUMOR_ANGIOGENESIS | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/GALIE_TUMOR_ANGIOGENESIS |
| GALI_TP53_TARGETS_APOPTOTIC_UP | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/GALI_TP53_TARGETS_APOPTOTIC_UP |
| GAUSSMANN_MLL_AF4_FUSION_TARGETS_B_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/GAUSSMANN_MLL_AF4_FUSION_TARGETS_B_DN |
| GAUTSCHI_SRC_SIGNALING | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/GAUTSCHI_SRC_SIGNALING |
| GROSS_HIF1A_TARGETS_UP | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/GROSS_HIF1A_TARGETS_UP |
| GROSS_HYPOXIA_VIA_HIF1A_ONLY | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/GROSS_HYPOXIA_VIA_HIF1A_ONLY |
| HOLLEMAN_DAUNORUBICIN_ALL_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLEMAN_DAUNORUBICIN_ALL_DN |
| IKEDA_MIR133_TARGETS_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/IKEDA_MIR133_TARGETS_DN |
| ISHIKAWA_STING_SIGNALING | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/ISHIKAWA_STING_SIGNALING |
| KANG_CISPLATIN_RESISTANCE_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/KANG_CISPLATIN_RESISTANCE_DN |
| KHETCHOUMIAN_TRIM24_TARGETS_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/KHETCHOUMIAN_TRIM24_TARGETS_DN |
| KOINUMA_COLON_CANCER_MSI_UP | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/KOINUMA_COLON_CANCER_MSI_UP |
| KONDO_HYPOXIA | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/KONDO_HYPOXIA |
| KREPPEL_CD99_TARGETS_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/KREPPEL_CD99_TARGETS_DN |
| LE_SKI_TARGETS_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/LE_SKI_TARGETS_DN |
| LINDGREN_BLADDER_CANCER_CLUSTER_2A_UP | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/LINDGREN_BLADDER_CANCER_CLUSTER_2A_UP |
| LIU_CDX2_TARGETS_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_CDX2_TARGETS_DN |
| LUCAS_HNF4A_TARGETS_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/LUCAS_HNF4A_TARGETS_DN |
| MARIADASON_RESPONSE_TO_BUTYRATE_CURCUMIN_SULINDAC_TSA_2 | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/MARIADASON_RESPONSE_TO_BUTYRATE_CURCUMIN_SULINDAC_TSA_2 |
| MATZUK_FERTILIZATION | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_FERTILIZATION |
| MATZUK_LUTEAL_GENES | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_LUTEAL_GENES |
| MIKKELSEN_DEDIFFERENTIATED_STATE_UP | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_DEDIFFERENTIATED_STATE_UP |
| MIKKELSEN_PLURIPOTENT_STATE_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_PLURIPOTENT_STATE_DN |
| MOOTHA_PYR | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/MOOTHA_PYR |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_18 | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_18 |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_7 | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_7 |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_9 | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_9 |
| NADELLA_PRKAR1A_TARGETS_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/NADELLA_PRKAR1A_TARGETS_DN |
| NAISHIRO_CTNNB1_TARGETS_WITH_LEF1_MOTIF | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/NAISHIRO_CTNNB1_TARGETS_WITH_LEF1_MOTIF |
| NAKAMURA_LUNG_CANCER | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAMURA_LUNG_CANCER |
| NEBEN_AML_WITH_FLT3_OR_NRAS_UP | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/NEBEN_AML_WITH_FLT3_OR_NRAS_UP |
| NIKOLSKY_BREAST_CANCER_20P13_AMPLICON | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_20P13_AMPLICON |
| NOUSHMEHR_GBM_GERMLINE_MUTATED | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/NOUSHMEHR_GBM_GERMLINE_MUTATED |
| OHM_EMBRYONIC_CARCINOMA_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/OHM_EMBRYONIC_CARCINOMA_DN |
| OXFORD_RALB_TARGETS_UP | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/OXFORD_RALB_TARGETS_UP |
| PHONG_TNF_TARGETS_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/PHONG_TNF_TARGETS_DN |
| RAMJAUN_APOPTOSIS_BY_TGFB1_VIA_MAPK1_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/RAMJAUN_APOPTOSIS_BY_TGFB1_VIA_MAPK1_DN |
| RAMJAUN_APOPTOSIS_BY_TGFB1_VIA_SMAD4_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/RAMJAUN_APOPTOSIS_BY_TGFB1_VIA_SMAD4_DN |
| RANKIN_ANGIOGENIC_TARGETS_OF_VHL_HIF2A_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/RANKIN_ANGIOGENIC_TARGETS_OF_VHL_HIF2A_DN |
| REICHERT_G1S_REGULATORS_AS_PI3K_TARGETS | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/REICHERT_G1S_REGULATORS_AS_PI3K_TARGETS |
| ROETH_TERT_TARGETS_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/ROETH_TERT_TARGETS_DN |
| SAMOLS_TARGETS_OF_KHSV_MIRNAS_UP | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/SAMOLS_TARGETS_OF_KHSV_MIRNAS_UP |
| SASAI_TARGETS_OF_CXCR6_AND_PTCH1_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/SASAI_TARGETS_OF_CXCR6_AND_PTCH1_DN |
| SA_G2_AND_M_PHASES | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/SA_G2_AND_M_PHASES |
| SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX5_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX5_DN |
| SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX6_UP | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX6_UP |
| SCHMIDT_POR_TARGETS_IN_LIMB_BUD_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHMIDT_POR_TARGETS_IN_LIMB_BUD_DN |
| SOUCEK_MYC_TARGETS | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/SOUCEK_MYC_TARGETS |
| STANHILL_HRAS_TRANSFROMATION_UP | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/STANHILL_HRAS_TRANSFROMATION_UP |
| TCGA_GLIOBLASTOMA_MUTATED | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/TCGA_GLIOBLASTOMA_MUTATED |
| TERAMOTO_OPN_TARGETS_CLUSTER_8 | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/TERAMOTO_OPN_TARGETS_CLUSTER_8 |
| THUM_MIR21_TARGETS_HEART_DISEASE_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/THUM_MIR21_TARGETS_HEART_DISEASE_DN |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_SUSTAINED_IN_MONOCYTE_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_SUSTAINED_IN_MONOCYTE_DN |
| TURJANSKI_MAPK7_TARGETS | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/TURJANSKI_MAPK7_TARGETS |
| TURJANSKI_MAPK8_AND_MAPK9_TARGETS | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/TURJANSKI_MAPK8_AND_MAPK9_TARGETS |
| WEBER_METHYLATED_ICP_IN_SPERM_UP | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/WEBER_METHYLATED_ICP_IN_SPERM_UP |
| WIEMANN_TELOMERE_SHORTENING_AND_CHRONIC_LIVER_DAMAGE_UP | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/WIEMANN_TELOMERE_SHORTENING_AND_CHRONIC_LIVER_DAMAGE_UP |
| YAGUE_PRETUMOR_DRUG_RESISTANCE_UP | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/YAGUE_PRETUMOR_DRUG_RESISTANCE_UP |
| YANG_BCL3_TARGETS_DN | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/YANG_BCL3_TARGETS_DN |
| ZHOU_CELL_CYCLE_GENES_IN_IR_RESPONSE_2HR | 8 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHOU_CELL_CYCLE_GENES_IN_IR_RESPONSE_2HR |
| ABBUD_LIF_SIGNALING_2_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/ABBUD_LIF_SIGNALING_2_DN |
| BAFNA_MUC4_TARGETS_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/BAFNA_MUC4_TARGETS_UP |
| BAKER_HEMATOPOESIS_STAT5_TARGETS | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/BAKER_HEMATOPOESIS_STAT5_TARGETS |
| BEGUM_TARGETS_OF_PAX3_FOXO1_FUSION_AND_PAX3 | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/BEGUM_TARGETS_OF_PAX3_FOXO1_FUSION_AND_PAX3 |
| BERGER_MBD2_TARGETS | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/BERGER_MBD2_TARGETS |
| BUDHU_LIVER_CANCER_METASTASIS_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/BUDHU_LIVER_CANCER_METASTASIS_DN |
| BUSA_SAM68_TARGETS_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/BUSA_SAM68_TARGETS_UP |
| CHESLER_BRAIN_D6MIT150_QTL_CIS | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/CHESLER_BRAIN_D6MIT150_QTL_CIS |
| DASU_IL6_SIGNALING_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/DASU_IL6_SIGNALING_DN |
| DER_IFN_ALPHA_RESPONSE_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/DER_IFN_ALPHA_RESPONSE_DN |
| FARDIN_HYPOXIA_9 | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/FARDIN_HYPOXIA_9 |
| FARMER_BREAST_CANCER_CLUSTER_8 | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/FARMER_BREAST_CANCER_CLUSTER_8 |
| FUNG_IL2_TARGETS_WITH_STAT5_BINDING_SITES | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/FUNG_IL2_TARGETS_WITH_STAT5_BINDING_SITES |
| GLINSKY_CANCER_DEATH_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/GLINSKY_CANCER_DEATH_UP |
| HEIDENBLAD_AMPLIFIED_IN_BONE_CANCER | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/HEIDENBLAD_AMPLIFIED_IN_BONE_CANCER |
| HERNANDEZ_ABERRANT_MITOSIS_BY_DOCETACEL_4NM_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/HERNANDEZ_ABERRANT_MITOSIS_BY_DOCETACEL_4NM_DN |
| HESSON_TUMOR_SUPPRESSOR_CLUSTER_3P21_3 | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/HESSON_TUMOR_SUPPRESSOR_CLUSTER_3P21_3 |
| HE_PTEN_TARGETS_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/HE_PTEN_TARGETS_DN |
| HOLLEMAN_DAUNORUBICIN_ALL_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/HOLLEMAN_DAUNORUBICIN_ALL_UP |
| IKEDA_MIR1_TARGETS_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/IKEDA_MIR1_TARGETS_DN |
| INGRAM_SHH_TARGETS | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/INGRAM_SHH_TARGETS |
| JOHANSSON_BRAIN_CANCER_EARLY_VS_LATE_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/JOHANSSON_BRAIN_CANCER_EARLY_VS_LATE_UP |
| JOHNSTONE_PARVB_TARGETS_1_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/JOHNSTONE_PARVB_TARGETS_1_UP |
| KOBAYASHI_EGFR_SIGNALING_6HR_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/KOBAYASHI_EGFR_SIGNALING_6HR_UP |
| KONDO_COLON_CANCER_HCP_WITH_H3K27ME3 | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/KONDO_COLON_CANCER_HCP_WITH_H3K27ME3 |
| KUROKAWA_LIVER_CANCER_EARLY_RECURRENCE_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/KUROKAWA_LIVER_CANCER_EARLY_RECURRENCE_DN |
| KUWANO_RNA_STABILIZED_BY_NO | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/KUWANO_RNA_STABILIZED_BY_NO |
| LIU_CMYB_TARGETS_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_CMYB_TARGETS_DN |
| LOPEZ_MESOTELIOMA_SURVIVAL_TIME_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/LOPEZ_MESOTELIOMA_SURVIVAL_TIME_DN |
| LOPEZ_MESOTHELIOMA_SURVIVAL_OVERALL_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/LOPEZ_MESOTHELIOMA_SURVIVAL_OVERALL_UP |
| LOPEZ_MESOTHELIOMA_SURVIVAL_WORST_VS_BEST_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/LOPEZ_MESOTHELIOMA_SURVIVAL_WORST_VS_BEST_DN |
| LUDWICZEK_TREATING_IRON_OVERLOAD | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/LUDWICZEK_TREATING_IRON_OVERLOAD |
| LY_AGING_OLD_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/LY_AGING_OLD_UP |
| MARSHALL_VIRAL_INFECTION_RESPONSE_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/MARSHALL_VIRAL_INFECTION_RESPONSE_UP |
| MATZUK_STEROIDOGENESIS | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/MATZUK_STEROIDOGENESIS |
| MCBRYAN_TERMINAL_END_BUD_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/MCBRYAN_TERMINAL_END_BUD_DN |
| MCCOLLUM_GELDANAMYCIN_RESISTANCE_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/MCCOLLUM_GELDANAMYCIN_RESISTANCE_DN |
| MIKHAYLOVA_OXIDATIVE_STRESS_RESPONSE_VIA_VHL_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKHAYLOVA_OXIDATIVE_STRESS_RESPONSE_VIA_VHL_UP |
| MIKI_COEXPRESSED_WITH_CYP19A1 | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKI_COEXPRESSED_WITH_CYP19A1 |
| MIKKELSEN_DEDIFFERENTIATED_STATE_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_DEDIFFERENTIATED_STATE_DN |
| MIKKELSEN_NPC_WITH_LCP_H3K27ME3 | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_NPC_WITH_LCP_H3K27ME3 |
| MOOTHA_ROS | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/MOOTHA_ROS |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_1 | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_1 |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_5 | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_5 |
| NAKAMURA_BRONCHIAL_AND_BRONCHIOLAR_EPITHELIA | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAMURA_BRONCHIAL_AND_BRONCHIOLAR_EPITHELIA |
| NIKOLSKY_BREAST_CANCER_19Q13.4_AMPLICON | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_19Q13.4_AMPLICON |
| PALOMERO_GSI_SENSITIVITY_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/PALOMERO_GSI_SENSITIVITY_UP |
| PETRETTO_BLOOD_PRESSURE_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/PETRETTO_BLOOD_PRESSURE_DN |
| PETRETTO_LEFT_VENTRICLE_MASS_QTL_CIS_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/PETRETTO_LEFT_VENTRICLE_MASS_QTL_CIS_DN |
| RAMJAUN_APOPTOSIS_BY_TGFB1_VIA_SMAD4_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/RAMJAUN_APOPTOSIS_BY_TGFB1_VIA_SMAD4_UP |
| SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX2_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX2_DN |
| SCHRAMM_INHBA_TARGETS_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHRAMM_INHBA_TARGETS_UP |
| SCHUHMACHER_MYC_TARGETS_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHUHMACHER_MYC_TARGETS_DN |
| SHARMA_PILOCYTIC_ASTROCYTOMA_LOCATION_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/SHARMA_PILOCYTIC_ASTROCYTOMA_LOCATION_DN |
| STONER_ESOPHAGEAL_CARCINOGENESIS_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/STONER_ESOPHAGEAL_CARCINOGENESIS_DN |
| ST_IL_13_PATHWAY | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_IL_13_PATHWAY |
| ST_INTERLEUKIN_13_PATHWAY | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_INTERLEUKIN_13_PATHWAY |
| ST_PAC1_RECEPTOR_PATHWAY | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/ST_PAC1_RECEPTOR_PATHWAY |
| TAKADA_GASTRIC_CANCER_COPY_NUMBER_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/TAKADA_GASTRIC_CANCER_COPY_NUMBER_UP |
| TERAMOTO_OPN_TARGETS_CLUSTER_5 | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/TERAMOTO_OPN_TARGETS_CLUSTER_5 |
| TESAR_ALK_TARGETS_EPISC_3D_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/TESAR_ALK_TARGETS_EPISC_3D_UP |
| TESAR_ALK_TARGETS_EPISC_4D_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/TESAR_ALK_TARGETS_EPISC_4D_UP |
| TESAR_ALK_TARGETS_HUMAN_ES_5D_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/TESAR_ALK_TARGETS_HUMAN_ES_5D_DN |
| TESAR_JAK_TARGETS_MOUSE_ES_D3_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/TESAR_JAK_TARGETS_MOUSE_ES_D3_UP |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_SUSTAINED_IN_GRANULOCYTE_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_SUSTAINED_IN_GRANULOCYTE_DN |
| TURASHVILI_BREAST_CARCINOMA_DUCTAL_VS_LOBULAR_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/TURASHVILI_BREAST_CARCINOMA_DUCTAL_VS_LOBULAR_DN |
| TURASHVILI_BREAST_NORMAL_DUCTAL_VS_LOBULAR_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/TURASHVILI_BREAST_NORMAL_DUCTAL_VS_LOBULAR_DN |
| WEBER_METHYLATED_LCP_IN_SPERM_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/WEBER_METHYLATED_LCP_IN_SPERM_DN |
| YAMASHITA_SILENCED_BY_METHYLATION | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/YAMASHITA_SILENCED_BY_METHYLATION |
| ZEILSTRA_CD44_TARGETS_DN | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/ZEILSTRA_CD44_TARGETS_DN |
| ZEILSTRA_CD44_TARGETS_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/ZEILSTRA_CD44_TARGETS_UP |
| ZHAN_EARLY_DIFFERENTIATION_GENES_UP | 7 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHAN_EARLY_DIFFERENTIATION_GENES_UP |
| BASSO_HAIRY_CELL_LEUKEMIA_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/BASSO_HAIRY_CELL_LEUKEMIA_UP |
| BIOCARTA_NEUROTRANSMITTERS_PATHWAY | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/BIOCARTA_NEUROTRANSMITTERS_PATHWAY |
| BUSA_SAM68_TARGETS_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/BUSA_SAM68_TARGETS_DN |
| BYSTRYKH_SCP2_QTL | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/BYSTRYKH_SCP2_QTL |
| CASTELLANO_HRAS_AND_NRAS_TARGETS_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/CASTELLANO_HRAS_AND_NRAS_TARGETS_UP |
| CASTELLANO_HRAS_TARGETS_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/CASTELLANO_HRAS_TARGETS_DN |
| CHENG_TAF7L_TARGETS | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/CHENG_TAF7L_TARGETS |
| CHEN_HOXA5_TARGETS_6HR_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEN_HOXA5_TARGETS_6HR_DN |
| CHESLER_BRAIN_QTL_TRANS | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/CHESLER_BRAIN_QTL_TRANS |
| CHOI_ATL_ACUTE_STAGE | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/CHOI_ATL_ACUTE_STAGE |
| CORRADETTI_MTOR_PATHWAY_REGULATORS_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/CORRADETTI_MTOR_PATHWAY_REGULATORS_DN |
| COURTOIS_SENESCENCE_TRIGGERS | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/COURTOIS_SENESCENCE_TRIGGERS |
| CROSBY_E2F4_TARGETS | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/CROSBY_E2F4_TARGETS |
| DARWICHE_PAPILLOMA_RISK_HIGH_VS_LOW_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/DARWICHE_PAPILLOMA_RISK_HIGH_VS_LOW_UP |
| DIERICK_SEROTONIN_FUNCTION_GENES | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/DIERICK_SEROTONIN_FUNCTION_GENES |
| DING_LUNG_CANCER_MUTATED_RECURRENTLY | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/DING_LUNG_CANCER_MUTATED_RECURRENTLY |
| DONATO_CELL_CYCLE_TRETINOIN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/DONATO_CELL_CYCLE_TRETINOIN |
| ELVIDGE_HIF2A_TARGETS_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/ELVIDGE_HIF2A_TARGETS_UP |
| FOURNIER_ACINAR_DEVELOPMENT_EARLY_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/FOURNIER_ACINAR_DEVELOPMENT_EARLY_DN |
| GALIE_TUMOR_STEMNESS_GENES | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/GALIE_TUMOR_STEMNESS_GENES |
| GALI_TP53_TARGETS_APOPTOTIC_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/GALI_TP53_TARGETS_APOPTOTIC_DN |
| GERHOLD_RESPONSE_TO_TZD_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/GERHOLD_RESPONSE_TO_TZD_UP |
| IGARASHI_ATF4_TARGETS_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/IGARASHI_ATF4_TARGETS_UP |
| ISHIDA_TARGETS_OF_SYT_SSX_FUSIONS | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/ISHIDA_TARGETS_OF_SYT_SSX_FUSIONS |
| IWANAGA_E2F1_TARGETS_NOT_INDUCED_BY_SERUM | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/IWANAGA_E2F1_TARGETS_NOT_INDUCED_BY_SERUM |
| KAPOSI_LIVER_CANCER_MET_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/KAPOSI_LIVER_CANCER_MET_DN |
| KASLER_HDAC7_TARGETS_2_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/KASLER_HDAC7_TARGETS_2_UP |
| KAUFFMANN_MELANOMA_RELAPSE_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/KAUFFMANN_MELANOMA_RELAPSE_DN |
| KIM_GLIS2_TARGETS_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_GLIS2_TARGETS_DN |
| KIM_PTEN_TARGETS_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/KIM_PTEN_TARGETS_DN |
| KORKOLA_CHORIOCARCINOMA | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/KORKOLA_CHORIOCARCINOMA |
| KORKOLA_CHORIOCARCINOMA_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/KORKOLA_CHORIOCARCINOMA_UP |
| KREPPEL_CD99_TARGETS_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/KREPPEL_CD99_TARGETS_UP |
| MAINA_HYPOXIA_VHL_TARGETS_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/MAINA_HYPOXIA_VHL_TARGETS_UP |
| MIKHAYLOVA_OXIDATIVE_STRESS_RESPONSE_VIA_VHL_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKHAYLOVA_OXIDATIVE_STRESS_RESPONSE_VIA_VHL_DN |
| MIKKELSEN_ES_LCP_WITH_H3K4ME3_AND_H3K27ME3 | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_ES_LCP_WITH_H3K4ME3_AND_H3K27ME3 |
| MIZUKAMI_HYPOXIA_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/MIZUKAMI_HYPOXIA_DN |
| MOTAMED_RESPONSE_TO_ANDROGEN_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/MOTAMED_RESPONSE_TO_ANDROGEN_DN |
| MOTAMED_RESPONSE_TO_ANDROGEN_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/MOTAMED_RESPONSE_TO_ANDROGEN_UP |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_14 | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_14 |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_6 | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_6 |
| OHASHI_AURKA_TARGETS | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/OHASHI_AURKA_TARGETS |
| OHM_EMBRYONIC_CARCINOMA_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/OHM_EMBRYONIC_CARCINOMA_UP |
| OKAMOTO_LIVER_CANCER_MULTICENTRIC_OCCURRENCE_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/OKAMOTO_LIVER_CANCER_MULTICENTRIC_OCCURRENCE_DN |
| PALOMERO_GSI_SENSITIVITY_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/PALOMERO_GSI_SENSITIVITY_DN |
| PARK_OSTEOBLAST_DIFFERENTIATION_BY_PHENYLAMIL_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/PARK_OSTEOBLAST_DIFFERENTIATION_BY_PHENYLAMIL_DN |
| PETRETTO_LEFT_VENTRICLE_MASS_QTL_CIS_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/PETRETTO_LEFT_VENTRICLE_MASS_QTL_CIS_UP |
| PLASARI_TGFB1_TARGETS_1HR_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/PLASARI_TGFB1_TARGETS_1HR_DN |
| RAMJAUN_APOPTOSIS_BY_TGFB1_VIA_MAPK1_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/RAMJAUN_APOPTOSIS_BY_TGFB1_VIA_MAPK1_UP |
| SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX2_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX2_UP |
| SMID_BREAST_CANCER_NORMAL_LIKE_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_NORMAL_LIKE_DN |
| SMID_BREAST_CANCER_RELAPSE_IN_LIVER_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_RELAPSE_IN_LIVER_UP |
| SMID_BREAST_CANCER_RELAPSE_IN_PLEURA_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_RELAPSE_IN_PLEURA_UP |
| TERAO_AOX4_TARGETS_HG_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/TERAO_AOX4_TARGETS_HG_DN |
| TESAR_ALK_AND_JAK_TARGETS_MOUSE_ES_D4_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/TESAR_ALK_AND_JAK_TARGETS_MOUSE_ES_D4_DN |
| TESAR_ALK_TARGETS_HUMAN_ES_4D_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/TESAR_ALK_TARGETS_HUMAN_ES_4D_DN |
| TRACEY_RESISTANCE_TO_IFNA2_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/TRACEY_RESISTANCE_TO_IFNA2_UP |
| TSAI_DNAJB4_TARGETS_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/TSAI_DNAJB4_TARGETS_DN |
| WALLACE_PROSTATE_CANCER_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/WALLACE_PROSTATE_CANCER_DN |
| WEBER_METHYLATED_HCP_IN_FIBROBLAST_UP | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/WEBER_METHYLATED_HCP_IN_FIBROBLAST_UP |
| WIEMANN_TELOMERE_SHORTENING_AND_CHRONIC_LIVER_DAMAGE_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/WIEMANN_TELOMERE_SHORTENING_AND_CHRONIC_LIVER_DAMAGE_DN |
| WILLIAMS_ESR1_TARGETS_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/WILLIAMS_ESR1_TARGETS_DN |
| YANG_MUC2_TARGETS_COLON_3MO_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/YANG_MUC2_TARGETS_COLON_3MO_DN |
| ZERBINI_RESPONSE_TO_SULINDAC_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/ZERBINI_RESPONSE_TO_SULINDAC_DN |
| ZIRN_TRETINOIN_RESPONSE_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/ZIRN_TRETINOIN_RESPONSE_DN |
| ZIRN_TRETINOIN_RESPONSE_WT1_DN | 6 | http://www.broadinstitute.org/gsea/msigdb/cards/ZIRN_TRETINOIN_RESPONSE_WT1_DN |
| ABDULRAHMAN_KIDNEY_CANCER_VHL_UP | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/ABDULRAHMAN_KIDNEY_CANCER_VHL_UP |
| ALONSO_METASTASIS_EMT_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/ALONSO_METASTASIS_EMT_DN |
| BAFNA_MUC4_TARGETS_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/BAFNA_MUC4_TARGETS_DN |
| BIERIE_INFLAMMATORY_RESPONSE_TGFB1 | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/BIERIE_INFLAMMATORY_RESPONSE_TGFB1 |
| BRUNEAU_SEPTATION_ATRIAL | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/BRUNEAU_SEPTATION_ATRIAL |
| CAFFAREL_RESPONSE_TO_THC_8HR_3_UP | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/CAFFAREL_RESPONSE_TO_THC_8HR_3_UP |
| CALVET_IRINOTECAN_SENSITIVE_VS_RESISTANT_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/CALVET_IRINOTECAN_SENSITIVE_VS_RESISTANT_DN |
| CALVET_IRINOTECAN_SENSITIVE_VS_RESISTANT_UP | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/CALVET_IRINOTECAN_SENSITIVE_VS_RESISTANT_UP |
| CALVET_IRINOTECAN_SENSITIVE_VS_REVERTED_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/CALVET_IRINOTECAN_SENSITIVE_VS_REVERTED_DN |
| CALVET_IRINOTECAN_SENSITIVE_VS_REVERTED_UP | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/CALVET_IRINOTECAN_SENSITIVE_VS_REVERTED_UP |
| CASTELLANO_HRAS_TARGETS_UP | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/CASTELLANO_HRAS_TARGETS_UP |
| CHEOK_RESPONSE_TO_MERCAPTOPURINE_AND_HD_MTX_UP | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/CHEOK_RESPONSE_TO_MERCAPTOPURINE_AND_HD_MTX_UP |
| FERRARI_RESPONSE_TO_FENRETINIDE_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/FERRARI_RESPONSE_TO_FENRETINIDE_DN |
| FIGUEROA_AML_METHYLATION_CLUSTER_7_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/FIGUEROA_AML_METHYLATION_CLUSTER_7_DN |
| FUJIWARA_PARK2_IN_LIVER_CANCER_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/FUJIWARA_PARK2_IN_LIVER_CANCER_DN |
| GAVIN_IL2_RESPONSIVE_FOXP3_TARGETS_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/GAVIN_IL2_RESPONSIVE_FOXP3_TARGETS_DN |
| IM_SREBF1A_TARGETS | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/IM_SREBF1A_TARGETS |
| INAMURA_LUNG_CANCER_SCC_SUBTYPES_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/INAMURA_LUNG_CANCER_SCC_SUBTYPES_DN |
| JONES_TCOF1_TARGETS | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/JONES_TCOF1_TARGETS |
| JU_AGING_TERC_TARGETS_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/JU_AGING_TERC_TARGETS_DN |
| KANG_GIST_WITH_PDGFRA_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/KANG_GIST_WITH_PDGFRA_DN |
| KERLEY_RESPONSE_TO_CISPLATIN_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/KERLEY_RESPONSE_TO_CISPLATIN_DN |
| LAU_APOPTOSIS_CDKN2A_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/LAU_APOPTOSIS_CDKN2A_DN |
| LIU_IL13_MEMORY_MODEL_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/LIU_IL13_MEMORY_MODEL_DN |
| LI_WILMS_TUMOR_ANAPLASTIC_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/LI_WILMS_TUMOR_ANAPLASTIC_DN |
| LU_TUMOR_ENDOTHELIAL_MARKERS_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/LU_TUMOR_ENDOTHELIAL_MARKERS_DN |
| MCGOWAN_RSP6_TARGETS_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/MCGOWAN_RSP6_TARGETS_DN |
| MIKKELSEN_IPS_LCP_WITH_H3K4ME3_AND_H3K27ME3 | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/MIKKELSEN_IPS_LCP_WITH_H3K4ME3_AND_H3K27ME3 |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_11 | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_11 |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_2 | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_2 |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_3 | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_3 |
| NAKAMURA_ALVEOLAR_EPITHELIUM | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/NAKAMURA_ALVEOLAR_EPITHELIUM |
| NICK_RESPONSE_TO_PROC_TREATMENT_UP | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/NICK_RESPONSE_TO_PROC_TREATMENT_UP |
| NIKOLSKY_BREAST_CANCER_19P13_AMPLICON | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/NIKOLSKY_BREAST_CANCER_19P13_AMPLICON |
| OZANNE_AP1_TARGETS_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/OZANNE_AP1_TARGETS_DN |
| PASTURAL_RIZ1_TARGETS_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/PASTURAL_RIZ1_TARGETS_DN |
| RAFFEL_VEGFA_TARGETS_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/RAFFEL_VEGFA_TARGETS_DN |
| RANKIN_ANGIOGENIC_TARGETS_OF_VHL_HIF2A_UP | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/RANKIN_ANGIOGENIC_TARGETS_OF_VHL_HIF2A_UP |
| REN_MIF_TARGETS_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/REN_MIF_TARGETS_DN |
| RODRIGUES_DCC_TARGETS_UP | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/RODRIGUES_DCC_TARGETS_UP |
| SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX3 | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX3 |
| SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX6_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/SCHAEFFER_PROSTATE_DEVELOPMENT_AND_CANCER_BOX6_DN |
| SHARMA_ASTROCYTOMA_WITH_NF1_SYNDROM | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/SHARMA_ASTROCYTOMA_WITH_NF1_SYNDROM |
| SMID_BREAST_CANCER_ERBB2_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/SMID_BREAST_CANCER_ERBB2_DN |
| SMIRNOV_CIRCULATING_ENDOTHELIOCYTES_IN_CANCER_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/SMIRNOV_CIRCULATING_ENDOTHELIOCYTES_IN_CANCER_DN |
| TERAMOTO_OPN_TARGETS_CLUSTER_3 | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/TERAMOTO_OPN_TARGETS_CLUSTER_3 |
| TESAR_ALK_AND_JAK_TARGETS_MOUSE_ES_D4_UP | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/TESAR_ALK_AND_JAK_TARGETS_MOUSE_ES_D4_UP |
| TESAR_ALK_TARGETS_HUMAN_ES_5D_UP | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/TESAR_ALK_TARGETS_HUMAN_ES_5D_UP |
| TOMIDA_LUNG_CANCER_POOR_SURVIVAL | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/TOMIDA_LUNG_CANCER_POOR_SURVIVAL |
| TURJANSKI_MAPK11_TARGETS | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/TURJANSKI_MAPK11_TARGETS |
| VALK_AML_WITH_T_8_21_TRANSLOCATION | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/VALK_AML_WITH_T_8_21_TRANSLOCATION |
| VANDESLUIS_NORMAL_EMBRYOS_UP | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/VANDESLUIS_NORMAL_EMBRYOS_UP |
| VERRECCHIA_RESPONSE_TO_TGFB1_C6 | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/VERRECCHIA_RESPONSE_TO_TGFB1_C6 |
| WANG_RESPONSE_TO_PACLITAXEL_VIA_MAPK8_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_RESPONSE_TO_PACLITAXEL_VIA_MAPK8_DN |
| WANG_THOC1_TARGETS_UP | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/WANG_THOC1_TARGETS_UP |
| WILSON_PROTEASES_AT_TUMOR_BONE_INTERFACE_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/WILSON_PROTEASES_AT_TUMOR_BONE_INTERFACE_DN |
| ZHENG_FOXP3_TARGETS_DN | 5 | http://www.broadinstitute.org/gsea/msigdb/cards/ZHENG_FOXP3_TARGETS_DN |
Format: gmt
Web label: MSigDB Pathways
Web label: key| (|count|)
| Filename | Size | md5 |
|---|---|---|
| c2.all.v4.0.symbols.gmt | 3.12 MB | 1bb7d2e82387558c16d4f32737c42b5b |
| genomic_resource.yaml | 398.0 B | c9edec7b94291b4b0091418b5302b2a4 |
| statistics/ |