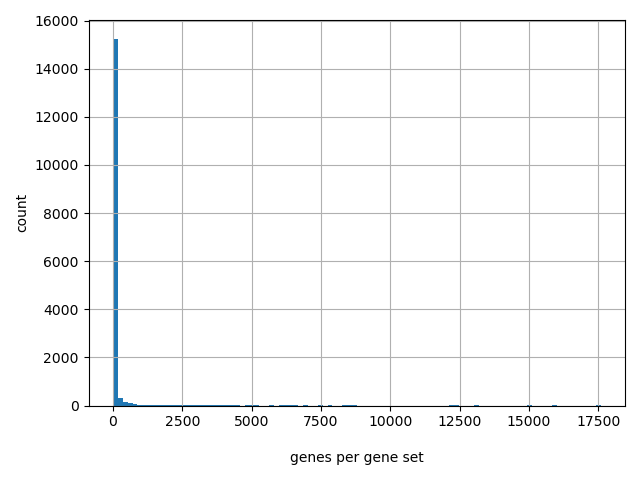
Count of genes per gene set

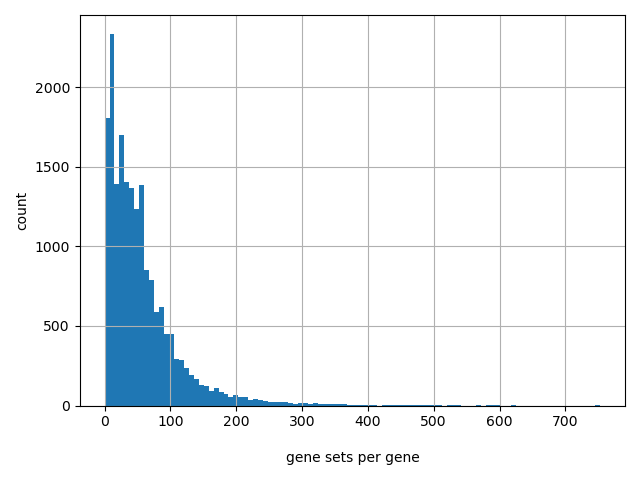
Count of gene sets per gene

| Id: | gene_properties/gene_sets/GO |
| Type: | gene_set_collection |
| Version: | 0 |
| Summary: |
GO (Gene Ontology) gene sets |
| Description: |
N/A
|
| Labels: |
Number of gene sets: 16065
Number of unique genes: 18829


| Gene Set | Gene Count | Description |
|---|---|---|
| GO:0005575 | 17587 | cellular_component |
| GO:0003674 | 15952 | molecular_function |
| GO:0008150 | 14998 | biological_process |
| GO:0044464 | 13144 | cell_part |
| GO:0009987 | 12312 | cellular_process |
| GO:0005488 | 12270 | binding |
| GO:0044424 | 12264 | intracellular_part |
| GO:0065007 | 8763 | biological_regulation |
| GO:0043226 | 8503 | organelle |
| GO:0043229 | 8486 | intracellular_organelle |
| GO:0008152 | 8384 | metabolic_process |
| GO:0050789 | 8301 | regulation_of_biological_process |
| GO:0044238 | 7810 | primary_metabolic_process |
| GO:0050794 | 7752 | regulation_of_cellular_process |
| GO:0043227 | 7521 | membrane-bounded_organelle |
| GO:0043231 | 7515 | intracellular_membrane-bounded_organelle |
| GO:0044237 | 7459 | cellular_metabolic_process |
| GO:0005515 | 7012 | protein_binding |
| GO:0044444 | 6651 | cytoplasmic_part |
| GO:0044422 | 6644 | organelle_part |
| GO:0044446 | 6559 | intracellular_organelle_part |
| GO:0043170 | 6361 | macromolecule_metabolic_process |
| GO:0044425 | 6239 | membrane_part |
| GO:0050896 | 6013 | response_to_stimulus |
| GO:0016020 | 5737 | membrane |
| GO:0003824 | 5667 | catalytic_activity |
| GO:0044260 | 5641 | cellular_macromolecule_metabolic_process |
| GO:0031224 | 5265 | intrinsic_to_membrane |
| GO:0016021 | 5149 | integral_to_membrane |
| GO:0005634 | 5011 | nucleus |
| GO:0019222 | 4784 | regulation_of_metabolic_process |
| GO:0006807 | 4572 | nitrogen_compound_metabolic_process |
| GO:0034641 | 4445 | cellular_nitrogen_compound_metabolic_process |
| GO:0051716 | 4364 | cellular_response_to_stimulus |
| GO:0031323 | 4355 | regulation_of_cellular_metabolic_process |
| GO:0080090 | 4265 | regulation_of_primary_metabolic_process |
| GO:0005737 | 4166 | cytoplasm |
| GO:0060255 | 4118 | regulation_of_macromolecule_metabolic_process |
| GO:0043167 | 4117 | ion_binding |
| GO:0043169 | 4106 | cation_binding |
| GO:0046872 | 4057 | metal_ion_binding |
| GO:0006139 | 4013 | nucleobase-containing_compound_metabolic_process |
| GO:0032991 | 3853 | macromolecular_complex |
| GO:0009058 | 3711 | biosynthetic_process |
| GO:0032502 | 3707 | developmental_process |
| GO:0044249 | 3567 | cellular_biosynthetic_process |
| GO:0007165 | 3554 | signal_transduction |
| GO:0005886 | 3547 | plasma_membrane |
| GO:0003676 | 3510 | nucleic_acid_binding |
| GO:0048518 | 3493 | positive_regulation_of_biological_process |
| GO:0090304 | 3483 | nucleic_acid_metabolic_process |
| GO:0051171 | 3436 | regulation_of_nitrogen_compound_metabolic_process |
| GO:0019219 | 3355 | regulation_of_nucleobase-containing_compound_metabolic_process |
| GO:0009889 | 3345 | regulation_of_biosynthetic_process |
| GO:0010468 | 3318 | regulation_of_gene_expression |
| GO:0031326 | 3317 | regulation_of_cellular_biosynthetic_process |
| GO:0019538 | 3247 | protein_metabolic_process |
| GO:0043234 | 3237 | protein_complex |
| GO:0071840 | 3223 | cellular_component_organization_or_biogenesis |
| GO:0016043 | 3187 | cellular_component_organization |
| GO:0010556 | 3175 | regulation_of_macromolecule_biosynthetic_process |
| GO:2000112 | 3114 | regulation_of_cellular_macromolecule_biosynthetic_process |
| GO:0048522 | 3073 | positive_regulation_of_cellular_process |
| GO:0044428 | 3024 | nuclear_part |
| GO:0051234 | 3003 | establishment_of_localization |
| GO:0016070 | 2998 | RNA_metabolic_process |
| GO:0051252 | 2957 | regulation_of_RNA_metabolic_process |
| GO:0006810 | 2950 | transport |
| GO:2001141 | 2899 | regulation_of_RNA_biosynthetic_process |
| GO:0006355 | 2882 | regulation_of_transcription,_DNA-dependent |
| GO:0048519 | 2873 | negative_regulation_of_biological_process |
| GO:0043228 | 2791 | non-membrane-bounded_organelle |
| GO:0043232 | 2791 | intracellular_non-membrane-bounded_organelle |
| GO:0009059 | 2707 | macromolecule_biosynthetic_process |
| GO:0036094 | 2680 | small_molecule_binding |
| GO:0034645 | 2672 | cellular_macromolecule_biosynthetic_process |
| GO:0071841 | 2629 | cellular_component_organization_or_biogenesis_at_cellular_level |
| GO:0048523 | 2624 | negative_regulation_of_cellular_process |
| GO:0032501 | 2606 | multicellular_organismal_process |
| GO:0042221 | 2604 | response_to_chemical_stimulus |
| GO:0071842 | 2593 | cellular_component_organization_at_cellular_level |
| GO:0016787 | 2500 | hydrolase_activity |
| GO:0000166 | 2490 | nucleotide_binding |
| GO:0044267 | 2481 | cellular_protein_metabolic_process |
| GO:0003677 | 2422 | DNA_binding |
| GO:0046914 | 2390 | transition_metal_ion_binding |
| GO:0032774 | 2332 | RNA_biosynthetic_process |
| GO:0006950 | 2325 | response_to_stress |
| GO:0048583 | 2256 | regulation_of_response_to_stimulus |
| GO:0005829 | 2253 | cytosol |
| GO:0048856 | 2242 | anatomical_structure_development |
| GO:0006351 | 2153 | transcription,_DNA-dependent |
| GO:0031090 | 2132 | organelle_membrane |
| GO:0065008 | 2109 | regulation_of_biological_quality |
| GO:0008270 | 2090 | zinc_ion_binding |
| GO:0043412 | 2071 | macromolecule_modification |
| GO:0044281 | 2006 | small_molecule_metabolic_process |
| GO:0007166 | 1983 | cell_surface_receptor_signaling_pathway |
| GO:0006464 | 1974 | cellular_protein_modification_process |
| GO:0036211 | 1974 | protein_modification_process |
| GO:0017076 | 1956 | purine_nucleotide_binding |
| GO:0032553 | 1944 | ribonucleotide_binding |
| GO:0032555 | 1943 | purine_ribonucleotide_binding |
| GO:0009893 | 1916 | positive_regulation_of_metabolic_process |
| GO:0035639 | 1911 | purine_ribonucleoside_triphosphate_binding |
| GO:0044459 | 1903 | plasma_membrane_part |
| GO:0016740 | 1885 | transferase_activity |
| GO:0023051 | 1841 | regulation_of_signaling |
| GO:0004872 | 1837 | receptor_activity |
| GO:0031325 | 1818 | positive_regulation_of_cellular_metabolic_process |
| GO:0048869 | 1812 | cellular_developmental_process |
| GO:0065009 | 1797 | regulation_of_molecular_function |
| GO:0010604 | 1782 | positive_regulation_of_macromolecule_metabolic_process |
| GO:0006996 | 1698 | organelle_organization |
| GO:0010033 | 1683 | response_to_organic_substance |
| GO:0051239 | 1664 | regulation_of_multicellular_organismal_process |
| GO:0004871 | 1661 | signal_transducer_activity |
| GO:0060089 | 1661 | molecular_transducer_activity |
| GO:0005576 | 1633 | extracellular_region |
| GO:0009966 | 1587 | regulation_of_signal_transduction |
| GO:0030554 | 1579 | adenyl_nucleotide_binding |
| GO:0009056 | 1575 | catabolic_process |
| GO:0032559 | 1569 | adenyl_ribonucleotide_binding |
| GO:0005524 | 1543 | ATP_binding |
| GO:0005730 | 1505 | nucleolus |
| GO:0005622 | 1446 | intracellular |
| GO:0050790 | 1408 | regulation_of_catalytic_activity |
| GO:0010646 | 1382 | regulation_of_cell_communication |
| GO:0002376 | 1368 | immune_system_process |
| GO:0038023 | 1352 | signaling_receptor_activity |
| GO:0051246 | 1341 | regulation_of_protein_metabolic_process |
| GO:0009892 | 1337 | negative_regulation_of_metabolic_process |
| GO:0050793 | 1317 | regulation_of_developmental_process |
| GO:0070887 | 1288 | cellular_response_to_chemical_stimulus |
| GO:0044430 | 1286 | cytoskeletal_part |
| GO:0051649 | 1286 | establishment_of_localization_in_cell |
| GO:0044248 | 1282 | cellular_catabolic_process |
| GO:0030154 | 1262 | cell_differentiation |
| GO:0004888 | 1259 | transmembrane_signaling_receptor_activity |
| GO:0032879 | 1256 | regulation_of_localization |
| GO:0022607 | 1253 | cellular_component_assembly |
| GO:0010605 | 1245 | negative_regulation_of_macromolecule_metabolic_process |
| GO:0031324 | 1245 | negative_regulation_of_cellular_metabolic_process |
| GO:0035556 | 1239 | intracellular_signal_transduction |
| GO:0009891 | 1225 | positive_regulation_of_biosynthetic_process |
| GO:0010941 | 1217 | regulation_of_cell_death |
| GO:0005215 | 1210 | transporter_activity |
| GO:0031328 | 1205 | positive_regulation_of_cellular_biosynthetic_process |
| GO:0005102 | 1191 | receptor_binding |
| GO:0032268 | 1188 | regulation_of_cellular_protein_metabolic_process |
| GO:0000267 | 1185 | cell_fraction |
| GO:0043067 | 1184 | regulation_of_programmed_cell_death |
| GO:0005739 | 1182 | mitochondrion |
| GO:0042981 | 1175 | regulation_of_apoptotic_process |
| GO:0051128 | 1169 | regulation_of_cellular_component_organization |
| GO:0009653 | 1156 | anatomical_structure_morphogenesis |
| GO:0048584 | 1154 | positive_regulation_of_response_to_stimulus |
| GO:0051173 | 1146 | positive_regulation_of_nitrogen_compound_metabolic_process |
| GO:0045935 | 1123 | positive_regulation_of_nucleobase-containing_compound_metabolic_process |
| GO:0042127 | 1119 | regulation_of_cell_proliferation |
| GO:0006357 | 1092 | regulation_of_transcription_from_RNA_polymerase_II_promoter |
| GO:0010557 | 1088 | positive_regulation_of_macromolecule_biosynthetic_process |
| GO:0031226 | 1076 | intrinsic_to_plasma_membrane |
| GO:0044093 | 1074 | positive_regulation_of_molecular_function |
| GO:0019899 | 1061 | enzyme_binding |
| GO:0022414 | 1051 | reproductive_process |
| GO:0010628 | 1037 | positive_regulation_of_gene_expression |
| GO:0071310 | 1036 | cellular_response_to_organic_substance |
| GO:0005887 | 1032 | integral_to_plasma_membrane |
| GO:0009605 | 1032 | response_to_external_stimulus |
| GO:0001071 | 1027 | nucleic_acid_binding_transcription_factor_activity |
| GO:0003700 | 1025 | sequence-specific_DNA_binding_transcription_factor_activity |
| GO:0045184 | 1023 | establishment_of_protein_localization |
| GO:2000026 | 1016 | regulation_of_multicellular_organismal_development |
| GO:0006629 | 1008 | lipid_metabolic_process |
| GO:0051254 | 1005 | positive_regulation_of_RNA_metabolic_process |
| GO:0015031 | 1003 | protein_transport |
| GO:0044421 | 993 | extracellular_region_part |
| GO:0016772 | 992 | transferase_activity,_transferring_phosphorus-containing_groups |
| GO:0022892 | 982 | substrate-specific_transporter_activity |
| GO:0030234 | 974 | enzyme_regulator_activity |
| GO:0003008 | 972 | system_process |
| GO:0033554 | 971 | cellular_response_to_stress |
| GO:0043933 | 969 | macromolecular_complex_subunit_organization |
| GO:0031399 | 966 | regulation_of_protein_modification_process |
| GO:0002682 | 958 | regulation_of_immune_system_process |
| GO:0045893 | 954 | positive_regulation_of_transcription,_DNA-dependent |
| GO:0046907 | 949 | intracellular_transport |
| GO:0022857 | 948 | transmembrane_transporter_activity |
| GO:0006952 | 945 | defense_response |
| GO:0040011 | 944 | locomotion |
| GO:0009890 | 933 | negative_regulation_of_biosynthetic_process |
| GO:0051049 | 930 | regulation_of_transport |
| GO:0031327 | 918 | negative_regulation_of_cellular_biosynthetic_process |
| GO:0045595 | 917 | regulation_of_cell_differentiation |
| GO:0005654 | 913 | nucleoplasm |
| GO:0042592 | 913 | homeostatic_process |
| GO:0016265 | 912 | death |
| GO:0007154 | 909 | cell_communication |
| GO:0008219 | 909 | cell_death |
| GO:0019220 | 909 | regulation_of_phosphate_metabolic_process |
| GO:0051174 | 909 | regulation_of_phosphorus_metabolic_process |
| GO:0005626 | 905 | insoluble_fraction |
| GO:0048513 | 898 | organ_development |
| GO:0003723 | 889 | RNA_binding |
| GO:0043085 | 885 | positive_regulation_of_catalytic_activity |
| GO:0009719 | 880 | response_to_endogenous_stimulus |
| GO:0005624 | 877 | membrane_fraction |
| GO:0004930 | 875 | G-protein_coupled_receptor_activity |
| GO:0071844 | 874 | cellular_component_assembly_at_cellular_level |
| GO:0010558 | 870 | negative_regulation_of_macromolecule_biosynthetic_process |
| GO:0006955 | 864 | immune_response |
| GO:0051172 | 862 | negative_regulation_of_nitrogen_compound_metabolic_process |
| GO:0045934 | 852 | negative_regulation_of_nucleobase-containing_compound_metabolic_process |
| GO:0016301 | 851 | kinase_activity |
| GO:0006508 | 850 | proteolysis |
| GO:0006793 | 849 | phosphorus_metabolic_process |
| GO:0006796 | 849 | phosphate-containing_compound_metabolic_process |
| GO:0022891 | 845 | substrate-specific_transmembrane_transporter_activity |
| GO:0042995 | 844 | cell_projection |
| GO:0022402 | 843 | cell_cycle_process |
| GO:0006928 | 842 | cellular_component_movement |
| GO:2000113 | 841 | negative_regulation_of_cellular_macromolecule_biosynthetic_process |
| GO:0065003 | 832 | macromolecular_complex_assembly |
| GO:0016817 | 829 | hydrolase_activity,_acting_on_acid_anhydrides |
| GO:0051247 | 829 | positive_regulation_of_protein_metabolic_process |
| GO:0016818 | 825 | hydrolase_activity,_acting_on_acid_anhydrides,_in_phosphorus-containing_anhydrides |
| GO:0042325 | 825 | regulation_of_phosphorylation |
| GO:0042802 | 824 | identical_protein_binding |
| GO:0044432 | 824 | endoplasmic_reticulum_part |
| GO:0016462 | 822 | pyrophosphatase_activity |
| GO:0005615 | 817 | extracellular_space |
| GO:0010629 | 817 | negative_regulation_of_gene_expression |
| GO:0044451 | 817 | nucleoplasm_part |
| GO:0051704 | 816 | multi-organism_process |
| GO:0016192 | 797 | vesicle-mediated_transport |
| GO:0046983 | 797 | protein_dimerization_activity |
| GO:0016788 | 790 | hydrolase_activity,_acting_on_ester_bonds |
| GO:0017111 | 789 | nucleoside-triphosphatase_activity |
| GO:0051253 | 777 | negative_regulation_of_RNA_metabolic_process |
| GO:0006082 | 774 | organic_acid_metabolic_process |
| GO:0001932 | 773 | regulation_of_protein_phosphorylation |
| GO:0042180 | 773 | cellular_ketone_metabolic_process |
| GO:0007155 | 770 | cell_adhesion |
| GO:0022610 | 770 | biological_adhesion |
| GO:0032270 | 758 | positive_regulation_of_cellular_protein_metabolic_process |
| GO:0016491 | 756 | oxidoreductase_activity |
| GO:0019752 | 756 | carboxylic_acid_metabolic_process |
| GO:0043436 | 756 | oxoacid_metabolic_process |
| GO:0008289 | 751 | lipid_binding |
| GO:0012501 | 751 | programmed_cell_death |
| GO:0045892 | 751 | negative_regulation_of_transcription,_DNA-dependent |
| GO:0016773 | 748 | phosphotransferase_activity,_alcohol_group_as_acceptor |
| GO:0051336 | 748 | regulation_of_hydrolase_activity |
| GO:0044429 | 747 | mitochondrial_part |
| GO:0006915 | 736 | apoptotic_process |
| GO:0044255 | 731 | cellular_lipid_metabolic_process |
| GO:0010647 | 725 | positive_regulation_of_cell_communication |
| GO:0023056 | 723 | positive_regulation_of_signaling |
| GO:0015075 | 722 | ion_transmembrane_transporter_activity |
| GO:0051726 | 721 | regulation_of_cell_cycle |
| GO:0009725 | 710 | response_to_hormone_stimulus |
| GO:0005975 | 709 | carbohydrate_metabolic_process |
| GO:0023052 | 707 | signaling |
| GO:0006811 | 706 | ion_transport |
| GO:0030054 | 706 | cell_junction |
| GO:0005794 | 705 | Golgi_apparatus |
| GO:0016310 | 698 | phosphorylation |
| GO:0071822 | 698 | protein_complex_subunit_organization |
| GO:0043565 | 696 | sequence-specific_DNA_binding |
| GO:0046483 | 694 | heterocycle_metabolic_process |
| GO:0007267 | 693 | cell-cell_signaling |
| GO:0031401 | 688 | positive_regulation_of_protein_modification_process |
| GO:0005509 | 687 | calcium_ion_binding |
| GO:0005856 | 684 | cytoskeleton |
| GO:0006259 | 682 | DNA_metabolic_process |
| GO:0009057 | 682 | macromolecule_catabolic_process |
| GO:0009967 | 682 | positive_regulation_of_signal_transduction |
| GO:0080134 | 681 | regulation_of_response_to_stress |
| GO:0048585 | 677 | negative_regulation_of_response_to_stimulus |
| GO:0005789 | 669 | endoplasmic_reticulum_membrane |
| GO:0044092 | 661 | negative_regulation_of_molecular_function |
| GO:0005198 | 659 | structural_molecule_activity |
| GO:0048878 | 656 | chemical_homeostasis |
| GO:0005783 | 651 | endoplasmic_reticulum |
| GO:0048731 | 651 | system_development |
| GO:0031982 | 648 | vesicle |
| GO:0002684 | 641 | positive_regulation_of_immune_system_process |
| GO:0045944 | 640 | positive_regulation_of_transcription_from_RNA_polymerase_II_promoter |
| GO:0050776 | 639 | regulation_of_immune_response |
| GO:0008233 | 638 | peptidase_activity |
| GO:0007167 | 636 | enzyme_linked_receptor_protein_signaling_pathway |
| GO:0010467 | 635 | gene_expression |
| GO:0051276 | 635 | chromosome_organization |
| GO:0006468 | 633 | protein_phosphorylation |
| GO:0004672 | 629 | protein_kinase_activity |
| GO:0051338 | 629 | regulation_of_transferase_activity |
| GO:0051094 | 626 | positive_regulation_of_developmental_process |
| GO:0006396 | 625 | RNA_processing |
| GO:0008092 | 617 | cytoskeletal_protein_binding |
| GO:0031410 | 616 | cytoplasmic_vesicle |
| GO:0055085 | 616 | transmembrane_transport |
| GO:0050877 | 615 | neurological_system_process |
| GO:0007010 | 614 | cytoskeleton_organization |
| GO:0008284 | 613 | positive_regulation_of_cell_proliferation |
| GO:0044431 | 613 | Golgi_apparatus_part |
| GO:0043549 | 610 | regulation_of_kinase_activity |
| GO:0070011 | 608 | peptidase_activity,_acting_on_L-amino_acid_peptides |
| GO:0006461 | 602 | protein_complex_assembly |
| GO:0060548 | 598 | negative_regulation_of_cell_death |
| GO:0048870 | 594 | cell_motility |
| GO:0044265 | 592 | cellular_macromolecule_catabolic_process |
| GO:0010648 | 588 | negative_regulation_of_cell_communication |
| GO:0010942 | 588 | positive_regulation_of_cell_death |
| GO:0016071 | 586 | mRNA_metabolic_process |
| GO:0009308 | 585 | amine_metabolic_process |
| GO:0023057 | 584 | negative_regulation_of_signaling |
| GO:0009628 | 582 | response_to_abiotic_stimulus |
| GO:0045859 | 582 | regulation_of_protein_kinase_activity |
| GO:0043069 | 579 | negative_regulation_of_programmed_cell_death |
| GO:0043068 | 574 | positive_regulation_of_programmed_cell_death |
| GO:0043066 | 573 | negative_regulation_of_apoptotic_process |
| GO:0030529 | 571 | ribonucleoprotein_complex |
| GO:0043065 | 569 | positive_regulation_of_apoptotic_process |
| GO:0007186 | 568 | G-protein_coupled_receptor_signaling_pathway |
| GO:0060341 | 568 | regulation_of_cellular_localization |
| GO:0007049 | 565 | cell_cycle |
| GO:0055086 | 564 | nucleobase-containing_small_molecule_metabolic_process |
| GO:0006974 | 563 | response_to_DNA_damage_stimulus |
| GO:0031988 | 563 | membrane-bounded_vesicle |
| GO:0048646 | 563 | anatomical_structure_formation_involved_in_morphogenesis |
| GO:0009607 | 560 | response_to_biotic_stimulus |
| GO:0010562 | 558 | positive_regulation_of_phosphorus_metabolic_process |
| GO:0045937 | 558 | positive_regulation_of_phosphate_metabolic_process |
| GO:0019725 | 555 | cellular_homeostasis |
| GO:0070647 | 555 | protein_modification_by_small_protein_conjugation_or_removal |
| GO:0034621 | 551 | cellular_macromolecular_complex_subunit_organization |
| GO:0050878 | 551 | regulation_of_body_fluid_levels |
| GO:0016023 | 550 | cytoplasmic_membrane-bounded_vesicle |
| GO:0042327 | 550 | positive_regulation_of_phosphorylation |
| GO:0044463 | 550 | cell_projection_part |
| GO:0042803 | 549 | protein_homodimerization_activity |
| GO:0006886 | 546 | intracellular_protein_transport |
| GO:0008324 | 544 | cation_transmembrane_transporter_activity |
| GO:0009888 | 544 | tissue_development |
| GO:0009968 | 543 | negative_regulation_of_signal_transduction |
| GO:0016477 | 543 | cell_migration |
| GO:0008283 | 540 | cell_proliferation |
| GO:0019904 | 538 | protein_domain_specific_binding |
| GO:0022603 | 538 | regulation_of_anatomical_structure_morphogenesis |
| GO:0001934 | 537 | positive_regulation_of_protein_phosphorylation |
| GO:0044427 | 536 | chromosomal_part |
| GO:0006935 | 526 | chemotaxis |
| GO:0042330 | 526 | taxis |
| GO:0006753 | 518 | nucleoside_phosphate_metabolic_process |
| GO:0009117 | 518 | nucleotide_metabolic_process |
| GO:0071495 | 518 | cellular_response_to_endogenous_stimulus |
| GO:0010627 | 517 | regulation_of_intracellular_protein_kinase_cascade |
| GO:0031974 | 512 | membrane-enclosed_lumen |
| GO:0008285 | 510 | negative_regulation_of_cell_proliferation |
| GO:0043086 | 507 | negative_regulation_of_catalytic_activity |
| GO:0005543 | 505 | phospholipid_binding |
| GO:0007169 | 505 | transmembrane_receptor_protein_tyrosine_kinase_signaling_pathway |
| GO:0007275 | 504 | multicellular_organismal_development |
| GO:0022403 | 504 | cell_cycle_phase |
| GO:0040008 | 501 | regulation_of_growth |
| GO:0001775 | 498 | cell_activation |
| GO:0016874 | 496 | ligase_activity |
| GO:0055114 | 494 | oxidation-reduction_process |
| GO:0006325 | 493 | chromatin_organization |
| GO:0050801 | 491 | ion_homeostasis |
| GO:0006812 | 489 | cation_transport |
| GO:0000988 | 487 | protein_binding_transcription_factor_activity |
| GO:0032446 | 487 | protein_modification_by_small_protein_conjugation |
| GO:0009894 | 485 | regulation_of_catabolic_process |
| GO:0044106 | 480 | cellular_amine_metabolic_process |
| GO:0009611 | 479 | response_to_wounding |
| GO:0034097 | 475 | response_to_cytokine_stimulus |
| GO:0051093 | 475 | negative_regulation_of_developmental_process |
| GO:0000139 | 474 | Golgi_membrane |
| GO:0000989 | 472 | transcription_factor_binding_transcription_factor_activity |
| GO:0043233 | 468 | organelle_lumen |
| GO:0055082 | 467 | cellular_chemical_homeostasis |
| GO:0007599 | 465 | hemostasis |
| GO:0045087 | 465 | innate_immune_response |
| GO:0031966 | 462 | mitochondrial_membrane |
| GO:0051240 | 462 | positive_regulation_of_multicellular_organismal_process |
| GO:0007596 | 461 | blood_coagulation |
| GO:0050817 | 461 | coagulation |
| GO:0016567 | 460 | protein_ubiquitination |
| GO:0022890 | 460 | inorganic_cation_transmembrane_transporter_activity |
| GO:0003712 | 457 | transcription_cofactor_activity |
| GO:0045597 | 457 | positive_regulation_of_cell_differentiation |
| GO:0050778 | 457 | positive_regulation_of_immune_response |
| GO:0060589 | 457 | nucleoside-triphosphatase_regulator_activity |
| GO:0033043 | 456 | regulation_of_organelle_organization |
| GO:0051130 | 456 | positive_regulation_of_cellular_component_organization |
| GO:0060284 | 456 | regulation_of_cell_development |
| GO:0048609 | 454 | multicellular_organismal_reproductive_process |
| GO:0018193 | 450 | peptidyl-amino_acid_modification |
| GO:0005815 | 449 | microtubule_organizing_center |
| GO:0048471 | 449 | perinuclear_region_of_cytoplasm |
| GO:0004674 | 448 | protein_serine/threonine_kinase_activity |
| GO:0051345 | 448 | positive_regulation_of_hydrolase_activity |
| GO:0030695 | 445 | GTPase_regulator_activity |
| GO:0000122 | 444 | negative_regulation_of_transcription_from_RNA_polymerase_II_promoter |
| GO:0051050 | 444 | positive_regulation_of_transport |
| GO:0006873 | 438 | cellular_ion_homeostasis |
| GO:0044057 | 437 | regulation_of_system_process |
| GO:0030030 | 436 | cell_projection_organization |
| GO:0072521 | 436 | purine-containing_compound_metabolic_process |
| GO:0008610 | 434 | lipid_biosynthetic_process |
| GO:0046903 | 430 | secretion |
| GO:0004984 | 429 | olfactory_receptor_activity |
| GO:0051248 | 428 | negative_regulation_of_protein_metabolic_process |
| GO:0007600 | 426 | sensory_perception |
| GO:0034622 | 425 | cellular_macromolecular_complex_assembly |
| GO:0030246 | 424 | carbohydrate_binding |
| GO:0031329 | 424 | regulation_of_cellular_catabolic_process |
| GO:0015267 | 423 | channel_activity |
| GO:0022803 | 423 | passive_transmembrane_transporter_activity |
| GO:0070013 | 423 | intracellular_organelle_lumen |
| GO:0007264 | 420 | small_GTPase_mediated_signal_transduction |
| GO:0040012 | 419 | regulation_of_locomotion |
| GO:0051347 | 419 | positive_regulation_of_transferase_activity |
| GO:0008047 | 417 | enzyme_activator_activity |
| GO:0051046 | 417 | regulation_of_secretion |
| GO:0007268 | 416 | synaptic_transmission |
| GO:0010564 | 415 | regulation_of_cell_cycle_process |
| GO:0019001 | 413 | guanyl_nucleotide_binding |
| GO:0032561 | 413 | guanyl_ribonucleotide_binding |
| GO:0051270 | 413 | regulation_of_cellular_component_movement |
| GO:0033674 | 409 | positive_regulation_of_kinase_activity |
| GO:0004175 | 407 | endopeptidase_activity |
| GO:0022838 | 403 | substrate-specific_channel_activity |
| GO:0043005 | 403 | neuron_projection |
| GO:0019900 | 402 | kinase_binding |
| GO:0005525 | 401 | GTP_binding |
| GO:0051960 | 400 | regulation_of_nervous_system_development |
| GO:0008134 | 399 | transcription_factor_binding |
| GO:0030001 | 397 | metal_ion_transport |
| GO:0071702 | 397 | organic_substance_transport |
| GO:0006091 | 395 | generation_of_precursor_metabolites_and_energy |
| GO:0006163 | 395 | purine_nucleotide_metabolic_process |
| GO:0031347 | 395 | regulation_of_defense_response |
| GO:0005216 | 393 | ion_channel_activity |
| GO:0006520 | 392 | cellular_amino_acid_metabolic_process |
| GO:0009887 | 392 | organ_morphogenesis |
| GO:0044271 | 392 | cellular_nitrogen_compound_biosynthetic_process |
| GO:2000145 | 389 | regulation_of_cell_motility |
| GO:0016568 | 388 | chromatin_modification |
| GO:0045860 | 388 | positive_regulation_of_protein_kinase_activity |
| GO:0007610 | 386 | behavior |
| GO:0032269 | 385 | negative_regulation_of_cellular_protein_metabolic_process |
| GO:0002253 | 383 | activation_of_immune_response |
| GO:0044419 | 382 | interspecies_interaction_between_organisms |
| GO:0006397 | 381 | mRNA_processing |
| GO:0032870 | 381 | cellular_response_to_hormone_stimulus |
| GO:0044433 | 381 | cytoplasmic_vesicle_part |
| GO:0071345 | 381 | cellular_response_to_cytokine_stimulus |
| GO:0048468 | 380 | cell_development |
| GO:0012502 | 376 | induction_of_programmed_cell_death |
| GO:0006281 | 374 | DNA_repair |
| GO:0043434 | 374 | response_to_peptide_hormone_stimulus |
| GO:0006917 | 373 | induction_of_apoptosis |
| GO:0010035 | 372 | response_to_inorganic_substance |
| GO:0016887 | 372 | ATPase_activity |
| GO:0045596 | 372 | negative_regulation_of_cell_differentiation |
| GO:0030334 | 370 | regulation_of_cell_migration |
| GO:0051707 | 370 | response_to_other_organism |
| GO:0042578 | 368 | phosphoric_ester_hydrolase_activity |
| GO:0055080 | 368 | cation_homeostasis |
| GO:0007389 | 367 | pattern_specification_process |
| GO:0046873 | 365 | metal_ion_transmembrane_transporter_activity |
| GO:0050767 | 365 | regulation_of_neurogenesis |
| GO:0032787 | 363 | monocarboxylic_acid_metabolic_process |
| GO:0005625 | 362 | soluble_fraction |
| GO:0032101 | 362 | regulation_of_response_to_external_stimulus |
| GO:0044456 | 360 | synapse_part |
| GO:0044262 | 359 | cellular_carbohydrate_metabolic_process |
| GO:0001817 | 354 | regulation_of_cytokine_production |
| GO:0003779 | 354 | actin_binding |
| GO:0010740 | 354 | positive_regulation_of_intracellular_protein_kinase_cascade |
| GO:0019866 | 354 | organelle_inner_membrane |
| GO:0032989 | 354 | cellular_component_morphogenesis |
| GO:0016032 | 353 | viral_reproduction |
| GO:0019901 | 352 | protein_kinase_binding |
| GO:0050865 | 352 | regulation_of_cell_activation |
| GO:0030029 | 351 | actin_filament-based_process |
| GO:0003006 | 349 | developmental_process_involved_in_reproduction |
| GO:0022804 | 349 | active_transmembrane_transporter_activity |
| GO:0032940 | 346 | secretion_by_cell |
| GO:0009991 | 345 | response_to_extracellular_stimulus |
| GO:0006140 | 344 | regulation_of_nucleotide_metabolic_process |
| GO:0007276 | 343 | gamete_generation |
| GO:0044270 | 342 | cellular_nitrogen_compound_catabolic_process |
| GO:0071900 | 342 | regulation_of_protein_serine/threonine_kinase_activity |
| GO:1900542 | 341 | regulation_of_purine_nucleotide_metabolic_process |
| GO:0002252 | 339 | immune_effector_process |
| GO:0005813 | 339 | centrosome |
| GO:0048610 | 339 | cellular_process_involved_in_reproduction |
| GO:0051179 | 339 | localization |
| GO:0042493 | 337 | response_to_drug |
| GO:0006605 | 336 | protein_targeting |
| GO:0007017 | 336 | microtubule-based_process |
| GO:0046700 | 336 | heterocycle_catabolic_process |
| GO:0006954 | 335 | inflammatory_response |
| GO:0007411 | 335 | axon_guidance |
| GO:0000975 | 334 | regulatory_region_DNA_binding |
| GO:0001067 | 334 | regulatory_region_nucleic_acid_binding |
| GO:0016337 | 334 | cell-cell_adhesion |
| GO:0016879 | 334 | ligase_activity,_forming_carbon-nitrogen_bonds |
| GO:0051301 | 333 | cell_division |
| GO:0005874 | 332 | microtubule |
| GO:0051603 | 332 | proteolysis_involved_in_cellular_protein_catabolic_process |
| GO:0007243 | 331 | intracellular_protein_kinase_cascade |
| GO:0009259 | 331 | ribonucleotide_metabolic_process |
| GO:0051090 | 330 | regulation_of_sequence-specific_DNA_binding_transcription_factor_activity |
| GO:0051241 | 330 | negative_regulation_of_multicellular_organismal_process |
| GO:0002694 | 329 | regulation_of_leukocyte_activation |
| GO:0005768 | 328 | endosome |
| GO:0005743 | 327 | mitochondrial_inner_membrane |
| GO:0015077 | 326 | monovalent_inorganic_cation_transmembrane_transporter_activity |
| GO:0044212 | 326 | transcription_regulatory_region_DNA_binding |
| GO:0000278 | 324 | mitotic_cell_cycle |
| GO:0031667 | 322 | response_to_nutrient_levels |
| GO:0043632 | 319 | modification-dependent_macromolecule_catabolic_process |
| GO:0032880 | 318 | regulation_of_protein_localization |
| GO:0004857 | 317 | enzyme_inhibitor_activity |
| GO:0019941 | 316 | modification-dependent_protein_catabolic_process |
| GO:0030003 | 316 | cellular_cation_homeostasis |
| GO:0044087 | 316 | regulation_of_cellular_component_biogenesis |
| GO:0048545 | 316 | response_to_steroid_hormone_stimulus |
| GO:0044440 | 314 | endosomal_part |
| GO:0048598 | 314 | embryonic_morphogenesis |
| GO:0061024 | 314 | membrane_organization |
| GO:0007283 | 313 | spermatogenesis |
| GO:0009150 | 313 | purine_ribonucleotide_metabolic_process |
| GO:0031012 | 313 | extracellular_matrix |
| GO:0048232 | 313 | male_gamete_generation |
| GO:0010608 | 312 | posttranscriptional_regulation_of_gene_expression |
| GO:0051259 | 312 | protein_oligomerization |
| GO:0009986 | 311 | cell_surface |
| GO:0034655 | 311 | nucleobase-containing_compound_catabolic_process |
| GO:0006511 | 310 | ubiquitin-dependent_protein_catabolic_process |
| GO:0016044 | 310 | cellular_membrane_organization |
| GO:0030036 | 309 | actin_cytoskeleton_organization |
| GO:0044283 | 308 | small_molecule_biosynthetic_process |
| GO:0006366 | 307 | transcription_from_RNA_polymerase_II_promoter |
| GO:0009141 | 306 | nucleoside_triphosphate_metabolic_process |
| GO:0009314 | 306 | response_to_radiation |
| GO:0045664 | 305 | regulation_of_neuron_differentiation |
| GO:0019221 | 304 | cytokine-mediated_signaling_pathway |
| GO:0031400 | 304 | negative_regulation_of_protein_modification_process |
| GO:0012506 | 303 | vesicle_membrane |
| GO:0051129 | 302 | negative_regulation_of_cellular_component_organization |
| GO:0007399 | 301 | nervous_system_development |
| GO:0055065 | 301 | metal_ion_homeostasis |
| GO:0016881 | 300 | acid-amino_acid_ligase_activity |
| GO:0010008 | 299 | endosome_membrane |
| GO:0022836 | 298 | gated_channel_activity |
| GO:0009790 | 296 | embryo_development |
| GO:0005083 | 292 | small_GTPase_regulator_activity |
| GO:0005667 | 292 | transcription_factor_complex |
| GO:0008380 | 292 | RNA_splicing |
| GO:0048729 | 292 | tissue_morphogenesis |
| GO:0009199 | 291 | ribonucleoside_triphosphate_metabolic_process |
| GO:0032403 | 291 | protein_complex_binding |
| GO:0034660 | 291 | ncRNA_metabolic_process |
| GO:0051249 | 291 | regulation_of_lymphocyte_activation |
| GO:0009144 | 290 | purine_nucleoside_triphosphate_metabolic_process |
| GO:0030659 | 290 | cytoplasmic_vesicle_membrane |
| GO:0042623 | 290 | ATPase_activity,_coupled |
| GO:0045321 | 290 | leukocyte_activation |
| GO:0001558 | 289 | regulation_of_cell_growth |
| GO:0043408 | 288 | regulation_of_MAPK_cascade |
| GO:0046982 | 288 | protein_heterodimerization_activity |
| GO:0042598 | 287 | vesicular_fraction |
| GO:0009205 | 286 | purine_ribonucleoside_triphosphate_metabolic_process |
| GO:0009166 | 285 | nucleotide_catabolic_process |
| GO:0033036 | 284 | macromolecule_localization |
| GO:0006875 | 283 | cellular_metal_ion_homeostasis |
| GO:0040007 | 283 | growth |
| GO:0005792 | 281 | microsome |
| GO:0016757 | 280 | transferase_activity,_transferring_glycosyl_groups |
| GO:0022604 | 280 | regulation_of_cell_morphogenesis |
| GO:0048037 | 280 | cofactor_binding |
| GO:0006412 | 279 | translation |
| GO:0070201 | 273 | regulation_of_establishment_of_protein_localization |
| GO:0051056 | 271 | regulation_of_small_GTPase_mediated_signal_transduction |
| GO:0072523 | 271 | purine-containing_compound_catabolic_process |
| GO:0005261 | 268 | cation_channel_activity |
| GO:0016791 | 268 | phosphatase_activity |
| GO:0005911 | 267 | cell-cell_junction |
| GO:0003713 | 266 | transcription_coactivator_activity |
| GO:0005096 | 266 | GTPase_activator_activity |
| GO:0006195 | 265 | purine_nucleotide_catabolic_process |
| GO:0016604 | 264 | nuclear_body |
| GO:0019787 | 264 | small_conjugating_protein_ligase_activity |
| GO:0005996 | 260 | monosaccharide_metabolic_process |
| GO:0006897 | 260 | endocytosis |
| GO:0080135 | 260 | regulation_of_cellular_response_to_stress |
| GO:0051493 | 259 | regulation_of_cytoskeleton_organization |
| GO:0070848 | 259 | response_to_growth_factor_stimulus |
| GO:0071156 | 258 | regulation_of_cell_cycle_arrest |
| GO:0003682 | 257 | chromatin_binding |
| GO:0006066 | 257 | alcohol_metabolic_process |
| GO:0009116 | 257 | nucleoside_metabolic_process |
| GO:0051186 | 257 | cofactor_metabolic_process |
| GO:0010038 | 256 | response_to_metal_ion |
| GO:0051051 | 256 | negative_regulation_of_transport |
| GO:0008104 | 255 | protein_localization |
| GO:0006631 | 254 | fatty_acid_metabolic_process |
| GO:0030155 | 254 | regulation_of_cell_adhesion |
| GO:0071375 | 254 | cellular_response_to_peptide_hormone_stimulus |
| GO:0019637 | 252 | organophosphate_metabolic_process |
| GO:0051223 | 252 | regulation_of_protein_transport |
| GO:0044297 | 251 | cell_body |
| GO:0045786 | 251 | negative_regulation_of_cell_cycle |
| GO:0032868 | 250 | response_to_insulin_stimulus |
| GO:0007346 | 249 | regulation_of_mitotic_cell_cycle |
| GO:0016053 | 249 | organic_acid_biosynthetic_process |
| GO:0046394 | 249 | carboxylic_acid_biosynthetic_process |
| GO:0052547 | 249 | regulation_of_peptidase_activity |
| GO:0004842 | 248 | ubiquitin-protein_ligase_activity |
| GO:0030811 | 246 | regulation_of_nucleotide_catabolic_process |
| GO:0033121 | 246 | regulation_of_purine_nucleotide_catabolic_process |
| GO:0050867 | 245 | positive_regulation_of_cell_activation |
| GO:0016746 | 244 | transferase_activity,_transferring_acyl_groups |
| GO:0008202 | 243 | steroid_metabolic_process |
| GO:0052548 | 242 | regulation_of_endopeptidase_activity |
| GO:0018130 | 241 | heterocycle_biosynthetic_process |
| GO:0051641 | 241 | cellular_localization |
| GO:0070482 | 241 | response_to_oxygen_levels |
| GO:0007584 | 240 | response_to_nutrient |
| GO:0006979 | 239 | response_to_oxidative_stress |
| GO:0046486 | 239 | glycerolipid_metabolic_process |
| GO:0071363 | 239 | cellular_response_to_growth_factor_stimulus |
| GO:0009261 | 238 | ribonucleotide_catabolic_process |
| GO:0002696 | 237 | positive_regulation_of_leukocyte_activation |
| GO:0003924 | 237 | GTPase_activity |
| GO:0031344 | 237 | regulation_of_cell_projection_organization |
| GO:0009154 | 236 | purine_ribonucleotide_catabolic_process |
| GO:0034762 | 236 | regulation_of_transmembrane_transport |
| GO:0009119 | 235 | ribonucleoside_metabolic_process |
| GO:0009143 | 235 | nucleoside_triphosphate_catabolic_process |
| GO:0043405 | 235 | regulation_of_MAP_kinase_activity |
| GO:0044454 | 234 | nuclear_chromosome_part |
| GO:0014070 | 233 | response_to_organic_cyclic_compound |
| GO:0046649 | 232 | lymphocyte_activation |
| GO:0006644 | 231 | phospholipid_metabolic_process |
| GO:0009146 | 231 | purine_nucleoside_triphosphate_catabolic_process |
| GO:0043025 | 231 | neuronal_cell_body |
| GO:0005506 | 230 | iron_ion_binding |
| GO:0006725 | 230 | cellular_aromatic_compound_metabolic_process |
| GO:0009203 | 230 | ribonucleoside_triphosphate_catabolic_process |
| GO:0009207 | 230 | purine_ribonucleoside_triphosphate_catabolic_process |
| GO:0016569 | 230 | covalent_chromatin_modification |
| GO:0043566 | 229 | structure-specific_DNA_binding |
| GO:0051346 | 229 | negative_regulation_of_hydrolase_activity |
| GO:0072507 | 229 | divalent_inorganic_cation_homeostasis |
| GO:0002009 | 228 | morphogenesis_of_an_epithelium |
| GO:0032844 | 228 | regulation_of_homeostatic_process |
| GO:0050863 | 228 | regulation_of_T_cell_activation |
| GO:0006916 | 227 | anti-apoptosis |
| GO:0016570 | 227 | histone_modification |
| GO:0032147 | 227 | activation_of_protein_kinase_activity |
| GO:0043269 | 227 | regulation_of_ion_transport |
| GO:0015672 | 226 | monovalent_inorganic_cation_transport |
| GO:0000981 | 225 | sequence-specific_DNA_binding_RNA_polymerase_II_transcription_factor_activity |
| GO:0070085 | 225 | glycosylation |
| GO:0001666 | 224 | response_to_hypoxia |
| GO:0003002 | 224 | regionalization |
| GO:0006457 | 224 | protein_folding |
| GO:0005773 | 223 | vacuole |
| GO:0006486 | 223 | protein_glycosylation |
| GO:0033124 | 223 | regulation_of_GTP_catabolic_process |
| GO:0043413 | 223 | macromolecule_glycosylation |
| GO:0051272 | 223 | positive_regulation_of_cellular_component_movement |
| GO:0000075 | 222 | cell_cycle_checkpoint |
| GO:0002764 | 221 | immune_response-regulating_signaling_pathway |
| GO:0006959 | 221 | humoral_immune_response |
| GO:0040017 | 221 | positive_regulation_of_locomotion |
| GO:0043087 | 221 | regulation_of_GTPase_activity |
| GO:0002237 | 220 | response_to_molecule_of_bacterial_origin |
| GO:0051251 | 219 | positive_regulation_of_lymphocyte_activation |
| GO:0042278 | 217 | purine_nucleoside_metabolic_process |
| GO:0055074 | 217 | calcium_ion_homeostasis |
| GO:0016042 | 216 | lipid_catabolic_process |
| GO:0072503 | 216 | cellular_divalent_inorganic_cation_homeostasis |
| GO:0044282 | 215 | small_molecule_catabolic_process |
| GO:0048011 | 215 | nerve_growth_factor_receptor_signaling_pathway |
| GO:0005126 | 214 | cytokine_receptor_binding |
| GO:0046128 | 214 | purine_ribonucleoside_metabolic_process |
| GO:2000147 | 214 | positive_regulation_of_cell_motility |
| GO:0005578 | 213 | proteinaceous_extracellular_matrix |
| GO:0034220 | 213 | ion_transmembrane_transport |
| GO:0016324 | 212 | apical_plasma_membrane |
| GO:0031644 | 212 | regulation_of_neurological_system_process |
| GO:0032990 | 212 | cell_part_morphogenesis |
| GO:0005125 | 211 | cytokine_activity |
| GO:0001525 | 210 | angiogenesis |
| GO:0016055 | 210 | Wnt_receptor_signaling_pathway |
| GO:0030335 | 210 | positive_regulation_of_cell_migration |
| GO:0010563 | 209 | negative_regulation_of_phosphorus_metabolic_process |
| GO:0045936 | 209 | negative_regulation_of_phosphate_metabolic_process |
| GO:0051260 | 209 | protein_homooligomerization |
| GO:0071902 | 209 | positive_regulation_of_protein_serine/threonine_kinase_activity |
| GO:0002757 | 208 | immune_response-activating_signal_transduction |
| GO:0003012 | 208 | muscle_system_process |
| GO:0022411 | 208 | cellular_component_disassembly |
| GO:0051047 | 208 | positive_regulation_of_secretion |
| GO:0070161 | 208 | anchoring_junction |
| GO:0032496 | 207 | response_to_lipopolysaccharide |
| GO:0060627 | 207 | regulation_of_vesicle-mediated_transport |
| GO:0071845 | 207 | cellular_component_disassembly_at_cellular_level |
| GO:0000785 | 206 | chromatin |
| GO:0006401 | 206 | RNA_catabolic_process |
| GO:0006874 | 206 | cellular_calcium_ion_homeostasis |
| GO:0010817 | 206 | regulation_of_hormone_levels |
| GO:0048285 | 206 | organelle_fission |
| GO:0061134 | 206 | peptidase_regulator_activity |
| GO:0009615 | 205 | response_to_virus |
| GO:0030168 | 205 | platelet_activation |
| GO:0044437 | 205 | vacuolar_part |
| GO:0097060 | 205 | synaptic_membrane |
| GO:0000226 | 204 | microtubule_cytoskeleton_organization |
| GO:0042326 | 204 | negative_regulation_of_phosphorylation |
| GO:0002697 | 203 | regulation_of_immune_effector_process |
| GO:0016741 | 203 | transferase_activity,_transferring_one-carbon_groups |
| GO:0019318 | 203 | hexose_metabolic_process |
| GO:0045088 | 203 | regulation_of_innate_immune_response |
| GO:0000323 | 202 | lytic_vacuole |
| GO:0005759 | 202 | mitochondrial_matrix |
| GO:0006732 | 202 | coenzyme_metabolic_process |
| GO:0034470 | 202 | ncRNA_processing |
| GO:0005764 | 201 | lysosome |
| GO:0031300 | 201 | intrinsic_to_organelle_membrane |
| GO:0043623 | 201 | cellular_protein_complex_assembly |
| GO:0050662 | 201 | coenzyme_binding |
| GO:0050678 | 201 | regulation_of_epithelial_cell_proliferation |
| GO:0072594 | 201 | establishment_of_protein_localization_to_organelle |
| GO:0034702 | 200 | ion_channel_complex |
| GO:0001871 | 199 | pattern_binding |
| GO:0019216 | 199 | regulation_of_lipid_metabolic_process |
| GO:0030247 | 199 | polysaccharide_binding |
| GO:0051969 | 199 | regulation_of_transmission_of_nerve_impulse |
| GO:0005882 | 198 | intermediate_filament |
| GO:0008168 | 198 | methyltransferase_activity |
| GO:0015291 | 197 | secondary_active_transmembrane_transporter_activity |
| GO:0031349 | 197 | positive_regulation_of_defense_response |
| GO:0017171 | 196 | serine_hydrolase_activity |
| GO:0071496 | 196 | cellular_response_to_external_stimulus |
| GO:0048858 | 195 | cell_projection_morphogenesis |
| GO:0006887 | 194 | exocytosis |
| GO:0010975 | 194 | regulation_of_neuron_projection_development |
| GO:0016758 | 194 | transferase_activity,_transferring_hexosyl_groups |
| GO:0032869 | 194 | cellular_response_to_insulin_stimulus |
| GO:0072376 | 194 | protein_activation_cascade |
| GO:0001664 | 193 | G-protein_coupled_receptor_binding |
| GO:0008236 | 193 | serine-type_peptidase_activity |
| GO:0009416 | 193 | response_to_light_stimulus |
| GO:0050900 | 193 | leukocyte_migration |
| GO:0051606 | 193 | detection_of_stimulus |
| GO:0010638 | 192 | positive_regulation_of_organelle_organization |
| GO:0044420 | 192 | extracellular_matrix_part |
| GO:0051052 | 192 | regulation_of_DNA_metabolic_process |
| GO:0005912 | 191 | adherens_junction |
| GO:0008237 | 191 | metallopeptidase_activity |
| GO:0045926 | 191 | negative_regulation_of_growth |
| GO:0001933 | 190 | negative_regulation_of_protein_phosphorylation |
| GO:0009792 | 190 | embryo_development_ending_in_birth_or_egg_hatching |
| GO:0000280 | 189 | nuclear_division |
| GO:0006260 | 189 | DNA_replication |
| GO:0007067 | 189 | mitosis |
| GO:0006310 | 188 | DNA_recombination |
| GO:0015980 | 186 | energy_derivation_by_oxidation_of_organic_compounds |
| GO:0016747 | 186 | transferase_activity,_transferring_acyl_groups_other_than_amino-acyl_groups |
| GO:0045121 | 186 | membrane_raft |
| GO:0050953 | 186 | sensory_perception_of_light_stimulus |
| GO:0051169 | 186 | nuclear_transport |
| GO:0070727 | 186 | cellular_macromolecule_localization |
| GO:0002521 | 185 | leukocyte_differentiation |
| GO:0010959 | 185 | regulation_of_metal_ion_transport |
| GO:0034654 | 185 | nucleobase-containing_compound_biosynthetic_process |
| GO:0043009 | 185 | chordate_embryonic_development |
| GO:0050804 | 185 | regulation_of_synaptic_transmission |
| GO:0001819 | 184 | positive_regulation_of_cytokine_production |
| GO:0048511 | 184 | rhythmic_process |
| GO:0001701 | 183 | in_utero_embryonic_development |
| GO:0006913 | 183 | nucleocytoplasmic_transport |
| GO:0009055 | 183 | electron_carrier_activity |
| GO:0003823 | 182 | antigen_binding |
| GO:0006417 | 182 | regulation_of_translation |
| GO:0034613 | 182 | cellular_protein_localization |
| GO:0000287 | 181 | magnesium_ion_binding |
| GO:0006352 | 181 | DNA-dependent_transcription,_initiation |
| GO:0006936 | 181 | muscle_contraction |
| GO:0043687 | 181 | post-translational_protein_modification |
| GO:0003714 | 180 | transcription_corepressor_activity |
| GO:0006402 | 180 | mRNA_catabolic_process |
| GO:0007568 | 180 | aging |
| GO:0009164 | 180 | nucleoside_catabolic_process |
| GO:0015629 | 180 | actin_cytoskeleton |
| GO:0016054 | 180 | organic_acid_catabolic_process |
| GO:0046395 | 180 | carboxylic_acid_catabolic_process |
| GO:0000375 | 179 | RNA_splicing,_via_transesterification_reactions |
| GO:0004518 | 179 | nuclease_activity |
| GO:0007601 | 179 | visual_perception |
| GO:0031253 | 179 | cell_projection_membrane |
| GO:0005244 | 178 | voltage-gated_ion_channel_activity |
| GO:0005539 | 178 | glycosaminoglycan_binding |
| GO:0022832 | 178 | voltage-gated_channel_activity |
| GO:0031301 | 178 | integral_to_organelle_membrane |
| GO:0032386 | 178 | regulation_of_intracellular_transport |
| GO:0045211 | 178 | postsynaptic_membrane |
| GO:0004721 | 176 | phosphoprotein_phosphatase_activity |
| GO:0016705 | 176 | oxidoreductase_activity,_acting_on_paired_donors,_with_incorporation_or_reduction_of_molecular_oxygen |
| GO:0043254 | 176 | regulation_of_protein_complex_assembly |
| GO:0050727 | 176 | regulation_of_inflammatory_response |
| GO:1901068 | 176 | guanosine-containing_compound_metabolic_process |
| GO:0006790 | 175 | sulfur_compound_metabolic_process |
| GO:0030425 | 175 | dendrite |
| GO:0031965 | 175 | nuclear_membrane |
| GO:0006956 | 174 | complement_activation |
| GO:0010498 | 174 | proteasomal_protein_catabolic_process |
| GO:0032970 | 174 | regulation_of_actin_filament-based_process |
| GO:0046883 | 174 | regulation_of_hormone_secretion |
| GO:0050870 | 174 | positive_regulation_of_T_cell_activation |
| GO:0090066 | 174 | regulation_of_anatomical_structure_size |
| GO:0030414 | 173 | peptidase_inhibitor_activity |
| GO:0043161 | 173 | proteasomal_ubiquitin-dependent_protein_catabolic_process |
| GO:0051091 | 173 | positive_regulation_of_sequence-specific_DNA_binding_transcription_factor_activity |
| GO:0072511 | 173 | divalent_inorganic_cation_transport |
| GO:0000377 | 172 | RNA_splicing,_via_transesterification_reactions_with_bulged_adenosine_as_nucleophile |
| GO:0000398 | 172 | nuclear_mRNA_splicing,_via_spliceosome |
| GO:0000956 | 172 | nuclear-transcribed_mRNA_catabolic_process |
| GO:0009165 | 172 | nucleotide_biosynthetic_process |
| GO:0016051 | 172 | carbohydrate_biosynthetic_process |
| GO:0003735 | 171 | structural_constituent_of_ribosome |
| GO:0005840 | 171 | ribosome |
| GO:0030182 | 171 | neuron_differentiation |
| GO:0031406 | 171 | carboxylic_acid_binding |
| GO:0043406 | 171 | positive_regulation_of_MAP_kinase_activity |
| GO:0061135 | 171 | endopeptidase_regulator_activity |
| GO:2000116 | 171 | regulation_of_cysteine-type_endopeptidase_activity |
| GO:0004252 | 170 | serine-type_endopeptidase_activity |
| GO:0010769 | 170 | regulation_of_cell_morphogenesis_involved_in_differentiation |
| GO:0030111 | 170 | regulation_of_Wnt_receptor_signaling_pathway |
| GO:0030522 | 170 | intracellular_receptor_mediated_signaling_pathway |
| GO:0032956 | 169 | regulation_of_actin_cytoskeleton_organization |
| GO:0070838 | 169 | divalent_metal_ion_transport |
| GO:0022415 | 168 | viral_reproductive_process |
| GO:0031396 | 168 | regulation_of_protein_ubiquitination |
| GO:0043122 | 168 | regulation_of_I-kappaB_kinase/NF-kappaB_cascade |
| GO:0043410 | 168 | positive_regulation_of_MAPK_cascade |
| GO:0048193 | 168 | Golgi_vesicle_transport |
| GO:0005085 | 167 | guanyl-nucleotide_exchange_factor_activity |
| GO:0030424 | 167 | axon |
| GO:0004866 | 166 | endopeptidase_inhibitor_activity |
| GO:0008083 | 166 | growth_factor_activity |
| GO:0009897 | 166 | external_side_of_plasma_membrane |
| GO:0016829 | 166 | lyase_activity |
| GO:0042454 | 166 | ribonucleoside_catabolic_process |
| GO:0043281 | 166 | regulation_of_cysteine-type_endopeptidase_activity_involved_in_apoptotic_process |
| GO:0045202 | 166 | synapse |
| GO:0007420 | 165 | brain_development |
| GO:0034330 | 165 | cell_junction_organization |
| GO:0043062 | 165 | extracellular_structure_organization |
| GO:0051098 | 165 | regulation_of_binding |
| GO:0000165 | 164 | MAPK_cascade |
| GO:0030198 | 164 | extracellular_matrix_organization |
| GO:0043235 | 164 | receptor_complex |
| GO:0046039 | 164 | GTP_metabolic_process |
| GO:0051348 | 164 | negative_regulation_of_transferase_activity |
| GO:0003690 | 163 | double-stranded_DNA_binding |
| GO:0015849 | 163 | organic_acid_transport |
| GO:0030141 | 162 | secretory_granule |
| GO:0033273 | 162 | response_to_vitamin |
| GO:0043627 | 162 | response_to_estrogen_stimulus |
| GO:0071705 | 162 | nitrogen_compound_transport |
| GO:0000209 | 161 | protein_polyubiquitination |
| GO:0005516 | 161 | calmodulin_binding |
| GO:0046942 | 161 | carboxylic_acid_transport |
| GO:0048812 | 161 | neuron_projection_morphogenesis |
| GO:0002683 | 160 | negative_regulation_of_immune_system_process |
| GO:0006958 | 160 | complement_activation,_classical_pathway |
| GO:0006650 | 159 | glycerophospholipid_metabolic_process |
| GO:0008022 | 159 | protein_C-terminus_binding |
| GO:0009617 | 159 | response_to_bacterium |
| GO:0010565 | 159 | regulation_of_cellular_ketone_metabolic_process |
| GO:0010639 | 159 | negative_regulation_of_organelle_organization |
| GO:0030855 | 159 | epithelial_cell_differentiation |
| GO:0060249 | 159 | anatomical_structure_homeostasis |
| GO:0005774 | 158 | vacuolar_membrane |
| GO:0006006 | 158 | glucose_metabolic_process |
| GO:0006152 | 158 | purine_nucleoside_catabolic_process |
| GO:0046130 | 158 | purine_ribonucleoside_catabolic_process |
| GO:0050890 | 158 | cognition |
| GO:0008234 | 157 | cysteine-type_peptidase_activity |
| GO:0032318 | 157 | regulation_of_Ras_GTPase_activity |
| GO:0046777 | 157 | protein_autophosphorylation |
| GO:0050730 | 156 | regulation_of_peptidyl-tyrosine_phosphorylation |
| GO:0015631 | 155 | tubulin_binding |
| GO:0048589 | 155 | developmental_growth |
| GO:0015931 | 154 | nucleobase-containing_compound_transport |
| GO:0033673 | 154 | negative_regulation_of_kinase_activity |
| GO:0000902 | 153 | cell_morphogenesis |
| GO:0001501 | 153 | skeletal_system_development |
| GO:0001763 | 153 | morphogenesis_of_a_branching_structure |
| GO:0004386 | 153 | helicase_activity |
| GO:0090068 | 153 | positive_regulation_of_cell_cycle_process |
| GO:0006367 | 152 | transcription_initiation_from_RNA_polymerase_II_promoter |
| GO:0016853 | 152 | isomerase_activity |
| GO:0019882 | 152 | antigen_processing_and_presentation |
| GO:0043523 | 152 | regulation_of_neuron_apoptotic_process |
| GO:0043547 | 152 | positive_regulation_of_GTPase_activity |
| GO:0070663 | 152 | regulation_of_leukocyte_proliferation |
| GO:0006869 | 151 | lipid_transport |
| GO:0035091 | 151 | phosphatidylinositol_binding |
| GO:0051020 | 151 | GTPase_binding |
| GO:1901069 | 151 | guanosine-containing_compound_catabolic_process |
| GO:0006612 | 150 | protein_targeting_to_membrane |
| GO:0009952 | 150 | anterior/posterior_pattern_specification |
| GO:0042110 | 150 | T_cell_activation |
| GO:0044455 | 150 | mitochondrial_membrane_part |
| GO:0051924 | 150 | regulation_of_calcium_ion_transport |
| GO:0004713 | 149 | protein_tyrosine_kinase_activity |
| GO:0005929 | 149 | cilium |
| GO:0006112 | 149 | energy_reserve_metabolic_process |
| GO:0006184 | 149 | GTP_catabolic_process |
| GO:0007507 | 149 | heart_development |
| GO:0008509 | 149 | anion_transmembrane_transporter_activity |
| GO:0042445 | 149 | hormone_metabolic_process |
| GO:0046906 | 149 | tetrapyrrole_binding |
| GO:0048534 | 149 | hemopoietic_or_lymphoid_organ_development |
| GO:2000241 | 149 | regulation_of_reproductive_process |
| GO:0000082 | 148 | G1/S_transition_of_mitotic_cell_cycle |
| GO:0000151 | 148 | ubiquitin_ligase_complex |
| GO:0009309 | 148 | amine_biosynthetic_process |
| GO:0016311 | 148 | dephosphorylation |
| GO:0030278 | 148 | regulation_of_ossification |
| GO:0046578 | 148 | regulation_of_Ras_protein_signal_transduction |
| GO:0090092 | 148 | regulation_of_transmembrane_receptor_protein_serine/threonine_kinase_signaling_pathway |
| GO:0008286 | 147 | insulin_receptor_signaling_pathway |
| GO:0008544 | 147 | epidermis_development |
| GO:0031668 | 147 | cellular_response_to_extracellular_stimulus |
| GO:0032103 | 147 | positive_regulation_of_response_to_external_stimulus |
| GO:0032944 | 147 | regulation_of_mononuclear_cell_proliferation |
| GO:0006805 | 146 | xenobiotic_metabolic_process |
| GO:0010243 | 146 | response_to_organic_nitrogen |
| GO:0016052 | 146 | carbohydrate_catabolic_process |
| GO:0019842 | 146 | vitamin_binding |
| GO:0022900 | 146 | electron_transport_chain |
| GO:0031960 | 146 | response_to_corticosteroid_stimulus |
| GO:0050670 | 146 | regulation_of_lymphocyte_proliferation |
| GO:0006469 | 145 | negative_regulation_of_protein_kinase_activity |
| GO:0010720 | 145 | positive_regulation_of_cell_development |
| GO:0030031 | 145 | cell_projection_assembly |
| GO:0030162 | 145 | regulation_of_proteolysis |
| GO:0051222 | 145 | positive_regulation_of_protein_transport |
| GO:0005694 | 144 | chromosome |
| GO:0007611 | 144 | learning_or_memory |
| GO:0031225 | 144 | anchored_to_membrane |
| GO:0002791 | 143 | regulation_of_peptide_secretion |
| GO:0007187 | 143 | G-protein_coupled_receptor_signaling_pathway,_coupled_to_cyclic_nucleotide_second_messenger |
| GO:0043123 | 143 | positive_regulation_of_I-kappaB_kinase/NF-kappaB_cascade |
| GO:0043176 | 143 | amine_binding |
| GO:0045765 | 143 | regulation_of_angiogenesis |
| GO:0051480 | 143 | cytosolic_calcium_ion_homeostasis |
| GO:0060090 | 143 | binding,_bridging |
| GO:0090087 | 143 | regulation_of_peptide_transport |
| GO:0006575 | 142 | cellular_modified_amino_acid_metabolic_process |
| GO:0007156 | 142 | homophilic_cell_adhesion |
| GO:0016607 | 142 | nuclear_speck |
| GO:0030135 | 142 | coated_vesicle |
| GO:0045927 | 142 | positive_regulation_of_growth |
| GO:0071824 | 142 | protein-DNA_complex_subunit_organization |
| GO:0000976 | 141 | transcription_regulatory_region_sequence-specific_DNA_binding |
| GO:0005681 | 141 | spliceosomal_complex |
| GO:0090276 | 141 | regulation_of_peptide_hormone_secretion |
| GO:0007018 | 140 | microtubule-based_movement |
| GO:0020037 | 140 | heme_binding |
| GO:0033157 | 140 | regulation_of_intracellular_protein_transport |
| GO:0042176 | 140 | regulation_of_protein_catabolic_process |
| GO:0042742 | 140 | defense_response_to_bacterium |
| GO:0044449 | 140 | contractile_fiber_part |
| GO:0005976 | 139 | polysaccharide_metabolic_process |
| GO:0007050 | 139 | cell_cycle_arrest |
| GO:0031625 | 139 | ubiquitin_protein_ligase_binding |
| GO:0044389 | 139 | small_conjugating_protein_ligase_binding |
| GO:0046822 | 139 | regulation_of_nucleocytoplasmic_transport |
| GO:0070302 | 139 | regulation_of_stress-activated_protein_kinase_signaling_cascade |
| GO:0030308 | 138 | negative_regulation_of_cell_growth |
| GO:0032872 | 138 | regulation_of_stress-activated_MAPK_cascade |
| GO:0034329 | 138 | cell_junction_assembly |
| GO:0042277 | 138 | peptide_binding |
| GO:0015276 | 137 | ligand-gated_ion_channel_activity |
| GO:0022834 | 137 | ligand-gated_channel_activity |
| GO:0045089 | 137 | positive_regulation_of_innate_immune_response |
| GO:0051384 | 137 | response_to_glucocorticoid_stimulus |
| GO:0002768 | 136 | immune_response-regulating_cell_surface_receptor_signaling_pathway |
| GO:0006816 | 136 | calcium_ion_transport |
| GO:0030055 | 136 | cell-substrate_junction |
| GO:0051188 | 136 | cofactor_biosynthetic_process |
| GO:0005765 | 135 | lysosomal_membrane |
| GO:0016197 | 135 | endosomal_transport |
| GO:0040013 | 135 | negative_regulation_of_locomotion |
| GO:0044344 | 135 | cellular_response_to_fibroblast_growth_factor_stimulus |
| GO:0071774 | 135 | response_to_fibroblast_growth_factor_stimulus |
| GO:0005769 | 134 | early_endosome |
| GO:0007626 | 134 | locomotory_behavior |
| GO:0034728 | 134 | nucleosome_organization |
| GO:0042391 | 134 | regulation_of_membrane_potential |
| GO:0044391 | 134 | ribosomal_subunit |
| GO:0050657 | 134 | nucleic_acid_transport |
| GO:0050658 | 134 | RNA_transport |
| GO:0051236 | 134 | establishment_of_RNA_localization |
| GO:0003774 | 133 | motor_activity |
| GO:0007005 | 133 | mitochondrion_organization |
| GO:0008201 | 133 | heparin_binding |
| GO:0015837 | 133 | amine_transport |
| GO:0016323 | 133 | basolateral_plasma_membrane |
| GO:0017038 | 133 | protein_import |
| GO:0035966 | 133 | response_to_topologically_incorrect_protein |
| GO:0006399 | 132 | tRNA_metabolic_process |
| GO:0022843 | 132 | voltage-gated_cation_channel_activity |
| GO:0030674 | 132 | protein_binding,_bridging |
| GO:0031267 | 132 | small_GTPase_binding |
| GO:0045017 | 132 | glycerolipid_biosynthetic_process |
| GO:0050795 | 132 | regulation_of_behavior |
| GO:0050796 | 132 | regulation_of_insulin_secretion |
| GO:0051082 | 132 | unfolded_protein_binding |
| GO:0007173 | 131 | epidermal_growth_factor_receptor_signaling_pathway |
| GO:0007265 | 131 | Ras_protein_signal_transduction |
| GO:0030136 | 131 | clathrin-coated_vesicle |
| GO:0038127 | 131 | ERBB_signaling_pathway |
| GO:0072522 | 131 | purine-containing_compound_biosynthetic_process |
| GO:0006470 | 130 | protein_dephosphorylation |
| GO:0016614 | 130 | oxidoreductase_activity,_acting_on_CH-OH_group_of_donors |
| GO:0016779 | 130 | nucleotidyltransferase_activity |
| GO:0071214 | 130 | cellular_response_to_abiotic_stimulus |
| GO:0005924 | 129 | cell-substrate_adherens_junction |
| GO:0008194 | 129 | UDP-glycosyltransferase_activity |
| GO:0010466 | 129 | negative_regulation_of_peptidase_activity |
| GO:0015293 | 129 | symporter_activity |
| GO:0016585 | 129 | chromatin_remodeling_complex |
| GO:0019867 | 129 | outer_membrane |
| GO:0031346 | 129 | positive_regulation_of_cell_projection_organization |
| GO:0034703 | 129 | cation_channel_complex |
| GO:0007204 | 128 | elevation_of_cytosolic_calcium_ion_concentration |
| GO:0008654 | 128 | phospholipid_biosynthetic_process |
| GO:0015079 | 128 | potassium_ion_transmembrane_transporter_activity |
| GO:0031398 | 128 | positive_regulation_of_protein_ubiquitination |
| GO:0072509 | 128 | divalent_inorganic_cation_transmembrane_transporter_activity |
| GO:0000187 | 127 | activation_of_MAPK_activity |
| GO:0009612 | 127 | response_to_mechanical_stimulus |
| GO:0061138 | 127 | morphogenesis_of_a_branching_epithelium |
| GO:0065004 | 127 | protein-DNA_complex_assembly |
| GO:0000086 | 126 | G2/M_transition_of_mitotic_cell_cycle |
| GO:0006986 | 126 | response_to_unfolded_protein |
| GO:0009896 | 126 | positive_regulation_of_catabolic_process |
| GO:0010951 | 126 | negative_regulation_of_endopeptidase_activity |
| GO:0016798 | 126 | hydrolase_activity,_acting_on_glycosyl_bonds |
| GO:0019207 | 126 | kinase_regulator_activity |
| GO:0033267 | 126 | axon_part |
| GO:0051271 | 126 | negative_regulation_of_cellular_component_movement |
| GO:0051427 | 126 | hormone_receptor_binding |
| GO:0002429 | 125 | immune_response-activating_cell_surface_receptor_signaling_pathway |
| GO:0006576 | 125 | cellular_biogenic_amine_metabolic_process |
| GO:0005925 | 124 | focal_adhesion |
| GO:0006820 | 124 | anion_transport |
| GO:0009310 | 124 | amine_catabolic_process |
| GO:0015081 | 124 | sodium_ion_transmembrane_transporter_activity |
| GO:0031968 | 124 | organelle_outer_membrane |
| GO:0009743 | 123 | response_to_carbohydrate_stimulus |
| GO:0017124 | 123 | SH3_domain_binding |
| GO:0019898 | 123 | extrinsic_to_membrane |
| GO:0031669 | 123 | cellular_response_to_nutrient_levels |
| GO:0046034 | 123 | ATP_metabolic_process |
| GO:0048754 | 123 | branching_morphogenesis_of_a_tube |
| GO:0001818 | 122 | negative_regulation_of_cytokine_production |
| GO:0006413 | 122 | translational_initiation |
| GO:0019717 | 122 | synaptosome |
| GO:0030100 | 122 | regulation_of_endocytosis |
| GO:0045637 | 122 | regulation_of_myeloid_cell_differentiation |
| GO:0046328 | 122 | regulation_of_JNK_cascade |
| GO:0051235 | 122 | maintenance_of_location |
| GO:2000027 | 122 | regulation_of_organ_morphogenesis |
| GO:0001653 | 121 | peptide_receptor_activity |
| GO:0003013 | 121 | circulatory_system_process |
| GO:0005099 | 121 | Ras_GTPase_activator_activity |
| GO:0005819 | 121 | spindle |
| GO:0008624 | 121 | induction_of_apoptosis_by_extracellular_signals |
| GO:0031967 | 121 | organelle_envelope |
| GO:0031975 | 121 | envelope |
| GO:0032102 | 121 | negative_regulation_of_response_to_external_stimulus |
| GO:0042770 | 121 | signal_transduction_in_response_to_DNA_damage |
| GO:0008528 | 120 | G-protein_coupled_peptide_receptor_activity |
| GO:0008543 | 120 | fibroblast_growth_factor_receptor_signaling_pathway |
| GO:0021700 | 120 | developmental_maturation |
| GO:0030099 | 120 | myeloid_cell_differentiation |
| GO:0032984 | 120 | macromolecular_complex_disassembly |
| GO:0043543 | 120 | protein_acylation |
| GO:0050679 | 120 | positive_regulation_of_epithelial_cell_proliferation |
| GO:0051028 | 120 | mRNA_transport |
| GO:2000146 | 120 | negative_regulation_of_cell_motility |
| GO:0000184 | 119 | nuclear-transcribed_mRNA_catabolic_process,_nonsense-mediated_decay |
| GO:0006164 | 119 | purine_nucleotide_biosynthetic_process |
| GO:0006766 | 119 | vitamin_metabolic_process |
| GO:0017016 | 119 | Ras_GTPase_binding |
| GO:0018904 | 119 | organic_ether_metabolic_process |
| GO:0030098 | 119 | lymphocyte_differentiation |
| GO:0034623 | 119 | cellular_macromolecular_complex_disassembly |
| GO:0045216 | 119 | cell-cell_junction_organization |
| GO:2000021 | 119 | regulation_of_ion_homeostasis |
| GO:0030336 | 118 | negative_regulation_of_cell_migration |
| GO:0044242 | 118 | cellular_lipid_catabolic_process |
| GO:0045785 | 118 | positive_regulation_of_cell_adhesion |
| GO:0050731 | 118 | positive_regulation_of_peptidyl-tyrosine_phosphorylation |
| GO:0016616 | 117 | oxidoreductase_activity,_acting_on_the_CH-OH_group_of_donors,_NAD_or_NADP_as_acceptor |
| GO:0018205 | 117 | peptidyl-lysine_modification |
| GO:0032259 | 117 | methylation |
| GO:0032409 | 117 | regulation_of_transporter_activity |
| GO:0032535 | 117 | regulation_of_cellular_component_size |
| GO:0052689 | 117 | carboxylic_ester_hydrolase_activity |
| GO:0060828 | 117 | regulation_of_canonical_Wnt_receptor_signaling_pathway |
| GO:0006643 | 116 | membrane_lipid_metabolic_process |
| GO:0008217 | 116 | regulation_of_blood_pressure |
| GO:0015103 | 116 | inorganic_anion_transmembrane_transporter_activity |
| GO:0016790 | 116 | thiolester_hydrolase_activity |
| GO:0016810 | 116 | hydrolase_activity,_acting_on_carbon-nitrogen_(but_not_peptide)_bonds |
| GO:0042306 | 116 | regulation_of_protein_import_into_nucleus |
| GO:0048608 | 116 | reproductive_structure_development |
| GO:0050851 | 116 | antigen_receptor-mediated_signaling_pathway |
| GO:1900180 | 116 | regulation_of_protein_localization_to_nucleus |
| GO:0006334 | 115 | nucleosome_assembly |
| GO:0006633 | 115 | fatty_acid_biosynthetic_process |
| GO:0007178 | 115 | transmembrane_receptor_protein_serine/threonine_kinase_signaling_pathway |
| GO:0015399 | 115 | primary_active_transmembrane_transporter_activity |
| GO:0015405 | 115 | P-P-bond-hydrolysis-driven_transmembrane_transporter_activity |
| GO:0030258 | 115 | lipid_modification |
| GO:0031589 | 115 | cell-substrate_adhesion |
| GO:0042579 | 115 | microbody |
| GO:0050769 | 115 | positive_regulation_of_neurogenesis |
| GO:0051048 | 115 | negative_regulation_of_secretion |
| GO:0005179 | 114 | hormone_activity |
| GO:0008026 | 114 | ATP-dependent_helicase_activity |
| GO:0010952 | 114 | positive_regulation_of_peptidase_activity |
| GO:0050708 | 114 | regulation_of_protein_secretion |
| GO:0051320 | 114 | S_phase |
| GO:0070035 | 114 | purine_NTP-dependent_helicase_activity |
| GO:0000084 | 113 | S_phase_of_mitotic_cell_cycle |
| GO:0000302 | 113 | response_to_reactive_oxygen_species |
| GO:0001822 | 113 | kidney_development |
| GO:0006662 | 113 | glycerol_ether_metabolic_process |
| GO:0007219 | 113 | Notch_signaling_pathway |
| GO:0014069 | 113 | postsynaptic_density |
| GO:0016298 | 113 | lipase_activity |
| GO:0005267 | 112 | potassium_channel_activity |
| GO:0005788 | 112 | endoplasmic_reticulum_lumen |
| GO:0007409 | 112 | axonogenesis |
| GO:0009063 | 112 | cellular_amino_acid_catabolic_process |
| GO:0004519 | 111 | endonuclease_activity |
| GO:0023061 | 111 | signal_release |
| GO:0034765 | 111 | regulation_of_ion_transmembrane_transport |
| GO:0050954 | 111 | sensory_perception_of_mechanical_stimulus |
| GO:0006694 | 110 | steroid_biosynthetic_process |
| GO:0010212 | 110 | response_to_ionizing_radiation |
| GO:0010927 | 110 | cellular_component_assembly_involved_in_morphogenesis |
| GO:0014075 | 110 | response_to_amine_stimulus |
| GO:0016072 | 110 | rRNA_metabolic_process |
| GO:0019887 | 110 | protein_kinase_regulator_activity |
| GO:0031099 | 110 | regeneration |
| GO:0035303 | 110 | regulation_of_dephosphorylation |
| GO:0070372 | 110 | regulation_of_ERK1_and_ERK2_cascade |
| GO:0000790 | 109 | nuclear_chromatin |
| GO:0002218 | 109 | activation_of_innate_immune_response |
| GO:0006665 | 109 | sphingolipid_metabolic_process |
| GO:0007188 | 109 | adenylate_cyclase-modulating_G-protein_coupled_receptor_signaling_pathway |
| GO:0007417 | 109 | central_nervous_system_development |
| GO:0009266 | 109 | response_to_temperature_stimulus |
| GO:0010950 | 109 | positive_regulation_of_endopeptidase_activity |
| GO:0015085 | 109 | calcium_ion_transmembrane_transporter_activity |
| GO:0016125 | 109 | sterol_metabolic_process |
| GO:0035257 | 109 | nuclear_hormone_receptor_binding |
| GO:0043433 | 109 | negative_regulation_of_sequence-specific_DNA_binding_transcription_factor_activity |
| GO:0007605 | 108 | sensory_perception_of_sound |
| GO:0019902 | 108 | phosphatase_binding |
| GO:0022904 | 108 | respiratory_electron_transport_chain |
| GO:0048002 | 108 | antigen_processing_and_presentation_of_peptide_antigen |
| GO:0050866 | 108 | negative_regulation_of_cell_activation |
| GO:0002758 | 107 | innate_immune_response-activating_signal_transduction |
| GO:0003705 | 107 | RNA_polymerase_II_distal_enhancer_sequence-specific_DNA_binding_transcription_factor_activity |
| GO:0004222 | 107 | metalloendopeptidase_activity |
| GO:0005635 | 107 | nuclear_envelope |
| GO:0007015 | 107 | actin_filament_organization |
| GO:0008238 | 107 | exopeptidase_activity |
| GO:0008652 | 107 | cellular_amino_acid_biosynthetic_process |
| GO:0015078 | 107 | hydrogen_ion_transmembrane_transporter_activity |
| GO:0019838 | 107 | growth_factor_binding |
| GO:0030027 | 107 | lamellipodium |
| GO:0033189 | 107 | response_to_vitamin_A |
| GO:0042692 | 107 | muscle_cell_differentiation |
| GO:0060191 | 107 | regulation_of_lipase_activity |
| GO:0005057 | 106 | receptor_signaling_protein_activity |
| GO:0006613 | 106 | cotranslational_protein_targeting_to_membrane |
| GO:0010721 | 106 | negative_regulation_of_cell_development |
| GO:0016820 | 106 | hydrolase_activity,_acting_on_acid_anhydrides,_catalyzing_transmembrane_movement_of_substances |
| GO:0045047 | 106 | protein_targeting_to_ER |
| GO:0072331 | 106 | signal_transduction_by_p53_class_mediator |
| GO:0072599 | 106 | establishment_of_protein_localization_in_endoplasmic_reticulum |
| GO:0001503 | 105 | ossification |
| GO:0004497 | 105 | monooxygenase_activity |
| GO:0004725 | 105 | protein_tyrosine_phosphatase_activity |
| GO:0006614 | 105 | SRP-dependent_cotranslational_protein_targeting_to_membrane |
| GO:0034976 | 105 | response_to_endoplasmic_reticulum_stress |
| GO:0043280 | 105 | positive_regulation_of_cysteine-type_endopeptidase_activity_involved_in_apoptotic_process |
| GO:0043492 | 105 | ATPase_activity,_coupled_to_movement_of_substances |
| GO:0045787 | 105 | positive_regulation_of_cell_cycle |
| GO:2001056 | 105 | positive_regulation_of_cysteine-type_endopeptidase_activity |
| GO:0006364 | 104 | rRNA_processing |
| GO:0006473 | 104 | protein_acetylation |
| GO:0008016 | 104 | regulation_of_heart_contraction |
| GO:0009636 | 104 | response_to_toxin |
| GO:0022898 | 104 | regulation_of_transmembrane_transporter_activity |
| GO:0034284 | 104 | response_to_monosaccharide_stimulus |
| GO:0042626 | 104 | ATPase_activity,_coupled_to_transmembrane_movement_of_substances |
| GO:0043414 | 104 | macromolecule_methylation |
| GO:0045165 | 104 | cell_fate_commitment |
| GO:0002221 | 103 | pattern_recognition_receptor_signaling_pathway |
| GO:0005741 | 103 | mitochondrial_outer_membrane |
| GO:0006414 | 103 | translational_elongation |
| GO:0030178 | 103 | negative_regulation_of_Wnt_receptor_signaling_pathway |
| GO:0071826 | 103 | ribonucleoprotein_complex_subunit_organization |
| GO:0090257 | 103 | regulation_of_muscle_system_process |
| GO:0000904 | 102 | cell_morphogenesis_involved_in_differentiation |
| GO:0009746 | 102 | response_to_hexose_stimulus |
| GO:0042129 | 102 | regulation_of_T_cell_proliferation |
| GO:0005875 | 101 | microtubule_associated_complex |
| GO:0008017 | 101 | microtubule_binding |
| GO:0008135 | 101 | translation_factor_activity,_nucleic_acid_binding |
| GO:0008203 | 101 | cholesterol_metabolic_process |
| GO:0008757 | 101 | S-adenosylmethionine-dependent_methyltransferase_activity |
| GO:0009123 | 101 | nucleoside_monophosphate_metabolic_process |
| GO:0015630 | 101 | microtubule_cytoskeleton |
| GO:0016410 | 101 | N-acyltransferase_activity |
| GO:0034341 | 101 | response_to_interferon-gamma |
| GO:0048732 | 101 | gland_development |
| GO:0070665 | 101 | positive_regulation_of_leukocyte_proliferation |
| GO:0097305 | 101 | response_to_alcohol |
| GO:0002474 | 100 | antigen_processing_and_presentation_of_peptide_antigen_via_MHC_class_I |
| GO:0004553 | 100 | hydrolase_activity,_hydrolyzing_O-glycosyl_compounds |
| GO:0005342 | 100 | organic_acid_transmembrane_transporter_activity |
| GO:0016651 | 100 | oxidoreductase_activity,_acting_on_NADH_or_NADPH |
| GO:0032355 | 100 | response_to_estradiol_stimulus |
| GO:0045471 | 100 | response_to_ethanol |
| GO:0045619 | 100 | regulation_of_lymphocyte_differentiation |
| GO:0048568 | 100 | embryonic_organ_development |
| GO:0051340 | 100 | regulation_of_ligase_activity |
| GO:0070851 | 100 | growth_factor_receptor_binding |
| GO:0002695 | 99 | negative_regulation_of_leukocyte_activation |
| GO:0002699 | 99 | positive_regulation_of_immune_effector_process |
| GO:0005923 | 99 | tight_junction |
| GO:0005938 | 99 | cell_cortex |
| GO:0008360 | 99 | regulation_of_cell_shape |
| GO:0009260 | 99 | ribonucleotide_biosynthetic_process |
| GO:0032271 | 99 | regulation_of_protein_polymerization |
| GO:0032412 | 99 | regulation_of_ion_transmembrane_transporter_activity |
| GO:0043241 | 99 | protein_complex_disassembly |
| GO:0044441 | 99 | cilium_part |
| GO:0045095 | 99 | keratin_filament |
| GO:0045185 | 99 | maintenance_of_protein_location |
| GO:0070160 | 99 | occluding_junction |
| GO:0008643 | 98 | carbohydrate_transport |
| GO:0009108 | 98 | coenzyme_biosynthetic_process |
| GO:0010810 | 98 | regulation_of_cell-substrate_adhesion |
| GO:0019932 | 98 | second-messenger-mediated_signaling |
| GO:0022618 | 98 | ribonucleoprotein_complex_assembly |
| GO:0030799 | 98 | regulation_of_cyclic_nucleotide_metabolic_process |
| GO:0032946 | 98 | positive_regulation_of_mononuclear_cell_proliferation |
| GO:0034754 | 98 | cellular_hormone_metabolic_process |
| GO:0043624 | 98 | cellular_protein_complex_disassembly |
| GO:0048638 | 98 | regulation_of_developmental_growth |
| GO:0050920 | 98 | regulation_of_chemotaxis |
| GO:0051092 | 98 | positive_regulation_of_NF-kappaB_transcription_factor_activity |
| GO:0060326 | 98 | cell_chemotaxis |
| GO:0061061 | 98 | muscle_structure_development |
| GO:0007606 | 97 | sensory_perception_of_chemical_stimulus |
| GO:0008081 | 97 | phosphoric_diester_hydrolase_activity |
| GO:0016407 | 97 | acetyltransferase_activity |
| GO:0030330 | 97 | DNA_damage_response,_signal_transduction_by_p53_class_mediator |
| GO:0030534 | 97 | adult_behavior |
| GO:0031647 | 97 | regulation_of_protein_stability |
| GO:0040029 | 97 | regulation_of_gene_expression,_epigenetic |
| GO:0046890 | 97 | regulation_of_lipid_biosynthetic_process |
| GO:0046943 | 97 | carboxylic_acid_transmembrane_transporter_activity |
| GO:0048469 | 97 | cell_maturation |
| GO:0050671 | 97 | positive_regulation_of_lymphocyte_proliferation |
| GO:0050770 | 97 | regulation_of_axonogenesis |
| GO:0051147 | 97 | regulation_of_muscle_cell_differentiation |
| GO:0000279 | 96 | M_phase |
| GO:0004620 | 96 | phospholipase_activity |
| GO:0004867 | 96 | serine-type_endopeptidase_inhibitor_activity |
| GO:0005802 | 96 | trans-Golgi_network |
| GO:0006813 | 96 | potassium_ion_transport |
| GO:0006865 | 96 | amino_acid_transport |
| GO:0007517 | 96 | muscle_organ_development |
| GO:0008406 | 96 | gonad_development |
| GO:0009749 | 96 | response_to_glucose_stimulus |
| GO:0030662 | 96 | coated_vesicle_membrane |
| GO:0031227 | 96 | intrinsic_to_endoplasmic_reticulum_membrane |
| GO:0048562 | 96 | embryonic_organ_morphogenesis |
| GO:0048666 | 96 | neuron_development |
| GO:0051438 | 96 | regulation_of_ubiquitin-protein_ligase_activity |
| GO:0051651 | 96 | maintenance_of_location_in_cell |
| GO:0071216 | 96 | cellular_response_to_biotic_stimulus |
| GO:0002703 | 95 | regulation_of_leukocyte_mediated_immunity |
| GO:0006275 | 95 | regulation_of_DNA_replication |
| GO:0006338 | 95 | chromatin_remodeling |
| GO:0008277 | 95 | regulation_of_G-protein_coupled_receptor_protein_signaling_pathway |
| GO:0031331 | 95 | positive_regulation_of_cellular_catabolic_process |
| GO:0032320 | 95 | positive_regulation_of_Ras_GTPase_activity |
| GO:0042393 | 95 | histone_binding |
| GO:0043900 | 95 | regulation_of_multi-organism_process |
| GO:0051054 | 95 | positive_regulation_of_DNA_metabolic_process |
| GO:0061448 | 95 | connective_tissue_development |
| GO:0000087 | 94 | M_phase_of_mitotic_cell_cycle |
| GO:0005581 | 94 | collagen |
| GO:0005777 | 94 | peroxisome |
| GO:0006839 | 94 | mitochondrial_transport |
| GO:0008629 | 94 | induction_of_apoptosis_by_intracellular_signals |
| GO:0010948 | 94 | negative_regulation_of_cell_cycle_process |
| GO:0016049 | 94 | cell_growth |
| GO:0032526 | 94 | response_to_retinoic_acid |
| GO:0032886 | 94 | regulation_of_microtubule-based_process |
| GO:0032993 | 94 | protein-DNA_complex |
| GO:0035239 | 94 | tube_morphogenesis |
| GO:0045834 | 94 | positive_regulation_of_lipid_metabolic_process |
| GO:0046474 | 94 | glycerophospholipid_biosynthetic_process |
| GO:0048634 | 94 | regulation_of_muscle_organ_development |
| GO:0050792 | 94 | regulation_of_viral_reproduction |
| GO:0051224 | 94 | negative_regulation_of_protein_transport |
| GO:0051592 | 94 | response_to_calcium_ion |
| GO:0005088 | 93 | Ras_guanyl-nucleotide_exchange_factor_activity |
| GO:0006487 | 93 | protein_N-linked_glycosylation |
| GO:0008565 | 93 | protein_transporter_activity |
| GO:0030808 | 93 | regulation_of_nucleotide_biosynthetic_process |
| GO:0034599 | 93 | cellular_response_to_oxidative_stress |
| GO:0035107 | 93 | appendage_morphogenesis |
| GO:0035108 | 93 | limb_morphogenesis |
| GO:0044445 | 93 | cytosolic_part |
| GO:0050906 | 93 | detection_of_stimulus_involved_in_sensory_perception |
| GO:0051495 | 93 | positive_regulation_of_cytoskeleton_organization |
| GO:1900371 | 93 | regulation_of_purine_nucleotide_biosynthetic_process |
| GO:0000041 | 92 | transition_metal_ion_transport |
| GO:0002819 | 92 | regulation_of_adaptive_immune_response |
| GO:0005262 | 92 | calcium_channel_activity |
| GO:0006475 | 92 | internal_protein_amino_acid_acetylation |
| GO:0006638 | 92 | neutral_lipid_metabolic_process |
| GO:0006898 | 92 | receptor-mediated_endocytosis |
| GO:0007218 | 92 | neuropeptide_signaling_pathway |
| GO:0010741 | 92 | negative_regulation_of_intracellular_protein_kinase_cascade |
| GO:0010921 | 92 | regulation_of_phosphatase_activity |
| GO:0016202 | 92 | regulation_of_striated_muscle_tissue_development |
| GO:0019058 | 92 | viral_infectious_cycle |
| GO:0030802 | 92 | regulation_of_cyclic_nucleotide_biosynthetic_process |
| GO:0032319 | 92 | regulation_of_Rho_GTPase_activity |
| GO:0035148 | 92 | tube_formation |
| GO:0042035 | 92 | regulation_of_cytokine_biosynthetic_process |
| GO:0046488 | 92 | phosphatidylinositol_metabolic_process |
| GO:0051168 | 92 | nuclear_export |
| GO:0071241 | 92 | cellular_response_to_inorganic_substance |
| GO:0000776 | 91 | kinetochore |
| GO:0002224 | 91 | toll-like_receptor_signaling_pathway |
| GO:0006639 | 91 | acylglycerol_metabolic_process |
| GO:0008144 | 91 | drug_binding |
| GO:0031072 | 91 | heat_shock_protein_binding |
| GO:0031397 | 91 | negative_regulation_of_protein_ubiquitination |
| GO:0043279 | 91 | response_to_alkaloid |
| GO:0001101 | 90 | response_to_acid |
| GO:0003729 | 90 | mRNA_binding |
| GO:0006109 | 90 | regulation_of_carbohydrate_metabolic_process |
| GO:0006984 | 90 | ER-nucleus_signaling_pathway |
| GO:0007281 | 90 | germ_cell_development |
| GO:0009411 | 90 | response_to_UV |
| GO:0009581 | 90 | detection_of_external_stimulus |
| GO:0009593 | 90 | detection_of_chemical_stimulus |
| GO:0016050 | 90 | vesicle_organization |
| GO:0022602 | 90 | ovulation_cycle_process |
| GO:0031098 | 90 | stress-activated_protein_kinase_signaling_cascade |
| GO:0031334 | 90 | positive_regulation_of_protein_complex_assembly |
| GO:0042113 | 90 | B_cell_activation |
| GO:0042470 | 90 | melanosome |
| GO:0048770 | 90 | pigment_granule |
| GO:0050852 | 90 | T_cell_receptor_signaling_pathway |
| GO:0051604 | 90 | protein_maturation |
| GO:0007423 | 89 | sensory_organ_development |
| GO:0018394 | 89 | peptidyl-lysine_acetylation |
| GO:0030832 | 89 | regulation_of_actin_filament_length |
| GO:0031123 | 89 | RNA_3'-end_processing |
| GO:0031256 | 89 | leading_edge_membrane |
| GO:0043393 | 89 | regulation_of_protein_binding |
| GO:0045667 | 89 | regulation_of_osteoblast_differentiation |
| GO:0046365 | 89 | monosaccharide_catabolic_process |
| GO:0007088 | 88 | regulation_of_mitosis |
| GO:0008064 | 88 | regulation_of_actin_polymerization_or_depolymerization |
| GO:0016597 | 88 | amino_acid_binding |
| GO:0018196 | 88 | peptidyl-asparagine_modification |
| GO:0033500 | 88 | carbohydrate_homeostasis |
| GO:0035023 | 88 | regulation_of_Rho_protein_signal_transduction |
| GO:0042593 | 88 | glucose_homeostasis |
| GO:0044450 | 88 | microtubule_organizing_center_part |
| GO:0051403 | 88 | stress-activated_MAPK_cascade |
| GO:0051783 | 88 | regulation_of_nuclear_division |
| GO:0004221 | 87 | ubiquitin_thiolesterase_activity |
| GO:0005770 | 87 | late_endosome |
| GO:0006302 | 87 | double-strand_break_repair |
| GO:0018279 | 87 | protein_N-linked_glycosylation_via_asparagine |
| GO:0018393 | 87 | internal_peptidyl-lysine_acetylation |
| GO:0030165 | 87 | PDZ_domain_binding |
| GO:0030307 | 87 | positive_regulation_of_cell_growth |
| GO:0033044 | 87 | regulation_of_chromosome_organization |
| GO:0034612 | 87 | response_to_tumor_necrosis_factor |
| GO:0048015 | 87 | phosphatidylinositol-mediated_signaling |
| GO:0048017 | 87 | inositol_lipid-mediated_signaling |
| GO:0048167 | 87 | regulation_of_synaptic_plasticity |
| GO:2000243 | 87 | positive_regulation_of_reproductive_process |
| GO:0001894 | 86 | tissue_homeostasis |
| GO:0006937 | 86 | regulation_of_muscle_contraction |
| GO:0010675 | 86 | regulation_of_cellular_carbohydrate_metabolic_process |
| GO:0015108 | 86 | chloride_transmembrane_transporter_activity |
| GO:0019884 | 86 | antigen_processing_and_presentation_of_exogenous_antigen |
| GO:0030097 | 86 | hemopoiesis |
| GO:0035967 | 86 | cellular_response_to_topologically_incorrect_protein |
| GO:0044448 | 86 | cell_cortex_part |
| GO:0048705 | 86 | skeletal_system_morphogenesis |
| GO:0051213 | 86 | dioxygenase_activity |
| GO:0051250 | 86 | negative_regulation_of_lymphocyte_activation |
| GO:0051351 | 86 | positive_regulation_of_ligase_activity |
| GO:0060759 | 86 | regulation_of_response_to_cytokine_stimulus |
| GO:0001505 | 85 | regulation_of_neurotransmitter_levels |
| GO:0001764 | 85 | neuron_migration |
| GO:0002822 | 85 | regulation_of_adaptive_immune_response_based_on_somatic_recombination_of_immune_receptors_built_from_immunoglobulin_superfamily_domains |
| GO:0006354 | 85 | DNA-dependent_transcription,_elongation |
| GO:0006415 | 85 | translational_termination |
| GO:0016573 | 85 | histone_acetylation |
| GO:0030814 | 85 | regulation_of_cAMP_metabolic_process |
| GO:2000602 | 85 | regulation_of_interphase_of_mitotic_cell_cycle |
| GO:0000079 | 84 | regulation_of_cyclin-dependent_protein_kinase_activity |
| GO:0000236 | 84 | mitotic_prometaphase |
| GO:0002478 | 84 | antigen_processing_and_presentation_of_exogenous_peptide_antigen |
| GO:0008080 | 84 | N-acetyltransferase_activity |
| GO:0019083 | 84 | viral_transcription |
| GO:0019199 | 84 | transmembrane_receptor_protein_kinase_activity |
| GO:0030426 | 84 | growth_cone |
| GO:0030427 | 84 | site_of_polarized_growth |
| GO:0031461 | 84 | cullin-RING_ubiquitin_ligase_complex |
| GO:0032507 | 84 | maintenance_of_protein_location_in_cell |
| GO:0043270 | 84 | positive_regulation_of_ion_transport |
| GO:0070374 | 84 | positive_regulation_of_ERK1_and_ERK2_cascade |
| GO:0071219 | 84 | cellular_response_to_molecule_of_bacterial_origin |
| GO:0071346 | 84 | cellular_response_to_interferon-gamma |
| GO:0071901 | 84 | negative_regulation_of_protein_serine/threonine_kinase_activity |
| GO:0001076 | 83 | RNA_polymerase_II_transcription_factor_binding_transcription_factor_activity |
| GO:0005275 | 83 | amine_transmembrane_transporter_activity |
| GO:0006919 | 83 | activation_of_cysteine-type_endopeptidase_activity_involved_in_apoptotic_process |
| GO:0009100 | 83 | glycoprotein_metabolic_process |
| GO:0009152 | 83 | purine_ribonucleotide_biosynthetic_process |
| GO:0016701 | 83 | oxidoreductase_activity,_acting_on_single_donors_with_incorporation_of_molecular_oxygen |
| GO:0030817 | 83 | regulation_of_cAMP_biosynthetic_process |
| GO:0042542 | 83 | response_to_hydrogen_peroxide |
| GO:0043524 | 83 | negative_regulation_of_neuron_apoptotic_process |
| GO:0044085 | 83 | cellular_component_biogenesis |
| GO:0047485 | 83 | protein_N-terminus_binding |
| GO:0048520 | 83 | positive_regulation_of_behavior |
| GO:0050660 | 83 | flavin_adenine_dinucleotide_binding |
| GO:0050864 | 83 | regulation_of_B_cell_activation |
| GO:0051302 | 83 | regulation_of_cell_division |
| GO:0055072 | 83 | iron_ion_homeostasis |
| GO:0097202 | 83 | activation_of_cysteine-type_endopeptidase_activity |
| GO:0006641 | 82 | triglyceride_metabolic_process |
| GO:0006836 | 82 | neurotransmitter_transport |
| GO:0007160 | 82 | cell-matrix_adhesion |
| GO:0007163 | 82 | establishment_or_maintenance_of_cell_polarity |
| GO:0015294 | 82 | solute:cation_symporter_activity |
| GO:0016702 | 82 | oxidoreductase_activity,_acting_on_single_donors_with_incorporation_of_molecular_oxygen,_incorporation_of_two_atoms_of_oxygen |
| GO:0030496 | 82 | midbody |
| GO:0032388 | 82 | positive_regulation_of_intracellular_transport |
| GO:0045580 | 82 | regulation_of_T_cell_differentiation |
| GO:0050768 | 82 | negative_regulation_of_neurogenesis |
| GO:0051443 | 82 | positive_regulation_of_ubiquitin-protein_ligase_activity |
| GO:0060193 | 82 | positive_regulation_of_lipase_activity |
| GO:0004540 | 81 | ribonuclease_activity |
| GO:0004896 | 81 | cytokine_receptor_activity |
| GO:0005253 | 81 | anion_channel_activity |
| GO:0006022 | 81 | aminoglycan_metabolic_process |
| GO:0006353 | 81 | DNA-dependent_transcription,_termination |
| GO:0006814 | 81 | sodium_ion_transport |
| GO:0010517 | 81 | regulation_of_phospholipase_activity |
| GO:0030968 | 81 | endoplasmic_reticulum_unfolded_protein_response |
| GO:0031175 | 81 | neuron_projection_development |
| GO:0034620 | 81 | cellular_response_to_unfolded_protein |
| GO:0045444 | 81 | fat_cell_differentiation |
| GO:0050808 | 81 | synapse_organization |
| GO:0070925 | 81 | organelle_assembly |
| GO:0071158 | 81 | positive_regulation_of_cell_cycle_arrest |
| GO:0071248 | 81 | cellular_response_to_metal_ion |
| GO:0001959 | 80 | regulation_of_cytokine-mediated_signaling_pathway |
| GO:0002576 | 80 | platelet_degranulation |
| GO:0002685 | 80 | regulation_of_leukocyte_migration |
| GO:0005201 | 80 | extracellular_matrix_structural_constituent |
| GO:0005249 | 80 | voltage-gated_potassium_channel_activity |
| GO:0006200 | 80 | ATP_catabolic_process |
| GO:0019897 | 80 | extrinsic_to_plasma_membrane |
| GO:0031145 | 80 | anaphase-promoting_complex-dependent_proteasomal_ubiquitin-dependent_protein_catabolic_process |
| GO:0042476 | 80 | odontogenesis |
| GO:0070507 | 80 | regulation_of_microtubule_cytoskeleton_organization |
| GO:0071013 | 80 | catalytic_step_2_spliceosome |
| GO:0000216 | 79 | M/G1_transition_of_mitotic_cell_cycle |
| GO:0000922 | 79 | spindle_pole |
| GO:0007059 | 79 | chromosome_segregation |
| GO:0007162 | 79 | negative_regulation_of_cell_adhesion |
| GO:0007612 | 79 | learning |
| GO:0016485 | 79 | protein_processing |
| GO:0018209 | 79 | peptidyl-serine_modification |
| GO:0030217 | 79 | T_cell_differentiation |
| GO:0031348 | 79 | negative_regulation_of_defense_response |
| GO:0031901 | 79 | early_endosome_membrane |
| GO:0042590 | 79 | antigen_processing_and_presentation_of_exogenous_peptide_antigen_via_MHC_class_I |
| GO:0050680 | 79 | negative_regulation_of_epithelial_cell_proliferation |
| GO:0051262 | 79 | protein_tetramerization |
| GO:0051494 | 79 | negative_regulation_of_cytoskeleton_organization |
| GO:0055088 | 79 | lipid_homeostasis |
| GO:0060070 | 79 | canonical_Wnt_receptor_signaling_pathway |
| GO:0071222 | 79 | cellular_response_to_lipopolysaccharide |
| GO:0001889 | 78 | liver_development |
| GO:0005178 | 78 | integrin_binding |
| GO:0006518 | 78 | peptide_metabolic_process |
| GO:0007631 | 78 | feeding_behavior |
| GO:0008033 | 78 | tRNA_processing |
| GO:0016363 | 78 | nuclear_matrix |
| GO:0017157 | 78 | regulation_of_exocytosis |
| GO:0030176 | 78 | integral_to_endoplasmic_reticulum_membrane |
| GO:0030326 | 78 | embryonic_limb_morphogenesis |
| GO:0030833 | 78 | regulation_of_actin_filament_polymerization |
| GO:0031294 | 78 | lymphocyte_costimulation |
| GO:0031295 | 78 | T_cell_costimulation |
| GO:0034142 | 78 | toll-like_receptor_4_signaling_pathway |
| GO:0035113 | 78 | embryonic_appendage_morphogenesis |
| GO:0051439 | 78 | regulation_of_ubiquitin-protein_ligase_activity_involved_in_mitotic_cell_cycle |
| GO:2001020 | 78 | regulation_of_response_to_DNA_damage_stimulus |
| GO:0001568 | 77 | blood_vessel_development |
| GO:0003777 | 77 | microtubule_motor_activity |
| GO:0005200 | 77 | structural_constituent_of_cytoskeleton |
| GO:0008063 | 77 | Toll_signaling_pathway |
| GO:0030595 | 77 | leukocyte_chemotaxis |
| GO:0044438 | 77 | microbody_part |
| GO:0044439 | 77 | peroxisomal_part |
| GO:0046930 | 77 | pore_complex |
| GO:0050714 | 77 | positive_regulation_of_protein_secretion |
| GO:0060537 | 77 | muscle_tissue_development |
| GO:0061041 | 77 | regulation_of_wound_healing |
| GO:0002706 | 76 | regulation_of_lymphocyte_mediated_immunity |
| GO:0009582 | 76 | detection_of_abiotic_stimulus |
| GO:0031902 | 76 | late_endosome_membrane |
| GO:0045766 | 76 | positive_regulation_of_angiogenesis |
| GO:0071103 | 76 | DNA_conformation_change |
| GO:0090316 | 76 | positive_regulation_of_intracellular_protein_transport |
| GO:0002479 | 75 | antigen_processing_and_presentation_of_exogenous_peptide_antigen_via_MHC_class_I,_TAP-dependent |
| GO:0005089 | 75 | Rho_guanyl-nucleotide_exchange_factor_activity |
| GO:0007093 | 75 | mitotic_cell_cycle_checkpoint |
| GO:0007565 | 75 | female_pregnancy |
| GO:0009895 | 75 | negative_regulation_of_catabolic_process |
| GO:0010827 | 75 | regulation_of_glucose_transport |
| GO:0012505 | 75 | endomembrane_system |
| GO:0015934 | 75 | large_ribosomal_subunit |
| GO:0019320 | 75 | hexose_catabolic_process |
| GO:0030324 | 75 | lung_development |
| GO:0031124 | 75 | mRNA_3'-end_processing |
| GO:0042594 | 75 | response_to_starvation |
| GO:0048524 | 75 | positive_regulation_of_viral_reproduction |
| GO:0000123 | 74 | histone_acetyltransferase_complex |
| GO:0001726 | 74 | ruffle |
| GO:0001936 | 74 | regulation_of_endothelial_cell_proliferation |
| GO:0002755 | 74 | MyD88-dependent_toll-like_receptor_signaling_pathway |
| GO:0002761 | 74 | regulation_of_myeloid_leukocyte_differentiation |
| GO:0002831 | 74 | regulation_of_response_to_biotic_stimulus |
| GO:0005230 | 74 | extracellular_ligand-gated_ion_channel_activity |
| GO:0005496 | 74 | steroid_binding |
| GO:0007229 | 74 | integrin-mediated_signaling_pathway |
| GO:0008021 | 74 | synaptic_vesicle |
| GO:0010518 | 74 | positive_regulation_of_phospholipase_activity |
| GO:0017015 | 74 | regulation_of_transforming_growth_factor_beta_receptor_signaling_pathway |
| GO:0033559 | 74 | unsaturated_fatty_acid_metabolic_process |
| GO:0033993 | 74 | response_to_lipid |
| GO:0051216 | 74 | cartilage_development |
| GO:0000910 | 73 | cytokinesis |
| GO:0003018 | 73 | vascular_process_in_circulatory_system |
| GO:0005604 | 73 | basement_membrane |
| GO:0009791 | 73 | post-embryonic_development |
| GO:0015698 | 73 | inorganic_anion_transport |
| GO:0030594 | 73 | neurotransmitter_receptor_activity |
| GO:0031570 | 73 | DNA_integrity_checkpoint |
| GO:0042303 | 73 | molting_cycle |
| GO:0042383 | 73 | sarcolemma |
| GO:0045732 | 73 | positive_regulation_of_protein_catabolic_process |
| GO:0048872 | 73 | homeostasis_of_number_of_cells |
| GO:0051015 | 73 | actin_filament_binding |
| GO:0051896 | 73 | regulation_of_protein_kinase_B_signaling_cascade |
| GO:0072593 | 73 | reactive_oxygen_species_metabolic_process |
| GO:0000786 | 72 | nucleosome |
| GO:0000982 | 72 | RNA_polymerase_II_core_promoter_proximal_region_sequence-specific_DNA_binding_transcription_factor_activity |
| GO:0005254 | 72 | chloride_channel_activity |
| GO:0008094 | 72 | DNA-dependent_ATPase_activity |
| GO:0016605 | 72 | PML_body |
| GO:0019903 | 72 | protein_phosphatase_binding |
| GO:0030518 | 72 | intracellular_steroid_hormone_receptor_signaling_pathway |
| GO:0030665 | 72 | clathrin_coated_vesicle_membrane |
| GO:0032069 | 72 | regulation_of_nuclease_activity |
| GO:0032580 | 72 | Golgi_cisterna_membrane |
| GO:0033238 | 72 | regulation_of_cellular_amine_metabolic_process |
| GO:0035295 | 72 | tube_development |
| GO:0045807 | 72 | positive_regulation_of_endocytosis |
| GO:0045995 | 72 | regulation_of_embryonic_development |
| GO:0050921 | 72 | positive_regulation_of_chemotaxis |
| GO:0051099 | 72 | positive_regulation_of_binding |
| GO:0051352 | 72 | negative_regulation_of_ligase_activity |
| GO:0051437 | 72 | positive_regulation_of_ubiquitin-protein_ligase_activity_involved_in_mitotic_cell_cycle |
| GO:0051444 | 72 | negative_regulation_of_ubiquitin-protein_ligase_activity |
| GO:0051591 | 72 | response_to_cAMP |
| GO:0070646 | 72 | protein_modification_by_small_protein_removal |
| GO:0072527 | 72 | pyrimidine-containing_compound_metabolic_process |
| GO:0090090 | 72 | negative_regulation_of_canonical_Wnt_receptor_signaling_pathway |
| GO:0002263 | 71 | cell_activation_involved_in_immune_response |
| GO:0002366 | 71 | leukocyte_activation_involved_in_immune_response |
| GO:0003697 | 71 | single-stranded_DNA_binding |
| GO:0006289 | 71 | nucleotide-excision_repair |
| GO:0006493 | 71 | protein_O-linked_glycosylation |
| GO:0006720 | 71 | isoprenoid_metabolic_process |
| GO:0009798 | 71 | axis_specification |
| GO:0019208 | 71 | phosphatase_regulator_activity |
| GO:0022407 | 71 | regulation_of_cell-cell_adhesion |
| GO:0030203 | 71 | glycosaminoglycan_metabolic_process |
| GO:0033365 | 71 | protein_localization_to_organelle |
| GO:0050707 | 71 | regulation_of_cytokine_secretion |
| GO:0090100 | 71 | positive_regulation_of_transmembrane_receptor_protein_serine/threonine_kinase_signaling_pathway |
| GO:0001012 | 70 | RNA_polymerase_II_regulatory_region_DNA_binding |
| GO:0004197 | 70 | cysteine-type_endopeptidase_activity |
| GO:0007126 | 70 | meiosis |
| GO:0008037 | 70 | cell_recognition |
| GO:0008076 | 70 | voltage-gated_potassium_channel_complex |
| GO:0008584 | 70 | male_gonad_development |
| GO:0009142 | 70 | nucleoside_triphosphate_biosynthetic_process |
| GO:0010906 | 70 | regulation_of_glucose_metabolic_process |
| GO:0019217 | 70 | regulation_of_fatty_acid_metabolic_process |
| GO:0022404 | 70 | molting_cycle_process |
| GO:0022405 | 70 | hair_cycle_process |
| GO:0030018 | 70 | Z_disc |
| GO:0030666 | 70 | endocytic_vesicle_membrane |
| GO:0031670 | 70 | cellular_response_to_nutrient |
| GO:0034134 | 70 | toll-like_receptor_2_signaling_pathway |
| GO:0034705 | 70 | potassium_channel_complex |
| GO:0050729 | 70 | positive_regulation_of_inflammatory_response |
| GO:0051170 | 70 | nuclear_import |
| GO:0061025 | 70 | membrane_fusion |
| GO:0090101 | 70 | negative_regulation_of_transmembrane_receptor_protein_serine/threonine_kinase_signaling_pathway |
| GO:0001667 | 69 | ameboidal_cell_migration |
| GO:0006690 | 69 | icosanoid_metabolic_process |
| GO:0007051 | 69 | spindle_organization |
| GO:0007179 | 69 | transforming_growth_factor_beta_receptor_signaling_pathway |
| GO:0009124 | 69 | nucleoside_monophosphate_biosynthetic_process |
| GO:0015171 | 69 | amino_acid_transmembrane_transporter_activity |
| GO:0032846 | 69 | positive_regulation_of_homeostatic_process |
| GO:0034130 | 69 | toll-like_receptor_1_signaling_pathway |
| GO:0042060 | 69 | wound_healing |
| GO:0042625 | 69 | ATPase_activity,_coupled_to_transmembrane_movement_of_ions |
| GO:0050818 | 69 | regulation_of_coagulation |
| GO:0060333 | 69 | interferon-gamma-mediated_signaling_pathway |
| GO:0072395 | 69 | signal_transduction_involved_in_cell_cycle_checkpoint |
| GO:0000777 | 68 | condensed_chromosome_kinetochore |
| GO:0000977 | 68 | RNA_polymerase_II_regulatory_region_sequence-specific_DNA_binding |
| GO:0002573 | 68 | myeloid_leukocyte_differentiation |
| GO:0006405 | 68 | RNA_export_from_nucleus |
| GO:0006479 | 68 | protein_methylation |
| GO:0007623 | 68 | circadian_rhythm |
| GO:0008213 | 68 | protein_alkylation |
| GO:0009311 | 68 | oligosaccharide_metabolic_process |
| GO:0009408 | 68 | response_to_heat |
| GO:0016247 | 68 | channel_regulator_activity |
| GO:0030133 | 68 | transport_vesicle |
| GO:0032680 | 68 | regulation_of_tumor_necrosis_factor_production |
| GO:0035770 | 68 | ribonucleoprotein_granule |
| GO:0043200 | 68 | response_to_amino_acid_stimulus |
| GO:0044272 | 68 | sulfur_compound_biosynthetic_process |
| GO:0045177 | 68 | apical_part_of_cell |
| GO:0046364 | 68 | monosaccharide_biosynthetic_process |
| GO:0046887 | 68 | positive_regulation_of_hormone_secretion |
| GO:0072401 | 68 | signal_transduction_involved_in_DNA_integrity_checkpoint |
| GO:0072404 | 68 | signal_transduction_involved_in_G1/S_transition_checkpoint |
| GO:0072422 | 68 | signal_transduction_involved_in_DNA_damage_checkpoint |
| GO:0000077 | 67 | DNA_damage_checkpoint |
| GO:0002756 | 67 | MyD88-independent_toll-like_receptor_signaling_pathway |
| GO:0004714 | 67 | transmembrane_receptor_protein_tyrosine_kinase_activity |
| GO:0006606 | 67 | protein_import_into_nucleus |
| GO:0006879 | 67 | cellular_iron_ion_homeostasis |
| GO:0006892 | 67 | post-Golgi_vesicle-mediated_transport |
| GO:0006977 | 67 | DNA_damage_response,_signal_transduction_by_p53_class_mediator_resulting_in_cell_cycle_arrest |
| GO:0010594 | 67 | regulation_of_endothelial_cell_migration |
| GO:0015718 | 67 | monocarboxylic_acid_transport |
| GO:0018212 | 67 | peptidyl-tyrosine_modification |
| GO:0019827 | 67 | stem_cell_maintenance |
| GO:0031345 | 67 | negative_regulation_of_cell_projection_organization |
| GO:0045981 | 67 | positive_regulation_of_nucleotide_metabolic_process |
| GO:0046519 | 67 | sphingoid_metabolic_process |
| GO:0046782 | 67 | regulation_of_viral_transcription |
| GO:0046824 | 67 | positive_regulation_of_nucleocytoplasmic_transport |
| GO:0050868 | 67 | negative_regulation_of_T_cell_activation |
| GO:0051100 | 67 | negative_regulation_of_binding |
| GO:0051341 | 67 | regulation_of_oxidoreductase_activity |
| GO:0051436 | 67 | negative_regulation_of_ubiquitin-protein_ligase_activity_involved_in_mitotic_cell_cycle |
| GO:0072413 | 67 | signal_transduction_involved_in_mitotic_cell_cycle_checkpoint |
| GO:0072431 | 67 | signal_transduction_involved_in_mitotic_cell_cycle_G1/S_transition_DNA_damage_checkpoint |
| GO:0072474 | 67 | signal_transduction_involved_in_mitotic_cell_cycle_G1/S_checkpoint |
| GO:1900544 | 67 | positive_regulation_of_purine_nucleotide_metabolic_process |
| GO:0001104 | 66 | RNA_polymerase_II_transcription_cofactor_activity |
| GO:0004527 | 66 | exonuclease_activity |
| GO:0006308 | 66 | DNA_catabolic_process |
| GO:0006944 | 66 | cellular_membrane_fusion |
| GO:0008276 | 66 | protein_methyltransferase_activity |
| GO:0009201 | 66 | ribonucleoside_triphosphate_biosynthetic_process |
| GO:0015749 | 66 | monosaccharide_transport |
| GO:0018105 | 66 | peptidyl-serine_phosphorylation |
| GO:0019218 | 66 | regulation_of_steroid_metabolic_process |
| GO:0030856 | 66 | regulation_of_epithelial_cell_differentiation |
| GO:0032075 | 66 | positive_regulation_of_nuclease_activity |
| GO:0032675 | 66 | regulation_of_interleukin-6_production |
| GO:0034340 | 66 | response_to_type_I_interferon |
| GO:0035258 | 66 | steroid_hormone_receptor_binding |
| GO:0042058 | 66 | regulation_of_epidermal_growth_factor_receptor_signaling_pathway |
| GO:0042102 | 66 | positive_regulation_of_T_cell_proliferation |
| GO:0042826 | 66 | histone_deacetylase_binding |
| GO:0043506 | 66 | regulation_of_JUN_kinase_activity |
| GO:0046425 | 66 | regulation_of_JAK-STAT_cascade |
| GO:0071356 | 66 | cellular_response_to_tumor_necrosis_factor |
| GO:0071843 | 66 | cellular_component_biogenesis_at_cellular_level |
| GO:0072329 | 66 | monocarboxylic_acid_catabolic_process |
| GO:0072372 | 66 | primary_cilium |
| GO:0000775 | 65 | chromosome,_centromeric_region |
| GO:0006368 | 65 | transcription_elongation_from_RNA_polymerase_II_promoter |
| GO:0007586 | 65 | digestion |
| GO:0007589 | 65 | body_fluid_secretion |
| GO:0007613 | 65 | memory |
| GO:0008170 | 65 | N-methyltransferase_activity |
| GO:0008344 | 65 | adult_locomotory_behavior |
| GO:0008645 | 65 | hexose_transport |
| GO:0009112 | 65 | nucleobase_metabolic_process |
| GO:0010469 | 65 | regulation_of_receptor_activity |
| GO:0015758 | 65 | glucose_transport |
| GO:0016459 | 65 | myosin_complex |
| GO:0018108 | 65 | peptidyl-tyrosine_phosphorylation |
| GO:0030177 | 65 | positive_regulation_of_Wnt_receptor_signaling_pathway |
| GO:0030801 | 65 | positive_regulation_of_cyclic_nucleotide_metabolic_process |
| GO:0032321 | 65 | positive_regulation_of_Rho_GTPase_activity |
| GO:0044264 | 65 | cellular_polysaccharide_metabolic_process |
| GO:0048660 | 65 | regulation_of_smooth_muscle_cell_proliferation |
| GO:0060337 | 65 | type_I_interferon-mediated_signaling_pathway |
| GO:0071357 | 65 | cellular_response_to_type_I_interferon |
| GO:0090305 | 65 | nucleic_acid_phosphodiester_bond_hydrolysis |
| GO:1900274 | 65 | regulation_of_phospholipase_C_activity |
| GO:0001654 | 64 | eye_development |
| GO:0002274 | 64 | myeloid_leukocyte_activation |
| GO:0002700 | 64 | regulation_of_production_of_molecular_mediator_of_immune_response |
| GO:0005319 | 64 | lipid_transporter_activity |
| GO:0006987 | 64 | activation_of_signaling_protein_activity_involved_in_unfolded_protein_response |
| GO:0010863 | 64 | positive_regulation_of_phospholipase_C_activity |
| GO:0014706 | 64 | striated_muscle_tissue_development |
| GO:0015297 | 64 | antiporter_activity |
| GO:0016209 | 64 | antioxidant_activity |
| GO:0030510 | 64 | regulation_of_BMP_signaling_pathway |
| GO:0030810 | 64 | positive_regulation_of_nucleotide_biosynthetic_process |
| GO:0034138 | 64 | toll-like_receptor_3_signaling_pathway |
| GO:0034440 | 64 | lipid_oxidation |
| GO:0035591 | 64 | signaling_adaptor_activity |
| GO:0042384 | 64 | cilium_assembly |
| GO:0050728 | 64 | negative_regulation_of_inflammatory_response |
| GO:0050821 | 64 | protein_stabilization |
| GO:0051101 | 64 | regulation_of_DNA_binding |
| GO:0051656 | 64 | establishment_of_organelle_localization |
| GO:0051899 | 64 | membrane_depolarization |
| GO:0051928 | 64 | positive_regulation_of_calcium_ion_transport |
| GO:0070167 | 64 | regulation_of_biomineral_tissue_development |
| GO:0070555 | 64 | response_to_interleukin-1 |
| GO:1900373 | 64 | positive_regulation_of_purine_nucleotide_biosynthetic_process |
| GO:0002250 | 63 | adaptive_immune_response |
| GO:0005932 | 63 | microtubule_basal_body |
| GO:0006767 | 63 | water-soluble_vitamin_metabolic_process |
| GO:0015711 | 63 | organic_anion_transport |
| GO:0019395 | 63 | fatty_acid_oxidation |
| GO:0030193 | 63 | regulation_of_blood_coagulation |
| GO:0030804 | 63 | positive_regulation_of_cyclic_nucleotide_biosynthetic_process |
| GO:0032479 | 63 | regulation_of_type_I_interferon_production |
| GO:0034708 | 63 | methyltransferase_complex |
| GO:0038024 | 63 | cargo_receptor_activity |
| GO:0043244 | 63 | regulation_of_protein_complex_disassembly |
| GO:0045666 | 63 | positive_regulation_of_neuron_differentiation |
| GO:1900046 | 63 | regulation_of_hemostasis |
| GO:0000502 | 62 | proteasome_complex |
| GO:0005097 | 62 | Rab_GTPase_activator_activity |
| GO:0006914 | 62 | autophagy |
| GO:0009145 | 62 | purine_nucleoside_triphosphate_biosynthetic_process |
| GO:0010923 | 62 | negative_regulation_of_phosphatase_activity |
| GO:0034614 | 62 | cellular_response_to_reactive_oxygen_species |
| GO:0034764 | 62 | positive_regulation_of_transmembrane_transport |
| GO:0035666 | 62 | TRIF-dependent_toll-like_receptor_signaling_pathway |
| GO:0042562 | 62 | hormone_binding |
| GO:0042990 | 62 | regulation_of_transcription_factor_import_into_nucleus |
| GO:0045621 | 62 | positive_regulation_of_lymphocyte_differentiation |
| GO:0046467 | 62 | membrane_lipid_biosynthetic_process |
| GO:0050907 | 62 | detection_of_chemical_stimulus_involved_in_sensory_perception |
| GO:0060560 | 62 | developmental_growth_involved_in_morphogenesis |
| GO:0000271 | 61 | polysaccharide_biosynthetic_process |
| GO:0005507 | 61 | copper_ion_binding |
| GO:0006304 | 61 | DNA_modification |
| GO:0006406 | 61 | mRNA_export_from_nucleus |
| GO:0006672 | 61 | ceramide_metabolic_process |
| GO:0006818 | 61 | hydrogen_transport |
| GO:0007043 | 61 | cell-cell_junction_assembly |
| GO:0009064 | 61 | glutamine_family_amino_acid_metabolic_process |
| GO:0009206 | 61 | purine_ribonucleoside_triphosphate_biosynthetic_process |
| GO:0016655 | 61 | oxidoreductase_activity,_acting_on_NADH_or_NADPH,_quinone_or_similar_compound_as_acceptor |
| GO:0016782 | 61 | transferase_activity,_transferring_sulfur-containing_groups |
| GO:0016835 | 61 | carbon-oxygen_lyase_activity |
| GO:0017053 | 61 | transcriptional_repressor_complex |
| GO:0019888 | 61 | protein_phosphatase_regulator_activity |
| GO:0019955 | 61 | cytokine_binding |
| GO:0030139 | 61 | endocytic_vesicle |
| GO:0035097 | 61 | histone_methyltransferase_complex |
| GO:0043407 | 61 | negative_regulation_of_MAP_kinase_activity |
| GO:0045598 | 61 | regulation_of_fat_cell_differentiation |
| GO:0048145 | 61 | regulation_of_fibroblast_proliferation |
| GO:2000117 | 61 | negative_regulation_of_cysteine-type_endopeptidase_activity |
| GO:0001948 | 60 | glycoprotein_binding |
| GO:0002687 | 60 | positive_regulation_of_leukocyte_migration |
| GO:0003743 | 60 | translation_initiation_factor_activity |
| GO:0005643 | 60 | nuclear_pore |
| GO:0006968 | 60 | cellular_defense_response |
| GO:0008013 | 60 | beta-catenin_binding |
| GO:0009913 | 60 | epidermal_cell_differentiation |
| GO:0015935 | 60 | small_ribosomal_subunit |
| GO:0016579 | 60 | protein_deubiquitination |
| GO:0030148 | 60 | sphingolipid_biosynthetic_process |
| GO:0031333 | 60 | negative_regulation_of_protein_complex_assembly |
| GO:0045454 | 60 | cell_redox_homeostasis |
| GO:0048771 | 60 | tissue_remodeling |
| GO:0051053 | 60 | negative_regulation_of_DNA_metabolic_process |
| GO:0051339 | 60 | regulation_of_lyase_activity |
| GO:0051701 | 60 | interaction_with_host |
| GO:0051781 | 60 | positive_regulation_of_cell_division |
| GO:0060021 | 60 | palate_development |
| GO:0006007 | 59 | glucose_catabolic_process |
| GO:0006733 | 59 | oxidoreduction_coenzyme_metabolic_process |
| GO:0008301 | 59 | DNA_binding,_bending |
| GO:0015833 | 59 | peptide_transport |
| GO:0015992 | 59 | proton_transport |
| GO:0021953 | 59 | central_nervous_system_neuron_differentiation |
| GO:0030170 | 59 | pyridoxal_phosphate_binding |
| GO:0032182 | 59 | small_conjugating_protein_binding |
| GO:0032649 | 59 | regulation_of_interferon-gamma_production |
| GO:0035821 | 59 | modification_of_morphology_or_physiology_of_other_organism |
| GO:0042108 | 59 | positive_regulation_of_cytokine_biosynthetic_process |
| GO:0045862 | 59 | positive_regulation_of_proteolysis |
| GO:0046332 | 59 | SMAD_binding |
| GO:0046634 | 59 | regulation_of_alpha-beta_T_cell_activation |
| GO:0048704 | 59 | embryonic_skeletal_system_morphogenesis |
| GO:0050773 | 59 | regulation_of_dendrite_development |
| GO:0051153 | 59 | regulation_of_striated_muscle_cell_differentiation |
| GO:0061136 | 59 | regulation_of_proteasomal_protein_catabolic_process |
| GO:0070279 | 59 | vitamin_B6_binding |
| GO:0071295 | 59 | cellular_response_to_vitamin |
| GO:0071383 | 59 | cellular_response_to_steroid_hormone_stimulus |
| GO:0001570 | 58 | vasculogenesis |
| GO:0001952 | 58 | regulation_of_cell-matrix_adhesion |
| GO:0002688 | 58 | regulation_of_leukocyte_chemotaxis |
| GO:0003014 | 58 | renal_system_process |
| GO:0005814 | 58 | centriole |
| GO:0007202 | 58 | activation_of_phospholipase_C_activity |
| GO:0007608 | 58 | sensory_perception_of_smell |
| GO:0009062 | 58 | fatty_acid_catabolic_process |
| GO:0009914 | 58 | hormone_transport |
| GO:0022613 | 58 | ribonucleoprotein_complex_biogenesis |
| GO:0030048 | 58 | actin_filament-based_movement |
| GO:0030500 | 58 | regulation_of_bone_mineralization |
| GO:0042379 | 58 | chemokine_receptor_binding |
| GO:0043154 | 58 | negative_regulation_of_cysteine-type_endopeptidase_activity_involved_in_apoptotic_process |
| GO:0043484 | 58 | regulation_of_RNA_splicing |
| GO:0051258 | 58 | protein_polymerization |
| GO:0051291 | 58 | protein_heterooligomerization |
| GO:0000228 | 57 | nuclear_chromosome |
| GO:0006521 | 57 | regulation_of_cellular_amino_acid_metabolic_process |
| GO:0009953 | 57 | dorsal/ventral_pattern_formation |
| GO:0014013 | 57 | regulation_of_gliogenesis |
| GO:0030509 | 57 | BMP_signaling_pathway |
| GO:0030816 | 57 | positive_regulation_of_cAMP_metabolic_process |
| GO:0030819 | 57 | positive_regulation_of_cAMP_biosynthetic_process |
| GO:0031279 | 57 | regulation_of_cyclase_activity |
| GO:0032200 | 57 | telomere_organization |
| GO:0032387 | 57 | negative_regulation_of_intracellular_transport |
| GO:0043038 | 57 | amino_acid_activation |
| GO:0043039 | 57 | tRNA_aminoacylation |
| GO:0050688 | 57 | regulation_of_defense_response_to_virus |
| GO:0060606 | 57 | tube_closure |
| GO:0071478 | 57 | cellular_response_to_radiation |
| GO:0000723 | 56 | telomere_maintenance |
| GO:0000792 | 56 | heterochromatin |
| GO:0001843 | 56 | neural_tube_closure |
| GO:0002526 | 56 | acute_inflammatory_response |
| GO:0003684 | 56 | damaged_DNA_binding |
| GO:0004521 | 56 | endoribonuclease_activity |
| GO:0005901 | 56 | caveola |
| GO:0005930 | 56 | axoneme |
| GO:0006941 | 56 | striated_muscle_contraction |
| GO:0016266 | 56 | O-glycan_processing |
| GO:0016765 | 56 | transferase_activity,_transferring_alkyl_or_aryl_(other_than_methyl)_groups |
| GO:0018958 | 56 | phenol-containing_compound_metabolic_process |
| GO:0032368 | 56 | regulation_of_lipid_transport |
| GO:0040014 | 56 | regulation_of_multicellular_organism_growth |
| GO:0042440 | 56 | pigment_metabolic_process |
| GO:0043130 | 56 | ubiquitin_binding |
| GO:0043255 | 56 | regulation_of_carbohydrate_biosynthetic_process |
| GO:0043507 | 56 | positive_regulation_of_JUN_kinase_activity |
| GO:0044309 | 56 | neuron_spine |
| GO:0045639 | 56 | positive_regulation_of_myeloid_cell_differentiation |
| GO:0045744 | 56 | negative_regulation_of_G-protein_coupled_receptor_protein_signaling_pathway |
| GO:0046620 | 56 | regulation_of_organ_growth |
| GO:0048029 | 56 | monosaccharide_binding |
| GO:0050905 | 56 | neuromuscular_process |
| GO:0051607 | 56 | defense_response_to_virus |
| GO:0004402 | 55 | histone_acetyltransferase_activity |
| GO:0006497 | 55 | protein_lipidation |
| GO:0006775 | 55 | fat-soluble_vitamin_metabolic_process |
| GO:0007254 | 55 | JNK_cascade |
| GO:0007569 | 55 | cell_aging |
| GO:0010976 | 55 | positive_regulation_of_neuron_projection_development |
| GO:0016627 | 55 | oxidoreductase_activity,_acting_on_the_CH-CH_group_of_donors |
| GO:0016667 | 55 | oxidoreductase_activity,_acting_on_a_sulfur_group_of_donors |
| GO:0016811 | 55 | hydrolase_activity,_acting_on_carbon-nitrogen_(but_not_peptide)_bonds,_in_linear_amides |
| GO:0019722 | 55 | calcium-mediated_signaling |
| GO:0031100 | 55 | organ_regeneration |
| GO:0031214 | 55 | biomineral_tissue_development |
| GO:0031983 | 55 | vesicle_lumen |
| GO:0033293 | 55 | monocarboxylic_acid_binding |
| GO:0034707 | 55 | chloride_channel_complex |
| GO:0042307 | 55 | positive_regulation_of_protein_import_into_nucleus |
| GO:0042632 | 55 | cholesterol_homeostasis |
| GO:0043112 | 55 | receptor_metabolic_process |
| GO:0043409 | 55 | negative_regulation_of_MAPK_cascade |
| GO:0045638 | 55 | negative_regulation_of_myeloid_cell_differentiation |
| GO:0045833 | 55 | negative_regulation_of_lipid_metabolic_process |
| GO:0048641 | 55 | regulation_of_skeletal_muscle_tissue_development |
| GO:0050434 | 55 | positive_regulation_of_viral_transcription |
| GO:0050871 | 55 | positive_regulation_of_B_cell_activation |
| GO:0051536 | 55 | iron-sulfur_cluster_binding |
| GO:0051540 | 55 | metal_cluster_binding |
| GO:0055092 | 55 | sterol_homeostasis |
| GO:0070304 | 55 | positive_regulation_of_stress-activated_protein_kinase_signaling_cascade |
| GO:0000186 | 54 | activation_of_MAPKK_activity |
| GO:0003707 | 54 | steroid_hormone_receptor_activity |
| GO:0006418 | 54 | tRNA_aminoacylation_for_protein_translation |
| GO:0006636 | 54 | unsaturated_fatty_acid_biosynthetic_process |
| GO:0006664 | 54 | glycolipid_metabolic_process |
| GO:0007033 | 54 | vacuole_organization |
| GO:0016525 | 54 | negative_regulation_of_angiogenesis |
| GO:0032874 | 54 | positive_regulation_of_stress-activated_MAPK_cascade |
| GO:0035150 | 54 | regulation_of_tube_size |
| GO:0043197 | 54 | dendritic_spine |
| GO:0043498 | 54 | cell_surface_binding |
| GO:0045761 | 54 | regulation_of_adenylate_cyclase_activity |
| GO:0046879 | 54 | hormone_secretion |
| GO:0051897 | 54 | positive_regulation_of_protein_kinase_B_signaling_cascade |
| GO:0090317 | 54 | negative_regulation_of_intracellular_protein_transport |
| GO:2000242 | 54 | negative_regulation_of_reproductive_process |
| GO:0006888 | 53 | ER_to_Golgi_vesicle-mediated_transport |
| GO:0006921 | 53 | cellular_component_disassembly_involved_in_apoptotic_process |
| GO:0009566 | 53 | fertilization |
| GO:0009880 | 53 | embryonic_pattern_specification |
| GO:0010811 | 53 | positive_regulation_of_cell-substrate_adhesion |
| GO:0016763 | 53 | transferase_activity,_transferring_pentosyl_groups |
| GO:0031056 | 53 | regulation_of_histone_modification |
| GO:0032088 | 53 | negative_regulation_of_NF-kappaB_transcription_factor_activity |
| GO:0035335 | 53 | peptidyl-tyrosine_dephosphorylation |
| GO:0042472 | 53 | inner_ear_morphogenesis |
| GO:0042641 | 53 | actomyosin |
| GO:0043467 | 53 | regulation_of_generation_of_precursor_metabolites_and_energy |
| GO:0045582 | 53 | positive_regulation_of_T_cell_differentiation |
| GO:0048514 | 53 | blood_vessel_morphogenesis |
| GO:0050839 | 53 | cell_adhesion_molecule_binding |
| GO:0050880 | 53 | regulation_of_blood_vessel_size |
| GO:0051149 | 53 | positive_regulation_of_muscle_cell_differentiation |
| GO:0060205 | 53 | cytoplasmic_membrane-bounded_vesicle_lumen |
| GO:0060429 | 53 | epithelium_development |
| GO:0070661 | 53 | leukocyte_proliferation |
| GO:0070664 | 53 | negative_regulation_of_leukocyte_proliferation |
| GO:0071260 | 53 | cellular_response_to_mechanical_stimulus |
| GO:0071407 | 53 | cellular_response_to_organic_cyclic_compound |
| GO:0071417 | 53 | cellular_response_to_organic_nitrogen |
| GO:0071453 | 53 | cellular_response_to_oxygen_levels |
| GO:0001938 | 52 | positive_regulation_of_endothelial_cell_proliferation |
| GO:0004702 | 52 | receptor_signaling_protein_serine/threonine_kinase_activity |
| GO:0005902 | 52 | microvillus |
| GO:0006213 | 52 | pyrimidine_nucleoside_metabolic_process |
| GO:0006458 | 52 | 'de_novo'_protein_folding |
| GO:0007269 | 52 | neurotransmitter_secretion |
| GO:0007286 | 52 | spermatid_development |
| GO:0008146 | 52 | sulfotransferase_activity |
| GO:0009187 | 52 | cyclic_nucleotide_metabolic_process |
| GO:0009451 | 52 | RNA_modification |
| GO:0015662 | 52 | ATPase_activity,_coupled_to_transmembrane_movement_of_ions,_phosphorylative_mechanism |
| GO:0019319 | 52 | hexose_biosynthetic_process |
| GO:0022625 | 52 | cytosolic_large_ribosomal_subunit |
| GO:0030117 | 52 | membrane_coat |
| GO:0031513 | 52 | nonmotile_primary_cilium |
| GO:0031646 | 52 | positive_regulation_of_neurological_system_process |
| GO:0032587 | 52 | ruffle_membrane |
| GO:0045665 | 52 | negative_regulation_of_neuron_differentiation |
| GO:0051640 | 52 | organelle_localization |
| GO:0051705 | 52 | behavioral_interaction_between_organisms |
| GO:0090183 | 52 | regulation_of_kidney_development |
| GO:0097285 | 52 | cell-type_specific_apoptotic_process |
| GO:0000288 | 51 | nuclear-transcribed_mRNA_catabolic_process,_deadenylation-dependent_decay |
| GO:0001085 | 51 | RNA_polymerase_II_transcription_factor_binding |
| GO:0002673 | 51 | regulation_of_acute_inflammatory_response |
| GO:0005778 | 51 | peroxisomal_membrane |
| GO:0006721 | 51 | terpenoid_metabolic_process |
| GO:0007272 | 51 | ensheathment_of_neurons |
| GO:0008366 | 51 | axon_ensheathment |
| GO:0030183 | 51 | B_cell_differentiation |
| GO:0030705 | 51 | cytoskeleton-dependent_intracellular_transport |
| GO:0030888 | 51 | regulation_of_B_cell_proliferation |
| GO:0030900 | 51 | forebrain_development |
| GO:0031228 | 51 | intrinsic_to_Golgi_membrane |
| GO:0031903 | 51 | microbody_membrane |
| GO:0034763 | 51 | negative_regulation_of_transmembrane_transport |
| GO:0042475 | 51 | odontogenesis_of_dentin-containing_tooth |
| GO:0045767 | 51 | regulation_of_anti-apoptosis |
| GO:0050673 | 51 | epithelial_cell_proliferation |
| GO:0050715 | 51 | positive_regulation_of_cytokine_secretion |
| GO:0051287 | 51 | NAD_binding |
| GO:0051289 | 51 | protein_homotetramerization |
| GO:0071418 | 51 | cellular_response_to_amine_stimulus |
| GO:0072655 | 51 | establishment_of_protein_localization_in_mitochondrion |
| GO:2000377 | 51 | regulation_of_reactive_oxygen_species_metabolic_process |
| GO:0001708 | 50 | cell_fate_specification |
| GO:0002690 | 50 | positive_regulation_of_leukocyte_chemotaxis |
| GO:0004812 | 50 | aminoacyl-tRNA_ligase_activity |
| GO:0004869 | 50 | cysteine-type_endopeptidase_inhibitor_activity |
| GO:0004879 | 50 | ligand-activated_sequence-specific_DNA_binding_RNA_polymerase_II_transcription_factor_activity |
| GO:0005070 | 50 | SH3/SH2_adaptor_activity |
| GO:0005796 | 50 | Golgi_lumen |
| GO:0006661 | 50 | phosphatidylinositol_biosynthetic_process |
| GO:0007189 | 50 | adenylate_cyclase-activating_G-protein_coupled_receptor_signaling_pathway |
| GO:0010506 | 50 | regulation_of_autophagy |
| GO:0014066 | 50 | regulation_of_phosphatidylinositol_3-kinase_cascade |
| GO:0016571 | 50 | histone_methylation |
| GO:0016875 | 50 | ligase_activity,_forming_carbon-oxygen_bonds |
| GO:0016876 | 50 | ligase_activity,_forming_aminoacyl-tRNA_and_related_compounds |
| GO:0030163 | 50 | protein_catabolic_process |
| GO:0030658 | 50 | transport_vesicle_membrane |
| GO:0031970 | 50 | organelle_envelope_lumen |
| GO:0032434 | 50 | regulation_of_proteasomal_ubiquitin-dependent_protein_catabolic_process |
| GO:0032943 | 50 | mononuclear_cell_proliferation |
| GO:0032945 | 50 | negative_regulation_of_mononuclear_cell_proliferation |
| GO:0034774 | 50 | secretory_granule_lumen |
| GO:0035085 | 50 | cilium_axoneme |
| GO:0042509 | 50 | regulation_of_tyrosine_phosphorylation_of_STAT_protein |
| GO:0042734 | 50 | presynaptic_membrane |
| GO:0043021 | 50 | ribonucleoprotein_complex_binding |
| GO:0043648 | 50 | dicarboxylic_acid_metabolic_process |
| GO:0046504 | 50 | glycerol_ether_biosynthetic_process |
| GO:0050672 | 50 | negative_regulation_of_lymphocyte_proliferation |
| GO:0055067 | 50 | monovalent_inorganic_cation_homeostasis |
| GO:0072524 | 50 | pyridine-containing_compound_metabolic_process |
| GO:0000118 | 49 | histone_deacetylase_complex |
| GO:0002460 | 49 | adaptive_immune_response_based_on_somatic_recombination_of_immune_receptors_built_from_immunoglobulin_superfamily_domains |
| GO:0005518 | 49 | collagen_binding |
| GO:0006446 | 49 | regulation_of_translational_initiation |
| GO:0006626 | 49 | protein_targeting_to_mitochondrion |
| GO:0007519 | 49 | skeletal_muscle_tissue_development |
| GO:0008009 | 49 | chemokine_activity |
| GO:0008287 | 49 | protein_serine/threonine_phosphatase_complex |
| GO:0015300 | 49 | solute:solute_antiporter_activity |
| GO:0015370 | 49 | solute:sodium_symporter_activity |
| GO:0016706 | 49 | oxidoreductase_activity,_acting_on_paired_donors,_with_incorporation_or_reduction_of_molecular_oxygen,_2-oxoglutarate_as_one_donor,_and_incorporation_of_one_atom_each_of_oxygen_into_both_donors |
| GO:0019205 | 49 | nucleobase-containing_compound_kinase_activity |
| GO:0019362 | 49 | pyridine_nucleotide_metabolic_process |
| GO:0019783 | 49 | small_conjugating_protein-specific_protease_activity |
| GO:0030216 | 49 | keratinocyte_differentiation |
| GO:0030667 | 49 | secretory_granule_membrane |
| GO:0031023 | 49 | microtubule_organizing_center_organization |
| GO:0031234 | 49 | extrinsic_to_internal_side_of_plasma_membrane |
| GO:0033555 | 49 | multicellular_organismal_response_to_stress |
| GO:0042054 | 49 | histone_methyltransferase_activity |
| GO:0046427 | 49 | positive_regulation_of_JAK-STAT_cascade |
| GO:0046456 | 49 | icosanoid_biosynthetic_process |
| GO:0046496 | 49 | nicotinamide_nucleotide_metabolic_process |
| GO:0051971 | 49 | positive_regulation_of_transmission_of_nerve_impulse |
| GO:0060491 | 49 | regulation_of_cell_projection_assembly |
| GO:0060688 | 49 | regulation_of_morphogenesis_of_a_branching_structure |
| GO:0071299 | 49 | cellular_response_to_vitamin_A |
| GO:0001228 | 48 | RNA_polymerase_II_transcription_regulatory_region_sequence-specific_DNA_binding_transcription_factor_activity_involved_in_positive_regulation_of_transcription |
| GO:0001649 | 48 | osteoblast_differentiation |
| GO:0002020 | 48 | protease_binding |
| GO:0002821 | 48 | positive_regulation_of_adaptive_immune_response |
| GO:0005231 | 48 | excitatory_extracellular_ligand-gated_ion_channel_activity |
| GO:0006687 | 48 | glycosphingolipid_metabolic_process |
| GO:0006754 | 48 | ATP_biosynthetic_process |
| GO:0006826 | 48 | iron_ion_transport |
| GO:0006970 | 48 | response_to_osmotic_stress |
| GO:0007200 | 48 | phospholipase_C-activating_G-protein_coupled_receptor_signaling_pathway |
| GO:0007266 | 48 | Rho_protein_signal_transduction |
| GO:0009267 | 48 | cellular_response_to_starvation |
| GO:0009306 | 48 | protein_secretion |
| GO:0016458 | 48 | gene_silencing |
| GO:0016860 | 48 | intramolecular_oxidoreductase_activity |
| GO:0017048 | 48 | Rho_GTPase_binding |
| GO:0030173 | 48 | integral_to_Golgi_membrane |
| GO:0031330 | 48 | negative_regulation_of_cellular_catabolic_process |
| GO:0032273 | 48 | positive_regulation_of_protein_polymerization |
| GO:0032432 | 48 | actin_filament_bundle |
| GO:0042439 | 48 | ethanolamine-containing_compound_metabolic_process |
| GO:0042446 | 48 | hormone_biosynthetic_process |
| GO:0042552 | 48 | myelination |
| GO:0043542 | 48 | endothelial_cell_migration |
| GO:0046651 | 48 | lymphocyte_proliferation |
| GO:0050777 | 48 | negative_regulation_of_immune_response |
| GO:0050810 | 48 | regulation_of_steroid_biosynthetic_process |
| GO:0051087 | 48 | chaperone_binding |
| GO:0051925 | 48 | regulation_of_calcium_ion_transport_via_voltage-gated_calcium_channel_activity |
| GO:0055001 | 48 | muscle_cell_development |
| GO:0071300 | 48 | cellular_response_to_retinoic_acid |
| GO:0071456 | 48 | cellular_response_to_hypoxia |
| GO:0000781 | 47 | chromosome,_telomeric_region |
| GO:0000793 | 47 | condensed_chromosome |
| GO:0002443 | 47 | leukocyte_mediated_immunity |
| GO:0002698 | 47 | negative_regulation_of_immune_effector_process |
| GO:0002705 | 47 | positive_regulation_of_leukocyte_mediated_immunity |
| GO:0002708 | 47 | positive_regulation_of_lymphocyte_mediated_immunity |
| GO:0002793 | 47 | positive_regulation_of_peptide_secretion |
| GO:0003954 | 47 | NADH_dehydrogenase_activity |
| GO:0004722 | 47 | protein_serine/threonine_phosphatase_activity |
| GO:0005044 | 47 | scavenger_receptor_activity |
| GO:0005793 | 47 | endoplasmic_reticulum-Golgi_intermediate_compartment |
| GO:0005905 | 47 | coated_pit |
| GO:0006073 | 47 | cellular_glucan_metabolic_process |
| GO:0006220 | 47 | pyrimidine_nucleotide_metabolic_process |
| GO:0008137 | 47 | NADH_dehydrogenase_(ubiquinone)_activity |
| GO:0009163 | 47 | nucleoside_biosynthetic_process |
| GO:0015030 | 47 | Cajal_body |
| GO:0016893 | 47 | endonuclease_activity,_active_with_either_ribo-_or_deoxyribonucleic_acids_and_producing_5'-phosphomonoesters |
| GO:0031093 | 47 | platelet_alpha_granule_lumen |
| GO:0043010 | 47 | camera-type_eye_development |
| GO:0043242 | 47 | negative_regulation_of_protein_complex_disassembly |
| GO:0044042 | 47 | glucan_metabolic_process |
| GO:0045111 | 47 | intermediate_filament_cytoskeleton |
| GO:0045669 | 47 | positive_regulation_of_osteoblast_differentiation |
| GO:0046330 | 47 | positive_regulation_of_JNK_cascade |
| GO:0046823 | 47 | negative_regulation_of_nucleocytoplasmic_transport |
| GO:0050136 | 47 | NADH_dehydrogenase_(quinone)_activity |
| GO:0051084 | 47 | 'de_novo'_posttranslational_protein_folding |
| GO:0051219 | 47 | phosphoprotein_binding |
| GO:0051817 | 47 | modification_of_morphology_or_physiology_of_other_organism_involved_in_symbiotic_interaction |
| GO:0051952 | 47 | regulation_of_amine_transport |
| GO:0060415 | 47 | muscle_tissue_morphogenesis |
| GO:0061387 | 47 | regulation_of_extent_of_cell_growth |
| GO:0072657 | 47 | protein_localization_in_membrane |
| GO:0000725 | 46 | recombinational_repair |
| GO:0001047 | 46 | core_promoter_binding |
| GO:0001077 | 46 | RNA_polymerase_II_core_promoter_proximal_region_sequence-specific_DNA_binding_transcription_factor_activity_involved_in_positive_regulation_of_transcription |
| GO:0001669 | 46 | acrosomal_vesicle |
| GO:0002039 | 46 | p53_binding |
| GO:0002824 | 46 | positive_regulation_of_adaptive_immune_response_based_on_somatic_recombination_of_immune_receptors_built_from_immunoglobulin_superfamily_domains |
| GO:0003678 | 46 | DNA_helicase_activity |
| GO:0005747 | 46 | mitochondrial_respiratory_chain_complex_I |
| GO:0005977 | 46 | glycogen_metabolic_process |
| GO:0006096 | 46 | glycolysis |
| GO:0006283 | 46 | transcription-coupled_nucleotide-excision_repair |
| GO:0006635 | 46 | fatty_acid_beta-oxidation |
| GO:0006749 | 46 | glutathione_metabolic_process |
| GO:0007498 | 46 | mesoderm_development |
| GO:0008015 | 46 | blood_circulation |
| GO:0008138 | 46 | protein_tyrosine/serine/threonine_phosphatase_activity |
| GO:0008306 | 46 | associative_learning |
| GO:0009312 | 46 | oligosaccharide_biosynthetic_process |
| GO:0010970 | 46 | microtubule-based_transport |
| GO:0016830 | 46 | carbon-carbon_lyase_activity |
| GO:0017148 | 46 | negative_regulation_of_translation |
| GO:0019210 | 46 | kinase_inhibitor_activity |
| GO:0019229 | 46 | regulation_of_vasoconstriction |
| GO:0019861 | 46 | flagellum |
| GO:0030964 | 46 | NADH_dehydrogenase_complex |
| GO:0032231 | 46 | regulation_of_actin_filament_bundle_assembly |
| GO:0032642 | 46 | regulation_of_chemokine_production |
| GO:0033135 | 46 | regulation_of_peptidyl-serine_phosphorylation |
| GO:0035264 | 46 | multicellular_organism_growth |
| GO:0035282 | 46 | segmentation |
| GO:0042044 | 46 | fluid_transport |
| GO:0042787 | 46 | protein_ubiquitination_involved_in_ubiquitin-dependent_protein_catabolic_process |
| GO:0045271 | 46 | respiratory_chain_complex_I |
| GO:0045727 | 46 | positive_regulation_of_translation |
| GO:0050661 | 46 | NADP_binding |
| GO:0050806 | 46 | positive_regulation_of_synaptic_transmission |
| GO:0050807 | 46 | regulation_of_synapse_organization |
| GO:0051146 | 46 | striated_muscle_cell_differentiation |
| GO:0061097 | 46 | regulation_of_protein_tyrosine_kinase_activity |
| GO:0090277 | 46 | positive_regulation_of_peptide_hormone_secretion |
| GO:0000724 | 45 | double-strand_break_repair_via_homologous_recombination |
| GO:0001725 | 45 | stress_fiber |
| GO:0001816 | 45 | cytokine_production |
| GO:0002790 | 45 | peptide_secretion |
| GO:0004715 | 45 | non-membrane_spanning_protein_tyrosine_kinase_activity |
| GO:0006282 | 45 | regulation_of_DNA_repair |
| GO:0007270 | 45 | neuron-neuron_synaptic_transmission |
| GO:0007416 | 45 | synapse_assembly |
| GO:0007588 | 45 | excretion |
| GO:0008307 | 45 | structural_constituent_of_muscle |
| GO:0010522 | 45 | regulation_of_calcium_ion_transport_into_cytosol |
| GO:0015179 | 45 | L-amino_acid_transmembrane_transporter_activity |
| GO:0030512 | 45 | negative_regulation_of_transforming_growth_factor_beta_receptor_signaling_pathway |
| GO:0030672 | 45 | synaptic_vesicle_membrane |
| GO:0032768 | 45 | regulation_of_monooxygenase_activity |
| GO:0032947 | 45 | protein_complex_scaffold |
| GO:0033762 | 45 | response_to_glucagon_stimulus |
| GO:0042611 | 45 | MHC_protein_complex |
| GO:0043588 | 45 | skin_development |
| GO:0045740 | 45 | positive_regulation_of_DNA_replication |
| GO:0046460 | 45 | neutral_lipid_biosynthetic_process |
| GO:0046463 | 45 | acylglycerol_biosynthetic_process |
| GO:0046635 | 45 | positive_regulation_of_alpha-beta_T_cell_activation |
| GO:0046888 | 45 | negative_regulation_of_hormone_secretion |
| GO:0046889 | 45 | positive_regulation_of_lipid_biosynthetic_process |
| GO:0051297 | 45 | centrosome_organization |
| GO:0090263 | 45 | positive_regulation_of_canonical_Wnt_receptor_signaling_pathway |
| GO:2001022 | 45 | positive_regulation_of_response_to_DNA_damage_stimulus |
| GO:0000272 | 44 | polysaccharide_catabolic_process |
| GO:0001541 | 44 | ovarian_follicle_development |
| GO:0002285 | 44 | lymphocyte_activation_involved_in_immune_response |
| GO:0003899 | 44 | DNA-directed_RNA_polymerase_activity |
| GO:0004843 | 44 | ubiquitin-specific_protease_activity |
| GO:0004860 | 44 | protein_kinase_inhibitor_activity |
| GO:0006084 | 44 | acetyl-CoA_metabolic_process |
| GO:0006120 | 44 | mitochondrial_electron_transport,_NADH_to_ubiquinone |
| GO:0006369 | 44 | termination_of_RNA_polymerase_II_transcription |
| GO:0006821 | 44 | chloride_transport |
| GO:0006909 | 44 | phagocytosis |
| GO:0006940 | 44 | regulation_of_smooth_muscle_contraction |
| GO:0007009 | 44 | plasma_membrane_organization |
| GO:0007034 | 44 | vacuolar_transport |
| GO:0009583 | 44 | detection_of_light_stimulus |
| GO:0017137 | 44 | Rab_GTPase_binding |
| GO:0030175 | 44 | filopodium |
| GO:0030521 | 44 | androgen_receptor_signaling_pathway |
| GO:0030545 | 44 | receptor_regulator_activity |
| GO:0031645 | 44 | negative_regulation_of_neurological_system_process |
| GO:0034062 | 44 | RNA_polymerase_activity |
| GO:0035872 | 44 | nucleotide-binding_domain,_leucine_rich_repeat_containing_receptor_signaling_pathway |
| GO:0043204 | 44 | perikaryon |
| GO:0045428 | 44 | regulation_of_nitric_oxide_biosynthetic_process |
| GO:0045685 | 44 | regulation_of_glial_cell_differentiation |
| GO:0060041 | 44 | retina_development_in_camera-type_eye |
| GO:2001233 | 44 | regulation_of_apoptotic_signaling_pathway |
| GO:0000096 | 43 | sulfur_amino_acid_metabolic_process |
| GO:0001704 | 43 | formation_of_primary_germ_layer |
| GO:0004180 | 43 | carboxypeptidase_activity |
| GO:0005080 | 43 | protein_kinase_C_binding |
| GO:0005758 | 43 | mitochondrial_intermembrane_space |
| GO:0005811 | 43 | lipid_particle |
| GO:0005913 | 43 | cell-cell_adherens_junction |
| GO:0006094 | 43 | gluconeogenesis |
| GO:0006333 | 43 | chromatin_assembly_or_disassembly |
| GO:0006833 | 43 | water_transport |
| GO:0007193 | 43 | adenylate_cyclase-inhibiting_G-protein_coupled_receptor_signaling_pathway |
| GO:0007259 | 43 | JAK-STAT_cascade |
| GO:0009799 | 43 | specification_of_symmetry |
| GO:0010001 | 43 | glial_cell_differentiation |
| GO:0010043 | 43 | response_to_zinc_ion |
| GO:0016836 | 43 | hydro-lyase_activity |
| GO:0019209 | 43 | kinase_activator_activity |
| GO:0030072 | 43 | peptide_hormone_secretion |
| GO:0031424 | 43 | keratinization |
| GO:0033613 | 43 | activating_transcription_factor_binding |
| GO:0034637 | 43 | cellular_carbohydrate_biosynthetic_process |
| GO:0043271 | 43 | negative_regulation_of_ion_transport |
| GO:0043966 | 43 | histone_H3_acetylation |
| GO:0044236 | 43 | multicellular_organismal_metabolic_process |
| GO:0046148 | 43 | pigment_biosynthetic_process |
| GO:0048168 | 43 | regulation_of_neuronal_synaptic_plasticity |
| GO:0048565 | 43 | digestive_tract_development |
| GO:0050772 | 43 | positive_regulation_of_axonogenesis |
| GO:0060562 | 43 | epithelial_tube_morphogenesis |
| GO:0061035 | 43 | regulation_of_cartilage_development |
| GO:0070301 | 43 | cellular_response_to_hydrogen_peroxide |
| GO:0071704 | 43 | organic_substance_metabolic_process |
| GO:2000106 | 43 | regulation_of_leukocyte_apoptotic_process |
| GO:0001709 | 42 | cell_fate_determination |
| GO:0003727 | 42 | single-stranded_RNA_binding |
| GO:0006261 | 42 | DNA-dependent_DNA_replication |
| GO:0009855 | 42 | determination_of_bilateral_symmetry |
| GO:0016101 | 42 | diterpenoid_metabolic_process |
| GO:0016278 | 42 | lysine_N-methyltransferase_activity |
| GO:0016279 | 42 | protein-lysine_N-methyltransferase_activity |
| GO:0016903 | 42 | oxidoreductase_activity,_acting_on_the_aldehyde_or_oxo_group_of_donors |
| GO:0019048 | 42 | virus-host_interaction |
| GO:0019233 | 42 | sensory_perception_of_pain |
| GO:0021782 | 42 | glial_cell_development |
| GO:0023021 | 42 | termination_of_signal_transduction |
| GO:0030374 | 42 | ligand-dependent_nuclear_receptor_transcription_coactivator_activity |
| GO:0030516 | 42 | regulation_of_axon_extension |
| GO:0031032 | 42 | actomyosin_structure_organization |
| GO:0031110 | 42 | regulation_of_microtubule_polymerization_or_depolymerization |
| GO:0032272 | 42 | negative_regulation_of_protein_polymerization |
| GO:0033013 | 42 | tetrapyrrole_metabolic_process |
| GO:0034061 | 42 | DNA_polymerase_activity |
| GO:0034332 | 42 | adherens_junction_organization |
| GO:0042157 | 42 | lipoprotein_metabolic_process |
| GO:0042308 | 42 | negative_regulation_of_protein_import_into_nucleus |
| GO:0042531 | 42 | positive_regulation_of_tyrosine_phosphorylation_of_STAT_protein |
| GO:0043028 | 42 | cysteine-type_endopeptidase_regulator_activity_involved_in_apoptotic_process |
| GO:0043195 | 42 | terminal_button |
| GO:0045768 | 42 | positive_regulation_of_anti-apoptosis |
| GO:0051318 | 42 | G1_phase |
| GO:0055008 | 42 | cardiac_muscle_tissue_morphogenesis |
| GO:0055037 | 42 | recycling_endosome |
| GO:0071229 | 42 | cellular_response_to_acid |
| GO:0000018 | 41 | regulation_of_DNA_recombination |
| GO:0001756 | 41 | somitogenesis |
| GO:0001947 | 41 | heart_looping |
| GO:0003725 | 41 | double-stranded_RNA_binding |
| GO:0004601 | 41 | peroxidase_activity |
| GO:0005876 | 41 | spindle_microtubule |
| GO:0006778 | 41 | porphyrin-containing_compound_metabolic_process |
| GO:0006939 | 41 | smooth_muscle_contraction |
| GO:0006953 | 41 | acute-phase_response |
| GO:0007224 | 41 | smoothened_signaling_pathway |
| GO:0008188 | 41 | neuropeptide_receptor_activity |
| GO:0008589 | 41 | regulation_of_smoothened_signaling_pathway |
| GO:0009755 | 41 | hormone-mediated_signaling_pathway |
| GO:0010464 | 41 | regulation_of_mesenchymal_cell_proliferation |
| GO:0016126 | 41 | sterol_biosynthetic_process |
| GO:0016684 | 41 | oxidoreductase_activity,_acting_on_peroxide_as_acceptor |
| GO:0016776 | 41 | phosphotransferase_activity,_phosphate_group_as_acceptor |
| GO:0016859 | 41 | cis-trans_isomerase_activity |
| GO:0030010 | 41 | establishment_of_cell_polarity |
| GO:0030863 | 41 | cortical_cytoskeleton |
| GO:0031018 | 41 | endocrine_pancreas_development |
| GO:0031576 | 41 | G2/M_transition_checkpoint |
| GO:0031594 | 41 | neuromuscular_junction |
| GO:0033143 | 41 | regulation_of_intracellular_steroid_hormone_receptor_signaling_pathway |
| GO:0034103 | 41 | regulation_of_tissue_remodeling |
| GO:0034704 | 41 | calcium_channel_complex |
| GO:0042059 | 41 | negative_regulation_of_epidermal_growth_factor_receptor_signaling_pathway |
| GO:0042398 | 41 | cellular_modified_amino_acid_biosynthetic_process |
| GO:0043178 | 41 | alcohol_binding |
| GO:0043967 | 41 | histone_H4_acetylation |
| GO:0045670 | 41 | regulation_of_osteoclast_differentiation |
| GO:0045913 | 41 | positive_regulation_of_carbohydrate_metabolic_process |
| GO:0046677 | 41 | response_to_antibiotic |
| GO:0048678 | 41 | response_to_axon_injury |
| GO:0048814 | 41 | regulation_of_dendrite_morphogenesis |
| GO:0050684 | 41 | regulation_of_mRNA_processing |
| GO:0050819 | 41 | negative_regulation_of_coagulation |
| GO:0050994 | 41 | regulation_of_lipid_catabolic_process |
| GO:0060078 | 41 | regulation_of_postsynaptic_membrane_potential |
| GO:2001252 | 41 | positive_regulation_of_chromosome_organization |
| GO:0001523 | 40 | retinoid_metabolic_process |
| GO:0001755 | 40 | neural_crest_cell_migration |
| GO:0001776 | 40 | leukocyte_homeostasis |
| GO:0001837 | 40 | epithelial_to_mesenchymal_transition |
| GO:0003755 | 40 | peptidyl-prolyl_cis-trans_isomerase_activity |
| GO:0004091 | 40 | carboxylesterase_activity |
| GO:0006383 | 40 | transcription_from_RNA_polymerase_III_promoter |
| GO:0007249 | 40 | I-kappaB_kinase/NF-kappaB_cascade |
| GO:0007595 | 40 | lactation |
| GO:0008206 | 40 | bile_acid_metabolic_process |
| GO:0008227 | 40 | G-protein_coupled_amine_receptor_activity |
| GO:0009161 | 40 | ribonucleoside_monophosphate_metabolic_process |
| GO:0009295 | 40 | nucleoid |
| GO:0014068 | 40 | positive_regulation_of_phosphatidylinositol_3-kinase_cascade |
| GO:0015918 | 40 | sterol_transport |
| GO:0018024 | 40 | histone-lysine_N-methyltransferase_activity |
| GO:0019432 | 40 | triglyceride_biosynthetic_process |
| GO:0030071 | 40 | regulation_of_mitotic_metaphase/anaphase_transition |
| GO:0030301 | 40 | cholesterol_transport |
| GO:0031526 | 40 | brush_border_membrane |
| GO:0032155 | 40 | cell_division_site_part |
| GO:0032813 | 40 | tumor_necrosis_factor_receptor_superfamily_binding |
| GO:0042401 | 40 | cellular_biogenic_amine_biosynthetic_process |
| GO:0046637 | 40 | regulation_of_alpha-beta_T_cell_differentiation |
| GO:0048146 | 40 | positive_regulation_of_fibroblast_proliferation |
| GO:0048306 | 40 | calcium-dependent_protein_binding |
| GO:0051588 | 40 | regulation_of_neurotransmitter_transport |
| GO:0051970 | 40 | negative_regulation_of_transmission_of_nerve_impulse |
| GO:0070252 | 40 | actin-mediated_cell_contraction |
| GO:0071347 | 40 | cellular_response_to_interleukin-1 |
| GO:0072528 | 40 | pyrimidine-containing_compound_biosynthetic_process |
| GO:0000080 | 39 | G1_phase_of_mitotic_cell_cycle |
| GO:0000149 | 39 | SNARE_binding |
| GO:0000932 | 39 | cytoplasmic_mRNA_processing_body |
| GO:0001508 | 39 | regulation_of_action_potential |
| GO:0001657 | 39 | ureteric_bud_development |
| GO:0001658 | 39 | branching_involved_in_ureteric_bud_morphogenesis |
| GO:0002028 | 39 | regulation_of_sodium_ion_transport |
| GO:0002449 | 39 | lymphocyte_mediated_immunity |
| GO:0004536 | 39 | deoxyribonuclease_activity |
| GO:0006144 | 39 | purine_base_metabolic_process |
| GO:0006637 | 39 | acyl-CoA_metabolic_process |
| GO:0006691 | 39 | leukotriene_metabolic_process |
| GO:0007041 | 39 | lysosomal_transport |
| GO:0007368 | 39 | determination_of_left/right_symmetry |
| GO:0007585 | 39 | respiratory_gaseous_exchange |
| GO:0008374 | 39 | O-acyltransferase_activity |
| GO:0008375 | 39 | acetylglucosaminyltransferase_activity |
| GO:0008630 | 39 | DNA_damage_response,_signal_transduction_resulting_in_induction_of_apoptosis |
| GO:0010595 | 39 | positive_regulation_of_endothelial_cell_migration |
| GO:0014823 | 39 | response_to_activity |
| GO:0015908 | 39 | fatty_acid_transport |
| GO:0019840 | 39 | isoprenoid_binding |
| GO:0030145 | 39 | manganese_ion_binding |
| GO:0032024 | 39 | positive_regulation_of_insulin_secretion |
| GO:0032663 | 39 | regulation_of_interleukin-2_production |
| GO:0035383 | 39 | thioester_metabolic_process |
| GO:0038032 | 39 | termination_of_G-protein_coupled_receptor_signaling_pathway |
| GO:0042130 | 39 | negative_regulation_of_T_cell_proliferation |
| GO:0042733 | 39 | embryonic_digit_morphogenesis |
| GO:0043449 | 39 | cellular_alkene_metabolic_process |
| GO:0043525 | 39 | positive_regulation_of_neuron_apoptotic_process |
| GO:0045778 | 39 | positive_regulation_of_ossification |
| GO:0045930 | 39 | negative_regulation_of_mitotic_cell_cycle |
| GO:0048259 | 39 | regulation_of_receptor-mediated_endocytosis |
| GO:0048639 | 39 | positive_regulation_of_developmental_growth |
| GO:0048661 | 39 | positive_regulation_of_smooth_muscle_cell_proliferation |
| GO:0048742 | 39 | regulation_of_skeletal_muscle_fiber_development |
| GO:0048839 | 39 | inner_ear_development |
| GO:0050681 | 39 | androgen_receptor_binding |
| GO:0051187 | 39 | cofactor_catabolic_process |
| GO:0051492 | 39 | regulation_of_stress_fiber_assembly |
| GO:0070001 | 39 | aspartic-type_peptidase_activity |
| GO:0070988 | 39 | demethylation |
| GO:1900673 | 39 | olefin_metabolic_process |
| GO:0002200 | 38 | somatic_diversification_of_immune_receptors |
| GO:0002718 | 38 | regulation_of_cytokine_production_involved_in_immune_response |
| GO:0004177 | 38 | aminopeptidase_activity |
| GO:0004190 | 38 | aspartic-type_endopeptidase_activity |
| GO:0006029 | 38 | proteoglycan_metabolic_process |
| GO:0006040 | 38 | amino_sugar_metabolic_process |
| GO:0006081 | 38 | cellular_aldehyde_metabolic_process |
| GO:0007205 | 38 | protein_kinase_C-activating_G-protein_coupled_receptor_signaling_pathway |
| GO:0007422 | 38 | peripheral_nervous_system_development |
| GO:0008156 | 38 | negative_regulation_of_DNA_replication |
| GO:0008235 | 38 | metalloexopeptidase_activity |
| GO:0009066 | 38 | aspartate_family_amino_acid_metabolic_process |
| GO:0010676 | 38 | positive_regulation_of_cellular_carbohydrate_metabolic_process |
| GO:0016709 | 38 | oxidoreductase_activity,_acting_on_paired_donors,_with_incorporation_or_reduction_of_molecular_oxygen,_NADH_or_NADPH_as_one_donor,_and_incorporation_of_one_atom_of_oxygen |
| GO:0017144 | 38 | drug_metabolic_process |
| GO:0019212 | 38 | phosphatase_inhibitor_activity |
| GO:0019751 | 38 | polyol_metabolic_process |
| GO:0021987 | 38 | cerebral_cortex_development |
| GO:0022008 | 38 | neurogenesis |
| GO:0022600 | 38 | digestive_system_process |
| GO:0030049 | 38 | muscle_filament_sliding |
| GO:0030119 | 38 | AP-type_membrane_coat_adaptor_complex |
| GO:0031623 | 38 | receptor_internalization |
| GO:0032655 | 38 | regulation_of_interleukin-12_production |
| GO:0032994 | 38 | protein-lipid_complex |
| GO:0033138 | 38 | positive_regulation_of_peptidyl-serine_phosphorylation |
| GO:0033275 | 38 | actin-myosin_filament_sliding |
| GO:0035136 | 38 | forelimb_morphogenesis |
| GO:0042558 | 38 | pteridine-containing_compound_metabolic_process |
| GO:0043487 | 38 | regulation_of_RNA_stability |
| GO:0046324 | 38 | regulation_of_glucose_import |
| GO:0048592 | 38 | eye_morphogenesis |
| GO:0050911 | 38 | detection_of_chemical_stimulus_involved_in_sensory_perception_of_smell |
| GO:0060401 | 38 | cytosolic_calcium_ion_transport |
| GO:0002027 | 37 | regulation_of_heart_rate |
| GO:0002053 | 37 | positive_regulation_of_mesenchymal_cell_proliferation |
| GO:0004629 | 37 | phospholipase_C_activity |
| GO:0006323 | 37 | DNA_packaging |
| GO:0006776 | 37 | vitamin_A_metabolic_process |
| GO:0006885 | 37 | regulation_of_pH |
| GO:0007030 | 37 | Golgi_organization |
| GO:0007159 | 37 | leukocyte_cell-cell_adhesion |
| GO:0007369 | 37 | gastrulation |
| GO:0010821 | 37 | regulation_of_mitochondrion_organization |
| GO:0019825 | 37 | oxygen_binding |
| GO:0030195 | 37 | negative_regulation_of_blood_coagulation |
| GO:0030218 | 37 | erythrocyte_differentiation |
| GO:0030286 | 37 | dynein_complex |
| GO:0030295 | 37 | protein_kinase_activator_activity |
| GO:0031341 | 37 | regulation_of_cell_killing |
| GO:0031519 | 37 | PcG_protein_complex |
| GO:0032092 | 37 | positive_regulation_of_protein_binding |
| GO:0032154 | 37 | cleavage_furrow |
| GO:0032652 | 37 | regulation_of_interleukin-1_production |
| GO:0032677 | 37 | regulation_of_interleukin-8_production |
| GO:0034358 | 37 | plasma_lipoprotein_particle |
| GO:0034968 | 37 | histone_lysine_methylation |
| GO:0035051 | 37 | cardiac_cell_differentiation |
| GO:0035137 | 37 | hindlimb_morphogenesis |
| GO:0035586 | 37 | purinergic_receptor_activity |
| GO:0042147 | 37 | retrograde_transport,_endosome_to_Golgi |
| GO:0042752 | 37 | regulation_of_circadian_rhythm |
| GO:0043954 | 37 | cellular_component_maintenance |
| GO:0044259 | 37 | multicellular_organismal_macromolecule_metabolic_process |
| GO:0045839 | 37 | negative_regulation_of_mitosis |
| GO:0046320 | 37 | regulation_of_fatty_acid_oxidation |
| GO:0048588 | 37 | developmental_cell_growth |
| GO:0050764 | 37 | regulation_of_phagocytosis |
| GO:0051349 | 37 | positive_regulation_of_lyase_activity |
| GO:0051784 | 37 | negative_regulation_of_nuclear_division |
| GO:0060402 | 37 | calcium_ion_transport_into_cytosol |
| GO:0071804 | 37 | cellular_potassium_ion_transport |
| GO:0071805 | 37 | potassium_ion_transmembrane_transport |
| GO:1900047 | 37 | negative_regulation_of_hemostasis |
| GO:0004864 | 36 | protein_phosphatase_inhibitor_activity |
| GO:0005100 | 36 | Rho_GTPase_activator_activity |
| GO:0005501 | 36 | retinoid_binding |
| GO:0005834 | 36 | heterotrimeric_G-protein_complex |
| GO:0005884 | 36 | actin_filament |
| GO:0006284 | 36 | base-excision_repair |
| GO:0006901 | 36 | vesicle_coating |
| GO:0007602 | 36 | phototransduction |
| GO:0009712 | 36 | catechol-containing_compound_metabolic_process |
| GO:0010332 | 36 | response_to_gamma_radiation |
| GO:0021766 | 36 | hippocampus_development |
| GO:0022627 | 36 | cytosolic_small_ribosomal_subunit |
| GO:0030199 | 36 | collagen_fibril_organization |
| GO:0030890 | 36 | positive_regulation_of_B_cell_proliferation |
| GO:0031532 | 36 | actin_cytoskeleton_reorganization |
| GO:0031577 | 36 | spindle_checkpoint |
| GO:0034311 | 36 | diol_metabolic_process |
| GO:0035064 | 36 | methylated_histone_residue_binding |
| GO:0042246 | 36 | tissue_regeneration |
| GO:0042645 | 36 | mitochondrial_nucleoid |
| GO:0043488 | 36 | regulation_of_mRNA_stability |
| GO:0043535 | 36 | regulation_of_blood_vessel_endothelial_cell_migration |
| GO:0044275 | 36 | cellular_carbohydrate_catabolic_process |
| GO:0046165 | 36 | alcohol_biosynthetic_process |
| GO:0050709 | 36 | negative_regulation_of_protein_secretion |
| GO:0050771 | 36 | negative_regulation_of_axonogenesis |
| GO:0050805 | 36 | negative_regulation_of_synaptic_transmission |
| GO:0050840 | 36 | extracellular_matrix_binding |
| GO:0050885 | 36 | neuromuscular_process_controlling_balance |
| GO:0051225 | 36 | spindle_assembly |
| GO:0060079 | 36 | regulation_of_excitatory_postsynaptic_membrane_potential |
| GO:0071230 | 36 | cellular_response_to_amino_acid_stimulus |
| GO:0071377 | 36 | cellular_response_to_glucagon_stimulus |
| GO:2000177 | 36 | regulation_of_neural_precursor_cell_proliferation |
| GO:0001944 | 35 | vasculature_development |
| GO:0002709 | 35 | regulation_of_T_cell_mediated_immunity |
| GO:0002712 | 35 | regulation_of_B_cell_mediated_immunity |
| GO:0004712 | 35 | protein_serine/threonine/tyrosine_kinase_activity |
| GO:0005272 | 35 | sodium_channel_activity |
| GO:0006584 | 35 | catecholamine_metabolic_process |
| GO:0007257 | 35 | activation_of_JUN_kinase_activity |
| GO:0007632 | 35 | visual_behavior |
| GO:0008637 | 35 | apoptotic_mitochondrial_changes |
| GO:0009156 | 35 | ribonucleoside_monophosphate_biosynthetic_process |
| GO:0009898 | 35 | internal_side_of_plasma_membrane |
| GO:0010833 | 35 | telomere_maintenance_via_telomere_lengthening |
| GO:0016469 | 35 | proton-transporting_two-sector_ATPase_complex |
| GO:0016592 | 35 | mediator_complex |
| GO:0016891 | 35 | endoribonuclease_activity,_producing_5'-phosphomonoesters |
| GO:0018210 | 35 | peptidyl-threonine_modification |
| GO:0022406 | 35 | membrane_docking |
| GO:0022616 | 35 | DNA_strand_elongation |
| GO:0030838 | 35 | positive_regulation_of_actin_filament_polymerization |
| GO:0031281 | 35 | positive_regulation_of_cyclase_activity |
| GO:0032104 | 35 | regulation_of_response_to_extracellular_stimulus |
| GO:0032107 | 35 | regulation_of_response_to_nutrient_levels |
| GO:0033628 | 35 | regulation_of_cell_adhesion_mediated_by_integrin |
| GO:0035176 | 35 | social_behavior |
| GO:0042254 | 35 | ribosome_biogenesis |
| GO:0043094 | 35 | cellular_metabolic_compound_salvage |
| GO:0043292 | 35 | contractile_fiber |
| GO:0045333 | 35 | cellular_respiration |
| GO:0046131 | 35 | pyrimidine_ribonucleoside_metabolic_process |
| GO:0050999 | 35 | regulation_of_nitric-oxide_synthase_activity |
| GO:0070588 | 35 | calcium_ion_transmembrane_transport |
| GO:0070936 | 35 | protein_K48-linked_ubiquitination |
| GO:2000045 | 35 | regulation_of_G1/S_transition_of_mitotic_cell_cycle |
| GO:0001159 | 34 | core_promoter_proximal_region_DNA_binding |
| GO:0001656 | 34 | metanephros_development |
| GO:0001906 | 34 | cell_killing |
| GO:0002260 | 34 | lymphocyte_homeostasis |
| GO:0002889 | 34 | regulation_of_immunoglobulin_mediated_immune_response |
| GO:0004003 | 34 | ATP-dependent_DNA_helicase_activity |
| GO:0005775 | 34 | vacuolar_lumen |
| GO:0005881 | 34 | cytoplasmic_microtubule |
| GO:0006695 | 34 | cholesterol_biosynthetic_process |
| GO:0008060 | 34 | ARF_GTPase_activator_activity |
| GO:0008207 | 34 | C21-steroid_hormone_metabolic_process |
| GO:0008593 | 34 | regulation_of_Notch_signaling_pathway |
| GO:0010717 | 34 | regulation_of_epithelial_to_mesenchymal_transition |
| GO:0016504 | 34 | peptidase_activator_activity |
| GO:0016814 | 34 | hydrolase_activity,_acting_on_carbon-nitrogen_(but_not_peptide)_bonds,_in_cyclic_amidines |
| GO:0017046 | 34 | peptide_hormone_binding |
| GO:0030837 | 34 | negative_regulation_of_actin_filament_polymerization |
| GO:0032411 | 34 | positive_regulation_of_transporter_activity |
| GO:0032651 | 34 | regulation_of_interleukin-1_beta_production |
| GO:0042177 | 34 | negative_regulation_of_protein_catabolic_process |
| GO:0042345 | 34 | regulation_of_NF-kappaB_import_into_nucleus |
| GO:0045682 | 34 | regulation_of_epidermis_development |
| GO:0050909 | 34 | sensory_perception_of_taste |
| GO:0070206 | 34 | protein_trimerization |
| GO:0070613 | 34 | regulation_of_protein_processing |
| GO:2001257 | 34 | regulation_of_cation_channel_activity |
| GO:0000987 | 33 | core_promoter_proximal_region_sequence-specific_DNA_binding |
| GO:0001046 | 33 | core_promoter_sequence-specific_DNA_binding |
| GO:0001942 | 33 | hair_follicle_development |
| GO:0002064 | 33 | epithelial_cell_development |
| GO:0002637 | 33 | regulation_of_immunoglobulin_production |
| GO:0002763 | 33 | positive_regulation_of_myeloid_leukocyte_differentiation |
| GO:0005245 | 33 | voltage-gated_calcium_channel_activity |
| GO:0005891 | 33 | voltage-gated_calcium_channel_complex |
| GO:0006023 | 33 | aminoglycan_biosynthetic_process |
| GO:0006221 | 33 | pyrimidine_nucleotide_biosynthetic_process |
| GO:0006271 | 33 | DNA_strand_elongation_involved_in_DNA_replication |
| GO:0006305 | 33 | DNA_alkylation |
| GO:0006306 | 33 | DNA_methylation |
| GO:0006513 | 33 | protein_monoubiquitination |
| GO:0006730 | 33 | one-carbon_metabolic_process |
| GO:0006942 | 33 | regulation_of_striated_muscle_contraction |
| GO:0007006 | 33 | mitochondrial_membrane_organization |
| GO:0007044 | 33 | cell-substrate_junction_assembly |
| GO:0007566 | 33 | embryo_implantation |
| GO:0008376 | 33 | acetylgalactosaminyltransferase_activity |
| GO:0008378 | 33 | galactosyltransferase_activity |
| GO:0008408 | 33 | 3'-5'_exonuclease_activity |
| GO:0009109 | 33 | coenzyme_catabolic_process |
| GO:0014015 | 33 | positive_regulation_of_gliogenesis |
| GO:0015036 | 33 | disulfide_oxidoreductase_activity |
| GO:0019438 | 33 | aromatic_compound_biosynthetic_process |
| GO:0030131 | 33 | clathrin_adaptor_complex |
| GO:0030433 | 33 | ER-associated_protein_catabolic_process |
| GO:0030971 | 33 | receptor_tyrosine_kinase_binding |
| GO:0031058 | 33 | positive_regulation_of_histone_modification |
| GO:0032312 | 33 | regulation_of_ARF_GTPase_activity |
| GO:0032436 | 33 | positive_regulation_of_proteasomal_ubiquitin-dependent_protein_catabolic_process |
| GO:0032481 | 33 | positive_regulation_of_type_I_interferon_production |
| GO:0032881 | 33 | regulation_of_polysaccharide_metabolic_process |
| GO:0033692 | 33 | cellular_polysaccharide_biosynthetic_process |
| GO:0035019 | 33 | somatic_stem_cell_maintenance |
| GO:0042169 | 33 | SH2_domain_binding |
| GO:0045429 | 33 | positive_regulation_of_nitric_oxide_biosynthetic_process |
| GO:0045762 | 33 | positive_regulation_of_adenylate_cyclase_activity |
| GO:0045840 | 33 | positive_regulation_of_mitosis |
| GO:0045931 | 33 | positive_regulation_of_mitotic_cell_cycle |
| GO:0046658 | 33 | anchored_to_plasma_membrane |
| GO:0046686 | 33 | response_to_cadmium_ion |
| GO:0048286 | 33 | lung_alveolus_development |
| GO:0048706 | 33 | embryonic_skeletal_system_development |
| GO:0050830 | 33 | defense_response_to_Gram-positive_bacterium |
| GO:0050848 | 33 | regulation_of_calcium-mediated_signaling |
| GO:0051055 | 33 | negative_regulation_of_lipid_biosynthetic_process |
| GO:0051785 | 33 | positive_regulation_of_nuclear_division |
| GO:0055038 | 33 | recycling_endosome_membrane |
| GO:0060271 | 33 | cilium_morphogenesis |
| GO:0071174 | 33 | mitotic_cell_cycle_spindle_checkpoint |
| GO:0071396 | 33 | cellular_response_to_lipid |
| GO:0071482 | 33 | cellular_response_to_light_stimulus |
| GO:0071559 | 33 | response_to_transforming_growth_factor_beta_stimulus |
| GO:0001614 | 32 | purinergic_nucleotide_receptor_activity |
| GO:0001707 | 32 | mesoderm_formation |
| GO:0001910 | 32 | regulation_of_leukocyte_mediated_cytotoxicity |
| GO:0002062 | 32 | chondrocyte_differentiation |
| GO:0002920 | 32 | regulation_of_humoral_immune_response |
| GO:0005782 | 32 | peroxisomal_matrix |
| GO:0005795 | 32 | Golgi_stack |
| GO:0006312 | 32 | mitotic_recombination |
| GO:0006488 | 32 | dolichol-linked_oligosaccharide_biosynthetic_process |
| GO:0006891 | 32 | intra-Golgi_vesicle-mediated_transport |
| GO:0007292 | 32 | female_gamete_generation |
| GO:0010389 | 32 | regulation_of_G2/M_transition_of_mitotic_cell_cycle |
| GO:0015909 | 32 | long-chain_fatty_acid_transport |
| GO:0016502 | 32 | nucleotide_receptor_activity |
| GO:0016831 | 32 | carboxy-lyase_activity |
| GO:0018107 | 32 | peptidyl-threonine_phosphorylation |
| GO:0019370 | 32 | leukotriene_biosynthetic_process |
| GO:0019748 | 32 | secondary_metabolic_process |
| GO:0019829 | 32 | cation-transporting_ATPase_activity |
| GO:0019843 | 32 | rRNA_binding |
| GO:0019905 | 32 | syntaxin_binding |
| GO:0022409 | 32 | positive_regulation_of_cell-cell_adhesion |
| GO:0030120 | 32 | vesicle_coat |
| GO:0030532 | 32 | small_nuclear_ribonucleoprotein_complex |
| GO:0031343 | 32 | positive_regulation_of_cell_killing |
| GO:0031572 | 32 | G2/M_transition_DNA_damage_checkpoint |
| GO:0031907 | 32 | microbody_lumen |
| GO:0032006 | 32 | regulation_of_TOR_signaling_cascade |
| GO:0032091 | 32 | negative_regulation_of_protein_binding |
| GO:0032233 | 32 | positive_regulation_of_actin_filament_bundle_assembly |
| GO:0032371 | 32 | regulation_of_sterol_transport |
| GO:0032374 | 32 | regulation_of_cholesterol_transport |
| GO:0032392 | 32 | DNA_geometric_change |
| GO:0032480 | 32 | negative_regulation_of_type_I_interferon_production |
| GO:0032755 | 32 | positive_regulation_of_interleukin-6_production |
| GO:0032963 | 32 | collagen_metabolic_process |
| GO:0043392 | 32 | negative_regulation_of_DNA_binding |
| GO:0043450 | 32 | alkene_biosynthetic_process |
| GO:0043621 | 32 | protein_self-association |
| GO:0045841 | 32 | negative_regulation_of_mitotic_metaphase/anaphase_transition |
| GO:0045921 | 32 | positive_regulation_of_exocytosis |
| GO:0046058 | 32 | cAMP_metabolic_process |
| GO:0046631 | 32 | alpha-beta_T_cell_activation |
| GO:0046834 | 32 | lipid_phosphorylation |
| GO:0048489 | 32 | synaptic_vesicle_transport |
| GO:0051353 | 32 | positive_regulation_of_oxidoreductase_activity |
| GO:0055010 | 32 | ventricular_cardiac_muscle_tissue_morphogenesis |
| GO:0055024 | 32 | regulation_of_cardiac_muscle_tissue_development |
| GO:0070491 | 32 | repressing_transcription_factor_binding |
| GO:0070830 | 32 | tight_junction_assembly |
| GO:0070997 | 32 | neuron_death |
| GO:0072529 | 32 | pyrimidine-containing_compound_catabolic_process |
| GO:1900674 | 32 | olefin_biosynthetic_process |
| GO:1901071 | 32 | glucosamine-containing_compound_metabolic_process |
| GO:2001251 | 32 | negative_regulation_of_chromosome_organization |
| GO:0002562 | 31 | somatic_diversification_of_immune_receptors_via_germline_recombination_within_a_single_locus |
| GO:0002753 | 31 | cytoplasmic_pattern_recognition_receptor_signaling_pathway |
| GO:0002762 | 31 | negative_regulation_of_myeloid_leukocyte_differentiation |
| GO:0004693 | 31 | cyclin-dependent_protein_kinase_activity |
| GO:0006024 | 31 | glycosaminoglycan_biosynthetic_process |
| GO:0006206 | 31 | pyrimidine_base_metabolic_process |
| GO:0006801 | 31 | superoxide_metabolic_process |
| GO:0006997 | 31 | nucleus_organization |
| GO:0007131 | 31 | reciprocal_meiotic_recombination |
| GO:0008154 | 31 | actin_polymerization_or_depolymerization |
| GO:0008633 | 31 | activation_of_pro-apoptotic_gene_products |
| GO:0009069 | 31 | serine_family_amino_acid_metabolic_process |
| GO:0009101 | 31 | glycoprotein_biosynthetic_process |
| GO:0009262 | 31 | deoxyribonucleotide_metabolic_process |
| GO:0009452 | 31 | 7-methylguanosine_RNA_capping |
| GO:0009948 | 31 | anterior/posterior_axis_specification |
| GO:0010883 | 31 | regulation_of_lipid_storage |
| GO:0015296 | 31 | anion:cation_symporter_activity |
| GO:0015914 | 31 | phospholipid_transport |
| GO:0015988 | 31 | energy_coupled_proton_transport,_against_electrochemical_gradient |
| GO:0015991 | 31 | ATP_hydrolysis_coupled_proton_transport |
| GO:0016234 | 31 | inclusion_body |
| GO:0016444 | 31 | somatic_cell_DNA_recombination |
| GO:0016675 | 31 | oxidoreductase_activity,_acting_on_a_heme_group_of_donors |
| GO:0019213 | 31 | deacetylase_activity |
| GO:0032760 | 31 | positive_regulation_of_tumor_necrosis_factor_production |
| GO:0032885 | 31 | regulation_of_polysaccharide_biosynthetic_process |
| GO:0032934 | 31 | sterol_binding |
| GO:0035094 | 31 | response_to_nicotine |
| GO:0035115 | 31 | embryonic_forelimb_morphogenesis |
| GO:0035825 | 31 | reciprocal_DNA_recombination |
| GO:0036260 | 31 | RNA_capping |
| GO:0042451 | 31 | purine_nucleoside_biosynthetic_process |
| GO:0042455 | 31 | ribonucleoside_biosynthetic_process |
| GO:0044306 | 31 | neuron_projection_terminus |
| GO:0045069 | 31 | regulation_of_viral_genome_replication |
| GO:0045646 | 31 | regulation_of_erythrocyte_differentiation |
| GO:0045668 | 31 | negative_regulation_of_osteoblast_differentiation |
| GO:0045776 | 31 | negative_regulation_of_blood_pressure |
| GO:0045861 | 31 | negative_regulation_of_proteolysis |
| GO:0046129 | 31 | purine_ribonucleoside_biosynthetic_process |
| GO:0046854 | 31 | phosphatidylinositol_phosphorylation |
| GO:0048284 | 31 | organelle_fusion |
| GO:0048538 | 31 | thymus_development |
| GO:0048844 | 31 | artery_morphogenesis |
| GO:0050931 | 31 | pigment_cell_differentiation |
| GO:0051017 | 31 | actin_filament_bundle_assembly |
| GO:0051963 | 31 | regulation_of_synapse_assembly |
| GO:0060338 | 31 | regulation_of_type_I_interferon-mediated_signaling_pathway |
| GO:0070169 | 31 | positive_regulation_of_biomineral_tissue_development |
| GO:0071173 | 31 | spindle_assembly_checkpoint |
| GO:0072175 | 31 | epithelial_tube_formation |
| GO:0000049 | 30 | tRNA_binding |
| GO:0001750 | 30 | photoreceptor_outer_segment |
| GO:0001838 | 30 | embryonic_epithelial_tube_formation |
| GO:0001974 | 30 | blood_vessel_remodeling |
| GO:0003007 | 30 | heart_morphogenesis |
| GO:0004129 | 30 | cytochrome-c_oxidase_activity |
| GO:0004435 | 30 | phosphatidylinositol_phospholipase_C_activity |
| GO:0006370 | 30 | 7-methylguanosine_mRNA_capping |
| GO:0006692 | 30 | prostanoid_metabolic_process |
| GO:0007040 | 30 | lysosome_organization |
| GO:0007094 | 30 | mitotic_cell_cycle_spindle_assembly_checkpoint |
| GO:0007190 | 30 | activation_of_adenylate_cyclase_activity |
| GO:0008180 | 30 | signalosome |
| GO:0008305 | 30 | integrin_complex |
| GO:0008542 | 30 | visual_learning |
| GO:0009190 | 30 | cyclic_nucleotide_biosynthetic_process |
| GO:0009620 | 30 | response_to_fungus |
| GO:0010977 | 30 | negative_regulation_of_neuron_projection_development |
| GO:0015002 | 30 | heme-copper_terminal_oxidase_activity |
| GO:0015020 | 30 | glucuronosyltransferase_activity |
| GO:0015682 | 30 | ferric_iron_transport |
| GO:0015695 | 30 | organic_cation_transport |
| GO:0016208 | 30 | AMP_binding |
| GO:0016229 | 30 | steroid_dehydrogenase_activity |
| GO:0016620 | 30 | oxidoreductase_activity,_acting_on_the_aldehyde_or_oxo_group_of_donors,_NAD_or_NADP_as_acceptor |
| GO:0016676 | 30 | oxidoreductase_activity,_acting_on_a_heme_group_of_donors,_oxygen_as_acceptor |
| GO:0017156 | 30 | calcium_ion-dependent_exocytosis |
| GO:0018149 | 30 | peptide_cross-linking |
| GO:0018410 | 30 | C-terminal_protein_amino_acid_modification |
| GO:0022617 | 30 | extracellular_matrix_disassembly |
| GO:0030073 | 30 | insulin_secretion |
| GO:0030261 | 30 | chromosome_condensation |
| GO:0030279 | 30 | negative_regulation_of_ossification |
| GO:0030318 | 30 | melanocyte_differentiation |
| GO:0030513 | 30 | positive_regulation_of_BMP_signaling_pathway |
| GO:0030514 | 30 | negative_regulation_of_BMP_signaling_pathway |
| GO:0030670 | 30 | phagocytic_vesicle_membrane |
| GO:0030901 | 30 | midbrain_development |
| GO:0032330 | 30 | regulation_of_chondrocyte_differentiation |
| GO:0032508 | 30 | DNA_duplex_unwinding |
| GO:0032729 | 30 | positive_regulation_of_interferon-gamma_production |
| GO:0032784 | 30 | regulation_of_DNA-dependent_transcription,_elongation |
| GO:0033017 | 30 | sarcoplasmic_reticulum_membrane |
| GO:0033261 | 30 | regulation_of_S_phase |
| GO:0033572 | 30 | transferrin_transport |
| GO:0042098 | 30 | T_cell_proliferation |
| GO:0042743 | 30 | hydrogen_peroxide_metabolic_process |
| GO:0042993 | 30 | positive_regulation_of_transcription_factor_import_into_nucleus |
| GO:0043198 | 30 | dendritic_shaft |
| GO:0043202 | 30 | lysosomal_lumen |
| GO:0043550 | 30 | regulation_of_lipid_kinase_activity |
| GO:0044325 | 30 | ion_channel_binding |
| GO:0045599 | 30 | negative_regulation_of_fat_cell_differentiation |
| GO:0045980 | 30 | negative_regulation_of_nucleotide_metabolic_process |
| GO:0046638 | 30 | positive_regulation_of_alpha-beta_T_cell_differentiation |
| GO:0046966 | 30 | thyroid_hormone_receptor_binding |
| GO:0051539 | 30 | 4_iron,_4_sulfur_cluster_binding |
| GO:0051865 | 30 | protein_autoubiquitination |
| GO:0051966 | 30 | regulation_of_synaptic_transmission,_glutamatergic |
| GO:0060393 | 30 | regulation_of_pathway-restricted_SMAD_protein_phosphorylation |
| GO:0060411 | 30 | cardiac_septum_morphogenesis |
| GO:0001569 | 29 | patterning_of_blood_vessels |
| GO:0003887 | 29 | DNA-directed_DNA_polymerase_activity |
| GO:0004520 | 29 | endodeoxyribonuclease_activity |
| GO:0004623 | 29 | phospholipase_A2_activity |
| GO:0005158 | 29 | insulin_receptor_binding |
| GO:0005544 | 29 | calcium-dependent_phospholipid_binding |
| GO:0005546 | 29 | phosphatidylinositol-4,5-bisphosphate_binding |
| GO:0005548 | 29 | phospholipid_transporter_activity |
| GO:0005798 | 29 | Golgi-associated_vesicle |
| GO:0006505 | 29 | GPI_anchor_metabolic_process |
| GO:0007338 | 29 | single_fertilization |
| GO:0008023 | 29 | transcription_elongation_factor_complex |
| GO:0008038 | 29 | neuron_recognition |
| GO:0008209 | 29 | androgen_metabolic_process |
| GO:0008328 | 29 | ionotropic_glutamate_receptor_complex |
| GO:0008334 | 29 | histone_mRNA_metabolic_process |
| GO:0009072 | 29 | aromatic_amino_acid_family_metabolic_process |
| GO:0009409 | 29 | response_to_cold |
| GO:0009925 | 29 | basal_plasma_membrane |
| GO:0010812 | 29 | negative_regulation_of_cell-substrate_adhesion |
| GO:0010828 | 29 | positive_regulation_of_glucose_transport |
| GO:0015459 | 29 | potassium_channel_regulator_activity |
| GO:0016328 | 29 | lateral_plasma_membrane |
| GO:0016445 | 29 | somatic_diversification_of_immunoglobulins |
| GO:0016712 | 29 | oxidoreductase_activity,_acting_on_paired_donors,_with_incorporation_or_reduction_of_molecular_oxygen,_reduced_flavin_or_flavoprotein_as_one_donor,_and_incorporation_of_one_atom_of_oxygen |
| GO:0016877 | 29 | ligase_activity,_forming_carbon-sulfur_bonds |
| GO:0019439 | 29 | aromatic_compound_catabolic_process |
| GO:0021885 | 29 | forebrain_cell_migration |
| GO:0030501 | 29 | positive_regulation_of_bone_mineralization |
| GO:0030546 | 29 | receptor_activator_activity |
| GO:0030551 | 29 | cyclic_nucleotide_binding |
| GO:0030593 | 29 | neutrophil_chemotaxis |
| GO:0030834 | 29 | regulation_of_actin_filament_depolymerization |
| GO:0033619 | 29 | membrane_protein_proteolysis |
| GO:0033865 | 29 | nucleoside_bisphosphate_metabolic_process |
| GO:0035116 | 29 | embryonic_hindlimb_morphogenesis |
| GO:0035690 | 29 | cellular_response_to_drug |
| GO:0040018 | 29 | positive_regulation_of_multicellular_organism_growth |
| GO:0042287 | 29 | MHC_protein_binding |
| GO:0042304 | 29 | regulation_of_fatty_acid_biosynthetic_process |
| GO:0042516 | 29 | regulation_of_tyrosine_phosphorylation_of_Stat3_protein |
| GO:0042992 | 29 | negative_regulation_of_transcription_factor_import_into_nucleus |
| GO:0043044 | 29 | ATP-dependent_chromatin_remodeling |
| GO:0043388 | 29 | positive_regulation_of_DNA_binding |
| GO:0043473 | 29 | pigmentation |
| GO:0045620 | 29 | negative_regulation_of_lymphocyte_differentiation |
| GO:0046520 | 29 | sphingoid_biosynthetic_process |
| GO:0046850 | 29 | regulation_of_bone_remodeling |
| GO:0046928 | 29 | regulation_of_neurotransmitter_secretion |
| GO:0048024 | 29 | regulation_of_nuclear_mRNA_splicing,_via_spliceosome |
| GO:0048278 | 29 | vesicle_docking |
| GO:0048675 | 29 | axon_extension |
| GO:0050732 | 29 | negative_regulation_of_peptidyl-tyrosine_phosphorylation |
| GO:0050853 | 29 | B_cell_receptor_signaling_pathway |
| GO:0051148 | 29 | negative_regulation_of_muscle_cell_differentiation |
| GO:0051279 | 29 | regulation_of_release_of_sequestered_calcium_ion_into_cytosol |
| GO:0051702 | 29 | interaction_with_symbiont |
| GO:0060761 | 29 | negative_regulation_of_response_to_cytokine_stimulus |
| GO:0061351 | 29 | neural_precursor_cell_proliferation |
| GO:0070873 | 29 | regulation_of_glycogen_metabolic_process |
| GO:0071560 | 29 | cellular_response_to_transforming_growth_factor_beta_stimulus |
| GO:1900076 | 29 | regulation_of_cellular_response_to_insulin_stimulus |
| GO:2000514 | 29 | regulation_of_CD4-positive,_alpha-beta_T_cell_activation |
| GO:2000756 | 29 | regulation_of_peptidyl-lysine_acetylation |
| GO:2001235 | 29 | positive_regulation_of_apoptotic_signaling_pathway |
| GO:0000578 | 28 | embryonic_axis_specification |
| GO:0001578 | 28 | microtubule_bundle_formation |
| GO:0001676 | 28 | long-chain_fatty_acid_metabolic_process |
| GO:0002792 | 28 | negative_regulation_of_peptide_secretion |
| GO:0003724 | 28 | RNA_helicase_activity |
| GO:0003746 | 28 | translation_elongation_factor_activity |
| GO:0005164 | 28 | tumor_necrosis_factor_receptor_binding |
| GO:0005791 | 28 | rough_endoplasmic_reticulum |
| GO:0006090 | 28 | pyruvate_metabolic_process |
| GO:0006779 | 28 | porphyrin-containing_compound_biosynthetic_process |
| GO:0007628 | 28 | adult_walking_behavior |
| GO:0008173 | 28 | RNA_methyltransferase_activity |
| GO:0009218 | 28 | pyrimidine_ribonucleotide_metabolic_process |
| GO:0010226 | 28 | response_to_lithium_ion |
| GO:0010524 | 28 | positive_regulation_of_calcium_ion_transport_into_cytosol |
| GO:0010907 | 28 | positive_regulation_of_glucose_metabolic_process |
| GO:0014014 | 28 | negative_regulation_of_gliogenesis |
| GO:0015035 | 28 | protein_disulfide_oxidoreductase_activity |
| GO:0016574 | 28 | histone_ubiquitination |
| GO:0016866 | 28 | intramolecular_transferase_activity |
| GO:0017022 | 28 | myosin_binding |
| GO:0019400 | 28 | alditol_metabolic_process |
| GO:0019674 | 28 | NAD_metabolic_process |
| GO:0030016 | 28 | myofibril |
| GO:0030282 | 28 | bone_mineralization |
| GO:0030317 | 28 | sperm_motility |
| GO:0030331 | 28 | estrogen_receptor_binding |
| GO:0030809 | 28 | negative_regulation_of_nucleotide_biosynthetic_process |
| GO:0030858 | 28 | positive_regulation_of_epithelial_cell_differentiation |
| GO:0031111 | 28 | negative_regulation_of_microtubule_polymerization_or_depolymerization |
| GO:0031663 | 28 | lipopolysaccharide-mediated_signaling_pathway |
| GO:0032410 | 28 | negative_regulation_of_transporter_activity |
| GO:0032570 | 28 | response_to_progesterone_stimulus |
| GO:0032592 | 28 | integral_to_mitochondrial_membrane |
| GO:0032720 | 28 | negative_regulation_of_tumor_necrosis_factor_production |
| GO:0033014 | 28 | tetrapyrrole_biosynthetic_process |
| GO:0033077 | 28 | T_cell_differentiation_in_thymus |
| GO:0034453 | 28 | microtubule_anchoring |
| GO:0034644 | 28 | cellular_response_to_UV |
| GO:0035601 | 28 | protein_deacylation |
| GO:0042168 | 28 | heme_metabolic_process |
| GO:0042461 | 28 | photoreceptor_cell_development |
| GO:0043526 | 28 | neuroprotection |
| GO:0044243 | 28 | multicellular_organismal_catabolic_process |
| GO:0046356 | 28 | acetyl-CoA_catabolic_process |
| GO:0046470 | 28 | phosphatidylcholine_metabolic_process |
| GO:0046626 | 28 | regulation_of_insulin_receptor_signaling_pathway |
| GO:0048260 | 28 | positive_regulation_of_receptor-mediated_endocytosis |
| GO:0048485 | 28 | sympathetic_nervous_system_development |
| GO:0048521 | 28 | negative_regulation_of_behavior |
| GO:0050433 | 28 | regulation_of_catecholamine_secretion |
| GO:0050690 | 28 | regulation_of_defense_response_to_virus_by_virus |
| GO:0050766 | 28 | positive_regulation_of_phagocytosis |
| GO:0051057 | 28 | positive_regulation_of_small_GTPase_mediated_signal_transduction |
| GO:0051282 | 28 | regulation_of_sequestering_of_calcium_ion |
| GO:0051385 | 28 | response_to_mineralocorticoid_stimulus |
| GO:0051496 | 28 | positive_regulation_of_stress_fiber_assembly |
| GO:0060048 | 28 | cardiac_muscle_contraction |
| GO:0060420 | 28 | regulation_of_heart_growth |
| GO:0070228 | 28 | regulation_of_lymphocyte_apoptotic_process |
| GO:0070888 | 28 | E-box_binding |
| GO:0071339 | 28 | MLL1_complex |
| GO:0071384 | 28 | cellular_response_to_corticosteroid_stimulus |
| GO:0071479 | 28 | cellular_response_to_ionizing_radiation |
| GO:0090199 | 28 | regulation_of_release_of_cytochrome_c_from_mitochondria |
| GO:0090287 | 28 | regulation_of_cellular_response_to_growth_factor_stimulus |
| GO:1900372 | 28 | negative_regulation_of_purine_nucleotide_biosynthetic_process |
| GO:1900543 | 28 | negative_regulation_of_purine_nucleotide_metabolic_process |
| GO:0000794 | 27 | condensed_nuclear_chromosome |
| GO:0001678 | 27 | cellular_glucose_homeostasis |
| GO:0001912 | 27 | positive_regulation_of_leukocyte_mediated_cytotoxicity |
| GO:0001975 | 27 | response_to_amphetamine |
| GO:0003151 | 27 | outflow_tract_morphogenesis |
| GO:0005520 | 27 | insulin-like_growth_factor_binding |
| GO:0005637 | 27 | nuclear_inner_membrane |
| GO:0005720 | 27 | nuclear_heterochromatin |
| GO:0005979 | 27 | regulation_of_glycogen_biosynthetic_process |
| GO:0006099 | 27 | tricarboxylic_acid_cycle |
| GO:0006476 | 27 | protein_deacetylation |
| GO:0006904 | 27 | vesicle_docking_involved_in_exocytosis |
| GO:0007492 | 27 | endoderm_development |
| GO:0009126 | 27 | purine_nucleoside_monophosphate_metabolic_process |
| GO:0010743 | 27 | regulation_of_macrophage_derived_foam_cell_differentiation |
| GO:0010962 | 27 | regulation_of_glucan_biosynthetic_process |
| GO:0014910 | 27 | regulation_of_smooth_muscle_cell_migration |
| GO:0015144 | 27 | carbohydrate_transmembrane_transporter_activity |
| GO:0016529 | 27 | sarcoplasmic_reticulum |
| GO:0016645 | 27 | oxidoreductase_activity,_acting_on_the_CH-NH_group_of_donors |
| GO:0019239 | 27 | deaminase_activity |
| GO:0021515 | 27 | cell_differentiation_in_spinal_cord |
| GO:0030800 | 27 | negative_regulation_of_cyclic_nucleotide_metabolic_process |
| GO:0030803 | 27 | negative_regulation_of_cyclic_nucleotide_biosynthetic_process |
| GO:0031069 | 27 | hair_follicle_morphogenesis |
| GO:0032414 | 27 | positive_regulation_of_ion_transmembrane_transporter_activity |
| GO:0032722 | 27 | positive_regulation_of_chemokine_production |
| GO:0032890 | 27 | regulation_of_organic_acid_transport |
| GO:0033209 | 27 | tumor_necrosis_factor-mediated_signaling_pathway |
| GO:0034121 | 27 | regulation_of_toll-like_receptor_signaling_pathway |
| GO:0035065 | 27 | regulation_of_histone_acetylation |
| GO:0035567 | 27 | non-canonical_Wnt_receptor_signaling_pathway |
| GO:0042036 | 27 | negative_regulation_of_cytokine_biosynthetic_process |
| GO:0043022 | 27 | ribosome_binding |
| GO:0043331 | 27 | response_to_dsRNA |
| GO:0043370 | 27 | regulation_of_CD4-positive,_alpha-beta_T_cell_differentiation |
| GO:0043470 | 27 | regulation_of_carbohydrate_catabolic_process |
| GO:0043471 | 27 | regulation_of_cellular_carbohydrate_catabolic_process |
| GO:0044452 | 27 | nucleolar_part |
| GO:0045103 | 27 | intermediate_filament-based_process |
| GO:0045777 | 27 | positive_regulation_of_blood_pressure |
| GO:0045923 | 27 | positive_regulation_of_fatty_acid_metabolic_process |
| GO:0046209 | 27 | nitric_oxide_metabolic_process |
| GO:0046632 | 27 | alpha-beta_T_cell_differentiation |
| GO:0046847 | 27 | filopodium_assembly |
| GO:0048663 | 27 | neuron_fate_commitment |
| GO:0048871 | 27 | multicellular_organismal_homeostasis |
| GO:0051004 | 27 | regulation_of_lipoprotein_lipase_activity |
| GO:0051058 | 27 | negative_regulation_of_small_GTPase_mediated_signal_transduction |
| GO:0051209 | 27 | release_of_sequestered_calcium_ion_into_cytosol |
| GO:0051283 | 27 | negative_regulation_of_sequestering_of_calcium_ion |
| GO:0051602 | 27 | response_to_electrical_stimulus |
| GO:0060325 | 27 | face_morphogenesis |
| GO:0060485 | 27 | mesenchyme_development |
| GO:0070423 | 27 | nucleotide-binding_oligomerization_domain_containing_signaling_pathway |
| GO:0070461 | 27 | SAGA-type_complex |
| GO:0071277 | 27 | cellular_response_to_calcium_ion |
| GO:0071385 | 27 | cellular_response_to_glucocorticoid_stimulus |
| GO:0071825 | 27 | protein-lipid_complex_subunit_organization |
| GO:0071827 | 27 | plasma_lipoprotein_particle_organization |
| GO:0072332 | 27 | signal_transduction_by_p53_class_mediator_resulting_in_induction_of_apoptosis |
| GO:0090278 | 27 | negative_regulation_of_peptide_hormone_secretion |
| GO:2000379 | 27 | positive_regulation_of_reactive_oxygen_species_metabolic_process |
| GO:0000291 | 26 | nuclear-transcribed_mRNA_catabolic_process,_exonucleolytic |
| GO:0000387 | 26 | spliceosomal_snRNP_assembly |
| GO:0000722 | 26 | telomere_maintenance_via_recombination |
| GO:0000737 | 26 | DNA_catabolic_process,_endonucleolytic |
| GO:0001190 | 26 | RNA_polymerase_II_transcription_factor_binding_transcription_factor_activity_involved_in_positive_regulation_of_transcription |
| GO:0002275 | 26 | myeloid_cell_activation_involved_in_immune_response |
| GO:0002711 | 26 | positive_regulation_of_T_cell_mediated_immunity |
| GO:0003950 | 26 | NAD+_ADP-ribosyltransferase_activity |
| GO:0004112 | 26 | cyclic-nucleotide_phosphodiesterase_activity |
| GO:0006071 | 26 | glycerol_metabolic_process |
| GO:0006506 | 26 | GPI_anchor_biosynthetic_process |
| GO:0007157 | 26 | heterophilic_cell-cell_adhesion |
| GO:0007212 | 26 | dopamine_receptor_signaling_pathway |
| GO:0008066 | 26 | glutamate_receptor_activity |
| GO:0008333 | 26 | endosome_to_lysosome_transport |
| GO:0008361 | 26 | regulation_of_cell_size |
| GO:0009167 | 26 | purine_ribonucleoside_monophosphate_metabolic_process |
| GO:0009220 | 26 | pyrimidine_ribonucleotide_biosynthetic_process |
| GO:0009595 | 26 | detection_of_biotic_stimulus |
| GO:0010470 | 26 | regulation_of_gastrulation |
| GO:0014072 | 26 | response_to_isoquinoline_alkaloid |
| GO:0015074 | 26 | DNA_integration |
| GO:0016331 | 26 | morphogenesis_of_embryonic_epithelium |
| GO:0016339 | 26 | calcium-dependent_cell-cell_adhesion |
| GO:0016358 | 26 | dendrite_development |
| GO:0017147 | 26 | Wnt-protein_binding |
| GO:0019003 | 26 | GDP_binding |
| GO:0021675 | 26 | nerve_development |
| GO:0022029 | 26 | telencephalon_cell_migration |
| GO:0022408 | 26 | negative_regulation_of_cell-cell_adhesion |
| GO:0030004 | 26 | cellular_monovalent_inorganic_cation_homeostasis |
| GO:0030315 | 26 | T-tubule |
| GO:0030815 | 26 | negative_regulation_of_cAMP_metabolic_process |
| GO:0030818 | 26 | negative_regulation_of_cAMP_biosynthetic_process |
| GO:0030880 | 26 | RNA_polymerase_complex |
| GO:0031016 | 26 | pancreas_development |
| GO:0031952 | 26 | regulation_of_protein_autophosphorylation |
| GO:0033116 | 26 | endoplasmic_reticulum-Golgi_intermediate_compartment_membrane |
| GO:0033574 | 26 | response_to_testosterone_stimulus |
| GO:0033764 | 26 | steroid_dehydrogenase_activity,_acting_on_the_CH-OH_group_of_donors,_NAD_or_NADP_as_acceptor |
| GO:0035326 | 26 | enhancer_binding |
| GO:0042312 | 26 | regulation_of_vasodilation |
| GO:0042462 | 26 | eye_photoreceptor_cell_development |
| GO:0042612 | 26 | MHC_class_I_protein_complex |
| GO:0042755 | 26 | eating_behavior |
| GO:0043278 | 26 | response_to_morphine |
| GO:0043486 | 26 | histone_exchange |
| GO:0043666 | 26 | regulation_of_phosphoprotein_phosphatase_activity |
| GO:0043928 | 26 | exonucleolytic_nuclear-transcribed_mRNA_catabolic_process_involved_in_deadenylation-dependent_decay |
| GO:0044423 | 26 | virion_part |
| GO:0044447 | 26 | axoneme_part |
| GO:0045104 | 26 | intermediate_filament_cytoskeleton_organization |
| GO:0045124 | 26 | regulation_of_bone_resorption |
| GO:0045739 | 26 | positive_regulation_of_DNA_repair |
| GO:0046326 | 26 | positive_regulation_of_glucose_import |
| GO:0046580 | 26 | negative_regulation_of_Ras_protein_signal_transduction |
| GO:0048008 | 26 | platelet-derived_growth_factor_receptor_signaling_pathway |
| GO:0048013 | 26 | ephrin_receptor_signaling_pathway |
| GO:0050854 | 26 | regulation_of_antigen_receptor-mediated_signaling_pathway |
| GO:0050869 | 26 | negative_regulation_of_B_cell_activation |
| GO:0050873 | 26 | brown_fat_cell_differentiation |
| GO:0051117 | 26 | ATPase_binding |
| GO:0051145 | 26 | smooth_muscle_cell_differentiation |
| GO:0051402 | 26 | neuron_apoptotic_process |
| GO:0051851 | 26 | modification_by_host_of_symbiont_morphology_or_physiology |
| GO:0051926 | 26 | negative_regulation_of_calcium_ion_transport |
| GO:0055007 | 26 | cardiac_muscle_cell_differentiation |
| GO:0061098 | 26 | positive_regulation_of_protein_tyrosine_kinase_activity |
| GO:0070979 | 26 | protein_K11-linked_ubiquitination |
| GO:0071322 | 26 | cellular_response_to_carbohydrate_stimulus |
| GO:2000107 | 26 | negative_regulation_of_leukocyte_apoptotic_process |
| GO:0000038 | 25 | very_long-chain_fatty_acid_metabolic_process |
| GO:0000152 | 25 | nuclear_ubiquitin_ligase_complex |
| GO:0000188 | 25 | inactivation_of_MAPK_activity |
| GO:0000289 | 25 | nuclear-transcribed_mRNA_poly(A)_tail_shortening |
| GO:0000428 | 25 | DNA-directed_RNA_polymerase_complex |
| GO:0001078 | 25 | RNA_polymerase_II_core_promoter_proximal_region_sequence-specific_DNA_binding_transcription_factor_activity_involved_in_negative_regulation_of_transcription |
| GO:0001102 | 25 | RNA_polymerase_II_activating_transcription_factor_binding |
| GO:0001227 | 25 | RNA_polymerase_II_transcription_regulatory_region_sequence-specific_DNA_binding_transcription_factor_activity_involved_in_negative_regulation_of_transcription |
| GO:0001540 | 25 | beta-amyloid_binding |
| GO:0001608 | 25 | G-protein_coupled_nucleotide_receptor_activity |
| GO:0001637 | 25 | G-protein_coupled_chemoattractant_receptor_activity |
| GO:0001954 | 25 | positive_regulation_of_cell-matrix_adhesion |
| GO:0001960 | 25 | negative_regulation_of_cytokine-mediated_signaling_pathway |
| GO:0003156 | 25 | regulation_of_organ_formation |
| GO:0004114 | 25 | 3',5'-cyclic-nucleotide_phosphodiesterase_activity |
| GO:0004364 | 25 | glutathione_transferase_activity |
| GO:0004950 | 25 | chemokine_receptor_activity |
| GO:0005109 | 25 | frizzled_binding |
| GO:0005504 | 25 | fatty_acid_binding |
| GO:0005801 | 25 | cis-Golgi_network |
| GO:0006026 | 25 | aminoglycan_catabolic_process |
| GO:0006041 | 25 | glucosamine_metabolic_process |
| GO:0006044 | 25 | N-acetylglucosamine_metabolic_process |
| GO:0006298 | 25 | mismatch_repair |
| GO:0006309 | 25 | apoptotic_DNA_fragmentation |
| GO:0006501 | 25 | C-terminal_protein_lipidation |
| GO:0006586 | 25 | indolalkylamine_metabolic_process |
| GO:0006760 | 25 | folic_acid-containing_compound_metabolic_process |
| GO:0007215 | 25 | glutamate_receptor_signaling_pathway |
| GO:0009125 | 25 | nucleoside_monophosphate_catabolic_process |
| GO:0009147 | 25 | pyrimidine_nucleoside_triphosphate_metabolic_process |
| GO:0009225 | 25 | nucleotide-sugar_metabolic_process |
| GO:0014073 | 25 | response_to_tropane |
| GO:0015298 | 25 | solute:cation_antiporter_activity |
| GO:0016628 | 25 | oxidoreductase_activity,_acting_on_the_CH-CH_group_of_donors,_NAD_or_NADP_as_acceptor |
| GO:0016755 | 25 | transferase_activity,_transferring_amino-acyl_groups |
| GO:0016769 | 25 | transferase_activity,_transferring_nitrogenous_groups |
| GO:0019005 | 25 | SCF_ubiquitin_ligase_complex |
| GO:0030017 | 25 | sarcomere |
| GO:0030835 | 25 | negative_regulation_of_actin_filament_depolymerization |
| GO:0032228 | 25 | regulation_of_synaptic_transmission,_GABAergic |
| GO:0032465 | 25 | regulation_of_cytokinesis |
| GO:0032589 | 25 | neuron_projection_membrane |
| GO:0032648 | 25 | regulation_of_interferon-beta_production |
| GO:0032855 | 25 | positive_regulation_of_Rac_GTPase_activity |
| GO:0034109 | 25 | homotypic_cell-cell_adhesion |
| GO:0034364 | 25 | high-density_lipoprotein_particle |
| GO:0042133 | 25 | neurotransmitter_metabolic_process |
| GO:0042220 | 25 | response_to_cocaine |
| GO:0042430 | 25 | indole-containing_compound_metabolic_process |
| GO:0043043 | 25 | peptide_biosynthetic_process |
| GO:0043489 | 25 | RNA_stabilization |
| GO:0043516 | 25 | regulation_of_DNA_damage_response,_signal_transduction_by_p53_class_mediator |
| GO:0043551 | 25 | regulation_of_phosphatidylinositol_3-kinase_activity |
| GO:0043679 | 25 | axon_terminus |
| GO:0043901 | 25 | negative_regulation_of_multi-organism_process |
| GO:0045028 | 25 | G-protein_coupled_purinergic_nucleotide_receptor_activity |
| GO:0045494 | 25 | photoreceptor_cell_maintenance |
| GO:0045933 | 25 | positive_regulation_of_muscle_contraction |
| GO:0046006 | 25 | regulation_of_activated_T_cell_proliferation |
| GO:0046164 | 25 | alcohol_catabolic_process |
| GO:0046503 | 25 | glycerolipid_catabolic_process |
| GO:0046579 | 25 | positive_regulation_of_Ras_protein_signal_transduction |
| GO:0046676 | 25 | negative_regulation_of_insulin_secretion |
| GO:0046688 | 25 | response_to_copper_ion |
| GO:0046915 | 25 | transition_metal_ion_transmembrane_transporter_activity |
| GO:0048010 | 25 | vascular_endothelial_growth_factor_receptor_signaling_pathway |
| GO:0048255 | 25 | mRNA_stabilization |
| GO:0048487 | 25 | beta-tubulin_binding |
| GO:0048662 | 25 | negative_regulation_of_smooth_muscle_cell_proliferation |
| GO:0048806 | 25 | genitalia_development |
| GO:0051489 | 25 | regulation_of_filopodium_assembly |
| GO:0051893 | 25 | regulation_of_focal_adhesion_assembly |
| GO:0055029 | 25 | nuclear_DNA-directed_RNA_polymerase_complex |
| GO:0060330 | 25 | regulation_of_response_to_interferon-gamma |
| GO:0060740 | 25 | prostate_gland_epithelium_morphogenesis |
| GO:0070098 | 25 | chemokine-mediated_signaling_pathway |
| GO:0072384 | 25 | organelle_transport_along_microtubule |
| GO:0090109 | 25 | regulation_of_cell-substrate_junction_assembly |
| GO:0090218 | 25 | positive_regulation_of_lipid_kinase_activity |
| GO:2001236 | 25 | regulation_of_extrinsic_apoptotic_signaling_pathway |
| GO:0001533 | 24 | cornified_envelope |
| GO:0001836 | 24 | release_of_cytochrome_c_from_mitochondria |
| GO:0001890 | 24 | placenta_development |
| GO:0002504 | 24 | antigen_processing_and_presentation_of_peptide_or_polysaccharide_antigen_via_MHC_class_II |
| GO:0002675 | 24 | positive_regulation_of_acute_inflammatory_response |
| GO:0002702 | 24 | positive_regulation_of_production_of_molecular_mediator_of_immune_response |
| GO:0003382 | 24 | epithelial_cell_morphogenesis |
| GO:0004181 | 24 | metallocarboxypeptidase_activity |
| GO:0005326 | 24 | neurotransmitter_transporter_activity |
| GO:0005343 | 24 | organic_acid:sodium_symporter_activity |
| GO:0005689 | 24 | U12-type_spliceosomal_complex |
| GO:0006360 | 24 | transcription_from_RNA_polymerase_I_promoter |
| GO:0006361 | 24 | transcription_initiation_from_RNA_polymerase_I_promoter |
| GO:0006471 | 24 | protein_ADP-ribosylation |
| GO:0006890 | 24 | retrograde_vesicle-mediated_transport,_Golgi_to_ER |
| GO:0007052 | 24 | mitotic_spindle_organization |
| GO:0008093 | 24 | cytoskeletal_adaptor_activity |
| GO:0009065 | 24 | glutamine_family_amino_acid_catabolic_process |
| GO:0009268 | 24 | response_to_pH |
| GO:0009394 | 24 | 2'-deoxyribonucleotide_metabolic_process |
| GO:0009395 | 24 | phospholipid_catabolic_process |
| GO:0009434 | 24 | microtubule-based_flagellum |
| GO:0010453 | 24 | regulation_of_cell_fate_commitment |
| GO:0010596 | 24 | negative_regulation_of_endothelial_cell_migration |
| GO:0010862 | 24 | positive_regulation_of_pathway-restricted_SMAD_protein_phosphorylation |
| GO:0016894 | 24 | endonuclease_activity,_active_with_either_ribo-_or_deoxyribonucleic_acids_and_producing_3'-phosphomonoesters |
| GO:0017158 | 24 | regulation_of_calcium_ion-dependent_exocytosis |
| GO:0018208 | 24 | peptidyl-proline_modification |
| GO:0021549 | 24 | cerebellum_development |
| GO:0021983 | 24 | pituitary_gland_development |
| GO:0030101 | 24 | natural_killer_cell_activation |
| GO:0030325 | 24 | adrenal_gland_development |
| GO:0031055 | 24 | chromatin_remodeling_at_centromere |
| GO:0031201 | 24 | SNARE_complex |
| GO:0032350 | 24 | regulation_of_hormone_metabolic_process |
| GO:0032370 | 24 | positive_regulation_of_lipid_transport |
| GO:0032451 | 24 | demethylase_activity |
| GO:0032653 | 24 | regulation_of_interleukin-10_production |
| GO:0032856 | 24 | activation_of_Ras_GTPase_activity |
| GO:0032873 | 24 | negative_regulation_of_stress-activated_MAPK_cascade |
| GO:0033003 | 24 | regulation_of_mast_cell_activation |
| GO:0033280 | 24 | response_to_vitamin_D |
| GO:0035329 | 24 | hippo_signaling_cascade |
| GO:0035337 | 24 | fatty-acyl-CoA_metabolic_process |
| GO:0042517 | 24 | positive_regulation_of_tyrosine_phosphorylation_of_Stat3_protein |
| GO:0042559 | 24 | pteridine-containing_compound_biosynthetic_process |
| GO:0043491 | 24 | protein_kinase_B_signaling_cascade |
| GO:0043499 | 24 | eukaryotic_cell_surface_binding |
| GO:0043534 | 24 | blood_vessel_endothelial_cell_migration |
| GO:0043552 | 24 | positive_regulation_of_phosphatidylinositol_3-kinase_activity |
| GO:0043631 | 24 | RNA_polyadenylation |
| GO:0045055 | 24 | regulated_secretory_pathway |
| GO:0045058 | 24 | T_cell_selection |
| GO:0045168 | 24 | cell-cell_signaling_involved_in_cell_fate_commitment |
| GO:0045182 | 24 | translation_regulator_activity |
| GO:0045581 | 24 | negative_regulation_of_T_cell_differentiation |
| GO:0045600 | 24 | positive_regulation_of_fat_cell_differentiation |
| GO:0045687 | 24 | positive_regulation_of_glial_cell_differentiation |
| GO:0045814 | 24 | negative_regulation_of_gene_expression,_epigenetic |
| GO:0045884 | 24 | regulation_of_survival_gene_product_expression |
| GO:0045912 | 24 | negative_regulation_of_carbohydrate_metabolic_process |
| GO:0046339 | 24 | diacylglycerol_metabolic_process |
| GO:0046513 | 24 | ceramide_biosynthetic_process |
| GO:0048265 | 24 | response_to_pain |
| GO:0048863 | 24 | stem_cell_differentiation |
| GO:0050850 | 24 | positive_regulation_of_calcium-mediated_signaling |
| GO:0051983 | 24 | regulation_of_chromosome_segregation |
| GO:0055002 | 24 | striated_muscle_cell_development |
| GO:0055021 | 24 | regulation_of_cardiac_muscle_tissue_growth |
| GO:0060334 | 24 | regulation_of_interferon-gamma-mediated_signaling_pathway |
| GO:0060397 | 24 | JAK-STAT_cascade_involved_in_growth_hormone_signaling_pathway |
| GO:0070303 | 24 | negative_regulation_of_stress-activated_protein_kinase_signaling_cascade |
| GO:0070330 | 24 | aromatase_activity |
| GO:0070542 | 24 | response_to_fatty_acid |
| GO:0072073 | 24 | kidney_epithelium_development |
| GO:0001158 | 23 | enhancer_sequence-specific_DNA_binding |
| GO:0001892 | 23 | embryonic_placenta_development |
| GO:0001937 | 23 | negative_regulation_of_endothelial_cell_proliferation |
| GO:0002286 | 23 | T_cell_activation_involved_in_immune_response |
| GO:0002886 | 23 | regulation_of_myeloid_leukocyte_mediated_immunity |
| GO:0003730 | 23 | mRNA_3'-UTR_binding |
| GO:0005104 | 23 | fibroblast_growth_factor_receptor_binding |
| GO:0005184 | 23 | neuropeptide_hormone_activity |
| GO:0005545 | 23 | 1-phosphatidylinositol_binding |
| GO:0006536 | 23 | glutamate_metabolic_process |
| GO:0006693 | 23 | prostaglandin_metabolic_process |
| GO:0006706 | 23 | steroid_catabolic_process |
| GO:0006739 | 23 | NADP_metabolic_process |
| GO:0006829 | 23 | zinc_ion_transport |
| GO:0007176 | 23 | regulation_of_epidermal_growth_factor-activated_receptor_activity |
| GO:0007530 | 23 | sex_determination |
| GO:0007622 | 23 | rhythmic_behavior |
| GO:0008483 | 23 | transaminase_activity |
| GO:0009081 | 23 | branched_chain_family_amino_acid_metabolic_process |
| GO:0009975 | 23 | cyclase_activity |
| GO:0010559 | 23 | regulation_of_glycoprotein_biosynthetic_process |
| GO:0010574 | 23 | regulation_of_vascular_endothelial_growth_factor_production |
| GO:0010677 | 23 | negative_regulation_of_cellular_carbohydrate_metabolic_process |
| GO:0010770 | 23 | positive_regulation_of_cell_morphogenesis_involved_in_differentiation |
| GO:0015485 | 23 | cholesterol_binding |
| GO:0016460 | 23 | myosin_II_complex |
| GO:0016796 | 23 | exonuclease_activity,_active_with_either_ribo-_or_deoxyribonucleic_acids_and_producing_5'-phosphomonoesters |
| GO:0016799 | 23 | hydrolase_activity,_hydrolyzing_N-glycosyl_compounds |
| GO:0019201 | 23 | nucleotide_kinase_activity |
| GO:0019935 | 23 | cyclic-nucleotide-mediated_signaling |
| GO:0021954 | 23 | central_nervous_system_neuron_development |
| GO:0022412 | 23 | cellular_process_involved_in_reproduction_in_multicellular_organism |
| GO:0030166 | 23 | proteoglycan_biosynthetic_process |
| GO:0030947 | 23 | regulation_of_vascular_endothelial_growth_factor_receptor_signaling_pathway |
| GO:0031102 | 23 | neuron_projection_regeneration |
| GO:0031128 | 23 | developmental_induction |
| GO:0031593 | 23 | polyubiquitin_binding |
| GO:0031941 | 23 | filamentous_actin |
| GO:0033177 | 23 | proton-transporting_two-sector_ATPase_complex,_proton-transporting_domain |
| GO:0034367 | 23 | macromolecular_complex_remodeling |
| GO:0034368 | 23 | protein-lipid_complex_remodeling |
| GO:0034369 | 23 | plasma_lipoprotein_particle_remodeling |
| GO:0034446 | 23 | substrate_adhesion-dependent_cell_spreading |
| GO:0035249 | 23 | synaptic_transmission,_glutamatergic |
| GO:0035254 | 23 | glutamate_receptor_binding |
| GO:0040036 | 23 | regulation_of_fibroblast_growth_factor_receptor_signaling_pathway |
| GO:0042402 | 23 | cellular_biogenic_amine_catabolic_process |
| GO:0043114 | 23 | regulation_of_vascular_permeability |
| GO:0043462 | 23 | regulation_of_ATPase_activity |
| GO:0043502 | 23 | regulation_of_muscle_adaptation |
| GO:0043603 | 23 | cellular_amide_metabolic_process |
| GO:0044246 | 23 | regulation_of_multicellular_organismal_metabolic_process |
| GO:0045296 | 23 | cadherin_binding |
| GO:0045806 | 23 | negative_regulation_of_endocytosis |
| GO:0045940 | 23 | positive_regulation_of_steroid_metabolic_process |
| GO:0046068 | 23 | cGMP_metabolic_process |
| GO:0046875 | 23 | ephrin_receptor_binding |
| GO:0048169 | 23 | regulation_of_long-term_neuronal_synaptic_plasticity |
| GO:0048536 | 23 | spleen_development |
| GO:0048640 | 23 | negative_regulation_of_developmental_growth |
| GO:0048747 | 23 | muscle_fiber_development |
| GO:0050912 | 23 | detection_of_chemical_stimulus_involved_in_sensory_perception_of_taste |
| GO:0051119 | 23 | sugar_transmembrane_transporter_activity |
| GO:0051350 | 23 | negative_regulation_of_lyase_activity |
| GO:0051412 | 23 | response_to_corticosterone_stimulus |
| GO:0051693 | 23 | actin_filament_capping |
| GO:0055093 | 23 | response_to_hyperoxia |
| GO:0060170 | 23 | cilium_membrane |
| GO:0070469 | 23 | respiratory_chain |
| GO:0070534 | 23 | protein_K63-linked_ubiquitination |
| GO:0072595 | 23 | maintenance_of_protein_localization_to_organelle |
| GO:1901019 | 23 | regulation_of_calcium_ion_transmembrane_transporter_activity |
| GO:0000045 | 22 | autophagic_vacuole_assembly |
| GO:0000060 | 22 | protein_import_into_nucleus,_translocation |
| GO:0000083 | 22 | regulation_of_transcription_involved_in_G1/S_phase_of_mitotic_cell_cycle |
| GO:0000313 | 22 | organellar_ribosome |
| GO:0001191 | 22 | RNA_polymerase_II_transcription_factor_binding_transcription_factor_activity_involved_in_negative_regulation_of_transcription |
| GO:0002088 | 22 | lens_development_in_camera-type_eye |
| GO:0002244 | 22 | hemopoietic_progenitor_cell_differentiation |
| GO:0002828 | 22 | regulation_of_type_2_immune_response |
| GO:0003073 | 22 | regulation_of_systemic_arterial_blood_pressure |
| GO:0004298 | 22 | threonine-type_endopeptidase_activity |
| GO:0005217 | 22 | intracellular_ligand-gated_ion_channel_activity |
| GO:0005242 | 22 | inward_rectifier_potassium_channel_activity |
| GO:0005484 | 22 | SNAP_receptor_activity |
| GO:0005761 | 22 | mitochondrial_ribosome |
| GO:0005871 | 22 | kinesin_complex |
| GO:0006270 | 22 | DNA-dependent_DNA_replication_initiation |
| GO:0006297 | 22 | nucleotide-excision_repair,_DNA_gap_filling |
| GO:0006336 | 22 | DNA_replication-independent_nucleosome_assembly |
| GO:0006611 | 22 | protein_export_from_nucleus |
| GO:0007090 | 22 | regulation_of_S_phase_of_mitotic_cell_cycle |
| GO:0007528 | 22 | neuromuscular_junction_development |
| GO:0008088 | 22 | axon_cargo_transport |
| GO:0008210 | 22 | estrogen_metabolic_process |
| GO:0008299 | 22 | isoprenoid_biosynthetic_process |
| GO:0008308 | 22 | voltage-gated_anion_channel_activity |
| GO:0009132 | 22 | nucleoside_diphosphate_metabolic_process |
| GO:0009988 | 22 | cell-cell_recognition |
| GO:0010165 | 22 | response_to_X-ray |
| GO:0010543 | 22 | regulation_of_platelet_activation |
| GO:0010632 | 22 | regulation_of_epithelial_cell_migration |
| GO:0010822 | 22 | positive_regulation_of_mitochondrion_organization |
| GO:0010876 | 22 | lipid_localization |
| GO:0012507 | 22 | ER_to_Golgi_transport_vesicle_membrane |
| GO:0015026 | 22 | coreceptor_activity |
| GO:0015175 | 22 | neutral_amino_acid_transmembrane_transporter_activity |
| GO:0016338 | 22 | calcium-independent_cell-cell_adhesion |
| GO:0016486 | 22 | peptide_hormone_processing |
| GO:0016505 | 22 | apoptotic_protease_activator_activity |
| GO:0016575 | 22 | histone_deacetylation |
| GO:0016849 | 22 | phosphorus-oxygen_lyase_activity |
| GO:0016917 | 22 | GABA_receptor_activity |
| GO:0016925 | 22 | protein_sumoylation |
| GO:0019724 | 22 | B_cell_mediated_immunity |
| GO:0019915 | 22 | lipid_storage |
| GO:0022829 | 22 | wide_pore_channel_activity |
| GO:0030057 | 22 | desmosome |
| GO:0031047 | 22 | gene_silencing_by_RNA |
| GO:0031114 | 22 | regulation_of_microtubule_depolymerization |
| GO:0031233 | 22 | intrinsic_to_external_side_of_plasma_membrane |
| GO:0031252 | 22 | cell_leading_edge |
| GO:0031280 | 22 | negative_regulation_of_cyclase_activity |
| GO:0031683 | 22 | G-protein_beta/gamma-subunit_complex_binding |
| GO:0032201 | 22 | telomere_maintenance_via_semi-conservative_replication |
| GO:0032369 | 22 | negative_regulation_of_lipid_transport |
| GO:0032732 | 22 | positive_regulation_of_interleukin-1_production |
| GO:0032845 | 22 | negative_regulation_of_homeostatic_process |
| GO:0034080 | 22 | CenH3-containing_nucleosome_assembly_at_centromere |
| GO:0034504 | 22 | protein_localization_to_nucleus |
| GO:0034698 | 22 | response_to_gonadotropin_stimulus |
| GO:0034724 | 22 | DNA_replication-independent_nucleosome_organization |
| GO:0042165 | 22 | neurotransmitter_binding |
| GO:0043531 | 22 | ADP_binding |
| GO:0043903 | 22 | regulation_of_symbiosis,_encompassing_mutualism_through_parasitism |
| GO:0045071 | 22 | negative_regulation_of_viral_genome_replication |
| GO:0045604 | 22 | regulation_of_epidermal_cell_differentiation |
| GO:0045622 | 22 | regulation_of_T-helper_cell_differentiation |
| GO:0045686 | 22 | negative_regulation_of_glial_cell_differentiation |
| GO:0045773 | 22 | positive_regulation_of_axon_extension |
| GO:0045907 | 22 | positive_regulation_of_vasoconstriction |
| GO:0048525 | 22 | negative_regulation_of_viral_reproduction |
| GO:0048701 | 22 | embryonic_cranial_skeleton_morphogenesis |
| GO:0048762 | 22 | mesenchymal_cell_differentiation |
| GO:0048813 | 22 | dendrite_morphogenesis |
| GO:0050710 | 22 | negative_regulation_of_cytokine_secretion |
| GO:0050820 | 22 | positive_regulation_of_coagulation |
| GO:0050926 | 22 | regulation_of_positive_chemotaxis |
| GO:0051059 | 22 | NF-kappaB_binding |
| GO:0051180 | 22 | vitamin_transport |
| GO:0051181 | 22 | cofactor_transport |
| GO:0051445 | 22 | regulation_of_meiotic_cell_cycle |
| GO:0060043 | 22 | regulation_of_cardiac_muscle_cell_proliferation |
| GO:0060674 | 22 | placenta_blood_vessel_development |
| GO:0060998 | 22 | regulation_of_dendritic_spine_development |
| GO:0061077 | 22 | chaperone-mediated_protein_folding |
| GO:0070003 | 22 | threonine-type_peptidase_activity |
| GO:0090207 | 22 | regulation_of_triglyceride_metabolic_process |
| GO:0090329 | 22 | regulation_of_DNA-dependent_DNA_replication |
| GO:1900449 | 22 | regulation_of_glutamate_receptor_signaling_pathway |
| GO:2000179 | 22 | positive_regulation_of_neural_precursor_cell_proliferation |
| GO:0000718 | 21 | nucleotide-excision_repair,_DNA_damage_removal |
| GO:0000979 | 21 | RNA_polymerase_II_core_promoter_sequence-specific_DNA_binding |
| GO:0001539 | 21 | ciliary_or_flagellar_motility |
| GO:0001972 | 21 | retinoic_acid_binding |
| GO:0004683 | 21 | calmodulin-dependent_protein_kinase_activity |
| GO:0004709 | 21 | MAP_kinase_kinase_kinase_activity |
| GO:0005048 | 21 | signal_sequence_binding |
| GO:0005154 | 21 | epidermal_growth_factor_receptor_binding |
| GO:0005227 | 21 | calcium_activated_cation_channel_activity |
| GO:0005540 | 21 | hyaluronic_acid_binding |
| GO:0005640 | 21 | nuclear_outer_membrane |
| GO:0005669 | 21 | transcription_factor_TFIID_complex |
| GO:0005680 | 21 | anaphase-promoting_complex |
| GO:0005839 | 21 | proteasome_core_complex |
| GO:0005903 | 21 | brush_border |
| GO:0005922 | 21 | connexon_complex |
| GO:0006278 | 21 | RNA-dependent_DNA_replication |
| GO:0006363 | 21 | termination_of_RNA_polymerase_I_transcription |
| GO:0006378 | 21 | mRNA_polyadenylation |
| GO:0006509 | 21 | membrane_protein_ectodomain_proteolysis |
| GO:0006699 | 21 | bile_acid_biosynthetic_process |
| GO:0007026 | 21 | negative_regulation_of_microtubule_depolymerization |
| GO:0007032 | 21 | endosome_organization |
| GO:0007062 | 21 | sister_chromatid_cohesion |
| GO:0007194 | 21 | negative_regulation_of_adenylate_cyclase_activity |
| GO:0007616 | 21 | long-term_memory |
| GO:0008028 | 21 | monocarboxylic_acid_transmembrane_transporter_activity |
| GO:0008186 | 21 | RNA-dependent_ATPase_activity |
| GO:0008373 | 21 | sialyltransferase_activity |
| GO:0009067 | 21 | aspartate_family_amino_acid_biosynthetic_process |
| GO:0009127 | 21 | purine_nucleoside_monophosphate_biosynthetic_process |
| GO:0009148 | 21 | pyrimidine_nucleoside_triphosphate_biosynthetic_process |
| GO:0009168 | 21 | purine_ribonucleoside_monophosphate_biosynthetic_process |
| GO:0009584 | 21 | detection_of_visible_light |
| GO:0010039 | 21 | response_to_iron_ion |
| GO:0010494 | 21 | cytoplasmic_stress_granule |
| GO:0010508 | 21 | positive_regulation_of_autophagy |
| GO:0010660 | 21 | regulation_of_muscle_cell_apoptotic_process |
| GO:0010712 | 21 | regulation_of_collagen_metabolic_process |
| GO:0015850 | 21 | organic_alcohol_transport |
| GO:0015985 | 21 | energy_coupled_proton_transport,_down_electrochemical_gradient |
| GO:0015986 | 21 | ATP_synthesis_coupled_proton_transport |
| GO:0016447 | 21 | somatic_recombination_of_immunoglobulin_gene_segments |
| GO:0019098 | 21 | reproductive_behavior |
| GO:0019835 | 21 | cytolysis |
| GO:0022839 | 21 | ion_gated_channel_activity |
| GO:0030032 | 21 | lamellipodium_assembly |
| GO:0030291 | 21 | protein_serine/threonine_kinase_inhibitor_activity |
| GO:0030574 | 21 | collagen_catabolic_process |
| GO:0030641 | 21 | regulation_of_cellular_pH |
| GO:0031418 | 21 | L-ascorbic_acid_binding |
| GO:0031640 | 21 | killing_of_cells_of_other_organism |
| GO:0032413 | 21 | negative_regulation_of_ion_transmembrane_transporter_activity |
| GO:0032735 | 21 | positive_regulation_of_interleukin-12_production |
| GO:0032925 | 21 | regulation_of_activin_receptor_signaling_pathway |
| GO:0033344 | 21 | cholesterol_efflux |
| GO:0034105 | 21 | positive_regulation_of_tissue_remodeling |
| GO:0034405 | 21 | response_to_fluid_shear_stress |
| GO:0035162 | 21 | embryonic_hemopoiesis |
| GO:0035250 | 21 | UDP-galactosyltransferase_activity |
| GO:0042346 | 21 | positive_regulation_of_NF-kappaB_import_into_nucleus |
| GO:0042417 | 21 | dopamine_metabolic_process |
| GO:0042474 | 21 | middle_ear_morphogenesis |
| GO:0042596 | 21 | fear_response |
| GO:0042813 | 21 | Wnt-activated_receptor_activity |
| GO:0043014 | 21 | alpha-tubulin_binding |
| GO:0043027 | 21 | cysteine-type_endopeptidase_inhibitor_activity_involved_in_apoptotic_process |
| GO:0043113 | 21 | receptor_clustering |
| GO:0043236 | 21 | laminin_binding |
| GO:0043266 | 21 | regulation_of_potassium_ion_transport |
| GO:0043300 | 21 | regulation_of_leukocyte_degranulation |
| GO:0043425 | 21 | bHLH_transcription_factor_binding |
| GO:0044003 | 21 | modification_by_symbiont_of_host_morphology_or_physiology |
| GO:0044060 | 21 | regulation_of_endocrine_process |
| GO:0044089 | 21 | positive_regulation_of_cellular_component_biogenesis |
| GO:0044349 | 21 | DNA_excision |
| GO:0045309 | 21 | protein_phosphorylated_amino_acid_binding |
| GO:0045661 | 21 | regulation_of_myoblast_differentiation |
| GO:0046466 | 21 | membrane_lipid_catabolic_process |
| GO:0046627 | 21 | negative_regulation_of_insulin_receptor_signaling_pathway |
| GO:0048484 | 21 | enteric_nervous_system_development |
| GO:0048512 | 21 | circadian_behavior |
| GO:0048710 | 21 | regulation_of_astrocyte_differentiation |
| GO:0048713 | 21 | regulation_of_oligodendrocyte_differentiation |
| GO:0050663 | 21 | cytokine_secretion |
| GO:0050691 | 21 | regulation_of_defense_response_to_virus_by_host |
| GO:0050918 | 21 | positive_chemotaxis |
| GO:0050922 | 21 | negative_regulation_of_chemotaxis |
| GO:0050927 | 21 | positive_regulation_of_positive_chemotaxis |
| GO:0060343 | 21 | trabecula_formation |
| GO:0061005 | 21 | cell_differentiation_involved_in_kidney_development |
| GO:0070328 | 21 | triglyceride_homeostasis |
| GO:0070371 | 21 | ERK1_and_ERK2_cascade |
| GO:0070603 | 21 | SWI/SNF-type_complex |
| GO:0071326 | 21 | cellular_response_to_monosaccharide_stimulus |
| GO:0071331 | 21 | cellular_response_to_hexose_stimulus |
| GO:0071333 | 21 | cellular_response_to_glucose_stimulus |
| GO:0071813 | 21 | lipoprotein_particle_binding |
| GO:0071814 | 21 | protein-lipid_complex_binding |
| GO:0072659 | 21 | protein_localization_in_plasma_membrane |
| GO:0090103 | 21 | cochlea_morphogenesis |
| GO:0090189 | 21 | regulation_of_branching_involved_in_ureteric_bud_morphogenesis |
| GO:0097190 | 21 | apoptotic_signaling_pathway |
| GO:1900077 | 21 | negative_regulation_of_cellular_response_to_insulin_stimulus |
| GO:2001242 | 21 | regulation_of_intrinsic_apoptotic_signaling_pathway |
| GO:0000062 | 20 | fatty-acyl-CoA_binding |
| GO:0000146 | 20 | microfilament_motor_activity |
| GO:0000178 | 20 | exosome_(RNase_complex) |
| GO:0000978 | 20 | RNA_polymerase_II_core_promoter_proximal_region_sequence-specific_DNA_binding |
| GO:0001659 | 20 | temperature_homeostasis |
| GO:0001772 | 20 | immunological_synapse |
| GO:0001958 | 20 | endochondral_ossification |
| GO:0001968 | 20 | fibronectin_binding |
| GO:0001990 | 20 | regulation_of_systemic_arterial_blood_pressure_by_hormone |
| GO:0002026 | 20 | regulation_of_the_force_of_heart_contraction |
| GO:0002312 | 20 | B_cell_activation_involved_in_immune_response |
| GO:0002715 | 20 | regulation_of_natural_killer_cell_mediated_immunity |
| GO:0002825 | 20 | regulation_of_T-helper_1_type_immune_response |
| GO:0003044 | 20 | regulation_of_systemic_arterial_blood_pressure_mediated_by_a_chemical_signal |
| GO:0004004 | 20 | ATP-dependent_RNA_helicase_activity |
| GO:0004532 | 20 | exoribonuclease_activity |
| GO:0004653 | 20 | polypeptide_N-acetylgalactosaminyltransferase_activity |
| GO:0005086 | 20 | ARF_guanyl-nucleotide_exchange_factor_activity |
| GO:0005753 | 20 | mitochondrial_proton-transporting_ATP_synthase_complex |
| GO:0005776 | 20 | autophagic_vacuole |
| GO:0006110 | 20 | regulation_of_glycolysis |
| GO:0006342 | 20 | chromatin_silencing |
| GO:0006482 | 20 | protein_demethylation |
| GO:0006595 | 20 | polyamine_metabolic_process |
| GO:0006700 | 20 | C21-steroid_hormone_biosynthetic_process |
| GO:0006783 | 20 | heme_biosynthetic_process |
| GO:0007220 | 20 | Notch_receptor_processing |
| GO:0007548 | 20 | sex_differentiation |
| GO:0008214 | 20 | protein_dealkylation |
| GO:0009060 | 20 | aerobic_respiration |
| GO:0009084 | 20 | glutamine_family_amino_acid_biosynthetic_process |
| GO:0009110 | 20 | vitamin_biosynthetic_process |
| GO:0014003 | 20 | oligodendrocyte_development |
| GO:0015301 | 20 | anion:anion_antiporter_activity |
| GO:0015844 | 20 | monoamine_transport |
| GO:0016878 | 20 | acid-thiol_ligase_activity |
| GO:0019894 | 20 | kinesin_binding |
| GO:0021510 | 20 | spinal_cord_development |
| GO:0030149 | 20 | sphingolipid_catabolic_process |
| GO:0030201 | 20 | heparan_sulfate_proteoglycan_metabolic_process |
| GO:0030511 | 20 | positive_regulation_of_transforming_growth_factor_beta_receptor_signaling_pathway |
| GO:0030552 | 20 | cAMP_binding |
| GO:0030864 | 20 | cortical_actin_cytoskeleton |
| GO:0031490 | 20 | chromatin_DNA_binding |
| GO:0032673 | 20 | regulation_of_interleukin-4_production |
| GO:0032731 | 20 | positive_regulation_of_interleukin-1_beta_production |
| GO:0032965 | 20 | regulation_of_collagen_biosynthetic_process |
| GO:0033205 | 20 | cell_cycle_cytokinesis |
| GO:0034361 | 20 | very-low-density_lipoprotein_particle |
| GO:0034385 | 20 | triglyceride-rich_lipoprotein_particle |
| GO:0034502 | 20 | protein_localization_to_chromosome |
| GO:0034694 | 20 | response_to_prostaglandin_stimulus |
| GO:0035088 | 20 | establishment_or_maintenance_of_apical/basal_cell_polarity |
| GO:0035336 | 20 | long-chain_fatty-acyl-CoA_metabolic_process |
| GO:0035384 | 20 | thioester_biosynthetic_process |
| GO:0036075 | 20 | replacement_ossification |
| GO:0040020 | 20 | regulation_of_meiosis |
| GO:0042107 | 20 | cytokine_metabolic_process |
| GO:0042116 | 20 | macrophage_activation |
| GO:0042311 | 20 | vasodilation |
| GO:0042481 | 20 | regulation_of_odontogenesis |
| GO:0042573 | 20 | retinoic_acid_metabolic_process |
| GO:0042605 | 20 | peptide_antigen_binding |
| GO:0042744 | 20 | hydrogen_peroxide_catabolic_process |
| GO:0042771 | 20 | DNA_damage_response,_signal_transduction_by_p53_class_mediator_resulting_in_induction_of_apoptosis |
| GO:0043030 | 20 | regulation_of_macrophage_activation |
| GO:0043124 | 20 | negative_regulation_of_I-kappaB_kinase/NF-kappaB_cascade |
| GO:0043536 | 20 | positive_regulation_of_blood_vessel_endothelial_cell_migration |
| GO:0043548 | 20 | phosphatidylinositol_3-kinase_binding |
| GO:0043574 | 20 | peroxisomal_transport |
| GO:0043647 | 20 | inositol_phosphate_metabolic_process |
| GO:0044058 | 20 | regulation_of_digestive_system_process |
| GO:0044062 | 20 | regulation_of_excretion |
| GO:0044269 | 20 | glycerol_ether_catabolic_process |
| GO:0044291 | 20 | cell-cell_contact_zone |
| GO:0045214 | 20 | sarcomere_organization |
| GO:0045259 | 20 | proton-transporting_ATP_synthase_complex |
| GO:0045335 | 20 | phagocytic_vesicle |
| GO:0045823 | 20 | positive_regulation_of_heart_contraction |
| GO:0045939 | 20 | negative_regulation_of_steroid_metabolic_process |
| GO:0046135 | 20 | pyrimidine_nucleoside_catabolic_process |
| GO:0046461 | 20 | neutral_lipid_catabolic_process |
| GO:0046464 | 20 | acylglycerol_catabolic_process |
| GO:0046605 | 20 | regulation_of_centrosome_cycle |
| GO:0046640 | 20 | regulation_of_alpha-beta_T_cell_proliferation |
| GO:0046717 | 20 | acid_secretion |
| GO:0046949 | 20 | fatty-acyl-CoA_biosynthetic_process |
| GO:0046961 | 20 | proton-transporting_ATPase_activity,_rotational_mechanism |
| GO:0048147 | 20 | negative_regulation_of_fibroblast_proliferation |
| GO:0048873 | 20 | homeostasis_of_number_of_cells_within_a_tissue |
| GO:0050699 | 20 | WW_domain_binding |
| GO:0050856 | 20 | regulation_of_T_cell_receptor_signaling_pathway |
| GO:0050886 | 20 | endocrine_process |
| GO:0050982 | 20 | detection_of_mechanical_stimulus |
| GO:0051220 | 20 | cytoplasmic_sequestering_of_protein |
| GO:0051354 | 20 | negative_regulation_of_oxidoreductase_activity |
| GO:0051648 | 20 | vesicle_localization |
| GO:0052312 | 20 | modulation_of_transcription_in_other_organism_involved_in_symbiotic_interaction |
| GO:0060444 | 20 | branching_involved_in_mammary_gland_duct_morphogenesis |
| GO:0060765 | 20 | regulation_of_androgen_receptor_signaling_pathway |
| GO:0061245 | 20 | establishment_or_maintenance_of_bipolar_cell_polarity |
| GO:0070373 | 20 | negative_regulation_of_ERK1_and_ERK2_cascade |
| GO:0070566 | 20 | adenylyltransferase_activity |
| GO:0070741 | 20 | response_to_interleukin-6 |
| GO:0071320 | 20 | cellular_response_to_cAMP |
| GO:0071616 | 20 | acyl-CoA_biosynthetic_process |
| GO:0080008 | 20 | CUL4-RING_ubiquitin_ligase_complex |
| GO:0090398 | 20 | cellular_senescence |
| GO:1901070 | 20 | guanosine-containing_compound_biosynthetic_process |
| GO:2001014 | 20 | regulation_of_skeletal_muscle_cell_differentiation |
| GO:0000097 | 19 | sulfur_amino_acid_biosynthetic_process |
| GO:0000159 | 19 | protein_phosphatase_type_2A_complex |
| GO:0000245 | 19 | spliceosomal_complex_assembly |
| GO:0000413 | 19 | protein_peptidyl-prolyl_isomerization |
| GO:0000819 | 19 | sister_chromatid_segregation |
| GO:0001105 | 19 | RNA_polymerase_II_transcription_coactivator_activity |
| GO:0001702 | 19 | gastrulation_with_mouth_forming_second |
| GO:0002090 | 19 | regulation_of_receptor_internalization |
| GO:0002720 | 19 | positive_regulation_of_cytokine_production_involved_in_immune_response |
| GO:0003179 | 19 | heart_valve_morphogenesis |
| GO:0004890 | 19 | GABA-A_receptor_activity |
| GO:0005234 | 19 | extracellular-glutamate-gated_ion_channel_activity |
| GO:0005246 | 19 | calcium_channel_regulator_activity |
| GO:0005328 | 19 | neurotransmitter:sodium_symporter_activity |
| GO:0006004 | 19 | fucose_metabolic_process |
| GO:0006103 | 19 | 2-oxoglutarate_metabolic_process |
| GO:0006111 | 19 | regulation_of_gluconeogenesis |
| GO:0006362 | 19 | transcription_elongation_from_RNA_polymerase_I_promoter |
| GO:0006625 | 19 | protein_targeting_to_peroxisome |
| GO:0007031 | 19 | peroxisome_organization |
| GO:0008045 | 19 | motor_axon_guidance |
| GO:0008200 | 19 | ion_channel_inhibitor_activity |
| GO:0008395 | 19 | steroid_hydroxylase_activity |
| GO:0008601 | 19 | protein_phosphatase_type_2A_regulator_activity |
| GO:0008656 | 19 | cysteine-type_endopeptidase_activator_activity_involved_in_apoptotic_process |
| GO:0009074 | 19 | aromatic_amino_acid_family_catabolic_process |
| GO:0009083 | 19 | branched_chain_family_amino_acid_catabolic_process |
| GO:0009208 | 19 | pyrimidine_ribonucleoside_triphosphate_metabolic_process |
| GO:0009251 | 19 | glucan_catabolic_process |
| GO:0009651 | 19 | response_to_salt_stress |
| GO:0009954 | 19 | proximal/distal_pattern_formation |
| GO:0010575 | 19 | positive_regulation_vascular_endothelial_growth_factor_production |
| GO:0010718 | 19 | positive_regulation_of_epithelial_to_mesenchymal_transition |
| GO:0010799 | 19 | regulation_of_peptidyl-threonine_phosphorylation |
| GO:0014031 | 19 | mesenchymal_cell_development |
| GO:0014065 | 19 | phosphatidylinositol_3-kinase_cascade |
| GO:0014074 | 19 | response_to_purine-containing_compound |
| GO:0015491 | 19 | cation:cation_antiporter_activity |
| GO:0015804 | 19 | neutral_amino_acid_transport |
| GO:0015807 | 19 | L-amino_acid_transport |
| GO:0016235 | 19 | aggresome |
| GO:0016248 | 19 | channel_inhibitor_activity |
| GO:0016409 | 19 | palmitoyltransferase_activity |
| GO:0016577 | 19 | histone_demethylation |
| GO:0016922 | 19 | ligand-dependent_nuclear_receptor_binding |
| GO:0017145 | 19 | stem_cell_division |
| GO:0018200 | 19 | peptidyl-glutamic_acid_modification |
| GO:0019200 | 19 | carbohydrate_kinase_activity |
| GO:0019433 | 19 | triglyceride_catabolic_process |
| GO:0019865 | 19 | immunoglobulin_binding |
| GO:0019933 | 19 | cAMP-mediated_signaling |
| GO:0021533 | 19 | cell_differentiation_in_hindbrain |
| GO:0021915 | 19 | neural_tube_development |
| GO:0021955 | 19 | central_nervous_system_neuron_axonogenesis |
| GO:0030159 | 19 | receptor_signaling_complex_scaffold_activity |
| GO:0030194 | 19 | positive_regulation_of_blood_coagulation |
| GO:0030204 | 19 | chondroitin_sulfate_metabolic_process |
| GO:0030316 | 19 | osteoclast_differentiation |
| GO:0030530 | 19 | heterogeneous_nuclear_ribonucleoprotein_complex |
| GO:0030865 | 19 | cortical_cytoskeleton_organization |
| GO:0030879 | 19 | mammary_gland_development |
| GO:0030902 | 19 | hindbrain_development |
| GO:0030983 | 19 | mismatched_DNA_binding |
| GO:0031113 | 19 | regulation_of_microtubule_polymerization |
| GO:0031497 | 19 | chromatin_assembly |
| GO:0031529 | 19 | ruffle_organization |
| GO:0031648 | 19 | protein_destabilization |
| GO:0032393 | 19 | MHC_class_I_receptor_activity |
| GO:0032757 | 19 | positive_regulation_of_interleukin-8_production |
| GO:0032862 | 19 | activation_of_Rho_GTPase_activity |
| GO:0033081 | 19 | regulation_of_T_cell_differentiation_in_thymus |
| GO:0033108 | 19 | mitochondrial_respiratory_chain_complex_assembly |
| GO:0033144 | 19 | negative_regulation_of_intracellular_steroid_hormone_receptor_signaling_pathway |
| GO:0033178 | 19 | proton-transporting_two-sector_ATPase_complex,_catalytic_domain |
| GO:0033198 | 19 | response_to_ATP |
| GO:0034381 | 19 | plasma_lipoprotein_particle_clearance |
| GO:0034605 | 19 | cellular_response_to_heat |
| GO:0035412 | 19 | regulation_of_catenin_import_into_nucleus |
| GO:0042269 | 19 | regulation_of_natural_killer_cell_mediated_cytotoxicity |
| GO:0042310 | 19 | vasoconstriction |
| GO:0042522 | 19 | regulation_of_tyrosine_phosphorylation_of_Stat5_protein |
| GO:0042613 | 19 | MHC_class_II_protein_complex |
| GO:0042730 | 19 | fibrinolysis |
| GO:0043015 | 19 | gamma-tubulin_binding |
| GO:0043168 | 19 | anion_binding |
| GO:0043500 | 19 | muscle_adaptation |
| GO:0043567 | 19 | regulation_of_insulin-like_growth_factor_receptor_signaling_pathway |
| GO:0043921 | 19 | modulation_by_host_of_viral_transcription |
| GO:0043984 | 19 | histone_H4-K16_acetylation |
| GO:0044247 | 19 | cellular_polysaccharide_catabolic_process |
| GO:0044442 | 19 | microtubule-based_flagellum_part |
| GO:0044460 | 19 | flagellum_part |
| GO:0045076 | 19 | regulation_of_interleukin-2_biosynthetic_process |
| GO:0045577 | 19 | regulation_of_B_cell_differentiation |
| GO:0045616 | 19 | regulation_of_keratinocyte_differentiation |
| GO:0045742 | 19 | positive_regulation_of_epidermal_growth_factor_receptor_signaling_pathway |
| GO:0045987 | 19 | positive_regulation_of_smooth_muscle_contraction |
| GO:0046329 | 19 | negative_regulation_of_JNK_cascade |
| GO:0046825 | 19 | regulation_of_protein_export_from_nucleus |
| GO:0048365 | 19 | Rac_GTPase_binding |
| GO:0050704 | 19 | regulation_of_interleukin-1_secretion |
| GO:0050892 | 19 | intestinal_absorption |
| GO:0050996 | 19 | positive_regulation_of_lipid_catabolic_process |
| GO:0051453 | 19 | regulation_of_intracellular_pH |
| GO:0051537 | 19 | 2_iron,_2_sulfur_cluster_binding |
| GO:0051668 | 19 | localization_within_membrane |
| GO:0051930 | 19 | regulation_of_sensory_perception_of_pain |
| GO:0051931 | 19 | regulation_of_sensory_perception |
| GO:0051954 | 19 | positive_regulation_of_amine_transport |
| GO:0051962 | 19 | positive_regulation_of_nervous_system_development |
| GO:0051965 | 19 | positive_regulation_of_synapse_assembly |
| GO:0052472 | 19 | modulation_by_host_of_symbiont_transcription |
| GO:0060349 | 19 | bone_morphogenesis |
| GO:0060479 | 19 | lung_cell_differentiation |
| GO:0070192 | 19 | chromosome_organization_involved_in_meiosis |
| GO:0070325 | 19 | lipoprotein_particle_receptor_binding |
| GO:0070723 | 19 | response_to_cholesterol |
| GO:0071634 | 19 | regulation_of_transforming_growth_factor_beta_production |
| GO:0071779 | 19 | G1/S_transition_checkpoint |
| GO:0072663 | 19 | establishment_of_protein_localization_to_peroxisome |
| GO:0072676 | 19 | lymphocyte_migration |
| GO:0090003 | 19 | regulation_of_establishment_of_protein_localization_in_plasma_membrane |
| GO:0090022 | 19 | regulation_of_neutrophil_chemotaxis |
| GO:0090190 | 19 | positive_regulation_of_branching_involved_in_ureteric_bud_morphogenesis |
| GO:0090200 | 19 | positive_regulation_of_release_of_cytochrome_c_from_mitochondria |
| GO:0097035 | 19 | regulation_of_membrane_lipid_distribution |
| GO:1900048 | 19 | positive_regulation_of_hemostasis |
| GO:2000401 | 19 | regulation_of_lymphocyte_migration |
| GO:2000516 | 19 | positive_regulation_of_CD4-positive,_alpha-beta_T_cell_activation |
| GO:0000314 | 18 | organellar_small_ribosomal_subunit |
| GO:0001502 | 18 | cartilage_condensation |
| GO:0001782 | 18 | B_cell_homeostasis |
| GO:0001917 | 18 | photoreceptor_inner_segment |
| GO:0002076 | 18 | osteoblast_development |
| GO:0002209 | 18 | behavioral_defense_response |
| GO:0002861 | 18 | regulation_of_inflammatory_response_to_antigenic_stimulus |
| GO:0003279 | 18 | cardiac_septum_development |
| GO:0004033 | 18 | aldo-keto_reductase_(NADP)_activity |
| GO:0004970 | 18 | ionotropic_glutamate_receptor_activity |
| GO:0005001 | 18 | transmembrane_receptor_protein_tyrosine_phosphatase_activity |
| GO:0005160 | 18 | transforming_growth_factor_beta_receptor_binding |
| GO:0005212 | 18 | structural_constituent_of_eye_lens |
| GO:0005542 | 18 | folic_acid_binding |
| GO:0005547 | 18 | phosphatidylinositol-3,4,5-trisphosphate_binding |
| GO:0005657 | 18 | replication_fork |
| GO:0005763 | 18 | mitochondrial_small_ribosomal_subunit |
| GO:0005771 | 18 | multivesicular_body |
| GO:0005844 | 18 | polysome |
| GO:0005859 | 18 | muscle_myosin_complex |
| GO:0005980 | 18 | glycogen_catabolic_process |
| GO:0006027 | 18 | glycosaminoglycan_catabolic_process |
| GO:0006241 | 18 | CTP_biosynthetic_process |
| GO:0006376 | 18 | mRNA_splice_site_selection |
| GO:0006385 | 18 | transcription_elongation_from_RNA_polymerase_III_promoter |
| GO:0006386 | 18 | termination_of_RNA_polymerase_III_transcription |
| GO:0006400 | 18 | tRNA_modification |
| GO:0006516 | 18 | glycoprotein_catabolic_process |
| GO:0006541 | 18 | glutamine_metabolic_process |
| GO:0006555 | 18 | methionine_metabolic_process |
| GO:0006656 | 18 | phosphatidylcholine_biosynthetic_process |
| GO:0007214 | 18 | gamma-aminobutyric_acid_signaling_pathway |
| GO:0007398 | 18 | ectoderm_development |
| GO:0007597 | 18 | blood_coagulation,_intrinsic_pathway |
| GO:0008320 | 18 | protein_transmembrane_transporter_activity |
| GO:0008484 | 18 | sulfuric_ester_hydrolase_activity |
| GO:0008527 | 18 | taste_receptor_activity |
| GO:0008603 | 18 | cAMP-dependent_protein_kinase_regulator_activity |
| GO:0009209 | 18 | pyrimidine_ribonucleoside_triphosphate_biosynthetic_process |
| GO:0009303 | 18 | rRNA_transcription |
| GO:0009950 | 18 | dorsal/ventral_axis_specification |
| GO:0010771 | 18 | negative_regulation_of_cell_morphogenesis_involved_in_differentiation |
| GO:0010869 | 18 | regulation_of_receptor_biosynthetic_process |
| GO:0010874 | 18 | regulation_of_cholesterol_efflux |
| GO:0010894 | 18 | negative_regulation_of_steroid_biosynthetic_process |
| GO:0014032 | 18 | neural_crest_cell_development |
| GO:0014704 | 18 | intercalated_disc |
| GO:0014743 | 18 | regulation_of_muscle_hypertrophy |
| GO:0015464 | 18 | acetylcholine_receptor_activity |
| GO:0015721 | 18 | bile_acid_and_bile_salt_transport |
| GO:0015949 | 18 | nucleobase-containing_small_molecule_interconversion |
| GO:0016064 | 18 | immunoglobulin_mediated_immune_response |
| GO:0016538 | 18 | cyclin-dependent_protein_kinase_regulator_activity |
| GO:0016638 | 18 | oxidoreductase_activity,_acting_on_the_CH-NH2_group_of_donors |
| GO:0016646 | 18 | oxidoreductase_activity,_acting_on_the_CH-NH_group_of_donors,_NAD_or_NADP_as_acceptor |
| GO:0016775 | 18 | phosphotransferase_activity,_nitrogenous_group_as_acceptor |
| GO:0017134 | 18 | fibroblast_growth_factor_binding |
| GO:0019198 | 18 | transmembrane_receptor_protein_phosphatase_activity |
| GO:0019321 | 18 | pentose_metabolic_process |
| GO:0022884 | 18 | macromolecule_transmembrane_transporter_activity |
| GO:0030239 | 18 | myofibril_assembly |
| GO:0030506 | 18 | ankyrin_binding |
| GO:0030539 | 18 | male_genitalia_development |
| GO:0030728 | 18 | ovulation |
| GO:0030878 | 18 | thyroid_gland_development |
| GO:0031362 | 18 | anchored_to_external_side_of_plasma_membrane |
| GO:0031575 | 18 | mitotic_cell_cycle_G1/S_transition_checkpoint |
| GO:0032012 | 18 | regulation_of_ARF_protein_signal_transduction |
| GO:0032420 | 18 | stereocilium |
| GO:0032452 | 18 | histone_demethylase_activity |
| GO:0032459 | 18 | regulation_of_protein_oligomerization |
| GO:0032660 | 18 | regulation_of_interleukin-17_production |
| GO:0032715 | 18 | negative_regulation_of_interleukin-6_production |
| GO:0032728 | 18 | positive_regulation_of_interferon-beta_production |
| GO:0032814 | 18 | regulation_of_natural_killer_cell_activation |
| GO:0032982 | 18 | myosin_filament |
| GO:0033146 | 18 | regulation_of_intracellular_estrogen_receptor_signaling_pathway |
| GO:0033558 | 18 | protein_deacetylase_activity |
| GO:0034394 | 18 | protein_localization_at_cell_surface |
| GO:0035338 | 18 | long-chain_fatty-acyl-CoA_biosynthetic_process |
| GO:0035813 | 18 | regulation_of_renal_sodium_excretion |
| GO:0035924 | 18 | cellular_response_to_vascular_endothelial_growth_factor_stimulus |
| GO:0042089 | 18 | cytokine_biosynthetic_process |
| GO:0042104 | 18 | positive_regulation_of_activated_T_cell_proliferation |
| GO:0042375 | 18 | quinone_cofactor_metabolic_process |
| GO:0042551 | 18 | neuron_maturation |
| GO:0042572 | 18 | retinol_metabolic_process |
| GO:0042974 | 18 | retinoic_acid_receptor_binding |
| GO:0043299 | 18 | leukocyte_degranulation |
| GO:0043620 | 18 | regulation_of_DNA-dependent_transcription_in_response_to_stress |
| GO:0044253 | 18 | positive_regulation_of_multicellular_organismal_metabolic_process |
| GO:0045061 | 18 | thymic_T_cell_selection |
| GO:0045453 | 18 | bone_resorption |
| GO:0045736 | 18 | negative_regulation_of_cyclin-dependent_protein_kinase_activity |
| GO:0045879 | 18 | negative_regulation_of_smoothened_signaling_pathway |
| GO:0046036 | 18 | CTP_metabolic_process |
| GO:0046434 | 18 | organophosphate_catabolic_process |
| GO:0046636 | 18 | negative_regulation_of_alpha-beta_T_cell_activation |
| GO:0046685 | 18 | response_to_arsenic-containing_substance |
| GO:0046885 | 18 | regulation_of_hormone_biosynthetic_process |
| GO:0048041 | 18 | focal_adhesion_assembly |
| GO:0048048 | 18 | embryonic_eye_morphogenesis |
| GO:0050706 | 18 | regulation_of_interleukin-1_beta_secretion |
| GO:0051293 | 18 | establishment_of_spindle_localization |
| GO:0051303 | 18 | establishment_of_chromosome_localization |
| GO:0051898 | 18 | negative_regulation_of_protein_kinase_B_signaling_cascade |
| GO:0055117 | 18 | regulation_of_cardiac_muscle_contraction |
| GO:0060135 | 18 | maternal_process_involved_in_female_pregnancy |
| GO:0060416 | 18 | response_to_growth_hormone_stimulus |
| GO:0060487 | 18 | lung_epithelial_cell_differentiation |
| GO:0060612 | 18 | adipose_tissue_development |
| GO:0070168 | 18 | negative_regulation_of_biomineral_tissue_development |
| GO:0072215 | 18 | regulation_of_metanephros_development |
| GO:0072341 | 18 | modified_amino_acid_binding |
| GO:0090181 | 18 | regulation_of_cholesterol_metabolic_process |
| GO:0090184 | 18 | positive_regulation_of_kidney_development |
| GO:0090342 | 18 | regulation_of_cell_aging |
| GO:2000191 | 18 | regulation_of_fatty_acid_transport |
| GO:2000273 | 18 | positive_regulation_of_receptor_activity |
| GO:2001258 | 18 | negative_regulation_of_cation_channel_activity |
| GO:0000030 | 17 | mannosyltransferase_activity |
| GO:0000299 | 17 | integral_to_membrane_of_membrane_fraction |
| GO:0000307 | 17 | cyclin-dependent_protein_kinase_holoenzyme_complex |
| GO:0000381 | 17 | regulation_of_alternative_nuclear_mRNA_splicing,_via_spliceosome |
| GO:0000784 | 17 | nuclear_chromosome,_telomeric_region |
| GO:0001106 | 17 | RNA_polymerase_II_transcription_corepressor_activity |
| GO:0001522 | 17 | pseudouridine_synthesis |
| GO:0001662 | 17 | behavioral_fear_response |
| GO:0001824 | 17 | blastocyst_development |
| GO:0001909 | 17 | leukocyte_mediated_cytotoxicity |
| GO:0001935 | 17 | endothelial_cell_proliferation |
| GO:0001953 | 17 | negative_regulation_of_cell-matrix_adhesion |
| GO:0002820 | 17 | negative_regulation_of_adaptive_immune_response |
| GO:0003206 | 17 | cardiac_chamber_morphogenesis |
| GO:0003254 | 17 | regulation_of_membrane_depolarization |
| GO:0003333 | 17 | amino_acid_transmembrane_transport |
| GO:0004407 | 17 | histone_deacetylase_activity |
| GO:0004675 | 17 | transmembrane_receptor_protein_serine/threonine_kinase_activity |
| GO:0004806 | 17 | triglyceride_lipase_activity |
| GO:0004835 | 17 | tubulin-tyrosine_ligase_activity |
| GO:0005003 | 17 | ephrin_receptor_activity |
| GO:0005024 | 17 | transforming_growth_factor_beta-activated_receptor_activity |
| GO:0005537 | 17 | mannose_binding |
| GO:0005921 | 17 | gap_junction |
| GO:0005978 | 17 | glycogen_biosynthetic_process |
| GO:0006000 | 17 | fructose_metabolic_process |
| GO:0006359 | 17 | regulation_of_transcription_from_RNA_polymerase_III_promoter |
| GO:0006544 | 17 | glycine_metabolic_process |
| GO:0006750 | 17 | glutathione_biosynthetic_process |
| GO:0006906 | 17 | vesicle_fusion |
| GO:0006972 | 17 | hyperosmotic_response |
| GO:0007029 | 17 | endoplasmic_reticulum_organization |
| GO:0007127 | 17 | meiosis_I |
| GO:0007250 | 17 | activation_of_NF-kappaB-inducing_kinase_activity |
| GO:0007274 | 17 | neuromuscular_synaptic_transmission |
| GO:0009214 | 17 | cyclic_nucleotide_catabolic_process |
| GO:0009247 | 17 | glycolipid_biosynthetic_process |
| GO:0009250 | 17 | glucan_biosynthetic_process |
| GO:0010390 | 17 | histone_monoubiquitination |
| GO:0010611 | 17 | regulation_of_cardiac_muscle_hypertrophy |
| GO:0010922 | 17 | positive_regulation_of_phosphatase_activity |
| GO:0015269 | 17 | calcium-activated_potassium_channel_activity |
| GO:0015299 | 17 | solute:hydrogen_antiporter_activity |
| GO:0015697 | 17 | quaternary_ammonium_group_transport |
| GO:0015800 | 17 | acidic_amino_acid_transport |
| GO:0015932 | 17 | nucleobase-containing_compound_transmembrane_transporter_activity |
| GO:0016241 | 17 | regulation_of_macroautophagy |
| GO:0016289 | 17 | CoA_hydrolase_activity |
| GO:0016780 | 17 | phosphotransferase_activity,_for_other_substituted_phosphate_groups |
| GO:0016892 | 17 | endoribonuclease_activity,_producing_3'-phosphomonoesters |
| GO:0019359 | 17 | nicotinamide_nucleotide_biosynthetic_process |
| GO:0019363 | 17 | pyridine_nucleotide_biosynthetic_process |
| GO:0021545 | 17 | cranial_nerve_development |
| GO:0021795 | 17 | cerebral_cortex_cell_migration |
| GO:0023019 | 17 | signal_transduction_involved_in_regulation_of_gene_expression |
| GO:0030041 | 17 | actin_filament_polymerization |
| GO:0030104 | 17 | water_homeostasis |
| GO:0030212 | 17 | hyaluronan_metabolic_process |
| GO:0030224 | 17 | monocyte_differentiation |
| GO:0030449 | 17 | regulation_of_complement_activation |
| GO:0030866 | 17 | cortical_actin_cytoskeleton_organization |
| GO:0031057 | 17 | negative_regulation_of_histone_modification |
| GO:0031060 | 17 | regulation_of_histone_methylation |
| GO:0031365 | 17 | N-terminal_protein_amino_acid_modification |
| GO:0032105 | 17 | negative_regulation_of_response_to_extracellular_stimulus |
| GO:0032108 | 17 | negative_regulation_of_response_to_nutrient_levels |
| GO:0032266 | 17 | phosphatidylinositol-3-phosphate_binding |
| GO:0032391 | 17 | photoreceptor_connecting_cilium |
| GO:0032689 | 17 | negative_regulation_of_interferon-gamma_production |
| GO:0032733 | 17 | positive_regulation_of_interleukin-10_production |
| GO:0032770 | 17 | positive_regulation_of_monooxygenase_activity |
| GO:0033006 | 17 | regulation_of_mast_cell_activation_involved_in_immune_response |
| GO:0033158 | 17 | regulation_of_protein_import_into_nucleus,_translocation |
| GO:0033630 | 17 | positive_regulation_of_cell_adhesion_mediated_by_integrin |
| GO:0033688 | 17 | regulation_of_osteoblast_proliferation |
| GO:0035710 | 17 | CD4-positive,_alpha-beta_T_cell_activation |
| GO:0035725 | 17 | sodium_ion_transmembrane_transport |
| GO:0035809 | 17 | regulation_of_urine_volume |
| GO:0042403 | 17 | thyroid_hormone_metabolic_process |
| GO:0042809 | 17 | vitamin_D_receptor_binding |
| GO:0043029 | 17 | T_cell_homeostasis |
| GO:0043073 | 17 | germ_cell_nucleus |
| GO:0043088 | 17 | regulation_of_Cdc42_GTPase_activity |
| GO:0043330 | 17 | response_to_exogenous_dsRNA |
| GO:0043372 | 17 | positive_regulation_of_CD4-positive,_alpha-beta_T_cell_differentiation |
| GO:0043691 | 17 | reverse_cholesterol_transport |
| GO:0044036 | 17 | cell_wall_macromolecule_metabolic_process |
| GO:0044144 | 17 | modulation_of_growth_of_symbiont_involved_in_interaction_with_host |
| GO:0044146 | 17 | negative_regulation_of_growth_of_symbiont_involved_in_interaction_with_host |
| GO:0044462 | 17 | external_encapsulating_structure_part |
| GO:0045191 | 17 | regulation_of_isotype_switching |
| GO:0045671 | 17 | negative_regulation_of_osteoclast_differentiation |
| GO:0045684 | 17 | positive_regulation_of_epidermis_development |
| GO:0045730 | 17 | respiratory_burst |
| GO:0045880 | 17 | positive_regulation_of_smoothened_signaling_pathway |
| GO:0045909 | 17 | positive_regulation_of_vasodilation |
| GO:0046697 | 17 | decidualization |
| GO:0046716 | 17 | muscle_cell_homeostasis |
| GO:0046839 | 17 | phospholipid_dephosphorylation |
| GO:0048477 | 17 | oogenesis |
| GO:0048557 | 17 | embryonic_digestive_tract_morphogenesis |
| GO:0048745 | 17 | smooth_muscle_tissue_development |
| GO:0048753 | 17 | pigment_granule_organization |
| GO:0050832 | 17 | defense_response_to_fungus |
| GO:0050995 | 17 | negative_regulation_of_lipid_catabolic_process |
| GO:0051018 | 17 | protein_kinase_A_binding |
| GO:0051019 | 17 | mitogen-activated_protein_kinase_binding |
| GO:0051155 | 17 | positive_regulation_of_striated_muscle_cell_differentiation |
| GO:0051183 | 17 | vitamin_transporter_activity |
| GO:0051491 | 17 | positive_regulation_of_filopodium_assembly |
| GO:0051568 | 17 | histone_H3-K4_methylation |
| GO:0051635 | 17 | bacterial_cell_surface_binding |
| GO:0051879 | 17 | Hsp90_protein_binding |
| GO:0052866 | 17 | phosphatidylinositol_phosphate_phosphatase_activity |
| GO:0060348 | 17 | bone_development |
| GO:0060395 | 17 | SMAD_protein_signal_transduction |
| GO:0060600 | 17 | dichotomous_subdivision_of_an_epithelial_terminal_unit |
| GO:0060644 | 17 | mammary_gland_epithelial_cell_differentiation |
| GO:0060760 | 17 | positive_regulation_of_response_to_cytokine_stimulus |
| GO:0060788 | 17 | ectodermal_placode_formation |
| GO:0070207 | 17 | protein_homotrimerization |
| GO:0070271 | 17 | protein_complex_biogenesis |
| GO:0070412 | 17 | R-SMAD_binding |
| GO:0071285 | 17 | cellular_response_to_lithium_ion |
| GO:0071312 | 17 | cellular_response_to_alkaloid |
| GO:0071514 | 17 | genetic_imprinting |
| GO:0071855 | 17 | neuropeptide_receptor_binding |
| GO:0072525 | 17 | pyridine-containing_compound_biosynthetic_process |
| GO:2000257 | 17 | regulation_of_protein_activation_cascade |
| GO:2000311 | 17 | regulation_of_alpha-amino-3-hydroxy-5-methyl-4-isoxazole_propionate_selective_glutamate_receptor_activity |
| GO:2000736 | 17 | regulation_of_stem_cell_differentiation |
| GO:0000070 | 16 | mitotic_sister_chromatid_segregation |
| GO:0000301 | 16 | retrograde_transport,_vesicle_recycling_within_Golgi |
| GO:0000421 | 16 | autophagic_vacuole_membrane |
| GO:0001914 | 16 | regulation_of_T_cell_mediated_cytotoxicity |
| GO:0001961 | 16 | positive_regulation_of_cytokine-mediated_signaling_pathway |
| GO:0002717 | 16 | positive_regulation_of_natural_killer_cell_mediated_immunity |
| GO:0003071 | 16 | renal_system_process_involved_in_regulation_of_systemic_arterial_blood_pressure |
| GO:0004198 | 16 | calcium-dependent_cysteine-type_endopeptidase_activity |
| GO:0004659 | 16 | prenyltransferase_activity |
| GO:0004697 | 16 | protein_kinase_C_activity |
| GO:0004708 | 16 | MAP_kinase_kinase_activity |
| GO:0004889 | 16 | acetylcholine-activated_cation-selective_channel_activity |
| GO:0005892 | 16 | acetylcholine-gated_channel_complex |
| GO:0006063 | 16 | uronic_acid_metabolic_process |
| GO:0006337 | 16 | nucleosome_disassembly |
| GO:0006525 | 16 | arginine_metabolic_process |
| GO:0006651 | 16 | diacylglycerol_biosynthetic_process |
| GO:0006893 | 16 | Golgi_to_plasma_membrane_transport |
| GO:0008329 | 16 | pattern_recognition_receptor_activity |
| GO:0009435 | 16 | NAD_biosynthetic_process |
| GO:0010714 | 16 | positive_regulation_of_collagen_metabolic_process |
| GO:0010830 | 16 | regulation_of_myotube_differentiation |
| GO:0014047 | 16 | glutamate_secretion |
| GO:0014059 | 16 | regulation_of_dopamine_secretion |
| GO:0014911 | 16 | positive_regulation_of_smooth_muscle_cell_migration |
| GO:0015813 | 16 | L-glutamate_transport |
| GO:0016010 | 16 | dystrophin-associated_glycoprotein_complex |
| GO:0016137 | 16 | glycoside_metabolic_process |
| GO:0016254 | 16 | preassembly_of_GPI_anchor_in_ER_membrane |
| GO:0016578 | 16 | histone_deubiquitination |
| GO:0016581 | 16 | NuRD_complex |
| GO:0016863 | 16 | intramolecular_oxidoreductase_activity,_transposing_C=C_bonds |
| GO:0016896 | 16 | exoribonuclease_activity,_producing_5'-phosphomonoesters |
| GO:0016998 | 16 | cell_wall_macromolecule_catabolic_process |
| GO:0017080 | 16 | sodium_channel_regulator_activity |
| GO:0019585 | 16 | glucuronate_metabolic_process |
| GO:0021537 | 16 | telencephalon_development |
| GO:0022410 | 16 | circadian_sleep/wake_cycle_process |
| GO:0030502 | 16 | negative_regulation_of_bone_mineralization |
| GO:0030517 | 16 | negative_regulation_of_axon_extension |
| GO:0030857 | 16 | negative_regulation_of_epithelial_cell_differentiation |
| GO:0030949 | 16 | positive_regulation_of_vascular_endothelial_growth_factor_receptor_signaling_pathway |
| GO:0031103 | 16 | axon_regeneration |
| GO:0031435 | 16 | mitogen-activated_protein_kinase_kinase_kinase_binding |
| GO:0031528 | 16 | microvillus_membrane |
| GO:0031674 | 16 | I_band |
| GO:0031984 | 16 | organelle_subcompartment |
| GO:0032095 | 16 | regulation_of_response_to_food |
| GO:0032332 | 16 | positive_regulation_of_chondrocyte_differentiation |
| GO:0032438 | 16 | melanosome_organization |
| GO:0032781 | 16 | positive_regulation_of_ATPase_activity |
| GO:0032786 | 16 | positive_regulation_of_DNA-dependent_transcription,_elongation |
| GO:0032816 | 16 | positive_regulation_of_natural_killer_cell_activation |
| GO:0032967 | 16 | positive_regulation_of_collagen_biosynthetic_process |
| GO:0032986 | 16 | protein-DNA_complex_disassembly |
| GO:0034199 | 16 | activation_of_protein_kinase_A_activity |
| GO:0034308 | 16 | primary_alcohol_metabolic_process |
| GO:0035035 | 16 | histone_acetyltransferase_binding |
| GO:0035121 | 16 | tail_morphogenesis |
| GO:0035637 | 16 | multicellular_organismal_signaling |
| GO:0035914 | 16 | skeletal_muscle_cell_differentiation |
| GO:0040001 | 16 | establishment_of_mitotic_spindle_localization |
| GO:0042056 | 16 | chemoattractant_activity |
| GO:0042119 | 16 | neutrophil_activation |
| GO:0042162 | 16 | telomeric_DNA_binding |
| GO:0042523 | 16 | positive_regulation_of_tyrosine_phosphorylation_of_Stat5_protein |
| GO:0042534 | 16 | regulation_of_tumor_necrosis_factor_biosynthetic_process |
| GO:0042659 | 16 | regulation_of_cell_fate_specification |
| GO:0042805 | 16 | actinin_binding |
| GO:0042994 | 16 | cytoplasmic_sequestering_of_transcription_factor |
| GO:0043034 | 16 | costamere |
| GO:0043101 | 16 | purine-containing_compound_salvage |
| GO:0043189 | 16 | H4/H2A_histone_acetyltransferase_complex |
| GO:0043304 | 16 | regulation_of_mast_cell_degranulation |
| GO:0043367 | 16 | CD4-positive,_alpha-beta_T_cell_differentiation |
| GO:0043394 | 16 | proteoglycan_binding |
| GO:0043496 | 16 | regulation_of_protein_homodimerization_activity |
| GO:0043537 | 16 | negative_regulation_of_blood_vessel_endothelial_cell_migration |
| GO:0044126 | 16 | regulation_of_growth_of_symbiont_in_host |
| GO:0044130 | 16 | negative_regulation_of_growth_of_symbiont_in_host |
| GO:0045072 | 16 | regulation_of_interferon-gamma_biosynthetic_process |
| GO:0045648 | 16 | positive_regulation_of_erythrocyte_differentiation |
| GO:0045672 | 16 | positive_regulation_of_osteoclast_differentiation |
| GO:0045747 | 16 | positive_regulation_of_Notch_signaling_pathway |
| GO:0045838 | 16 | positive_regulation_of_membrane_potential |
| GO:0045922 | 16 | negative_regulation_of_fatty_acid_metabolic_process |
| GO:0045932 | 16 | negative_regulation_of_muscle_contraction |
| GO:0046134 | 16 | pyrimidine_nucleoside_biosynthetic_process |
| GO:0046653 | 16 | tetrahydrofolate_metabolic_process |
| GO:0046933 | 16 | hydrogen_ion_transporting_ATP_synthase_activity,_rotational_mechanism |
| GO:0048384 | 16 | retinoic_acid_receptor_signaling_pathway |
| GO:0048566 | 16 | embryonic_digestive_tract_development |
| GO:0048635 | 16 | negative_regulation_of_muscle_organ_development |
| GO:0048665 | 16 | neuron_fate_specification |
| GO:0050716 | 16 | positive_regulation_of_interleukin-1_secretion |
| GO:0050718 | 16 | positive_regulation_of_interleukin-1_beta_secretion |
| GO:0050974 | 16 | detection_of_mechanical_stimulus_involved_in_sensory_perception |
| GO:0051043 | 16 | regulation_of_membrane_protein_ectodomain_proteolysis |
| GO:0051184 | 16 | cofactor_transporter_activity |
| GO:0051953 | 16 | negative_regulation_of_amine_transport |
| GO:0055081 | 16 | anion_homeostasis |
| GO:0055102 | 16 | lipase_inhibitor_activity |
| GO:0060045 | 16 | positive_regulation_of_cardiac_muscle_cell_proliferation |
| GO:0060412 | 16 | ventricular_septum_morphogenesis |
| GO:0060441 | 16 | epithelial_tube_branching_involved_in_lung_morphogenesis |
| GO:0060706 | 16 | cell_differentiation_involved_in_embryonic_placenta_development |
| GO:0060968 | 16 | regulation_of_gene_silencing |
| GO:0061001 | 16 | regulation_of_dendritic_spine_morphogenesis |
| GO:0070076 | 16 | histone_lysine_demethylation |
| GO:0070229 | 16 | negative_regulation_of_lymphocyte_apoptotic_process |
| GO:0070232 | 16 | regulation_of_T_cell_apoptotic_process |
| GO:0071556 | 16 | integral_to_lumenal_side_of_endoplasmic_reticulum_membrane |
| GO:0090150 | 16 | establishment_of_protein_localization_in_membrane |
| GO:0090208 | 16 | positive_regulation_of_triglyceride_metabolic_process |
| GO:0090311 | 16 | regulation_of_protein_deacetylation |
| GO:2000142 | 16 | regulation_of_DNA-dependent_transcription,_initiation |
| GO:2000758 | 16 | positive_regulation_of_peptidyl-lysine_acetylation |
| GO:2001021 | 16 | negative_regulation_of_response_to_DNA_damage_stimulus |
| GO:2001234 | 16 | negative_regulation_of_apoptotic_signaling_pathway |
| GO:0000012 | 15 | single_strand_break_repair |
| GO:0000266 | 15 | mitochondrial_fission |
| GO:0000315 | 15 | organellar_large_ribosomal_subunit |
| GO:0000930 | 15 | gamma-tubulin_complex |
| GO:0000959 | 15 | mitochondrial_RNA_metabolic_process |
| GO:0000980 | 15 | RNA_polymerase_II_distal_enhancer_sequence-specific_DNA_binding |
| GO:0001504 | 15 | neurotransmitter_uptake |
| GO:0001516 | 15 | prostaglandin_biosynthetic_process |
| GO:0001556 | 15 | oocyte_maturation |
| GO:0001784 | 15 | phosphotyrosine_binding |
| GO:0001786 | 15 | phosphatidylserine_binding |
| GO:0001882 | 15 | nucleoside_binding |
| GO:0001919 | 15 | regulation_of_receptor_recycling |
| GO:0002040 | 15 | sprouting_angiogenesis |
| GO:0002437 | 15 | inflammatory_response_to_antigenic_stimulus |
| GO:0002686 | 15 | negative_regulation_of_leukocyte_migration |
| GO:0002823 | 15 | negative_regulation_of_adaptive_immune_response_based_on_somatic_recombination_of_immune_receptors_built_from_immunoglobulin_superfamily_domains |
| GO:0003009 | 15 | skeletal_muscle_contraction |
| GO:0003208 | 15 | cardiac_ventricle_morphogenesis |
| GO:0003995 | 15 | acyl-CoA_dehydrogenase_activity |
| GO:0004012 | 15 | phospholipid-translocating_ATPase_activity |
| GO:0004089 | 15 | carbonate_dehydratase_activity |
| GO:0004143 | 15 | diacylglycerol_kinase_activity |
| GO:0004523 | 15 | ribonuclease_H_activity |
| GO:0004602 | 15 | glutathione_peroxidase_activity |
| GO:0004707 | 15 | MAP_kinase_activity |
| GO:0005247 | 15 | voltage-gated_chloride_channel_activity |
| GO:0005248 | 15 | voltage-gated_sodium_channel_activity |
| GO:0005523 | 15 | tropomyosin_binding |
| GO:0005527 | 15 | macrolide_binding |
| GO:0005528 | 15 | FK506_binding |
| GO:0005740 | 15 | mitochondrial_envelope |
| GO:0005762 | 15 | mitochondrial_large_ribosomal_subunit |
| GO:0005852 | 15 | eukaryotic_translation_initiation_factor_3_complex |
| GO:0006013 | 15 | mannose_metabolic_process |
| GO:0006171 | 15 | cAMP_biosynthetic_process |
| GO:0006182 | 15 | cGMP_biosynthetic_process |
| GO:0006198 | 15 | cAMP_catabolic_process |
| GO:0006577 | 15 | betaine_metabolic_process |
| GO:0006688 | 15 | glycosphingolipid_biosynthetic_process |
| GO:0006825 | 15 | copper_ion_transport |
| GO:0006884 | 15 | cell_volume_homeostasis |
| GO:0007271 | 15 | synaptic_transmission,_cholinergic |
| GO:0007413 | 15 | axonal_fasciculation |
| GO:0008211 | 15 | glucocorticoid_metabolic_process |
| GO:0008353 | 15 | RNA_polymerase_II_carboxy-terminal_domain_kinase_activity |
| GO:0008430 | 15 | selenium_binding |
| GO:0008625 | 15 | induction_of_apoptosis_via_death_domain_receptors |
| GO:0009068 | 15 | aspartate_family_amino_acid_catabolic_process |
| GO:0009086 | 15 | methionine_biosynthetic_process |
| GO:0009219 | 15 | pyrimidine_deoxyribonucleotide_metabolic_process |
| GO:0009264 | 15 | deoxyribonucleotide_catabolic_process |
| GO:0010634 | 15 | positive_regulation_of_epithelial_cell_migration |
| GO:0010824 | 15 | regulation_of_centrosome_duplication |
| GO:0010893 | 15 | positive_regulation_of_steroid_biosynthetic_process |
| GO:0014048 | 15 | regulation_of_glutamate_secretion |
| GO:0015145 | 15 | monosaccharide_transmembrane_transporter_activity |
| GO:0015278 | 15 | calcium-release_channel_activity |
| GO:0015669 | 15 | gas_transport |
| GO:0015701 | 15 | bicarbonate_transport |
| GO:0015923 | 15 | mannosidase_activity |
| GO:0016079 | 15 | synaptic_vesicle_exocytosis |
| GO:0016408 | 15 | C-acyltransferase_activity |
| GO:0016411 | 15 | acylglycerol_O-acyltransferase_activity |
| GO:0016514 | 15 | SWI/SNF_complex |
| GO:0017069 | 15 | snRNA_binding |
| GO:0030260 | 15 | entry_into_host_cell |
| GO:0030520 | 15 | intracellular_estrogen_receptor_signaling_pathway |
| GO:0031112 | 15 | positive_regulation_of_microtubule_polymerization_or_depolymerization |
| GO:0031146 | 15 | SCF-dependent_proteasomal_ubiquitin-dependent_protein_catabolic_process |
| GO:0031290 | 15 | retinal_ganglion_cell_axon_guidance |
| GO:0031293 | 15 | membrane_protein_intracellular_domain_proteolysis |
| GO:0031306 | 15 | intrinsic_to_mitochondrial_outer_membrane |
| GO:0031998 | 15 | regulation_of_fatty_acid_beta-oxidation |
| GO:0032225 | 15 | regulation_of_synaptic_transmission,_dopaminergic |
| GO:0032300 | 15 | mismatch_repair_complex |
| GO:0032461 | 15 | positive_regulation_of_protein_oligomerization |
| GO:0032467 | 15 | positive_regulation_of_cytokinesis |
| GO:0032494 | 15 | response_to_peptidoglycan |
| GO:0032838 | 15 | cell_projection_cytoplasm |
| GO:0032892 | 15 | positive_regulation_of_organic_acid_transport |
| GO:0032924 | 15 | activin_receptor_signaling_pathway |
| GO:0033119 | 15 | negative_regulation_of_RNA_splicing |
| GO:0033151 | 15 | V(D)J_recombination |
| GO:0033549 | 15 | MAP_kinase_phosphatase_activity |
| GO:0033875 | 15 | ribonucleoside_bisphosphate_metabolic_process |
| GO:0034032 | 15 | purine_nucleoside_bisphosphate_metabolic_process |
| GO:0034695 | 15 | response_to_prostaglandin_E_stimulus |
| GO:0034979 | 15 | NAD-dependent_protein_deacetylase_activity |
| GO:0035066 | 15 | positive_regulation_of_histone_acetylation |
| GO:0035265 | 15 | organ_growth |
| GO:0035267 | 15 | NuA4_histone_acetyltransferase_complex |
| GO:0035510 | 15 | DNA_dealkylation |
| GO:0042554 | 15 | superoxide_anion_generation |
| GO:0042749 | 15 | regulation_of_circadian_sleep/wake_cycle |
| GO:0042772 | 15 | DNA_damage_response,_signal_transduction_resulting_in_transcription |
| GO:0042776 | 15 | mitochondrial_ATP_synthesis_coupled_proton_transport |
| GO:0042982 | 15 | amyloid_precursor_protein_metabolic_process |
| GO:0043401 | 15 | steroid_hormone_mediated_signaling_pathway |
| GO:0043576 | 15 | regulation_of_respiratory_gaseous_exchange |
| GO:0043981 | 15 | histone_H4-K5_acetylation |
| GO:0043982 | 15 | histone_H4-K8_acetylation |
| GO:0044409 | 15 | entry_into_host |
| GO:0045022 | 15 | early_endosome_to_late_endosome_transport |
| GO:0045120 | 15 | pronucleus |
| GO:0045187 | 15 | regulation_of_circadian_sleep/wake_cycle,_sleep |
| GO:0045445 | 15 | myoblast_differentiation |
| GO:0045446 | 15 | endothelial_cell_differentiation |
| GO:0045737 | 15 | positive_regulation_of_cyclin-dependent_protein_kinase_activity |
| GO:0045745 | 15 | positive_regulation_of_G-protein_coupled_receptor_protein_signaling_pathway |
| GO:0045746 | 15 | negative_regulation_of_Notch_signaling_pathway |
| GO:0045815 | 15 | positive_regulation_of_gene_expression,_epigenetic |
| GO:0045851 | 15 | pH_reduction |
| GO:0045947 | 15 | negative_regulation_of_translational_initiation |
| GO:0045954 | 15 | positive_regulation_of_natural_killer_cell_mediated_cytotoxicity |
| GO:0046112 | 15 | nucleobase_biosynthetic_process |
| GO:0046457 | 15 | prostanoid_biosynthetic_process |
| GO:0046622 | 15 | positive_regulation_of_organ_growth |
| GO:0046641 | 15 | positive_regulation_of_alpha-beta_T_cell_proliferation |
| GO:0048020 | 15 | CCR_chemokine_receptor_binding |
| GO:0048208 | 15 | COPII_vesicle_coating |
| GO:0048546 | 15 | digestive_tract_morphogenesis |
| GO:0048645 | 15 | organ_formation |
| GO:0048709 | 15 | oligodendrocyte_differentiation |
| GO:0048846 | 15 | axon_extension_involved_in_axon_guidance |
| GO:0050253 | 15 | retinyl-palmitate_esterase_activity |
| GO:0050664 | 15 | oxidoreductase_activity,_acting_on_NADH_or_NADPH,_oxygen_as_acceptor |
| GO:0050829 | 15 | defense_response_to_Gram-negative_bacterium |
| GO:0050997 | 15 | quaternary_ammonium_group_binding |
| GO:0051150 | 15 | regulation_of_smooth_muscle_cell_differentiation |
| GO:0051721 | 15 | protein_phosphatase_2A_binding |
| GO:0051806 | 15 | entry_into_cell_of_other_organism_involved_in_symbiotic_interaction |
| GO:0051828 | 15 | entry_into_other_organism_involved_in_symbiotic_interaction |
| GO:0051861 | 15 | glycolipid_binding |
| GO:0051923 | 15 | sulfation |
| GO:0052126 | 15 | movement_in_host_environment |
| GO:0052192 | 15 | movement_in_environment_of_other_organism_involved_in_symbiotic_interaction |
| GO:0052742 | 15 | phosphatidylinositol_kinase_activity |
| GO:0052745 | 15 | inositol_phosphate_phosphatase_activity |
| GO:0055006 | 15 | cardiac_cell_development |
| GO:0055013 | 15 | cardiac_muscle_cell_development |
| GO:0055070 | 15 | copper_ion_homeostasis |
| GO:0060081 | 15 | membrane_hyperpolarization |
| GO:0060231 | 15 | mesenchymal_to_epithelial_transition |
| GO:0060716 | 15 | labyrinthine_layer_blood_vessel_development |
| GO:0060749 | 15 | mammary_gland_alveolus_development |
| GO:0061178 | 15 | regulation_of_insulin_secretion_involved_in_cellular_response_to_glucose_stimulus |
| GO:0070063 | 15 | RNA_polymerase_binding |
| GO:0070064 | 15 | proline-rich_region_binding |
| GO:0070536 | 15 | protein_K63-linked_deubiquitination |
| GO:0070875 | 15 | positive_regulation_of_glycogen_metabolic_process |
| GO:0071359 | 15 | cellular_response_to_dsRNA |
| GO:0071391 | 15 | cellular_response_to_estrogen_stimulus |
| GO:0071526 | 15 | semaphorin-plexin_signaling_pathway |
| GO:0071695 | 15 | anatomical_structure_maturation |
| GO:0071837 | 15 | HMG_box_domain_binding |
| GO:0071897 | 15 | DNA_biosynthetic_process |
| GO:0072600 | 15 | establishment_of_protein_localization_in_Golgi |
| GO:0090002 | 15 | establishment_of_protein_localization_in_plasma_membrane |
| GO:0090023 | 15 | positive_regulation_of_neutrophil_chemotaxis |
| GO:2000108 | 15 | positive_regulation_of_leukocyte_apoptotic_process |
| GO:2000278 | 15 | regulation_of_DNA_biosynthetic_process |
| GO:2000677 | 15 | regulation_of_transcription_regulatory_region_DNA_binding |
| GO:2000779 | 15 | regulation_of_double-strand_break_repair |
| GO:2001238 | 15 | positive_regulation_of_extrinsic_apoptotic_signaling_pathway |
| GO:2001244 | 15 | positive_regulation_of_intrinsic_apoptotic_signaling_pathway |
| GO:0000042 | 14 | protein_targeting_to_Golgi |
| GO:0000132 | 14 | establishment_of_mitotic_spindle_orientation |
| GO:0000175 | 14 | 3'-5'-exoribonuclease_activity |
| GO:0000217 | 14 | DNA_secondary_structure_binding |
| GO:0000242 | 14 | pericentriolar_material |
| GO:0000305 | 14 | response_to_oxygen_radical |
| GO:0000803 | 14 | sex_chromosome |
| GO:0001518 | 14 | voltage-gated_sodium_channel_complex |
| GO:0001655 | 14 | urogenital_system_development |
| GO:0001673 | 14 | male_germ_cell_nucleus |
| GO:0001759 | 14 | organ_induction |
| GO:0001916 | 14 | positive_regulation_of_T_cell_mediated_cytotoxicity |
| GO:0002042 | 14 | cell_migration_involved_in_sprouting_angiogenesis |
| GO:0002052 | 14 | positive_regulation_of_neuroblast_proliferation |
| GO:0002070 | 14 | epithelial_cell_maturation |
| GO:0002092 | 14 | positive_regulation_of_receptor_internalization |
| GO:0002204 | 14 | somatic_recombination_of_immunoglobulin_genes_involved_in_immune_response |
| GO:0002208 | 14 | somatic_diversification_of_immunoglobulins_involved_in_immune_response |
| GO:0002407 | 14 | dendritic_cell_chemotaxis |
| GO:0002704 | 14 | negative_regulation_of_leukocyte_mediated_immunity |
| GO:0002707 | 14 | negative_regulation_of_lymphocyte_mediated_immunity |
| GO:0002714 | 14 | positive_regulation_of_B_cell_mediated_immunity |
| GO:0002891 | 14 | positive_regulation_of_immunoglobulin_mediated_immune_response |
| GO:0003964 | 14 | RNA-directed_DNA_polymerase_activity |
| GO:0004622 | 14 | lysophospholipase_activity |
| GO:0004673 | 14 | protein_histidine_kinase_activity |
| GO:0004745 | 14 | retinol_dehydrogenase_activity |
| GO:0005035 | 14 | death_receptor_activity |
| GO:0005159 | 14 | insulin-like_growth_factor_receptor_binding |
| GO:0005310 | 14 | dicarboxylic_acid_transmembrane_transporter_activity |
| GO:0005385 | 14 | zinc_ion_transmembrane_transporter_activity |
| GO:0005605 | 14 | basal_lamina |
| GO:0005665 | 14 | DNA-directed_RNA_polymerase_II,_core_complex |
| GO:0005671 | 14 | Ada2/Gcn5/Ada3_transcription_activator_complex |
| GO:0005746 | 14 | mitochondrial_respiratory_chain |
| GO:0005779 | 14 | integral_to_peroxisomal_membrane |
| GO:0005849 | 14 | mRNA_cleavage_factor_complex |
| GO:0006067 | 14 | ethanol_metabolic_process |
| GO:0006301 | 14 | postreplication_repair |
| GO:0006349 | 14 | regulation_of_gene_expression_by_genetic_imprinting |
| GO:0006568 | 14 | tryptophan_metabolic_process |
| GO:0006590 | 14 | thyroid_hormone_generation |
| GO:0006607 | 14 | NLS-bearing_substrate_import_into_nucleus |
| GO:0006702 | 14 | androgen_biosynthetic_process |
| GO:0006809 | 14 | nitric_oxide_biosynthetic_process |
| GO:0006949 | 14 | syncytium_formation |
| GO:0006978 | 14 | DNA_damage_response,_signal_transduction_by_p53_class_mediator_resulting_in_transcription_of_p21_class_mediator |
| GO:0007164 | 14 | establishment_of_tissue_polarity |
| GO:0007263 | 14 | nitric_oxide_mediated_signal_transduction |
| GO:0008105 | 14 | asymmetric_protein_localization |
| GO:0008171 | 14 | O-methyltransferase_activity |
| GO:0008198 | 14 | ferrous_iron_binding |
| GO:0008417 | 14 | fucosyltransferase_activity |
| GO:0009070 | 14 | serine_family_amino_acid_biosynthetic_process |
| GO:0009071 | 14 | serine_family_amino_acid_catabolic_process |
| GO:0009129 | 14 | pyrimidine_nucleoside_monophosphate_metabolic_process |
| GO:0009698 | 14 | phenylpropanoid_metabolic_process |
| GO:0009881 | 14 | photoreceptor_activity |
| GO:0009982 | 14 | pseudouridine_synthase_activity |
| GO:0010181 | 14 | FMN_binding |
| GO:0010257 | 14 | NADH_dehydrogenase_complex_assembly |
| GO:0010288 | 14 | response_to_lead_ion |
| GO:0010324 | 14 | membrane_invagination |
| GO:0010507 | 14 | negative_regulation_of_autophagy |
| GO:0010623 | 14 | developmental_programmed_cell_death |
| GO:0010656 | 14 | negative_regulation_of_muscle_cell_apoptotic_process |
| GO:0010744 | 14 | positive_regulation_of_macrophage_derived_foam_cell_differentiation |
| GO:0010765 | 14 | positive_regulation_of_sodium_ion_transport |
| GO:0010866 | 14 | regulation_of_triglyceride_biosynthetic_process |
| GO:0010884 | 14 | positive_regulation_of_lipid_storage |
| GO:0010888 | 14 | negative_regulation_of_lipid_storage |
| GO:0014821 | 14 | phasic_smooth_muscle_contraction |
| GO:0015114 | 14 | phosphate_ion_transmembrane_transporter_activity |
| GO:0015116 | 14 | sulfate_transmembrane_transporter_activity |
| GO:0015149 | 14 | hexose_transmembrane_transporter_activity |
| GO:0015238 | 14 | drug_transmembrane_transporter_activity |
| GO:0015248 | 14 | sterol_transporter_activity |
| GO:0015295 | 14 | solute:hydrogen_symporter_activity |
| GO:0015645 | 14 | fatty_acid_ligase_activity |
| GO:0015929 | 14 | hexosaminidase_activity |
| GO:0015936 | 14 | coenzyme_A_metabolic_process |
| GO:0016073 | 14 | snRNA_metabolic_process |
| GO:0016572 | 14 | histone_phosphorylation |
| GO:0016641 | 14 | oxidoreductase_activity,_acting_on_the_CH-NH2_group_of_donors,_oxygen_as_acceptor |
| GO:0016805 | 14 | dipeptidase_activity |
| GO:0016846 | 14 | carbon-sulfur_lyase_activity |
| GO:0016861 | 14 | intramolecular_oxidoreductase_activity,_interconverting_aldoses_and_ketoses |
| GO:0017017 | 14 | MAP_kinase_tyrosine/serine/threonine_phosphatase_activity |
| GO:0017085 | 14 | response_to_insecticide |
| GO:0017091 | 14 | AU-rich_element_binding |
| GO:0017136 | 14 | NAD-dependent_histone_deacetylase_activity |
| GO:0018195 | 14 | peptidyl-arginine_modification |
| GO:0018198 | 14 | peptidyl-cysteine_modification |
| GO:0019028 | 14 | viral_capsid |
| GO:0019079 | 14 | viral_genome_replication |
| GO:0030126 | 14 | COPI_vesicle_coat |
| GO:0030228 | 14 | lipoprotein_particle_receptor_activity |
| GO:0030275 | 14 | LRR_domain_binding |
| GO:0030332 | 14 | cyclin_binding |
| GO:0030544 | 14 | Hsp70_protein_binding |
| GO:0030547 | 14 | receptor_inhibitor_activity |
| GO:0030675 | 14 | Rac_GTPase_activator_activity |
| GO:0030684 | 14 | preribosome |
| GO:0030823 | 14 | regulation_of_cGMP_metabolic_process |
| GO:0031000 | 14 | response_to_caffeine |
| GO:0031011 | 14 | Ino80_complex |
| GO:0031122 | 14 | cytoplasmic_microtubule_organization |
| GO:0031231 | 14 | intrinsic_to_peroxisomal_membrane |
| GO:0031235 | 14 | intrinsic_to_internal_side_of_plasma_membrane |
| GO:0031258 | 14 | lamellipodium_membrane |
| GO:0031307 | 14 | integral_to_mitochondrial_outer_membrane |
| GO:0031430 | 14 | M_band |
| GO:0031985 | 14 | Golgi_cisterna |
| GO:0032007 | 14 | negative_regulation_of_TOR_signaling_cascade |
| GO:0032008 | 14 | positive_regulation_of_TOR_signaling_cascade |
| GO:0032098 | 14 | regulation_of_appetite |
| GO:0032106 | 14 | positive_regulation_of_response_to_extracellular_stimulus |
| GO:0032109 | 14 | positive_regulation_of_response_to_nutrient_levels |
| GO:0032314 | 14 | regulation_of_Rac_GTPase_activity |
| GO:0032365 | 14 | intracellular_lipid_transport |
| GO:0032404 | 14 | mismatch_repair_complex_binding |
| GO:0032839 | 14 | dendrite_cytoplasm |
| GO:0032897 | 14 | negative_regulation_of_viral_transcription |
| GO:0032981 | 14 | mitochondrial_respiratory_chain_complex_I_assembly |
| GO:0033202 | 14 | DNA_helicase_complex |
| GO:0033276 | 14 | transcription_factor_TFTC_complex |
| GO:0033327 | 14 | Leydig_cell_differentiation |
| GO:0034035 | 14 | purine_ribonucleoside_bisphosphate_metabolic_process |
| GO:0034110 | 14 | regulation_of_homotypic_cell-cell_adhesion |
| GO:0034185 | 14 | apolipoprotein_binding |
| GO:0034204 | 14 | lipid_translocation |
| GO:0034362 | 14 | low-density_lipoprotein_particle |
| GO:0034375 | 14 | high-density_lipoprotein_particle_remodeling |
| GO:0034706 | 14 | sodium_channel_complex |
| GO:0035036 | 14 | sperm-egg_recognition |
| GO:0035050 | 14 | embryonic_heart_tube_development |
| GO:0035255 | 14 | ionotropic_glutamate_receptor_binding |
| GO:0035304 | 14 | regulation_of_protein_dephosphorylation |
| GO:0042100 | 14 | B_cell_proliferation |
| GO:0042136 | 14 | neurotransmitter_biosynthetic_process |
| GO:0042288 | 14 | MHC_class_I_protein_binding |
| GO:0042448 | 14 | progesterone_metabolic_process |
| GO:0043173 | 14 | nucleotide_salvage |
| GO:0043174 | 14 | nucleoside_salvage |
| GO:0043243 | 14 | positive_regulation_of_protein_complex_disassembly |
| GO:0043616 | 14 | keratinocyte_proliferation |
| GO:0045190 | 14 | isotype_switching |
| GO:0045332 | 14 | phospholipid_translocation |
| GO:0045408 | 14 | regulation_of_interleukin-6_biosynthetic_process |
| GO:0045723 | 14 | positive_regulation_of_fatty_acid_biosynthetic_process |
| GO:0045843 | 14 | negative_regulation_of_striated_muscle_tissue_development |
| GO:0045885 | 14 | positive_regulation_of_survival_gene_product_expression |
| GO:0045911 | 14 | positive_regulation_of_DNA_recombination |
| GO:0046051 | 14 | UTP_metabolic_process |
| GO:0048009 | 14 | insulin-like_growth_factor_receptor_signaling_pathway |
| GO:0048018 | 14 | receptor_agonist_activity |
| GO:0048019 | 14 | receptor_antagonist_activity |
| GO:0048103 | 14 | somatic_stem_cell_division |
| GO:0048593 | 14 | camera-type_eye_morphogenesis |
| GO:0048730 | 14 | epidermis_morphogenesis |
| GO:0048738 | 14 | cardiac_muscle_tissue_development |
| GO:0050427 | 14 | 3'-phosphoadenosine_5'-phosphosulfate_metabolic_process |
| GO:0050750 | 14 | low-density_lipoprotein_particle_receptor_binding |
| GO:0051193 | 14 | regulation_of_cofactor_metabolic_process |
| GO:0051196 | 14 | regulation_of_coenzyme_metabolic_process |
| GO:0051281 | 14 | positive_regulation_of_release_of_sequestered_calcium_ion_into_cytosol |
| GO:0051294 | 14 | establishment_of_spindle_orientation |
| GO:0051310 | 14 | metaphase_plate_congression |
| GO:0051457 | 14 | maintenance_of_protein_location_in_nucleus |
| GO:0051881 | 14 | regulation_of_mitochondrial_membrane_potential |
| GO:0051917 | 14 | regulation_of_fibrinolysis |
| GO:0051968 | 14 | positive_regulation_of_synaptic_transmission,_glutamatergic |
| GO:0055003 | 14 | cardiac_myofibril_assembly |
| GO:0060026 | 14 | convergent_extension |
| GO:0060071 | 14 | Wnt_receptor_signaling_pathway,_planar_cell_polarity_pathway |
| GO:0060260 | 14 | regulation_of_transcription_initiation_from_RNA_polymerase_II_promoter |
| GO:0060317 | 14 | cardiac_epithelial_to_mesenchymal_transition |
| GO:0061099 | 14 | negative_regulation_of_protein_tyrosine_kinase_activity |
| GO:0061333 | 14 | renal_tubule_morphogenesis |
| GO:0070849 | 14 | response_to_epidermal_growth_factor_stimulus |
| GO:0071044 | 14 | histone_mRNA_catabolic_process |
| GO:0071203 | 14 | WASH_complex |
| GO:0071354 | 14 | cellular_response_to_interleukin-6 |
| GO:0071371 | 14 | cellular_response_to_gonadotropin_stimulus |
| GO:0072530 | 14 | purine-containing_compound_transmembrane_transport |
| GO:0090025 | 14 | regulation_of_monocyte_chemotaxis |
| GO:0090175 | 14 | regulation_of_establishment_of_planar_polarity |
| GO:0097031 | 14 | mitochondrial_respiratory_chain_complex_I_biogenesis |
| GO:0097193 | 14 | intrinsic_apoptotic_signaling_pathway |
| GO:0097346 | 14 | INO80-type_complex |
| GO:2000178 | 14 | negative_regulation_of_neural_precursor_cell_proliferation |
| GO:2000403 | 14 | positive_regulation_of_lymphocyte_migration |
| GO:2000649 | 14 | regulation_of_sodium_ion_transmembrane_transporter_activity |
| GO:0000155 | 13 | two-component_sensor_activity |
| GO:0000303 | 13 | response_to_superoxide |
| GO:0001562 | 13 | response_to_protozoan |
| GO:0001573 | 13 | ganglioside_metabolic_process |
| GO:0001706 | 13 | endoderm_formation |
| GO:0001736 | 13 | establishment_of_planar_polarity |
| GO:0001829 | 13 | trophectodermal_cell_differentiation |
| GO:0002548 | 13 | monocyte_chemotaxis |
| GO:0002566 | 13 | somatic_diversification_of_immune_receptors_via_somatic_mutation |
| GO:0002724 | 13 | regulation_of_T_cell_cytokine_production |
| GO:0003281 | 13 | ventricular_septum_development |
| GO:0003401 | 13 | axis_elongation |
| GO:0003785 | 13 | actin_monomer_binding |
| GO:0004522 | 13 | pancreatic_ribonuclease_activity |
| GO:0004550 | 13 | nucleoside_diphosphate_kinase_activity |
| GO:0004859 | 13 | phospholipase_inhibitor_activity |
| GO:0004953 | 13 | icosanoid_receptor_activity |
| GO:0004954 | 13 | prostanoid_receptor_activity |
| GO:0004993 | 13 | serotonin_receptor_activity |
| GO:0005095 | 13 | GTPase_inhibitor_activity |
| GO:0005123 | 13 | death_receptor_binding |
| GO:0005149 | 13 | interleukin-1_receptor_binding |
| GO:0005161 | 13 | platelet-derived_growth_factor_receptor_binding |
| GO:0005344 | 13 | oxygen_transporter_activity |
| GO:0005372 | 13 | water_transmembrane_transporter_activity |
| GO:0005452 | 13 | inorganic_anion_exchanger_activity |
| GO:0005583 | 13 | fibrillar_collagen |
| GO:0005614 | 13 | interstitial_matrix |
| GO:0005732 | 13 | small_nucleolar_ribonucleoprotein_complex |
| GO:0005858 | 13 | axonemal_dynein_complex |
| GO:0006183 | 13 | GTP_biosynthetic_process |
| GO:0006228 | 13 | UTP_biosynthetic_process |
| GO:0006677 | 13 | glycosylceramide_metabolic_process |
| GO:0006684 | 13 | sphingomyelin_metabolic_process |
| GO:0006957 | 13 | complement_activation,_alternative_pathway |
| GO:0007076 | 13 | mitotic_chromosome_condensation |
| GO:0007140 | 13 | male_meiosis |
| GO:0007141 | 13 | male_meiosis_I |
| GO:0007210 | 13 | serotonin_receptor_signaling_pathway |
| GO:0007339 | 13 | binding_of_sperm_to_zona_pellucida |
| GO:0007340 | 13 | acrosome_reaction |
| GO:0007379 | 13 | segment_specification |
| GO:0007512 | 13 | adult_heart_development |
| GO:0008347 | 13 | glial_cell_migration |
| GO:0008514 | 13 | organic_anion_transmembrane_transporter_activity |
| GO:0008585 | 13 | female_gonad_development |
| GO:0009130 | 13 | pyrimidine_nucleoside_monophosphate_biosynthetic_process |
| GO:0009713 | 13 | catechol-containing_compound_biosynthetic_process |
| GO:0010042 | 13 | response_to_manganese_ion |
| GO:0010224 | 13 | response_to_UV-B |
| GO:0010460 | 13 | positive_regulation_of_heart_rate |
| GO:0010463 | 13 | mesenchymal_cell_proliferation |
| GO:0010485 | 13 | H4_histone_acetyltransferase_activity |
| GO:0010591 | 13 | regulation_of_lamellipodium_assembly |
| GO:0010719 | 13 | negative_regulation_of_epithelial_to_mesenchymal_transition |
| GO:0010745 | 13 | negative_regulation_of_macrophage_derived_foam_cell_differentiation |
| GO:0010800 | 13 | positive_regulation_of_peptidyl-threonine_phosphorylation |
| GO:0010823 | 13 | negative_regulation_of_mitochondrion_organization |
| GO:0010885 | 13 | regulation_of_cholesterol_storage |
| GO:0015101 | 13 | organic_cation_transmembrane_transporter_activity |
| GO:0015172 | 13 | acidic_amino_acid_transmembrane_transporter_activity |
| GO:0016307 | 13 | phosphatidylinositol_phosphate_kinase_activity |
| GO:0016446 | 13 | somatic_hypermutation_of_immunoglobulin_genes |
| GO:0016591 | 13 | DNA-directed_RNA_polymerase_II,_holoenzyme |
| GO:0016594 | 13 | glycine_binding |
| GO:0016854 | 13 | racemase_and_epimerase_activity |
| GO:0016888 | 13 | endodeoxyribonuclease_activity,_producing_5'-phosphomonoesters |
| GO:0016918 | 13 | retinal_binding |
| GO:0017075 | 13 | syntaxin-1_binding |
| GO:0017127 | 13 | cholesterol_transporter_activity |
| GO:0018022 | 13 | peptidyl-lysine_methylation |
| GO:0018298 | 13 | protein-chromophore_linkage |
| GO:0019002 | 13 | GMP_binding |
| GO:0019031 | 13 | viral_envelope |
| GO:0019104 | 13 | DNA_N-glycosylase_activity |
| GO:0019228 | 13 | regulation_of_action_potential_in_neuron |
| GO:0019369 | 13 | arachidonic_acid_metabolic_process |
| GO:0021522 | 13 | spinal_cord_motor_neuron_differentiation |
| GO:0021542 | 13 | dentate_gyrus_development |
| GO:0021772 | 13 | olfactory_bulb_development |
| GO:0021952 | 13 | central_nervous_system_projection_neuron_axonogenesis |
| GO:0022612 | 13 | gland_morphogenesis |
| GO:0030118 | 13 | clathrin_coat |
| GO:0030169 | 13 | low-density_lipoprotein_particle_binding |
| GO:0030225 | 13 | macrophage_differentiation |
| GO:0030238 | 13 | male_sex_determination |
| GO:0030288 | 13 | outer_membrane-bounded_periplasmic_space |
| GO:0030660 | 13 | Golgi-associated_vesicle_membrane |
| GO:0030852 | 13 | regulation_of_granulocyte_differentiation |
| GO:0030889 | 13 | negative_regulation_of_B_cell_proliferation |
| GO:0030914 | 13 | STAGA_complex |
| GO:0031143 | 13 | pseudopodium |
| GO:0031463 | 13 | Cul3-RING_ubiquitin_ligase_complex |
| GO:0031954 | 13 | positive_regulation_of_protein_autophosphorylation |
| GO:0032026 | 13 | response_to_magnesium_ion |
| GO:0032129 | 13 | histone_deacetylase_activity_(H3-K9_specific) |
| GO:0032204 | 13 | regulation_of_telomere_maintenance |
| GO:0032309 | 13 | icosanoid_secretion |
| GO:0032647 | 13 | regulation_of_interferon-alpha_production |
| GO:0032793 | 13 | positive_regulation_of_CREB_transcription_factor_activity |
| GO:0033598 | 13 | mammary_gland_epithelial_cell_proliferation |
| GO:0033599 | 13 | regulation_of_mammary_gland_epithelial_cell_proliferation |
| GO:0034104 | 13 | negative_regulation_of_tissue_remodeling |
| GO:0034122 | 13 | negative_regulation_of_toll-like_receptor_signaling_pathway |
| GO:0034312 | 13 | diol_biosynthetic_process |
| GO:0034661 | 13 | ncRNA_catabolic_process |
| GO:0035173 | 13 | histone_kinase_activity |
| GO:0042026 | 13 | protein_refolding |
| GO:0042101 | 13 | T_cell_receptor_complex |
| GO:0042166 | 13 | acetylcholine_binding |
| GO:0042301 | 13 | phosphate_ion_binding |
| GO:0042423 | 13 | catecholamine_biosynthetic_process |
| GO:0042537 | 13 | benzene-containing_compound_metabolic_process |
| GO:0042597 | 13 | periplasmic_space |
| GO:0042737 | 13 | drug_catabolic_process |
| GO:0042800 | 13 | histone_methyltransferase_activity_(H3-K4_specific) |
| GO:0042953 | 13 | lipoprotein_transport |
| GO:0043046 | 13 | DNA_methylation_involved_in_gamete_generation |
| GO:0043129 | 13 | surfactant_homeostasis |
| GO:0043186 | 13 | P_granule |
| GO:0043205 | 13 | fibril |
| GO:0043383 | 13 | negative_T_cell_selection |
| GO:0043618 | 13 | regulation_of_transcription_from_RNA_polymerase_II_promoter_in_response_to_stress |
| GO:0044257 | 13 | cellular_protein_catabolic_process |
| GO:0045026 | 13 | plasma_membrane_fusion |
| GO:0045073 | 13 | regulation_of_chemokine_biosynthetic_process |
| GO:0045086 | 13 | positive_regulation_of_interleukin-2_biosynthetic_process |
| GO:0045263 | 13 | proton-transporting_ATP_synthase_complex,_coupling_factor_F(o) |
| GO:0045576 | 13 | mast_cell_activation |
| GO:0045601 | 13 | regulation_of_endothelial_cell_differentiation |
| GO:0045725 | 13 | positive_regulation_of_glycogen_biosynthetic_process |
| GO:0045822 | 13 | negative_regulation_of_heart_contraction |
| GO:0046341 | 13 | CDP-diacylglycerol_metabolic_process |
| GO:0046415 | 13 | urate_metabolic_process |
| GO:0046689 | 13 | response_to_mercury_ion |
| GO:0046965 | 13 | retinoid_X_receptor_binding |
| GO:0046969 | 13 | NAD-dependent_histone_deacetylase_activity_(H3-K9_specific) |
| GO:0047617 | 13 | acyl-CoA_hydrolase_activity |
| GO:0048038 | 13 | quinone_binding |
| GO:0048200 | 13 | Golgi_transport_vesicle_coating |
| GO:0048205 | 13 | COPI_coating_of_Golgi_vesicle |
| GO:0048247 | 13 | lymphocyte_chemotaxis |
| GO:0048385 | 13 | regulation_of_retinoic_acid_receptor_signaling_pathway |
| GO:0048596 | 13 | embryonic_camera-type_eye_morphogenesis |
| GO:0048741 | 13 | skeletal_muscle_fiber_development |
| GO:0048857 | 13 | neural_nucleus_development |
| GO:0048875 | 13 | chemical_homeostasis_within_a_tissue |
| GO:0050482 | 13 | arachidonic_acid_secretion |
| GO:0050775 | 13 | positive_regulation_of_dendrite_morphogenesis |
| GO:0050930 | 13 | induction_of_positive_chemotaxis |
| GO:0051298 | 13 | centrosome_duplication |
| GO:0051324 | 13 | prophase |
| GO:0051428 | 13 | peptide_hormone_receptor_binding |
| GO:0051452 | 13 | intracellular_pH_reduction |
| GO:0051569 | 13 | regulation_of_histone_H3-K4_methylation |
| GO:0060037 | 13 | pharyngeal_system_development |
| GO:0060076 | 13 | excitatory_synapse |
| GO:0060122 | 13 | inner_ear_receptor_stereocilium_organization |
| GO:0060134 | 13 | prepulse_inhibition |
| GO:0060251 | 13 | regulation_of_glial_cell_proliferation |
| GO:0060291 | 13 | long-term_synaptic_potentiation |
| GO:0060314 | 13 | regulation_of_ryanodine-sensitive_calcium-release_channel_activity |
| GO:0060389 | 13 | pathway-restricted_SMAD_protein_phosphorylation |
| GO:0060442 | 13 | branching_involved_in_prostate_gland_morphogenesis |
| GO:0060445 | 13 | branching_involved_in_salivary_gland_morphogenesis |
| GO:0060795 | 13 | cell_fate_commitment_involved_in_formation_of_primary_germ_layer |
| GO:0061326 | 13 | renal_tubule_development |
| GO:0070633 | 13 | transepithelial_transport |
| GO:0070670 | 13 | response_to_interleukin-4 |
| GO:0070717 | 13 | poly-purine_tract_binding |
| GO:0070742 | 13 | C2H2_zinc_finger_domain_binding |
| GO:0071294 | 13 | cellular_response_to_zinc_ion |
| GO:0071425 | 13 | hemopoietic_stem_cell_proliferation |
| GO:0071715 | 13 | icosanoid_transport |
| GO:0071775 | 13 | regulation_of_cell_cycle_cytokinesis |
| GO:0071777 | 13 | positive_regulation_of_cell_cycle_cytokinesis |
| GO:0071941 | 13 | nitrogen_cycle_metabolic_process |
| GO:0071944 | 13 | cell_periphery |
| GO:0090030 | 13 | regulation_of_steroid_hormone_biosynthetic_process |
| GO:1900115 | 13 | extracellular_regulation_of_signal_transduction |
| GO:1900116 | 13 | extracellular_negative_regulation_of_signal_transduction |
| GO:1901021 | 13 | positive_regulation_of_calcium_ion_transmembrane_transporter_activity |
| GO:2000249 | 13 | regulation_of_actin_cytoskeleton_reorganization |
| GO:2000351 | 13 | regulation_of_endothelial_cell_apoptotic_process |
| GO:2000696 | 13 | regulation_of_epithelial_cell_differentiation_involved_in_kidney_development |
| GO:2001259 | 13 | positive_regulation_of_cation_channel_activity |
| GO:0000726 | 12 | non-recombinational_repair |
| GO:0000768 | 12 | syncytium_formation_by_plasma_membrane_fusion |
| GO:0000779 | 12 | condensed_chromosome,_centromeric_region |
| GO:0000782 | 12 | telomere_cap_complex |
| GO:0000783 | 12 | nuclear_telomere_cap_complex |
| GO:0000791 | 12 | euchromatin |
| GO:0000795 | 12 | synaptonemal_complex |
| GO:0000940 | 12 | condensed_chromosome_outer_kinetochore |
| GO:0001530 | 12 | lipopolysaccharide_binding |
| GO:0001823 | 12 | mesonephros_development |
| GO:0001833 | 12 | inner_cell_mass_cell_proliferation |
| GO:0001841 | 12 | neural_tube_formation |
| GO:0001891 | 12 | phagocytic_cup |
| GO:0001946 | 12 | lymphangiogenesis |
| GO:0001963 | 12 | synaptic_transmission,_dopaminergic |
| GO:0001965 | 12 | G-protein_alpha-subunit_binding |
| GO:0001967 | 12 | suckling_behavior |
| GO:0001977 | 12 | renal_system_process_involved_in_regulation_of_blood_volume |
| GO:0002011 | 12 | morphogenesis_of_an_epithelial_sheet |
| GO:0002089 | 12 | lens_morphogenesis_in_camera-type_eye |
| GO:0002102 | 12 | podosome |
| GO:0002230 | 12 | positive_regulation_of_defense_response_to_virus_by_host |
| GO:0002440 | 12 | production_of_molecular_mediator_of_immune_response |
| GO:0002832 | 12 | negative_regulation_of_response_to_biotic_stimulus |
| GO:0002902 | 12 | regulation_of_B_cell_apoptotic_process |
| GO:0003081 | 12 | regulation_of_systemic_arterial_blood_pressure_by_renin-angiotensin |
| GO:0003207 | 12 | cardiac_chamber_formation |
| GO:0003906 | 12 | DNA-(apurinic_or_apyrimidinic_site)_lyase_activity |
| GO:0004065 | 12 | arylsulfatase_activity |
| GO:0004549 | 12 | tRNA-specific_ribonuclease_activity |
| GO:0004955 | 12 | prostaglandin_receptor_activity |
| GO:0005031 | 12 | tumor_necrosis_factor-activated_receptor_activity |
| GO:0005112 | 12 | Notch_binding |
| GO:0005251 | 12 | delayed_rectifier_potassium_channel_activity |
| GO:0005313 | 12 | L-glutamate_transmembrane_transporter_activity |
| GO:0005355 | 12 | glucose_transmembrane_transporter_activity |
| GO:0005416 | 12 | cation:amino_acid_symporter_activity |
| GO:0005833 | 12 | hemoglobin_complex |
| GO:0005845 | 12 | mRNA_cap_binding_complex |
| GO:0005868 | 12 | cytoplasmic_dynein_complex |
| GO:0006303 | 12 | double-strand_break_repair_via_nonhomologous_end_joining |
| GO:0006379 | 12 | mRNA_cleavage |
| GO:0006740 | 12 | NADPH_regeneration |
| GO:0006903 | 12 | vesicle_targeting |
| GO:0006911 | 12 | phagocytosis,_engulfment |
| GO:0007007 | 12 | inner_mitochondrial_membrane_organization |
| GO:0007020 | 12 | microtubule_nucleation |
| GO:0007096 | 12 | regulation_of_exit_from_mitosis |
| GO:0007158 | 12 | neuron_cell-cell_adhesion |
| GO:0007213 | 12 | G-protein_coupled_acetylcholine_receptor_signaling_pathway |
| GO:0007405 | 12 | neuroblast_proliferation |
| GO:0007494 | 12 | midgut_development |
| GO:0007625 | 12 | grooming_behavior |
| GO:0008106 | 12 | alcohol_dehydrogenase_(NADP+)_activity |
| GO:0008195 | 12 | phosphatidate_phosphatase_activity |
| GO:0008199 | 12 | ferric_iron_binding |
| GO:0008242 | 12 | omega_peptidase_activity |
| GO:0008252 | 12 | nucleotidase_activity |
| GO:0008278 | 12 | cohesin_complex |
| GO:0008340 | 12 | determination_of_adult_lifespan |
| GO:0008536 | 12 | Ran_GTPase_binding |
| GO:0008556 | 12 | potassium-transporting_ATPase_activity |
| GO:0009113 | 12 | purine_base_biosynthetic_process |
| GO:0009200 | 12 | deoxyribonucleoside_triphosphate_metabolic_process |
| GO:0009396 | 12 | folic_acid-containing_compound_biosynthetic_process |
| GO:0009629 | 12 | response_to_gravity |
| GO:0009812 | 12 | flavonoid_metabolic_process |
| GO:0010155 | 12 | regulation_of_proton_transport |
| GO:0010259 | 12 | multicellular_organismal_aging |
| GO:0010510 | 12 | regulation_of_acetyl-CoA_biosynthetic_process_from_pyruvate |
| GO:0010576 | 12 | metalloenzyme_regulator_activity |
| GO:0010669 | 12 | epithelial_structure_maintenance |
| GO:0010837 | 12 | regulation_of_keratinocyte_proliferation |
| GO:0010842 | 12 | retina_layer_formation |
| GO:0010972 | 12 | negative_regulation_of_G2/M_transition_of_mitotic_cell_cycle |
| GO:0010984 | 12 | regulation_of_lipoprotein_particle_clearance |
| GO:0014888 | 12 | striated_muscle_adaptation |
| GO:0015012 | 12 | heparan_sulfate_proteoglycan_biosynthetic_process |
| GO:0015250 | 12 | water_channel_activity |
| GO:0015838 | 12 | betaine_transport |
| GO:0015879 | 12 | carnitine_transport |
| GO:0016024 | 12 | CDP-diacylglycerol_biosynthetic_process |
| GO:0016045 | 12 | detection_of_bacterium |
| GO:0016180 | 12 | snRNA_processing |
| GO:0016493 | 12 | C-C_chemokine_receptor_activity |
| GO:0016601 | 12 | Rac_protein_signal_transduction |
| GO:0016813 | 12 | hydrolase_activity,_acting_on_carbon-nitrogen_(but_not_peptide)_bonds,_in_linear_amidines |
| GO:0016862 | 12 | intramolecular_oxidoreductase_activity,_interconverting_keto-_and_enol-groups |
| GO:0019059 | 12 | initiation_of_viral_infection |
| GO:0019627 | 12 | urea_metabolic_process |
| GO:0019841 | 12 | retinol_binding |
| GO:0019956 | 12 | chemokine_binding |
| GO:0021527 | 12 | spinal_cord_association_neuron_differentiation |
| GO:0021904 | 12 | dorsal/ventral_neural_tube_patterning |
| GO:0030140 | 12 | trans-Golgi_network_transport_vesicle |
| GO:0030296 | 12 | protein_tyrosine_kinase_activator_activity |
| GO:0030553 | 12 | cGMP_binding |
| GO:0030851 | 12 | granulocyte_differentiation |
| GO:0030897 | 12 | HOPS_complex |
| GO:0031116 | 12 | positive_regulation_of_microtubule_polymerization |
| GO:0031514 | 12 | motile_cilium |
| GO:0031581 | 12 | hemidesmosome_assembly |
| GO:0031638 | 12 | zymogen_activation |
| GO:0031650 | 12 | regulation_of_heat_generation |
| GO:0031652 | 12 | positive_regulation_of_heat_generation |
| GO:0031996 | 12 | thioesterase_binding |
| GO:0032039 | 12 | integrator_complex |
| GO:0032148 | 12 | activation_of_protein_kinase_B_activity |
| GO:0032281 | 12 | alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic_acid_selective_glutamate_receptor_complex |
| GO:0032303 | 12 | regulation_of_icosanoid_secretion |
| GO:0032352 | 12 | positive_regulation_of_hormone_metabolic_process |
| GO:0032372 | 12 | negative_regulation_of_sterol_transport |
| GO:0032373 | 12 | positive_regulation_of_sterol_transport |
| GO:0032375 | 12 | negative_regulation_of_cholesterol_transport |
| GO:0032376 | 12 | positive_regulation_of_cholesterol_transport |
| GO:0032435 | 12 | negative_regulation_of_proteasomal_ubiquitin-dependent_protein_catabolic_process |
| GO:0032674 | 12 | regulation_of_interleukin-5_production |
| GO:0032695 | 12 | negative_regulation_of_interleukin-12_production |
| GO:0032922 | 12 | circadian_regulation_of_gene_expression |
| GO:0033057 | 12 | multicellular_organismal_reproductive_behavior |
| GO:0033268 | 12 | node_of_Ranvier |
| GO:0033522 | 12 | histone_H2A_ubiquitination |
| GO:0034243 | 12 | regulation_of_transcription_elongation_from_RNA_polymerase_II_promoter |
| GO:0034377 | 12 | plasma_lipoprotein_particle_assembly |
| GO:0034483 | 12 | heparan_sulfate_sulfotransferase_activity |
| GO:0034518 | 12 | RNA_cap_binding_complex |
| GO:0035004 | 12 | phosphatidylinositol_3-kinase_activity |
| GO:0035102 | 12 | PRC1_complex |
| GO:0035112 | 12 | genitalia_morphogenesis |
| GO:0035810 | 12 | positive_regulation_of_urine_volume |
| GO:0036038 | 12 | TCTN-B9D_complex |
| GO:0042074 | 12 | cell_migration_involved_in_gastrulation |
| GO:0042149 | 12 | cellular_response_to_glucose_starvation |
| GO:0042274 | 12 | ribosomal_small_subunit_biogenesis |
| GO:0042490 | 12 | mechanoreceptor_differentiation |
| GO:0042510 | 12 | regulation_of_tyrosine_phosphorylation_of_Stat1_protein |
| GO:0042627 | 12 | chylomicron |
| GO:0042634 | 12 | regulation_of_hair_cycle |
| GO:0043001 | 12 | Golgi_to_plasma_membrane_protein_transport |
| GO:0043209 | 12 | myelin_sheath |
| GO:0043252 | 12 | sodium-independent_organic_anion_transport |
| GO:0043296 | 12 | apical_junction_complex |
| GO:0043325 | 12 | phosphatidylinositol-3,4-bisphosphate_binding |
| GO:0043395 | 12 | heparan_sulfate_proteoglycan_binding |
| GO:0043517 | 12 | positive_regulation_of_DNA_damage_response,_signal_transduction_by_p53_class_mediator |
| GO:0043560 | 12 | insulin_receptor_substrate_binding |
| GO:0043604 | 12 | amide_biosynthetic_process |
| GO:0043922 | 12 | negative_regulation_by_host_of_viral_transcription |
| GO:0043968 | 12 | histone_H2A_acetylation |
| GO:0044273 | 12 | sulfur_compound_catabolic_process |
| GO:0044319 | 12 | wound_healing,_spreading_of_cells |
| GO:0045005 | 12 | maintenance_of_fidelity_involved_in_DNA-dependent_DNA_replication |
| GO:0045060 | 12 | negative_thymic_T_cell_selection |
| GO:0045078 | 12 | positive_regulation_of_interferon-gamma_biosynthetic_process |
| GO:0045197 | 12 | establishment_or_maintenance_of_epithelial_cell_apical/basal_polarity |
| GO:0045295 | 12 | gamma-catenin_binding |
| GO:0045346 | 12 | regulation_of_MHC_class_II_biosynthetic_process |
| GO:0045414 | 12 | regulation_of_interleukin-8_biosynthetic_process |
| GO:0045426 | 12 | quinone_cofactor_biosynthetic_process |
| GO:0045540 | 12 | regulation_of_cholesterol_biosynthetic_process |
| GO:0045624 | 12 | positive_regulation_of_T-helper_cell_differentiation |
| GO:0045780 | 12 | positive_regulation_of_bone_resorption |
| GO:0045824 | 12 | negative_regulation_of_innate_immune_response |
| GO:0045830 | 12 | positive_regulation_of_isotype_switching |
| GO:0046527 | 12 | glucosyltransferase_activity |
| GO:0046625 | 12 | sphingolipid_binding |
| GO:0046852 | 12 | positive_regulation_of_bone_remodeling |
| GO:0046856 | 12 | phosphatidylinositol_dephosphorylation |
| GO:0048016 | 12 | inositol_phosphate-mediated_signaling |
| GO:0048070 | 12 | regulation_of_developmental_pigmentation |
| GO:0048185 | 12 | activin_binding |
| GO:0048488 | 12 | synaptic_vesicle_endocytosis |
| GO:0048679 | 12 | regulation_of_axon_regeneration |
| GO:0048699 | 12 | generation_of_neurons |
| GO:0048703 | 12 | embryonic_viscerocranium_morphogenesis |
| GO:0048715 | 12 | negative_regulation_of_oligodendrocyte_differentiation |
| GO:0048736 | 12 | appendage_development |
| GO:0048854 | 12 | brain_morphogenesis |
| GO:0050431 | 12 | transforming_growth_factor_beta_binding |
| GO:0050858 | 12 | negative_regulation_of_antigen_receptor-mediated_signaling_pathway |
| GO:0050872 | 12 | white_fat_cell_differentiation |
| GO:0050908 | 12 | detection_of_light_stimulus_involved_in_visual_perception |
| GO:0050919 | 12 | negative_chemotaxis |
| GO:0050962 | 12 | detection_of_light_stimulus_involved_in_sensory_perception |
| GO:0051000 | 12 | positive_regulation_of_nitric-oxide_synthase_activity |
| GO:0051010 | 12 | microtubule_plus-end_binding |
| GO:0051044 | 12 | positive_regulation_of_membrane_protein_ectodomain_proteolysis |
| GO:0051154 | 12 | negative_regulation_of_striated_muscle_cell_differentiation |
| GO:0051233 | 12 | spindle_midzone |
| GO:0051322 | 12 | anaphase |
| GO:0051393 | 12 | alpha-actinin_binding |
| GO:0051414 | 12 | response_to_cortisol_stimulus |
| GO:0051482 | 12 | elevation_of_cytosolic_calcium_ion_concentration_involved_in_phospholipase_C-activating_G-protein_coupled_signaling_pathway |
| GO:0051580 | 12 | regulation_of_neurotransmitter_uptake |
| GO:0051769 | 12 | regulation_of_nitric-oxide_synthase_biosynthetic_process |
| GO:0055069 | 12 | zinc_ion_homeostasis |
| GO:0060004 | 12 | reflex |
| GO:0060173 | 12 | limb_development |
| GO:0060347 | 12 | heart_trabecula_formation |
| GO:0060351 | 12 | cartilage_development_involved_in_endochondral_bone_morphogenesis |
| GO:0060571 | 12 | morphogenesis_of_an_epithelial_fold |
| GO:0060590 | 12 | ATPase_regulator_activity |
| GO:0060736 | 12 | prostate_gland_growth |
| GO:0065005 | 12 | protein-lipid_complex_assembly |
| GO:0070193 | 12 | synaptonemal_complex_organization |
| GO:0070293 | 12 | renal_absorption |
| GO:0070403 | 12 | NAD+_binding |
| GO:0070527 | 12 | platelet_aggregation |
| GO:0070570 | 12 | regulation_of_neuron_projection_regeneration |
| GO:0070672 | 12 | response_to_interleukin-15 |
| GO:0071108 | 12 | protein_K48-linked_deubiquitination |
| GO:0071276 | 12 | cellular_response_to_cadmium_ion |
| GO:0071548 | 12 | response_to_dexamethasone_stimulus |
| GO:0071565 | 12 | nBAF_complex |
| GO:0071636 | 12 | positive_regulation_of_transforming_growth_factor_beta_production |
| GO:0071780 | 12 | mitotic_cell_cycle_G2/M_transition_checkpoint |
| GO:0071889 | 12 | 14-3-3_protein_binding |
| GO:0071956 | 12 | cellular_component_maintenance_at_cellular_level |
| GO:0072009 | 12 | nephron_epithelium_development |
| GO:0072080 | 12 | nephron_tubule_development |
| GO:0080111 | 12 | DNA_demethylation |
| GO:0090004 | 12 | positive_regulation_of_establishment_of_protein_localization_in_plasma_membrane |
| GO:0090079 | 12 | translation_regulator_activity,_nucleic_acid_binding |
| GO:0090322 | 12 | regulation_of_superoxide_metabolic_process |
| GO:0097028 | 12 | dendritic_cell_differentiation |
| GO:2000104 | 12 | negative_regulation_of_DNA-dependent_DNA_replication |
| GO:2000193 | 12 | positive_regulation_of_fatty_acid_transport |
| GO:0000050 | 11 | urea_cycle |
| GO:0001773 | 11 | myeloid_dendritic_cell_activation |
| GO:0002031 | 11 | G-protein_coupled_receptor_internalization |
| GO:0002063 | 11 | chondrocyte_development |
| GO:0002065 | 11 | columnar/cuboidal_epithelial_cell_differentiation |
| GO:0002080 | 11 | acrosomal_membrane |
| GO:0002281 | 11 | macrophage_activation_involved_in_immune_response |
| GO:0002287 | 11 | alpha-beta_T_cell_activation_involved_in_immune_response |
| GO:0002292 | 11 | T_cell_differentiation_involved_in_immune_response |
| GO:0002293 | 11 | alpha-beta_T_cell_differentiation_involved_in_immune_response |
| GO:0002456 | 11 | T_cell_mediated_immunity |
| GO:0002577 | 11 | regulation_of_antigen_processing_and_presentation |
| GO:0002643 | 11 | regulation_of_tolerance_induction |
| GO:0002701 | 11 | negative_regulation_of_production_of_molecular_mediator_of_immune_response |
| GO:0002719 | 11 | negative_regulation_of_cytokine_production_involved_in_immune_response |
| GO:0002922 | 11 | positive_regulation_of_humoral_immune_response |
| GO:0003211 | 11 | cardiac_ventricle_formation |
| GO:0003840 | 11 | gamma-glutamyltransferase_activity |
| GO:0003993 | 11 | acid_phosphatase_activity |
| GO:0004016 | 11 | adenylate_cyclase_activity |
| GO:0004115 | 11 | 3',5'-cyclic-AMP_phosphodiesterase_activity |
| GO:0004303 | 11 | estradiol_17-beta-dehydrogenase_activity |
| GO:0004385 | 11 | guanylate_kinase_activity |
| GO:0004861 | 11 | cyclin-dependent_protein_kinase_inhibitor_activity |
| GO:0005041 | 11 | low-density_lipoprotein_receptor_activity |
| GO:0005487 | 11 | nucleocytoplasmic_transporter_activity |
| GO:0005675 | 11 | holo_TFIIH_complex |
| GO:0005719 | 11 | nuclear_euchromatin |
| GO:0005721 | 11 | centromeric_heterochromatin |
| GO:0005916 | 11 | fascia_adherens |
| GO:0006012 | 11 | galactose_metabolic_process |
| GO:0006069 | 11 | ethanol_oxidation |
| GO:0006098 | 11 | pentose-phosphate_shunt |
| GO:0006107 | 11 | oxaloacetate_metabolic_process |
| GO:0006188 | 11 | IMP_biosynthetic_process |
| GO:0006569 | 11 | tryptophan_catabolic_process |
| GO:0006596 | 11 | polyamine_biosynthetic_process |
| GO:0006599 | 11 | phosphagen_metabolic_process |
| GO:0006707 | 11 | cholesterol_catabolic_process |
| GO:0006878 | 11 | cellular_copper_ion_homeostasis |
| GO:0006882 | 11 | cellular_zinc_ion_homeostasis |
| GO:0007004 | 11 | telomere_maintenance_via_telomerase |
| GO:0007080 | 11 | mitotic_metaphase_plate_congression |
| GO:0007091 | 11 | mitotic_metaphase/anaphase_transition |
| GO:0007130 | 11 | synaptonemal_complex_assembly |
| GO:0007168 | 11 | receptor_guanylyl_cyclase_signaling_pathway |
| GO:0007223 | 11 | Wnt_receptor_signaling_pathway,_calcium_modulating_pathway |
| GO:0007252 | 11 | I-kappaB_phosphorylation |
| GO:0007520 | 11 | myoblast_fusion |
| GO:0007603 | 11 | phototransduction,_visible_light |
| GO:0008175 | 11 | tRNA_methyltransferase_activity |
| GO:0008191 | 11 | metalloendopeptidase_inhibitor_activity |
| GO:0008239 | 11 | dipeptidyl-peptidase_activity |
| GO:0008271 | 11 | secondary_active_sulfate_transmembrane_transporter_activity |
| GO:0008595 | 11 | anterior/posterior_axis_specification,_embryo |
| GO:0008631 | 11 | induction_of_apoptosis_by_oxidative_stress |
| GO:0009263 | 11 | deoxyribonucleotide_biosynthetic_process |
| GO:0010269 | 11 | response_to_selenium_ion |
| GO:0010560 | 11 | positive_regulation_of_glycoprotein_biosynthetic_process |
| GO:0010662 | 11 | regulation_of_striated_muscle_cell_apoptotic_process |
| GO:0010870 | 11 | positive_regulation_of_receptor_biosynthetic_process |
| GO:0010875 | 11 | positive_regulation_of_cholesterol_efflux |
| GO:0010939 | 11 | regulation_of_necrotic_cell_death |
| GO:0010955 | 11 | negative_regulation_of_protein_processing |
| GO:0014902 | 11 | myotube_differentiation |
| GO:0015450 | 11 | P-P-bond-hydrolysis-driven_protein_transmembrane_transporter_activity |
| GO:0015665 | 11 | alcohol_transmembrane_transporter_activity |
| GO:0015893 | 11 | drug_transport |
| GO:0015939 | 11 | pantothenate_metabolic_process |
| GO:0016127 | 11 | sterol_catabolic_process |
| GO:0016175 | 11 | superoxide-generating_NADPH_oxidase_activity |
| GO:0016226 | 11 | iron-sulfur_cluster_assembly |
| GO:0016303 | 11 | 1-phosphatidylinositol-3-kinase_activity |
| GO:0016500 | 11 | protein-hormone_receptor_activity |
| GO:0016558 | 11 | protein_import_into_peroxisome_matrix |
| GO:0016580 | 11 | Sin3_complex |
| GO:0016634 | 11 | oxidoreductase_activity,_acting_on_the_CH-CH_group_of_donors,_oxygen_as_acceptor |
| GO:0016840 | 11 | carbon-nitrogen_lyase_activity |
| GO:0016868 | 11 | intramolecular_transferase_activity,_phosphotransferases |
| GO:0016884 | 11 | carbon-nitrogen_ligase_activity,_with_glutamine_as_amido-N-donor |
| GO:0017025 | 11 | TBP-class_protein_binding |
| GO:0017110 | 11 | nucleoside-diphosphatase_activity |
| GO:0017187 | 11 | peptidyl-glutamic_acid_carboxylation |
| GO:0018202 | 11 | peptidyl-histidine_modification |
| GO:0018214 | 11 | protein_carboxylation |
| GO:0018345 | 11 | protein_palmitoylation |
| GO:0019377 | 11 | glycolipid_catabolic_process |
| GO:0021854 | 11 | hypothalamus_development |
| GO:0030276 | 11 | clathrin_binding |
| GO:0030431 | 11 | sleep |
| GO:0030507 | 11 | spectrin_binding |
| GO:0030742 | 11 | GTP-dependent_protein_binding |
| GO:0031062 | 11 | positive_regulation_of_histone_methylation |
| GO:0031063 | 11 | regulation_of_histone_deacetylation |
| GO:0031078 | 11 | histone_deacetylase_activity_(H3-K14_specific) |
| GO:0031091 | 11 | platelet_alpha_granule |
| GO:0031105 | 11 | septin_complex |
| GO:0031163 | 11 | metallo-sulfur_cluster_assembly |
| GO:0031210 | 11 | phosphatidylcholine_binding |
| GO:0031369 | 11 | translation_initiation_factor_binding |
| GO:0031432 | 11 | titin_binding |
| GO:0031434 | 11 | mitogen-activated_protein_kinase_kinase_binding |
| GO:0031527 | 11 | filopodium_membrane |
| GO:0031904 | 11 | endosome_lumen |
| GO:0032041 | 11 | NAD-dependent_histone_deacetylase_activity_(H3-K14_specific) |
| GO:0032094 | 11 | response_to_food |
| GO:0032230 | 11 | positive_regulation_of_synaptic_transmission,_GABAergic |
| GO:0032305 | 11 | positive_regulation_of_icosanoid_secretion |
| GO:0032331 | 11 | negative_regulation_of_chondrocyte_differentiation |
| GO:0032395 | 11 | MHC_class_II_receptor_activity |
| GO:0032401 | 11 | establishment_of_melanosome_localization |
| GO:0032407 | 11 | MutSalpha_complex_binding |
| GO:0032462 | 11 | regulation_of_protein_homooligomerization |
| GO:0032656 | 11 | regulation_of_interleukin-13_production |
| GO:0032692 | 11 | negative_regulation_of_interleukin-1_production |
| GO:0032740 | 11 | positive_regulation_of_interleukin-17_production |
| GO:0032753 | 11 | positive_regulation_of_interleukin-4_production |
| GO:0032769 | 11 | negative_regulation_of_monooxygenase_activity |
| GO:0032836 | 11 | glomerular_basement_membrane_development |
| GO:0033002 | 11 | muscle_cell_proliferation |
| GO:0033005 | 11 | positive_regulation_of_mast_cell_activation |
| GO:0033032 | 11 | regulation_of_myeloid_cell_apoptotic_process |
| GO:0033363 | 11 | secretory_granule_organization |
| GO:0033540 | 11 | fatty_acid_beta-oxidation_using_acyl-CoA_oxidase |
| GO:0033605 | 11 | positive_regulation_of_catecholamine_secretion |
| GO:0034123 | 11 | positive_regulation_of_toll-like_receptor_signaling_pathway |
| GO:0034260 | 11 | negative_regulation_of_GTPase_activity |
| GO:0034374 | 11 | low-density_lipoprotein_particle_remodeling |
| GO:0034739 | 11 | histone_deacetylase_activity_(H4-K16_specific) |
| GO:0035145 | 11 | exon-exon_junction_complex |
| GO:0035587 | 11 | purinergic_receptor_signaling_pathway |
| GO:0035631 | 11 | CD40_receptor_complex |
| GO:0035815 | 11 | positive_regulation_of_renal_sodium_excretion |
| GO:0036092 | 11 | phosphatidylinositol-3-phosphate_biosynthetic_process |
| GO:0040019 | 11 | positive_regulation_of_embryonic_development |
| GO:0040023 | 11 | establishment_of_nucleus_localization |
| GO:0040037 | 11 | negative_regulation_of_fibroblast_growth_factor_receptor_signaling_pathway |
| GO:0042043 | 11 | neurexin_family_protein_binding |
| GO:0042069 | 11 | regulation_of_catecholamine_metabolic_process |
| GO:0042273 | 11 | ribosomal_large_subunit_biogenesis |
| GO:0042347 | 11 | negative_regulation_of_NF-kappaB_import_into_nucleus |
| GO:0042354 | 11 | L-fucose_metabolic_process |
| GO:0042359 | 11 | vitamin_D_metabolic_process |
| GO:0042436 | 11 | indole-containing_compound_catabolic_process |
| GO:0042574 | 11 | retinal_metabolic_process |
| GO:0042640 | 11 | anagen |
| GO:0042738 | 11 | exogenous_drug_catabolic_process |
| GO:0042832 | 11 | defense_response_to_protozoan |
| GO:0042987 | 11 | amyloid_precursor_protein_catabolic_process |
| GO:0043090 | 11 | amino_acid_import |
| GO:0043175 | 11 | RNA_polymerase_core_enzyme_binding |
| GO:0043240 | 11 | Fanconi_anaemia_nuclear_complex |
| GO:0043249 | 11 | erythrocyte_maturation |
| GO:0043274 | 11 | phospholipase_binding |
| GO:0043277 | 11 | apoptotic_cell_clearance |
| GO:0043288 | 11 | apocarotenoid_metabolic_process |
| GO:0043368 | 11 | positive_T_cell_selection |
| GO:0043518 | 11 | negative_regulation_of_DNA_damage_response,_signal_transduction_by_p53_class_mediator |
| GO:0045502 | 11 | dynein_binding |
| GO:0045649 | 11 | regulation_of_macrophage_differentiation |
| GO:0045652 | 11 | regulation_of_megakaryocyte_differentiation |
| GO:0045663 | 11 | positive_regulation_of_myoblast_differentiation |
| GO:0045749 | 11 | negative_regulation_of_S_phase_of_mitotic_cell_cycle |
| GO:0045910 | 11 | negative_regulation_of_DNA_recombination |
| GO:0045920 | 11 | negative_regulation_of_exocytosis |
| GO:0046040 | 11 | IMP_metabolic_process |
| GO:0046218 | 11 | indolalkylamine_catabolic_process |
| GO:0046321 | 11 | positive_regulation_of_fatty_acid_oxidation |
| GO:0046459 | 11 | short-chain_fatty_acid_metabolic_process |
| GO:0046639 | 11 | negative_regulation_of_alpha-beta_T_cell_differentiation |
| GO:0046718 | 11 | viral_entry_into_host_cell |
| GO:0046788 | 11 | egress_of_virus_within_host_cell |
| GO:0046827 | 11 | positive_regulation_of_protein_export_from_nucleus |
| GO:0046849 | 11 | bone_remodeling |
| GO:0046851 | 11 | negative_regulation_of_bone_remodeling |
| GO:0046902 | 11 | regulation_of_mitochondrial_membrane_permeability |
| GO:0046970 | 11 | NAD-dependent_histone_deacetylase_activity_(H4-K16_specific) |
| GO:0047496 | 11 | vesicle_transport_along_microtubule |
| GO:0047555 | 11 | 3',5'-cyclic-GMP_phosphodiesterase_activity |
| GO:0048246 | 11 | macrophage_chemotaxis |
| GO:0048407 | 11 | platelet-derived_growth_factor_binding |
| GO:0048535 | 11 | lymph_node_development |
| GO:0048551 | 11 | metalloenzyme_inhibitor_activity |
| GO:0048786 | 11 | presynaptic_active_zone |
| GO:0048821 | 11 | erythrocyte_development |
| GO:0048841 | 11 | regulation_of_axon_extension_involved_in_axon_guidance |
| GO:0048937 | 11 | lateral_line_nerve_glial_cell_development |
| GO:0050321 | 11 | tau-protein_kinase_activity |
| GO:0050685 | 11 | positive_regulation_of_mRNA_processing |
| GO:0050686 | 11 | negative_regulation_of_mRNA_processing |
| GO:0050780 | 11 | dopamine_receptor_binding |
| GO:0050860 | 11 | negative_regulation_of_T_cell_receptor_signaling_pathway |
| GO:0050891 | 11 | multicellular_organismal_water_homeostasis |
| GO:0051023 | 11 | regulation_of_immunoglobulin_secretion |
| GO:0051085 | 11 | chaperone_mediated_protein_folding_requiring_cofactor |
| GO:0051131 | 11 | chaperone-mediated_protein_complex_assembly |
| GO:0051205 | 11 | protein_insertion_into_membrane |
| GO:0051313 | 11 | attachment_of_spindle_microtubules_to_chromosome |
| GO:0051650 | 11 | establishment_of_vesicle_localization |
| GO:0051894 | 11 | positive_regulation_of_focal_adhesion_assembly |
| GO:0051905 | 11 | establishment_of_pigment_granule_localization |
| GO:0051927 | 11 | negative_regulation_of_calcium_ion_transport_via_voltage-gated_calcium_channel_activity |
| GO:0051972 | 11 | regulation_of_telomerase_activity |
| GO:0052695 | 11 | cellular_glucuronidation |
| GO:0055094 | 11 | response_to_lipoprotein_stimulus |
| GO:0060033 | 11 | anatomical_structure_regression |
| GO:0060039 | 11 | pericardium_development |
| GO:0060065 | 11 | uterus_development |
| GO:0060113 | 11 | inner_ear_receptor_cell_differentiation |
| GO:0060174 | 11 | limb_bud_formation |
| GO:0060425 | 11 | lung_morphogenesis |
| GO:0060536 | 11 | cartilage_morphogenesis |
| GO:0060638 | 11 | mesenchymal-epithelial_cell_signaling |
| GO:0060693 | 11 | regulation_of_branching_involved_in_salivary_gland_morphogenesis |
| GO:0060742 | 11 | epithelial_cell_differentiation_involved_in_prostate_gland_development |
| GO:0060766 | 11 | negative_regulation_of_androgen_receptor_signaling_pathway |
| GO:0061036 | 11 | positive_regulation_of_cartilage_development |
| GO:0061311 | 11 | cell_surface_receptor_signaling_pathway_involved_in_heart_development |
| GO:0070208 | 11 | protein_heterotrimerization |
| GO:0070243 | 11 | regulation_of_thymocyte_apoptotic_process |
| GO:0070410 | 11 | co-SMAD_binding |
| GO:0070411 | 11 | I-SMAD_binding |
| GO:0070584 | 11 | mitochondrion_morphogenesis |
| GO:0070822 | 11 | Sin3-type_complex |
| GO:0071157 | 11 | negative_regulation_of_cell_cycle_arrest |
| GO:0071364 | 11 | cellular_response_to_epidermal_growth_factor_stimulus |
| GO:0071545 | 11 | inositol_phosphate_catabolic_process |
| GO:0071564 | 11 | npBAF_complex |
| GO:0071887 | 11 | leukocyte_apoptotic_process |
| GO:0072182 | 11 | regulation_of_nephron_tubule_epithelial_cell_differentiation |
| GO:0072661 | 11 | protein_targeting_to_plasma_membrane |
| GO:0072686 | 11 | mitotic_spindle |
| GO:0080025 | 11 | phosphatidylinositol-3,5-bisphosphate_binding |
| GO:0085029 | 11 | extracellular_matrix_assembly |
| GO:0090136 | 11 | epithelial_cell-cell_adhesion |
| GO:0090162 | 11 | establishment_of_epithelial_cell_polarity |
| GO:0090192 | 11 | regulation_of_glomerulus_development |
| GO:0090279 | 11 | regulation_of_calcium_ion_import |
| GO:0090312 | 11 | positive_regulation_of_protein_deacetylation |
| GO:0097061 | 11 | dendritic_spine_organization |
| GO:0097066 | 11 | response_to_thyroid_hormone_stimulus |
| GO:2000058 | 11 | regulation_of_protein_ubiquitination_involved_in_ubiquitin-dependent_protein_catabolic_process |
| GO:2000272 | 11 | negative_regulation_of_receptor_activity |
| GO:2000380 | 11 | regulation_of_mesoderm_development |
| GO:2000404 | 11 | regulation_of_T_cell_migration |
| GO:2000463 | 11 | positive_regulation_of_excitatory_postsynaptic_membrane_potential |
| GO:2000826 | 11 | regulation_of_heart_morphogenesis |
| GO:2001237 | 11 | negative_regulation_of_extrinsic_apoptotic_signaling_pathway |
| GO:0000003 | 10 | reproduction |
| GO:0000052 | 10 | citrulline_metabolic_process |
| GO:0000076 | 10 | DNA_replication_checkpoint |
| GO:0000085 | 10 | G2_phase_of_mitotic_cell_cycle |
| GO:0000090 | 10 | mitotic_anaphase |
| GO:0000731 | 10 | DNA_synthesis_involved_in_DNA_repair |
| GO:0000780 | 10 | condensed_nuclear_chromosome,_centromeric_region |
| GO:0001098 | 10 | basal_transcription_machinery_binding |
| GO:0001099 | 10 | basal_RNA_polymerase_II_transcription_machinery_binding |
| GO:0001510 | 10 | RNA_methylation |
| GO:0002544 | 10 | chronic_inflammatory_response |
| GO:0002645 | 10 | positive_regulation_of_tolerance_induction |
| GO:0002710 | 10 | negative_regulation_of_T_cell_mediated_immunity |
| GO:0002833 | 10 | positive_regulation_of_response_to_biotic_stimulus |
| GO:0002863 | 10 | positive_regulation_of_inflammatory_response_to_antigenic_stimulus |
| GO:0002864 | 10 | regulation_of_acute_inflammatory_response_to_antigenic_stimulus |
| GO:0002923 | 10 | regulation_of_humoral_immune_response_mediated_by_circulating_immunoglobulin |
| GO:0003084 | 10 | positive_regulation_of_systemic_arterial_blood_pressure |
| GO:0003148 | 10 | outflow_tract_septum_morphogenesis |
| GO:0003323 | 10 | type_B_pancreatic_cell_development |
| GO:0003756 | 10 | protein_disulfide_isomerase_activity |
| GO:0003796 | 10 | lysozyme_activity |
| GO:0004000 | 10 | adenosine_deaminase_activity |
| GO:0004029 | 10 | aldehyde_dehydrogenase_(NAD)_activity |
| GO:0004467 | 10 | long-chain_fatty_acid-CoA_ligase_activity |
| GO:0004526 | 10 | ribonuclease_P_activity |
| GO:0004983 | 10 | neuropeptide_Y_receptor_activity |
| GO:0004985 | 10 | opioid_receptor_activity |
| GO:0005072 | 10 | transforming_growth_factor_beta_receptor,_cytoplasmic_mediator_activity |
| GO:0005092 | 10 | GDP-dissociation_inhibitor_activity |
| GO:0005218 | 10 | intracellular_ligand-gated_calcium_channel_activity |
| GO:0005243 | 10 | gap_junction_channel_activity |
| GO:0005283 | 10 | sodium:amino_acid_symporter_activity |
| GO:0005391 | 10 | sodium:potassium-exchanging_ATPase_activity |
| GO:0005451 | 10 | monovalent_cation:hydrogen_antiporter_activity |
| GO:0005678 | 10 | chromatin_assembly_complex |
| GO:0005790 | 10 | smooth_endoplasmic_reticulum |
| GO:0005952 | 10 | cAMP-dependent_protein_kinase_complex |
| GO:0006020 | 10 | inositol_metabolic_process |
| GO:0006047 | 10 | UDP-N-acetylglucosamine_metabolic_process |
| GO:0006105 | 10 | succinate_metabolic_process |
| GO:0006222 | 10 | UMP_biosynthetic_process |
| GO:0006356 | 10 | regulation_of_transcription_from_RNA_polymerase_I_promoter |
| GO:0006465 | 10 | signal_peptide_processing |
| GO:0006560 | 10 | proline_metabolic_process |
| GO:0006570 | 10 | tyrosine_metabolic_process |
| GO:0006582 | 10 | melanin_metabolic_process |
| GO:0006600 | 10 | creatine_metabolic_process |
| GO:0006670 | 10 | sphingosine_metabolic_process |
| GO:0006704 | 10 | glucocorticoid_biosynthetic_process |
| GO:0006743 | 10 | ubiquinone_metabolic_process |
| GO:0006895 | 10 | Golgi_to_endosome_transport |
| GO:0006983 | 10 | ER_overload_response |
| GO:0006998 | 10 | nuclear_envelope_organization |
| GO:0007016 | 10 | cytoskeletal_anchoring_at_plasma_membrane |
| GO:0007063 | 10 | regulation_of_sister_chromatid_cohesion |
| GO:0007128 | 10 | meiotic_prophase_I |
| GO:0007171 | 10 | activation_of_transmembrane_receptor_protein_tyrosine_kinase_activity |
| GO:0007342 | 10 | fusion_of_sperm_to_egg_plasma_membrane |
| GO:0007567 | 10 | parturition |
| GO:0007635 | 10 | chemosensory_behavior |
| GO:0008143 | 10 | poly(A)_RNA_binding |
| GO:0008158 | 10 | hedgehog_receptor_activity |
| GO:0008187 | 10 | poly-pyrimidine_tract_binding |
| GO:0008250 | 10 | oligosaccharyltransferase_complex |
| GO:0008253 | 10 | 5'-nucleotidase_activity |
| GO:0008318 | 10 | protein_prenyltransferase_activity |
| GO:0008385 | 10 | IkappaB_kinase_complex |
| GO:0008535 | 10 | respiratory_chain_complex_IV_assembly |
| GO:0008635 | 10 | activation_of_cysteine-type_endopeptidase_activity_involved_in_apoptotic_process_by_cytochrome_c |
| GO:0008641 | 10 | small_protein_activating_enzyme_activity |
| GO:0008655 | 10 | pyrimidine-containing_compound_salvage |
| GO:0009173 | 10 | pyrimidine_ribonucleoside_monophosphate_metabolic_process |
| GO:0009174 | 10 | pyrimidine_ribonucleoside_monophosphate_biosynthetic_process |
| GO:0009185 | 10 | ribonucleoside_diphosphate_metabolic_process |
| GO:0009299 | 10 | mRNA_transcription |
| GO:0009437 | 10 | carnitine_metabolic_process |
| GO:0009635 | 10 | response_to_herbicide |
| GO:0009650 | 10 | UV_protection |
| GO:0010092 | 10 | specification_of_organ_identity |
| GO:0010454 | 10 | negative_regulation_of_cell_fate_commitment |
| GO:0010613 | 10 | positive_regulation_of_cardiac_muscle_hypertrophy |
| GO:0010664 | 10 | negative_regulation_of_striated_muscle_cell_apoptotic_process |
| GO:0010665 | 10 | regulation_of_cardiac_muscle_cell_apoptotic_process |
| GO:0010738 | 10 | regulation_of_protein_kinase_A_signaling_cascade |
| GO:0010829 | 10 | negative_regulation_of_glucose_transport |
| GO:0010867 | 10 | positive_regulation_of_triglyceride_biosynthetic_process |
| GO:0010935 | 10 | regulation_of_macrophage_cytokine_production |
| GO:0014061 | 10 | regulation_of_norepinephrine_secretion |
| GO:0014742 | 10 | positive_regulation_of_muscle_hypertrophy |
| GO:0014812 | 10 | muscle_cell_migration |
| GO:0015125 | 10 | bile_acid_transmembrane_transporter_activity |
| GO:0015197 | 10 | peptide_transporter_activity |
| GO:0015643 | 10 | toxin_binding |
| GO:0015671 | 10 | oxygen_transport |
| GO:0015693 | 10 | magnesium_ion_transport |
| GO:0015924 | 10 | mannosyl-oligosaccharide_mannosidase_activity |
| GO:0015926 | 10 | glucosidase_activity |
| GO:0015937 | 10 | coenzyme_A_biosynthetic_process |
| GO:0016090 | 10 | prenol_metabolic_process |
| GO:0016239 | 10 | positive_regulation_of_macroautophagy |
| GO:0016272 | 10 | prefoldin_complex |
| GO:0016471 | 10 | vacuolar_proton-transporting_V-type_ATPase_complex |
| GO:0016679 | 10 | oxidoreductase_activity,_acting_on_diphenols_and_related_substances_as_donors |
| GO:0016722 | 10 | oxidoreductase_activity,_oxidizing_metal_ions |
| GO:0016725 | 10 | oxidoreductase_activity,_acting_on_CH_or_CH2_groups |
| GO:0016801 | 10 | hydrolase_activity,_acting_on_ether_bonds |
| GO:0016812 | 10 | hydrolase_activity,_acting_on_carbon-nitrogen_(but_not_peptide)_bonds,_in_cyclic_amides |
| GO:0016864 | 10 | intramolecular_oxidoreductase_activity,_transposing_S-S_bonds |
| GO:0017119 | 10 | Golgi_transport_complex |
| GO:0017166 | 10 | vinculin_binding |
| GO:0019012 | 10 | virion |
| GO:0019203 | 10 | carbohydrate_phosphatase_activity |
| GO:0019206 | 10 | nucleoside_kinase_activity |
| GO:0019430 | 10 | removal_of_superoxide_radicals |
| GO:0019789 | 10 | SUMO_ligase_activity |
| GO:0019852 | 10 | L-ascorbic_acid_metabolic_process |
| GO:0019864 | 10 | IgG_binding |
| GO:0021602 | 10 | cranial_nerve_morphogenesis |
| GO:0021702 | 10 | cerebellar_Purkinje_cell_differentiation |
| GO:0021846 | 10 | cell_proliferation_in_forebrain |
| GO:0030002 | 10 | cellular_anion_homeostasis |
| GO:0030125 | 10 | clathrin_vesicle_coat |
| GO:0030206 | 10 | chondroitin_sulfate_biosynthetic_process |
| GO:0030220 | 10 | platelet_formation |
| GO:0030371 | 10 | translation_repressor_activity |
| GO:0030673 | 10 | axolemma |
| GO:0030826 | 10 | regulation_of_cGMP_biosynthetic_process |
| GO:0030934 | 10 | anchoring_collagen |
| GO:0031080 | 10 | nuclear_pore_outer_ring |
| GO:0031109 | 10 | microtubule_polymerization_or_depolymerization |
| GO:0031272 | 10 | regulation_of_pseudopodium_assembly |
| GO:0031312 | 10 | extrinsic_to_organelle_membrane |
| GO:0031420 | 10 | alkali_metal_ion_binding |
| GO:0031503 | 10 | protein_complex_localization |
| GO:0031620 | 10 | regulation_of_fever_generation |
| GO:0031622 | 10 | positive_regulation_of_fever_generation |
| GO:0031641 | 10 | regulation_of_myelination |
| GO:0031690 | 10 | adrenergic_receptor_binding |
| GO:0031935 | 10 | regulation_of_chromatin_silencing |
| GO:0032096 | 10 | negative_regulation_of_response_to_food |
| GO:0032232 | 10 | negative_regulation_of_actin_filament_bundle_assembly |
| GO:0032354 | 10 | response_to_follicle-stimulating_hormone_stimulus |
| GO:0032402 | 10 | melanosome_transport |
| GO:0032456 | 10 | endocytic_recycling |
| GO:0032743 | 10 | positive_regulation_of_interleukin-2_production |
| GO:0032835 | 10 | glomerulus_development |
| GO:0032878 | 10 | regulation_of_establishment_or_maintenance_of_cell_polarity |
| GO:0032968 | 10 | positive_regulation_of_transcription_elongation_from_RNA_polymerase_II_promoter |
| GO:0033145 | 10 | positive_regulation_of_intracellular_steroid_hormone_receptor_signaling_pathway |
| GO:0033160 | 10 | positive_regulation_of_protein_import_into_nucleus,_translocation |
| GO:0033176 | 10 | proton-transporting_V-type_ATPase_complex |
| GO:0033180 | 10 | proton-transporting_V-type_ATPase,_V1_domain |
| GO:0033194 | 10 | response_to_hydroperoxide |
| GO:0033197 | 10 | response_to_vitamin_E |
| GO:0033233 | 10 | regulation_of_protein_sumoylation |
| GO:0033612 | 10 | receptor_serine/threonine_kinase_binding |
| GO:0033627 | 10 | cell_adhesion_mediated_by_integrin |
| GO:0033700 | 10 | phospholipid_efflux |
| GO:0034114 | 10 | regulation_of_heterotypic_cell-cell_adhesion |
| GO:0034391 | 10 | regulation_of_smooth_muscle_cell_apoptotic_process |
| GO:0034593 | 10 | phosphatidylinositol_bisphosphate_phosphatase_activity |
| GO:0034616 | 10 | response_to_laminar_fluid_shear_stress |
| GO:0035020 | 10 | regulation_of_Rac_protein_signal_transduction |
| GO:0035098 | 10 | ESC/E(Z)_complex |
| GO:0035240 | 10 | dopamine_binding |
| GO:0035251 | 10 | UDP-glucosyltransferase_activity |
| GO:0035253 | 10 | ciliary_rootlet |
| GO:0035306 | 10 | positive_regulation_of_dephosphorylation |
| GO:0035313 | 10 | wound_healing,_spreading_of_epidermal_cells |
| GO:0036065 | 10 | fucosylation |
| GO:0042053 | 10 | regulation_of_dopamine_metabolic_process |
| GO:0042255 | 10 | ribosome_assembly |
| GO:0042362 | 10 | fat-soluble_vitamin_biosynthetic_process |
| GO:0042364 | 10 | water-soluble_vitamin_biosynthetic_process |
| GO:0042535 | 10 | positive_regulation_of_tumor_necrosis_factor_biosynthetic_process |
| GO:0042698 | 10 | ovulation_cycle |
| GO:0043011 | 10 | myeloid_dendritic_cell_differentiation |
| GO:0043032 | 10 | positive_regulation_of_macrophage_activation |
| GO:0043092 | 10 | L-amino_acid_import |
| GO:0043116 | 10 | negative_regulation_of_vascular_permeability |
| GO:0043171 | 10 | peptide_catabolic_process |
| GO:0043256 | 10 | laminin_complex |
| GO:0043302 | 10 | positive_regulation_of_leukocyte_degranulation |
| GO:0043403 | 10 | skeletal_muscle_tissue_regeneration |
| GO:0043555 | 10 | regulation_of_translation_in_response_to_stress |
| GO:0043923 | 10 | positive_regulation_by_host_of_viral_transcription |
| GO:0043949 | 10 | regulation_of_cAMP-mediated_signaling |
| GO:0044070 | 10 | regulation_of_anion_transport |
| GO:0044550 | 10 | secondary_metabolite_biosynthetic_process |
| GO:0045019 | 10 | negative_regulation_of_nitric_oxide_biosynthetic_process |
| GO:0045070 | 10 | positive_regulation_of_viral_genome_replication |
| GO:0045080 | 10 | positive_regulation_of_chemokine_biosynthetic_process |
| GO:0045109 | 10 | intermediate_filament_organization |
| GO:0045176 | 10 | apical_protein_localization |
| GO:0045606 | 10 | positive_regulation_of_epidermal_cell_differentiation |
| GO:0045717 | 10 | negative_regulation_of_fatty_acid_biosynthetic_process |
| GO:0045741 | 10 | positive_regulation_of_epidermal_growth_factor-activated_receptor_activity |
| GO:0045844 | 10 | positive_regulation_of_striated_muscle_tissue_development |
| GO:0046049 | 10 | UMP_metabolic_process |
| GO:0046113 | 10 | nucleobase_catabolic_process |
| GO:0046348 | 10 | amino_sugar_catabolic_process |
| GO:0046426 | 10 | negative_regulation_of_JAK-STAT_cascade |
| GO:0046501 | 10 | protoporphyrinogen_IX_metabolic_process |
| GO:0046838 | 10 | phosphorylated_carbohydrate_dephosphorylation |
| GO:0046855 | 10 | inositol_phosphate_dephosphorylation |
| GO:0048025 | 10 | negative_regulation_of_nuclear_mRNA_splicing,_via_spliceosome |
| GO:0048266 | 10 | behavioral_response_to_pain |
| GO:0048268 | 10 | clathrin_coat_assembly |
| GO:0048302 | 10 | regulation_of_isotype_switching_to_IgG_isotypes |
| GO:0048599 | 10 | oocyte_development |
| GO:0048617 | 10 | embryonic_foregut_morphogenesis |
| GO:0048636 | 10 | positive_regulation_of_muscle_organ_development |
| GO:0048708 | 10 | astrocyte_differentiation |
| GO:0048712 | 10 | negative_regulation_of_astrocyte_differentiation |
| GO:0050308 | 10 | sugar-phosphatase_activity |
| GO:0050435 | 10 | beta-amyloid_metabolic_process |
| GO:0050746 | 10 | regulation_of_lipoprotein_metabolic_process |
| GO:0050857 | 10 | positive_regulation_of_antigen_receptor-mediated_signaling_pathway |
| GO:0050879 | 10 | multicellular_organismal_movement |
| GO:0050881 | 10 | musculoskeletal_movement |
| GO:0050910 | 10 | detection_of_mechanical_stimulus_involved_in_sensory_perception_of_sound |
| GO:0051261 | 10 | protein_depolymerization |
| GO:0051304 | 10 | chromosome_separation |
| GO:0051319 | 10 | G2_phase |
| GO:0051323 | 10 | metaphase |
| GO:0051378 | 10 | serotonin_binding |
| GO:0051593 | 10 | response_to_folic_acid |
| GO:0051597 | 10 | response_to_methylmercury |
| GO:0051646 | 10 | mitochondrion_localization |
| GO:0051797 | 10 | regulation_of_hair_follicle_development |
| GO:0051900 | 10 | regulation_of_mitochondrial_depolarization |
| GO:0051904 | 10 | pigment_granule_transport |
| GO:0051918 | 10 | negative_regulation_of_fibrinolysis |
| GO:0051937 | 10 | catecholamine_transport |
| GO:0051955 | 10 | regulation_of_amino_acid_transport |
| GO:0052173 | 10 | response_to_defenses_of_other_organism_involved_in_symbiotic_interaction |
| GO:0052200 | 10 | response_to_host_defenses |
| GO:0055015 | 10 | ventricular_cardiac_muscle_cell_development |
| GO:0055062 | 10 | phosphate_ion_homeostasis |
| GO:0055078 | 10 | sodium_ion_homeostasis |
| GO:0060056 | 10 | mammary_gland_involution |
| GO:0060080 | 10 | regulation_of_inhibitory_postsynaptic_membrane_potential |
| GO:0060192 | 10 | negative_regulation_of_lipase_activity |
| GO:0060216 | 10 | definitive_hemopoiesis |
| GO:0060292 | 10 | long_term_synaptic_depression |
| GO:0060322 | 10 | head_development |
| GO:0060538 | 10 | skeletal_muscle_organ_development |
| GO:0060602 | 10 | branch_elongation_of_an_epithelium |
| GO:0060768 | 10 | regulation_of_epithelial_cell_proliferation_involved_in_prostate_gland_development |
| GO:0060850 | 10 | regulation_of_transcription_involved_in_cell_fate_commitment |
| GO:0060992 | 10 | response_to_fungicide |
| GO:0060999 | 10 | positive_regulation_of_dendritic_spine_development |
| GO:0070016 | 10 | armadillo_repeat_domain_binding |
| GO:0070059 | 10 | intrinsic_apoptotic_signaling_pathway_in_response_to_endoplasmic_reticulum_stress |
| GO:0070402 | 10 | NADPH_binding |
| GO:0070498 | 10 | interleukin-1-mediated_signaling_pathway |
| GO:0070587 | 10 | regulation_of_cell-cell_adhesion_involved_in_gastrulation |
| GO:0070989 | 10 | oxidative_demethylation |
| GO:0071236 | 10 | cellular_response_to_antibiotic |
| GO:0071379 | 10 | cellular_response_to_prostaglandin_stimulus |
| GO:0071397 | 10 | cellular_response_to_cholesterol |
| GO:0071398 | 10 | cellular_response_to_fatty_acid |
| GO:0071450 | 10 | cellular_response_to_oxygen_radical |
| GO:0071451 | 10 | cellular_response_to_superoxide |
| GO:0071470 | 10 | cellular_response_to_osmotic_stress |
| GO:0071542 | 10 | dopaminergic_neuron_differentiation |
| GO:0071549 | 10 | cellular_response_to_dexamethasone_stimulus |
| GO:0071622 | 10 | regulation_of_granulocyte_chemotaxis |
| GO:0071675 | 10 | regulation_of_mononuclear_cell_migration |
| GO:0071731 | 10 | response_to_nitric_oxide |
| GO:0071867 | 10 | response_to_monoamine_stimulus |
| GO:0071868 | 10 | cellular_response_to_monoamine_stimulus |
| GO:0071869 | 10 | response_to_catecholamine_stimulus |
| GO:0071870 | 10 | cellular_response_to_catecholamine_stimulus |
| GO:0072078 | 10 | nephron_tubule_morphogenesis |
| GO:0072088 | 10 | nephron_epithelium_morphogenesis |
| GO:0072091 | 10 | regulation_of_stem_cell_proliferation |
| GO:0072506 | 10 | trivalent_inorganic_anion_homeostasis |
| GO:0072531 | 10 | pyrimidine-containing_compound_transmembrane_transport |
| GO:0075136 | 10 | response_to_host |
| GO:0090026 | 10 | positive_regulation_of_monocyte_chemotaxis |
| GO:0090049 | 10 | regulation_of_cell_migration_involved_in_sprouting_angiogenesis |
| GO:0090307 | 10 | spindle_assembly_involved_in_mitosis |
| GO:0090399 | 10 | replicative_senescence |
| GO:0097067 | 10 | cellular_response_to_thyroid_hormone_stimulus |
| GO:0097094 | 10 | craniofacial_suture_morphogenesis |
| GO:0097150 | 10 | neuronal_stem_cell_maintenance |
| GO:2000378 | 10 | negative_regulation_of_reactive_oxygen_species_metabolic_process |
| GO:2000406 | 10 | positive_regulation_of_T_cell_migration |
| GO:2000772 | 10 | regulation_of_cellular_senescence |
| GO:0000098 | 9 | sulfur_amino_acid_catabolic_process |
| GO:0000145 | 9 | exocyst |
| GO:0000338 | 9 | protein_deneddylation |
| GO:0000808 | 9 | origin_recognition_complex |
| GO:0000993 | 9 | RNA_polymerase_II_core_binding |
| GO:0001527 | 9 | microfibril |
| GO:0001542 | 9 | ovulation_from_ovarian_follicle |
| GO:0001580 | 9 | detection_of_chemical_stimulus_involved_in_sensory_perception_of_bitter_taste |
| GO:0001675 | 9 | acrosome_assembly |
| GO:0001741 | 9 | XY_body |
| GO:0001774 | 9 | microglial_cell_activation |
| GO:0001885 | 9 | endothelial_cell_development |
| GO:0002294 | 9 | CD4-positive,_alpha-beta_T_cell_differentiation_involved_in_immune_response |
| GO:0002320 | 9 | lymphoid_progenitor_cell_differentiation |
| GO:0002360 | 9 | T_cell_lineage_commitment |
| GO:0002444 | 9 | myeloid_leukocyte_mediated_immunity |
| GO:0002467 | 9 | germinal_center_formation |
| GO:0002480 | 9 | antigen_processing_and_presentation_of_exogenous_peptide_antigen_via_MHC_class_I,_TAP-independent |
| GO:0002495 | 9 | antigen_processing_and_presentation_of_peptide_antigen_via_MHC_class_II |
| GO:0002579 | 9 | positive_regulation_of_antigen_processing_and_presentation |
| GO:0002634 | 9 | regulation_of_germinal_center_formation |
| GO:0002664 | 9 | regulation_of_T_cell_tolerance_induction |
| GO:0002674 | 9 | negative_regulation_of_acute_inflammatory_response |
| GO:0002676 | 9 | regulation_of_chronic_inflammatory_response |
| GO:0002726 | 9 | positive_regulation_of_T_cell_cytokine_production |
| GO:0002827 | 9 | positive_regulation_of_T-helper_1_type_immune_response |
| GO:0002834 | 9 | regulation_of_response_to_tumor_cell |
| GO:0002836 | 9 | positive_regulation_of_response_to_tumor_cell |
| GO:0002837 | 9 | regulation_of_immune_response_to_tumor_cell |
| GO:0002839 | 9 | positive_regulation_of_immune_response_to_tumor_cell |
| GO:0003016 | 9 | respiratory_system_process |
| GO:0003337 | 9 | mesenchymal_to_epithelial_transition_involved_in_metanephros_morphogenesis |
| GO:0003810 | 9 | protein-glutamine_gamma-glutamyltransferase_activity |
| GO:0003841 | 9 | 1-acylglycerol-3-phosphate_O-acyltransferase_activity |
| GO:0004022 | 9 | alcohol_dehydrogenase_(NAD)_activity |
| GO:0004383 | 9 | guanylate_cyclase_activity |
| GO:0004576 | 9 | oligosaccharyl_transferase_activity |
| GO:0004690 | 9 | cyclic_nucleotide-dependent_protein_kinase_activity |
| GO:0004716 | 9 | receptor_signaling_protein_tyrosine_kinase_activity |
| GO:0004935 | 9 | adrenergic_receptor_activity |
| GO:0005005 | 9 | transmembrane-ephrin_receptor_activity |
| GO:0005221 | 9 | intracellular_cyclic_nucleotide_activated_cation_channel_activity |
| GO:0005337 | 9 | nucleoside_transmembrane_transporter_activity |
| GO:0005351 | 9 | sugar:hydrogen_symporter_activity |
| GO:0005388 | 9 | calcium-transporting_ATPase_activity |
| GO:0005402 | 9 | cation:sugar_symporter_activity |
| GO:0005513 | 9 | detection_of_calcium_ion |
| GO:0005664 | 9 | nuclear_origin_of_replication_recognition_complex |
| GO:0005744 | 9 | mitochondrial_inner_membrane_presequence_translocase_complex |
| GO:0005847 | 9 | mRNA_cleavage_and_polyadenylation_specificity_factor_complex |
| GO:0005883 | 9 | neurofilament |
| GO:0006014 | 9 | D-ribose_metabolic_process |
| GO:0006119 | 9 | oxidative_phosphorylation |
| GO:0006265 | 9 | DNA_topological_change |
| GO:0006266 | 9 | DNA_ligation |
| GO:0006390 | 9 | transcription_from_mitochondrial_promoter |
| GO:0006474 | 9 | N-terminal_protein_amino_acid_acetylation |
| GO:0006515 | 9 | misfolded_or_incompletely_synthesized_protein_catabolic_process |
| GO:0006527 | 9 | arginine_catabolic_process |
| GO:0006553 | 9 | lysine_metabolic_process |
| GO:0006554 | 9 | lysine_catabolic_process |
| GO:0006558 | 9 | L-phenylalanine_metabolic_process |
| GO:0006559 | 9 | L-phenylalanine_catabolic_process |
| GO:0006580 | 9 | ethanolamine_metabolic_process |
| GO:0006621 | 9 | protein_retention_in_ER_lumen |
| GO:0006622 | 9 | protein_targeting_to_lysosome |
| GO:0006623 | 9 | protein_targeting_to_vacuole |
| GO:0006646 | 9 | phosphatidylethanolamine_biosynthetic_process |
| GO:0006703 | 9 | estrogen_biosynthetic_process |
| GO:0006734 | 9 | NADH_metabolic_process |
| GO:0006744 | 9 | ubiquinone_biosynthetic_process |
| GO:0006782 | 9 | protoporphyrinogen_IX_biosynthetic_process |
| GO:0006910 | 9 | phagocytosis,_recognition |
| GO:0007191 | 9 | adenylate_cyclase-activating_dopamine_receptor_signaling_pathway |
| GO:0008091 | 9 | spectrin |
| GO:0008121 | 9 | ubiquinol-cytochrome-c_reductase_activity |
| GO:0008139 | 9 | nuclear_localization_sequence_binding |
| GO:0008409 | 9 | 5'-3'_exonuclease_activity |
| GO:0008608 | 9 | attachment_of_spindle_microtubules_to_kinetochore |
| GO:0008653 | 9 | lipopolysaccharide_metabolic_process |
| GO:0009151 | 9 | purine_deoxyribonucleotide_metabolic_process |
| GO:0009404 | 9 | toxin_metabolic_process |
| GO:0009415 | 9 | response_to_water |
| GO:0009649 | 9 | entrainment_of_circadian_clock |
| GO:0009744 | 9 | response_to_sucrose_stimulus |
| GO:0009820 | 9 | alkaloid_metabolic_process |
| GO:0009886 | 9 | post-embryonic_morphogenesis |
| GO:0010388 | 9 | cullin_deneddylation |
| GO:0010544 | 9 | negative_regulation_of_platelet_activation |
| GO:0010569 | 9 | regulation_of_double-strand_break_repair_via_homologous_recombination |
| GO:0010640 | 9 | regulation_of_platelet-derived_growth_factor_receptor_signaling_pathway |
| GO:0010667 | 9 | negative_regulation_of_cardiac_muscle_cell_apoptotic_process |
| GO:0010692 | 9 | regulation_of_alkaline_phosphatase_activity |
| GO:0010758 | 9 | regulation_of_macrophage_chemotaxis |
| GO:0010762 | 9 | regulation_of_fibroblast_migration |
| GO:0010803 | 9 | regulation_of_tumor_necrosis_factor-mediated_signaling_pathway |
| GO:0010872 | 9 | regulation_of_cholesterol_esterification |
| GO:0010880 | 9 | regulation_of_release_of_sequestered_calcium_ion_into_cytosol_by_sarcoplasmic_reticulum |
| GO:0010889 | 9 | regulation_of_sequestering_of_triglyceride |
| GO:0010919 | 9 | regulation_of_inositol_phosphate_biosynthetic_process |
| GO:0010944 | 9 | negative_regulation_of_transcription_by_competitive_promoter_binding |
| GO:0010954 | 9 | positive_regulation_of_protein_processing |
| GO:0014002 | 9 | astrocyte_development |
| GO:0014701 | 9 | junctional_sarcoplasmic_reticulum_membrane |
| GO:0014829 | 9 | vascular_smooth_muscle_contraction |
| GO:0014912 | 9 | negative_regulation_of_smooth_muscle_cell_migration |
| GO:0015245 | 9 | fatty_acid_transporter_activity |
| GO:0015271 | 9 | outward_rectifier_potassium_channel_activity |
| GO:0015280 | 9 | ligand-gated_sodium_channel_activity |
| GO:0015385 | 9 | sodium:hydrogen_antiporter_activity |
| GO:0015851 | 9 | nucleobase_transport |
| GO:0016273 | 9 | arginine_N-methyltransferase_activity |
| GO:0016274 | 9 | protein-arginine_N-methyltransferase_activity |
| GO:0016281 | 9 | eukaryotic_translation_initiation_factor_4F_complex |
| GO:0016290 | 9 | palmitoyl-CoA_hydrolase_activity |
| GO:0016322 | 9 | neuron_remodeling |
| GO:0016595 | 9 | glutamate_binding |
| GO:0016624 | 9 | oxidoreductase_activity,_acting_on_the_aldehyde_or_oxo_group_of_donors,_disulfide_as_acceptor |
| GO:0016668 | 9 | oxidoreductase_activity,_acting_on_a_sulfur_group_of_donors,_NAD_or_NADP_as_acceptor |
| GO:0016671 | 9 | oxidoreductase_activity,_acting_on_a_sulfur_group_of_donors,_disulfide_as_acceptor |
| GO:0016681 | 9 | oxidoreductase_activity,_acting_on_diphenols_and_related_substances_as_donors,_cytochrome_as_acceptor |
| GO:0016713 | 9 | oxidoreductase_activity,_acting_on_paired_donors,_with_incorporation_or_reduction_of_molecular_oxygen,_reduced_iron-sulfur_protein_as_one_donor,_and_incorporation_of_one_atom_of_oxygen |
| GO:0016999 | 9 | antibiotic_metabolic_process |
| GO:0017049 | 9 | GTP-Rho_binding |
| GO:0017153 | 9 | sodium:dicarboxylate_symporter_activity |
| GO:0018065 | 9 | protein-cofactor_linkage |
| GO:0018216 | 9 | peptidyl-arginine_methylation |
| GO:0019317 | 9 | fucose_catabolic_process |
| GO:0019322 | 9 | pentose_biosynthetic_process |
| GO:0019509 | 9 | L-methionine_salvage_from_methylthioadenosine |
| GO:0019953 | 9 | sexual_reproduction |
| GO:0019992 | 9 | diacylglycerol_binding |
| GO:0020027 | 9 | hemoglobin_metabolic_process |
| GO:0021670 | 9 | lateral_ventricle_development |
| GO:0021794 | 9 | thalamus_development |
| GO:0021884 | 9 | forebrain_neuron_development |
| GO:0021895 | 9 | cerebral_cortex_neuron_differentiation |
| GO:0030122 | 9 | AP-2_adaptor_complex |
| GO:0030127 | 9 | COPII_vesicle_coat |
| GO:0030515 | 9 | snoRNA_binding |
| GO:0030825 | 9 | positive_regulation_of_cGMP_metabolic_process |
| GO:0030898 | 9 | actin-dependent_ATPase_activity |
| GO:0030903 | 9 | notochord_development |
| GO:0031017 | 9 | exocrine_pancreas_development |
| GO:0031076 | 9 | embryonic_camera-type_eye_development |
| GO:0031082 | 9 | BLOC_complex |
| GO:0031083 | 9 | BLOC-1_complex |
| GO:0031274 | 9 | positive_regulation_of_pseudopodium_assembly |
| GO:0031304 | 9 | intrinsic_to_mitochondrial_inner_membrane |
| GO:0031305 | 9 | integral_to_mitochondrial_inner_membrane |
| GO:0031419 | 9 | cobalamin_binding |
| GO:0031440 | 9 | regulation_of_mRNA_3'-end_processing |
| GO:0031571 | 9 | mitotic_cell_cycle_G1/S_transition_DNA_damage_checkpoint |
| GO:0031943 | 9 | regulation_of_glucocorticoid_metabolic_process |
| GO:0032042 | 9 | mitochondrial_DNA_metabolic_process |
| GO:0032099 | 9 | negative_regulation_of_appetite |
| GO:0032205 | 9 | negative_regulation_of_telomere_maintenance |
| GO:0032234 | 9 | regulation_of_calcium_ion_transport_via_store-operated_calcium_channel_activity |
| GO:0032279 | 9 | asymmetric_synapse |
| GO:0032389 | 9 | MutLalpha_complex |
| GO:0032588 | 9 | trans-Golgi_network_membrane |
| GO:0032645 | 9 | regulation_of_granulocyte_macrophage_colony-stimulating_factor_production |
| GO:0032691 | 9 | negative_regulation_of_interleukin-1_beta_production |
| GO:0032703 | 9 | negative_regulation_of_interleukin-2_production |
| GO:0032794 | 9 | GTPase_activating_protein_binding |
| GO:0032863 | 9 | activation_of_Rac_GTPase_activity |
| GO:0032891 | 9 | negative_regulation_of_organic_acid_transport |
| GO:0032926 | 9 | negative_regulation_of_activin_receptor_signaling_pathway |
| GO:0033008 | 9 | positive_regulation_of_mast_cell_activation_involved_in_immune_response |
| GO:0033137 | 9 | negative_regulation_of_peptidyl-serine_phosphorylation |
| GO:0033148 | 9 | positive_regulation_of_intracellular_estrogen_receptor_signaling_pathway |
| GO:0033391 | 9 | chromatoid_body |
| GO:0033523 | 9 | histone_H2B_ubiquitination |
| GO:0033591 | 9 | response_to_L-ascorbic_acid |
| GO:0034045 | 9 | pre-autophagosomal_structure_membrane |
| GO:0034143 | 9 | regulation_of_toll-like_receptor_4_signaling_pathway |
| GO:0034285 | 9 | response_to_disaccharide_stimulus |
| GO:0034370 | 9 | triglyceride-rich_lipoprotein_particle_remodeling |
| GO:0034451 | 9 | centriolar_satellite |
| GO:0034464 | 9 | BBSome |
| GO:0034587 | 9 | piRNA_metabolic_process |
| GO:0035082 | 9 | axoneme_assembly |
| GO:0035086 | 9 | flagellar_axoneme |
| GO:0035259 | 9 | glucocorticoid_receptor_binding |
| GO:0035307 | 9 | positive_regulation_of_protein_dephosphorylation |
| GO:0035437 | 9 | maintenance_of_protein_localization_in_endoplasmic_reticulum |
| GO:0035850 | 9 | epithelial_cell_differentiation_involved_in_kidney_development |
| GO:0036002 | 9 | pre-mRNA_binding |
| GO:0038084 | 9 | vascular_endothelial_growth_factor_signaling_pathway |
| GO:0040034 | 9 | regulation_of_development,_heterochronic |
| GO:0042088 | 9 | T-helper_1_type_immune_response |
| GO:0042093 | 9 | T-helper_cell_differentiation |
| GO:0042135 | 9 | neurotransmitter_catabolic_process |
| GO:0042355 | 9 | L-fucose_catabolic_process |
| GO:0042428 | 9 | serotonin_metabolic_process |
| GO:0042438 | 9 | melanin_biosynthetic_process |
| GO:0042555 | 9 | MCM_complex |
| GO:0042575 | 9 | DNA_polymerase_complex |
| GO:0042581 | 9 | specific_granule |
| GO:0042589 | 9 | zymogen_granule_membrane |
| GO:0042599 | 9 | lamellar_body |
| GO:0042923 | 9 | neuropeptide_binding |
| GO:0042975 | 9 | peroxisome_proliferator_activated_receptor_binding |
| GO:0043020 | 9 | NADPH_oxidase_complex |
| GO:0043089 | 9 | positive_regulation_of_Cdc42_GTPase_activity |
| GO:0043097 | 9 | pyrimidine_nucleoside_salvage |
| GO:0043102 | 9 | amino_acid_salvage |
| GO:0043194 | 9 | axon_initial_segment |
| GO:0043230 | 9 | extracellular_organelle |
| GO:0043267 | 9 | negative_regulation_of_potassium_ion_transport |
| GO:0043306 | 9 | positive_regulation_of_mast_cell_degranulation |
| GO:0043508 | 9 | negative_regulation_of_JUN_kinase_activity |
| GO:0043568 | 9 | positive_regulation_of_insulin-like_growth_factor_receptor_signaling_pathway |
| GO:0043569 | 9 | negative_regulation_of_insulin-like_growth_factor_receptor_signaling_pathway |
| GO:0043583 | 9 | ear_development |
| GO:0043586 | 9 | tongue_development |
| GO:0043855 | 9 | cyclic_nucleotide-gated_ion_channel_activity |
| GO:0043995 | 9 | histone_acetyltransferase_activity_(H4-K5_specific) |
| GO:0043996 | 9 | histone_acetyltransferase_activity_(H4-K8_specific) |
| GO:0044065 | 9 | regulation_of_respiratory_system_process |
| GO:0044364 | 9 | disruption_of_cells_of_other_organism |
| GO:0045059 | 9 | positive_thymic_T_cell_selection |
| GO:0045125 | 9 | bioactive_lipid_receptor_activity |
| GO:0045161 | 9 | neuronal_ion_channel_clustering |
| GO:0045579 | 9 | positive_regulation_of_B_cell_differentiation |
| GO:0045603 | 9 | positive_regulation_of_endothelial_cell_differentiation |
| GO:0045618 | 9 | positive_regulation_of_keratinocyte_differentiation |
| GO:0045625 | 9 | regulation_of_T-helper_1_cell_differentiation |
| GO:0045628 | 9 | regulation_of_T-helper_2_cell_differentiation |
| GO:0045655 | 9 | regulation_of_monocyte_differentiation |
| GO:0045662 | 9 | negative_regulation_of_myoblast_differentiation |
| GO:0045779 | 9 | negative_regulation_of_bone_resorption |
| GO:0045792 | 9 | negative_regulation_of_cell_size |
| GO:0045821 | 9 | positive_regulation_of_glycolysis |
| GO:0045948 | 9 | positive_regulation_of_translational_initiation |
| GO:0045956 | 9 | positive_regulation_of_calcium_ion-dependent_exocytosis |
| GO:0045986 | 9 | negative_regulation_of_smooth_muscle_contraction |
| GO:0046033 | 9 | AMP_metabolic_process |
| GO:0046174 | 9 | polyol_catabolic_process |
| GO:0046335 | 9 | ethanolamine_biosynthetic_process |
| GO:0046337 | 9 | phosphatidylethanolamine_metabolic_process |
| GO:0046521 | 9 | sphingoid_catabolic_process |
| GO:0046581 | 9 | intercellular_canaliculus |
| GO:0046621 | 9 | negative_regulation_of_organ_growth |
| GO:0046643 | 9 | regulation_of_gamma-delta_T_cell_activation |
| GO:0046645 | 9 | positive_regulation_of_gamma-delta_T_cell_activation |
| GO:0046655 | 9 | folic_acid_metabolic_process |
| GO:0046870 | 9 | cadmium_ion_binding |
| GO:0046886 | 9 | positive_regulation_of_hormone_biosynthetic_process |
| GO:0046972 | 9 | histone_acetyltransferase_activity_(H4-K16_specific) |
| GO:0048066 | 9 | developmental_pigmentation |
| GO:0048188 | 9 | Set1C/COMPASS_complex |
| GO:0048387 | 9 | negative_regulation_of_retinoic_acid_receptor_signaling_pathway |
| GO:0048505 | 9 | regulation_of_timing_of_cell_differentiation |
| GO:0048531 | 9 | beta-1,3-galactosyltransferase_activity |
| GO:0048532 | 9 | anatomical_structure_arrangement |
| GO:0048711 | 9 | positive_regulation_of_astrocyte_differentiation |
| GO:0048714 | 9 | positive_regulation_of_oligodendrocyte_differentiation |
| GO:0048853 | 9 | forebrain_morphogenesis |
| GO:0048935 | 9 | peripheral_nervous_system_neuron_development |
| GO:0050665 | 9 | hydrogen_peroxide_biosynthetic_process |
| GO:0050667 | 9 | homocysteine_metabolic_process |
| GO:0050901 | 9 | leukocyte_tethering_or_rolling |
| GO:0050935 | 9 | iridophore_differentiation |
| GO:0051006 | 9 | positive_regulation_of_lipoprotein_lipase_activity |
| GO:0051238 | 9 | sequestering_of_metal_ion |
| GO:0051383 | 9 | kinetochore_organization |
| GO:0051770 | 9 | positive_regulation_of_nitric-oxide_synthase_biosynthetic_process |
| GO:0051818 | 9 | disruption_of_cells_of_other_organism_involved_in_symbiotic_interaction |
| GO:0051883 | 9 | killing_of_cells_in_other_organism_involved_in_symbiotic_interaction |
| GO:0051890 | 9 | regulation_of_cardioblast_differentiation |
| GO:0055089 | 9 | fatty_acid_homeostasis |
| GO:0055091 | 9 | phospholipid_homeostasis |
| GO:0060046 | 9 | regulation_of_acrosome_reaction |
| GO:0060123 | 9 | regulation_of_growth_hormone_secretion |
| GO:0060263 | 9 | regulation_of_respiratory_burst |
| GO:0060324 | 9 | face_development |
| GO:0060363 | 9 | cranial_suture_morphogenesis |
| GO:0060390 | 9 | regulation_of_SMAD_protein_import_into_nucleus |
| GO:0060413 | 9 | atrial_septum_morphogenesis |
| GO:0060484 | 9 | lung-associated_mesenchyme_development |
| GO:0060539 | 9 | diaphragm_development |
| GO:0060840 | 9 | artery_development |
| GO:0060969 | 9 | negative_regulation_of_gene_silencing |
| GO:0060997 | 9 | dendritic_spine_morphogenesis |
| GO:0061029 | 9 | eyelid_development_in_camera-type_eye |
| GO:0061303 | 9 | cornea_development_in_camera-type_eye |
| GO:0065010 | 9 | extracellular_membrane-bounded_organelle |
| GO:0070189 | 9 | kynurenine_metabolic_process |
| GO:0070227 | 9 | lymphocyte_apoptotic_process |
| GO:0070230 | 9 | positive_regulation_of_lymphocyte_apoptotic_process |
| GO:0070233 | 9 | negative_regulation_of_T_cell_apoptotic_process |
| GO:0070696 | 9 | transmembrane_receptor_protein_serine/threonine_kinase_binding |
| GO:0070874 | 9 | negative_regulation_of_glycogen_metabolic_process |
| GO:0070935 | 9 | 3'-UTR-mediated_mRNA_stabilization |
| GO:0071265 | 9 | L-methionine_biosynthetic_process |
| GO:0071267 | 9 | L-methionine_salvage |
| GO:0071350 | 9 | cellular_response_to_interleukin-15 |
| GO:0071392 | 9 | cellular_response_to_estradiol_stimulus |
| GO:0071577 | 9 | zinc_ion_transmembrane_transport |
| GO:0071772 | 9 | response_to_BMP_stimulus |
| GO:0071773 | 9 | cellular_response_to_BMP_stimulus |
| GO:0072044 | 9 | collecting_duct_development |
| GO:0072079 | 9 | nephron_tubule_formation |
| GO:0072111 | 9 | cell_proliferation_involved_in_kidney_development |
| GO:0072163 | 9 | mesonephric_epithelium_development |
| GO:0072349 | 9 | modified_amino_acid_transmembrane_transporter_activity |
| GO:0072666 | 9 | establishment_of_protein_localization_to_vacuole |
| GO:0086010 | 9 | membrane_depolarization_involved_in_regulation_of_action_potential |
| GO:0086012 | 9 | membrane_depolarization_involved_in_regulation_of_cardiac_muscle_cell_action_potential |
| GO:0090009 | 9 | primitive_streak_formation |
| GO:0090075 | 9 | relaxation_of_muscle |
| GO:0090102 | 9 | cochlea_development |
| GO:0090201 | 9 | negative_regulation_of_release_of_cytochrome_c_from_mitochondria |
| GO:0090288 | 9 | negative_regulation_of_cellular_response_to_growth_factor_stimulus |
| GO:0090344 | 9 | negative_regulation_of_cell_aging |
| GO:0097090 | 9 | presynaptic_membrane_organization |
| GO:1901072 | 9 | glucosamine-containing_compound_catabolic_process |
| GO:2000008 | 9 | regulation_of_protein_localization_at_cell_surface |
| GO:2000310 | 9 | regulation_of_N-methyl-D-aspartate_selective_glutamate_receptor_activity |
| GO:2000352 | 9 | negative_regulation_of_endothelial_cell_apoptotic_process |
| GO:2000515 | 9 | negative_regulation_of_CD4-positive,_alpha-beta_T_cell_activation |
| GO:2000757 | 9 | negative_regulation_of_peptidyl-lysine_acetylation |
| GO:0000109 | 8 | nucleotide-excision_repair_complex |
| GO:0000239 | 8 | pachytene |
| GO:0000276 | 8 | mitochondrial_proton-transporting_ATP_synthase_complex,_coupling_factor_F(o) |
| GO:0000339 | 8 | RNA_cap_binding |
| GO:0000346 | 8 | transcription_export_complex |
| GO:0000460 | 8 | maturation_of_5.8S_rRNA |
| GO:0000506 | 8 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase_(GPI-GnT)_complex |
| GO:0000738 | 8 | DNA_catabolic_process,_exonucleolytic |
| GO:0000983 | 8 | RNA_polymerase_II_core_promoter_sequence-specific_DNA_binding_transcription_factor_activity |
| GO:0001553 | 8 | luteinization |
| GO:0001671 | 8 | ATPase_activator_activity |
| GO:0001895 | 8 | retina_homeostasis |
| GO:0001921 | 8 | positive_regulation_of_receptor_recycling |
| GO:0001931 | 8 | uropod |
| GO:0002087 | 8 | regulation_of_respiratory_gaseous_exchange_by_neurological_system_process |
| GO:0002262 | 8 | myeloid_cell_homeostasis |
| GO:0002279 | 8 | mast_cell_activation_involved_in_immune_response |
| GO:0002283 | 8 | neutrophil_activation_involved_in_immune_response |
| GO:0002335 | 8 | mature_B_cell_differentiation |
| GO:0002367 | 8 | cytokine_production_involved_in_immune_response |
| GO:0002532 | 8 | production_of_molecular_mediator_involved_in_inflammatory_response |
| GO:0002604 | 8 | regulation_of_dendritic_cell_antigen_processing_and_presentation |
| GO:0002666 | 8 | positive_regulation_of_T_cell_tolerance_induction |
| GO:0002862 | 8 | negative_regulation_of_inflammatory_response_to_antigenic_stimulus |
| GO:0002883 | 8 | regulation_of_hypersensitivity |
| GO:0002903 | 8 | negative_regulation_of_B_cell_apoptotic_process |
| GO:0003015 | 8 | heart_process |
| GO:0003085 | 8 | negative_regulation_of_systemic_arterial_blood_pressure |
| GO:0003097 | 8 | renal_water_transport |
| GO:0003197 | 8 | endocardial_cushion_development |
| GO:0003339 | 8 | regulation_of_mesenchymal_to_epithelial_transition_involved_in_metanephros_morphogenesis |
| GO:0003341 | 8 | cilium_movement |
| GO:0003857 | 8 | 3-hydroxyacyl-CoA_dehydrogenase_activity |
| GO:0004017 | 8 | adenylate_kinase_activity |
| GO:0004126 | 8 | cytidine_deaminase_activity |
| GO:0004128 | 8 | cytochrome-b5_reductase_activity |
| GO:0004312 | 8 | fatty_acid_synthase_activity |
| GO:0004529 | 8 | exodeoxyribonuclease_activity |
| GO:0004551 | 8 | nucleotide_diphosphatase_activity |
| GO:0004571 | 8 | mannosyl-oligosaccharide_1,2-alpha-mannosidase_activity |
| GO:0004579 | 8 | dolichyl-diphosphooligosaccharide-protein_glycotransferase_activity |
| GO:0004726 | 8 | non-membrane_spanning_protein_tyrosine_phosphatase_activity |
| GO:0004887 | 8 | thyroid_hormone_receptor_activity |
| GO:0004982 | 8 | N-formyl_peptide_receptor_activity |
| GO:0005021 | 8 | vascular_endothelial_growth_factor-activated_receptor_activity |
| GO:0005068 | 8 | transmembrane_receptor_protein_tyrosine_kinase_adaptor_activity |
| GO:0005132 | 8 | interferon-alpha/beta_receptor_binding |
| GO:0005338 | 8 | nucleotide-sugar_transmembrane_transporter_activity |
| GO:0005436 | 8 | sodium:phosphate_symporter_activity |
| GO:0005521 | 8 | lamin_binding |
| GO:0005522 | 8 | profilin_binding |
| GO:0005536 | 8 | glucose_binding |
| GO:0005577 | 8 | fibrinogen_complex |
| GO:0005838 | 8 | proteasome_regulatory_particle |
| GO:0005861 | 8 | troponin_complex |
| GO:0005915 | 8 | zonula_adherens |
| GO:0005942 | 8 | phosphatidylinositol_3-kinase_complex |
| GO:0006108 | 8 | malate_metabolic_process |
| GO:0006208 | 8 | pyrimidine_base_catabolic_process |
| GO:0006216 | 8 | cytidine_catabolic_process |
| GO:0006244 | 8 | pyrimidine_nucleotide_catabolic_process |
| GO:0006264 | 8 | mitochondrial_DNA_replication |
| GO:0006268 | 8 | DNA_unwinding_involved_in_replication |
| GO:0006285 | 8 | base-excision_repair,_AP_site_formation |
| GO:0006403 | 8 | RNA_localization |
| GO:0006491 | 8 | N-glycan_processing |
| GO:0006546 | 8 | glycine_catabolic_process |
| GO:0006563 | 8 | L-serine_metabolic_process |
| GO:0006573 | 8 | valine_metabolic_process |
| GO:0006835 | 8 | dicarboxylic_acid_transport |
| GO:0006853 | 8 | carnitine_shuttle |
| GO:0006857 | 8 | oligopeptide_transport |
| GO:0007095 | 8 | mitotic_cell_cycle_G2/M_transition_DNA_damage_checkpoint |
| GO:0007099 | 8 | centriole_replication |
| GO:0007172 | 8 | signal_complex_assembly |
| GO:0007175 | 8 | negative_regulation_of_epidermal_growth_factor-activated_receptor_activity |
| GO:0007183 | 8 | SMAD_protein_complex_assembly |
| GO:0007199 | 8 | G-protein_coupled_receptor_signaling_pathway_coupled_to_cGMP_nucleotide_second_messenger |
| GO:0007217 | 8 | tachykinin_receptor_signaling_pathway |
| GO:0007260 | 8 | tyrosine_phosphorylation_of_STAT_protein |
| GO:0007617 | 8 | mating_behavior |
| GO:0008089 | 8 | anterograde_axon_cargo_transport |
| GO:0008140 | 8 | cAMP_response_element_binding_protein_binding |
| GO:0008157 | 8 | protein_phosphatase_1_binding |
| GO:0008266 | 8 | poly(U)_RNA_binding |
| GO:0008272 | 8 | sulfate_transport |
| GO:0008290 | 8 | F-actin_capping_protein_complex |
| GO:0008354 | 8 | germ_cell_migration |
| GO:0008432 | 8 | JUN_kinase_binding |
| GO:0008499 | 8 | UDP-galactose:beta-N-acetylglucosamine_beta-1,3-galactosyltransferase_activity |
| GO:0008553 | 8 | hydrogen-exporting_ATPase_activity,_phosphorylative_mechanism |
| GO:0008634 | 8 | negative_regulation_of_survival_gene_product_expression |
| GO:0008889 | 8 | glycerophosphodiester_phosphodiesterase_activity |
| GO:0009103 | 8 | lipopolysaccharide_biosynthetic_process |
| GO:0009135 | 8 | purine_nucleoside_diphosphate_metabolic_process |
| GO:0009179 | 8 | purine_ribonucleoside_diphosphate_metabolic_process |
| GO:0009265 | 8 | 2'-deoxyribonucleotide_biosynthetic_process |
| GO:0009750 | 8 | response_to_fructose_stimulus |
| GO:0009972 | 8 | cytidine_deamination |
| GO:0010171 | 8 | body_morphogenesis |
| GO:0010225 | 8 | response_to_UV-C |
| GO:0010310 | 8 | regulation_of_hydrogen_peroxide_metabolic_process |
| GO:0010755 | 8 | regulation_of_plasminogen_activation |
| GO:0010819 | 8 | regulation_of_T_cell_chemotaxis |
| GO:0010820 | 8 | positive_regulation_of_T_cell_chemotaxis |
| GO:0010832 | 8 | negative_regulation_of_myotube_differentiation |
| GO:0010851 | 8 | cyclase_regulator_activity |
| GO:0010896 | 8 | regulation_of_triglyceride_catabolic_process |
| GO:0014049 | 8 | positive_regulation_of_glutamate_secretion |
| GO:0014067 | 8 | negative_regulation_of_phosphatidylinositol_3-kinase_cascade |
| GO:0014741 | 8 | negative_regulation_of_muscle_hypertrophy |
| GO:0014850 | 8 | response_to_muscle_activity |
| GO:0014855 | 8 | striated_muscle_cell_proliferation |
| GO:0014896 | 8 | muscle_hypertrophy |
| GO:0015277 | 8 | kainate_selective_glutamate_receptor_activity |
| GO:0015288 | 8 | porin_activity |
| GO:0015377 | 8 | cation:chloride_symporter_activity |
| GO:0015884 | 8 | folic_acid_transport |
| GO:0016075 | 8 | rRNA_catabolic_process |
| GO:0016342 | 8 | catenin_complex |
| GO:0016405 | 8 | CoA-ligase_activity |
| GO:0016442 | 8 | RNA-induced_silencing_complex |
| GO:0016494 | 8 | C-X-C_chemokine_receptor_activity |
| GO:0016540 | 8 | protein_autoprocessing |
| GO:0016653 | 8 | oxidoreductase_activity,_acting_on_NADH_or_NADPH,_heme_protein_as_acceptor |
| GO:0016742 | 8 | hydroxymethyl-,_formyl-_and_related_transferase_activity |
| GO:0016832 | 8 | aldehyde-lyase_activity |
| GO:0016857 | 8 | racemase_and_epimerase_activity,_acting_on_carbohydrates_and_derivatives |
| GO:0016895 | 8 | exodeoxyribonuclease_activity,_producing_5'-phosphomonoesters |
| GO:0017146 | 8 | N-methyl-D-aspartate_selective_glutamate_receptor_complex |
| GO:0017154 | 8 | semaphorin_receptor_activity |
| GO:0017169 | 8 | CDP-alcohol_phosphatidyltransferase_activity |
| GO:0018342 | 8 | protein_prenylation |
| GO:0019047 | 8 | provirus_integration |
| GO:0019226 | 8 | transmission_of_nerve_impulse |
| GO:0019405 | 8 | alditol_catabolic_process |
| GO:0019673 | 8 | GDP-mannose_metabolic_process |
| GO:0019773 | 8 | proteasome_core_complex,_alpha-subunit_complex |
| GO:0019800 | 8 | peptide_cross-linking_via_chondroitin_4-sulfate_glycosaminoglycan |
| GO:0019883 | 8 | antigen_processing_and_presentation_of_endogenous_antigen |
| GO:0021680 | 8 | cerebellar_Purkinje_cell_layer_development |
| GO:0021756 | 8 | striatum_development |
| GO:0021799 | 8 | cerebral_cortex_radially_oriented_cell_migration |
| GO:0022010 | 8 | central_nervous_system_myelination |
| GO:0022011 | 8 | myelination_in_peripheral_nervous_system |
| GO:0023014 | 8 | signal_transduction_by_phosphorylation |
| GO:0030056 | 8 | hemidesmosome |
| GO:0030202 | 8 | heparin_metabolic_process |
| GO:0030277 | 8 | maintenance_of_gastrointestinal_epithelium |
| GO:0030280 | 8 | structural_constituent_of_epidermis |
| GO:0030299 | 8 | intestinal_cholesterol_absorption |
| GO:0030432 | 8 | peristalsis |
| GO:0030497 | 8 | fatty_acid_elongation |
| GO:0030647 | 8 | aminoglycoside_antibiotic_metabolic_process |
| GO:0030676 | 8 | Rac_guanyl-nucleotide_exchange_factor_activity |
| GO:0031065 | 8 | positive_regulation_of_histone_deacetylation |
| GO:0031095 | 8 | platelet_dense_tubular_network_membrane |
| GO:0031332 | 8 | RNAi_effector_complex |
| GO:0031371 | 8 | ubiquitin_conjugating_enzyme_complex |
| GO:0031491 | 8 | nucleosome_binding |
| GO:0031543 | 8 | peptidyl-proline_dioxygenase_activity |
| GO:0031579 | 8 | membrane_raft_organization |
| GO:0031624 | 8 | ubiquitin_conjugating_enzyme_binding |
| GO:0031657 | 8 | regulation_of_cyclin-dependent_protein_kinase_activity_involved_in_G1/S |
| GO:0031672 | 8 | A_band |
| GO:0031701 | 8 | angiotensin_receptor_binding |
| GO:0031929 | 8 | TOR_signaling_cascade |
| GO:0031953 | 8 | negative_regulation_of_protein_autophosphorylation |
| GO:0031958 | 8 | corticosteroid_receptor_signaling_pathway |
| GO:0032000 | 8 | positive_regulation_of_fatty_acid_beta-oxidation |
| GO:0032040 | 8 | small-subunit_processome |
| GO:0032229 | 8 | negative_regulation_of_synaptic_transmission,_GABAergic |
| GO:0032261 | 8 | purine_nucleotide_salvage |
| GO:0032276 | 8 | regulation_of_gonadotropin_secretion |
| GO:0032291 | 8 | axon_ensheathment_in_central_nervous_system |
| GO:0032292 | 8 | peripheral_nervous_system_axon_ensheathment |
| GO:0032306 | 8 | regulation_of_prostaglandin_secretion |
| GO:0032308 | 8 | positive_regulation_of_prostaglandin_secretion |
| GO:0032351 | 8 | negative_regulation_of_hormone_metabolic_process |
| GO:0032429 | 8 | regulation_of_phospholipase_A2_activity |
| GO:0032453 | 8 | histone_demethylase_activity_(H3-K4_specific) |
| GO:0032464 | 8 | positive_regulation_of_protein_homooligomerization |
| GO:0032469 | 8 | endoplasmic_reticulum_calcium_ion_homeostasis |
| GO:0032490 | 8 | detection_of_molecule_of_bacterial_origin |
| GO:0032495 | 8 | response_to_muramyl_dipeptide |
| GO:0032525 | 8 | somite_rostral/caudal_axis_specification |
| GO:0032700 | 8 | negative_regulation_of_interleukin-17_production |
| GO:0032727 | 8 | positive_regulation_of_interferon-alpha_production |
| GO:0032928 | 8 | regulation_of_superoxide_anion_generation |
| GO:0033083 | 8 | regulation_of_immature_T_cell_proliferation |
| GO:0033162 | 8 | melanosome_membrane |
| GO:0033235 | 8 | positive_regulation_of_protein_sumoylation |
| GO:0033240 | 8 | positive_regulation_of_cellular_amine_metabolic_process |
| GO:0033483 | 8 | gas_homeostasis |
| GO:0033690 | 8 | positive_regulation_of_osteoblast_proliferation |
| GO:0034310 | 8 | primary_alcohol_catabolic_process |
| GO:0034331 | 8 | cell_junction_maintenance |
| GO:0034366 | 8 | spherical_high-density_lipoprotein_particle |
| GO:0034383 | 8 | low-density_lipoprotein_particle_clearance |
| GO:0034450 | 8 | ubiquitin-ubiquitin_ligase_activity |
| GO:0034595 | 8 | phosphatidylinositol_phosphate_5-phosphatase_activity |
| GO:0034969 | 8 | histone_arginine_methylation |
| GO:0035024 | 8 | negative_regulation_of_Rho_protein_signal_transduction |
| GO:0035025 | 8 | positive_regulation_of_Rho_protein_signal_transduction |
| GO:0035067 | 8 | negative_regulation_of_histone_acetylation |
| GO:0035198 | 8 | miRNA_binding |
| GO:0035235 | 8 | ionotropic_glutamate_receptor_signaling_pathway |
| GO:0035246 | 8 | peptidyl-arginine_N-methylation |
| GO:0035327 | 8 | transcriptionally_active_chromatin |
| GO:0035413 | 8 | positive_regulation_of_catenin_import_into_nucleus |
| GO:0035414 | 8 | negative_regulation_of_catenin_import_into_nucleus |
| GO:0035455 | 8 | response_to_interferon-alpha |
| GO:0035590 | 8 | purinergic_nucleotide_receptor_signaling_pathway |
| GO:0035869 | 8 | ciliary_transition_zone |
| GO:0036128 | 8 | CatSper_complex |
| GO:0042447 | 8 | hormone_catabolic_process |
| GO:0042473 | 8 | outer_ear_morphogenesis |
| GO:0042511 | 8 | positive_regulation_of_tyrosine_phosphorylation_of_Stat1_protein |
| GO:0042753 | 8 | positive_regulation_of_circadian_rhythm |
| GO:0042756 | 8 | drinking_behavior |
| GO:0042769 | 8 | DNA_damage_response,_detection_of_DNA_damage |
| GO:0042834 | 8 | peptidoglycan_binding |
| GO:0042886 | 8 | amide_transport |
| GO:0042989 | 8 | sequestering_of_actin_monomers |
| GO:0043121 | 8 | neurotrophin_binding |
| GO:0043149 | 8 | stress_fiber_assembly |
| GO:0043206 | 8 | fibril_organization |
| GO:0043208 | 8 | glycosphingolipid_binding |
| GO:0043295 | 8 | glutathione_binding |
| GO:0043303 | 8 | mast_cell_degranulation |
| GO:0043371 | 8 | negative_regulation_of_CD4-positive,_alpha-beta_T_cell_differentiation |
| GO:0043422 | 8 | protein_kinase_B_binding |
| GO:0043522 | 8 | leucine_zipper_domain_binding |
| GO:0043587 | 8 | tongue_morphogenesis |
| GO:0044030 | 8 | regulation_of_DNA_methylation |
| GO:0044320 | 8 | cellular_response_to_leptin_stimulus |
| GO:0044321 | 8 | response_to_leptin_stimulus |
| GO:0044390 | 8 | small_protein_conjugating_enzyme_binding |
| GO:0044548 | 8 | S100_protein_binding |
| GO:0044597 | 8 | daunorubicin_metabolic_process |
| GO:0044598 | 8 | doxorubicin_metabolic_process |
| GO:0045010 | 8 | actin_nucleation |
| GO:0045075 | 8 | regulation_of_interleukin-12_biosynthetic_process |
| GO:0045217 | 8 | cell-cell_junction_maintenance |
| GO:0045294 | 8 | alpha-catenin_binding |
| GO:0045348 | 8 | positive_regulation_of_MHC_class_II_biosynthetic_process |
| GO:0045357 | 8 | regulation_of_interferon-beta_biosynthetic_process |
| GO:0045416 | 8 | positive_regulation_of_interleukin-8_biosynthetic_process |
| GO:0045589 | 8 | regulation_of_regulatory_T_cell_differentiation |
| GO:0045623 | 8 | negative_regulation_of_T-helper_cell_differentiation |
| GO:0045647 | 8 | negative_regulation_of_erythrocyte_differentiation |
| GO:0045760 | 8 | positive_regulation_of_action_potential |
| GO:0046030 | 8 | inositol_trisphosphate_phosphatase_activity |
| GO:0046085 | 8 | adenosine_metabolic_process |
| GO:0046087 | 8 | cytidine_metabolic_process |
| GO:0046133 | 8 | pyrimidine_ribonucleoside_catabolic_process |
| GO:0046173 | 8 | polyol_biosynthetic_process |
| GO:0046479 | 8 | glycosphingolipid_catabolic_process |
| GO:0046487 | 8 | glyoxylate_metabolic_process |
| GO:0046514 | 8 | ceramide_catabolic_process |
| GO:0046549 | 8 | retinal_cone_cell_development |
| GO:0046599 | 8 | regulation_of_centriole_replication |
| GO:0046784 | 8 | intronless_viral_mRNA_export_from_host_nucleus |
| GO:0046920 | 8 | alpha-(1->3)-fucosyltransferase_activity |
| GO:0046939 | 8 | nucleotide_phosphorylation |
| GO:0047498 | 8 | calcium-dependent_phospholipase_A2_activity |
| GO:0047760 | 8 | butyrate-CoA_ligase_activity |
| GO:0048172 | 8 | regulation_of_short-term_neuronal_synaptic_plasticity |
| GO:0048245 | 8 | eosinophil_chemotaxis |
| GO:0048261 | 8 | negative_regulation_of_receptor-mediated_endocytosis |
| GO:0048311 | 8 | mitochondrion_distribution |
| GO:0048537 | 8 | mucosal-associated_lymphoid_tissue_development |
| GO:0048643 | 8 | positive_regulation_of_skeletal_muscle_tissue_development |
| GO:0048644 | 8 | muscle_organ_morphogenesis |
| GO:0048670 | 8 | regulation_of_collateral_sprouting |
| GO:0048843 | 8 | negative_regulation_of_axon_extension_involved_in_axon_guidance |
| GO:0050291 | 8 | sphingosine_N-acyltransferase_activity |
| GO:0050700 | 8 | CARD_domain_binding |
| GO:0050811 | 8 | GABA_receptor_binding |
| GO:0050884 | 8 | neuromuscular_process_controlling_posture |
| GO:0050998 | 8 | nitric-oxide_synthase_binding |
| GO:0051095 | 8 | regulation_of_helicase_activity |
| GO:0051151 | 8 | negative_regulation_of_smooth_muscle_cell_differentiation |
| GO:0051382 | 8 | kinetochore_assembly |
| GO:0051386 | 8 | regulation_of_nerve_growth_factor_receptor_signaling_pathway |
| GO:0051400 | 8 | BH_domain_binding |
| GO:0051481 | 8 | reduction_of_cytosolic_calcium_ion_concentration |
| GO:0051532 | 8 | regulation_of_NFAT_protein_import_into_nucleus |
| GO:0051571 | 8 | positive_regulation_of_histone_H3-K4_methylation |
| GO:0051589 | 8 | negative_regulation_of_neurotransmitter_transport |
| GO:0051654 | 8 | establishment_of_mitochondrion_localization |
| GO:0051657 | 8 | maintenance_of_organelle_location |
| GO:0051766 | 8 | inositol_trisphosphate_kinase_activity |
| GO:0051787 | 8 | misfolded_protein_binding |
| GO:0051920 | 8 | peroxiredoxin_activity |
| GO:0051929 | 8 | positive_regulation_of_calcium_ion_transport_via_voltage-gated_calcium_channel_activity |
| GO:0051938 | 8 | L-glutamate_import |
| GO:0051967 | 8 | negative_regulation_of_synaptic_transmission,_glutamatergic |
| GO:0051988 | 8 | regulation_of_attachment_of_spindle_microtubules_to_kinetochore |
| GO:0060009 | 8 | Sertoli_cell_development |
| GO:0060038 | 8 | cardiac_muscle_cell_proliferation |
| GO:0060047 | 8 | heart_contraction |
| GO:0060052 | 8 | neurofilament_cytoskeleton_organization |
| GO:0060055 | 8 | angiogenesis_involved_in_wound_healing |
| GO:0060059 | 8 | embryonic_retina_morphogenesis_in_camera-type_eye |
| GO:0060068 | 8 | vagina_development |
| GO:0060119 | 8 | inner_ear_receptor_cell_development |
| GO:0060158 | 8 | phospholipase_C-activating_dopamine_receptor_signaling_pathway |
| GO:0060206 | 8 | estrous_cycle_phase |
| GO:0060229 | 8 | lipase_activator_activity |
| GO:0060253 | 8 | negative_regulation_of_glial_cell_proliferation |
| GO:0060272 | 8 | embryonic_skeletal_joint_morphogenesis |
| GO:0060346 | 8 | bone_trabecula_formation |
| GO:0060384 | 8 | innervation |
| GO:0060391 | 8 | positive_regulation_of_SMAD_protein_import_into_nucleus |
| GO:0060396 | 8 | growth_hormone_receptor_signaling_pathway |
| GO:0060426 | 8 | lung_vasculature_development |
| GO:0060457 | 8 | negative_regulation_of_digestive_system_process |
| GO:0060527 | 8 | prostate_epithelial_cord_arborization_involved_in_prostate_glandular_acinus_morphogenesis |
| GO:0060561 | 8 | apoptotic_process_involved_in_morphogenesis |
| GO:0060601 | 8 | lateral_sprouting_from_an_epithelium |
| GO:0060670 | 8 | branching_involved_in_labyrinthine_layer_morphogenesis |
| GO:0060678 | 8 | dichotomous_subdivision_of_terminal_units_involved_in_ureteric_bud_branching |
| GO:0060687 | 8 | regulation_of_branching_involved_in_prostate_gland_morphogenesis |
| GO:0060707 | 8 | trophoblast_giant_cell_differentiation |
| GO:0060711 | 8 | labyrinthine_layer_development |
| GO:0060972 | 8 | left/right_pattern_formation |
| GO:0061081 | 8 | positive_regulation_of_myeloid_leukocyte_cytokine_production_involved_in_immune_response |
| GO:0061162 | 8 | establishment_of_monopolar_cell_polarity |
| GO:0061202 | 8 | clathrin_sculpted_gamma-aminobutyric_acid_transport_vesicle_membrane |
| GO:0061337 | 8 | cardiac_conduction |
| GO:0061339 | 8 | establishment_or_maintenance_of_monopolar_cell_polarity |
| GO:0070008 | 8 | serine-type_exopeptidase_activity |
| GO:0070234 | 8 | positive_regulation_of_T_cell_apoptotic_process |
| GO:0070306 | 8 | lens_fiber_cell_differentiation |
| GO:0070513 | 8 | death_domain_binding |
| GO:0070577 | 8 | histone_acetyl-lysine_binding |
| GO:0070628 | 8 | proteasome_binding |
| GO:0070652 | 8 | HAUS_complex |
| GO:0070688 | 8 | MLL5-L_complex |
| GO:0071305 | 8 | cellular_response_to_vitamin_D |
| GO:0071353 | 8 | cellular_response_to_interleukin-4 |
| GO:0071378 | 8 | cellular_response_to_growth_hormone_stimulus |
| GO:0071467 | 8 | cellular_response_to_pH |
| GO:0071498 | 8 | cellular_response_to_fluid_shear_stress |
| GO:0071850 | 8 | mitotic_cell_cycle_arrest |
| GO:0072015 | 8 | glomerular_visceral_epithelial_cell_development |
| GO:0072164 | 8 | mesonephric_tubule_development |
| GO:0072202 | 8 | cell_differentiation_involved_in_metanephros_development |
| GO:0072203 | 8 | cell_proliferation_involved_in_metanephros_development |
| GO:0072310 | 8 | glomerular_epithelial_cell_development |
| GO:0072337 | 8 | modified_amino_acid_transport |
| GO:0072677 | 8 | eosinophil_migration |
| GO:0090231 | 8 | regulation_of_spindle_checkpoint |
| GO:0097105 | 8 | presynaptic_membrane_assembly |
| GO:0097354 | 8 | prenylation |
| GO:1900750 | 8 | oligopeptide_binding |
| GO:2000036 | 8 | regulation_of_stem_cell_maintenance |
| GO:2000109 | 8 | regulation_of_macrophage_apoptotic_process |
| GO:2000678 | 8 | negative_regulation_of_transcription_regulatory_region_DNA_binding |
| GO:2000725 | 8 | regulation_of_cardiac_muscle_cell_differentiation |
| GO:2001015 | 8 | negative_regulation_of_skeletal_muscle_cell_differentiation |
| GO:0000002 | 7 | mitochondrial_genome_maintenance |
| GO:0000183 | 7 | chromatin_silencing_at_rDNA |
| GO:0000185 | 7 | activation_of_MAPKKK_activity |
| GO:0000270 | 7 | peptidoglycan_metabolic_process |
| GO:0000281 | 7 | cytokinesis_after_mitosis |
| GO:0000380 | 7 | alternative_nuclear_mRNA_splicing,_via_spliceosome |
| GO:0000712 | 7 | resolution_of_meiotic_recombination_intermediates |
| GO:0000931 | 7 | gamma-tubulin_large_complex |
| GO:0001103 | 7 | RNA_polymerase_II_repressing_transcription_factor_binding |
| GO:0001574 | 7 | ganglioside_biosynthetic_process |
| GO:0001609 | 7 | G-protein_coupled_adenosine_receptor_activity |
| GO:0001758 | 7 | retinal_dehydrogenase_activity |
| GO:0001779 | 7 | natural_killer_cell_differentiation |
| GO:0001848 | 7 | complement_binding |
| GO:0001867 | 7 | complement_activation,_lectin_pathway |
| GO:0001878 | 7 | response_to_yeast |
| GO:0001886 | 7 | endothelial_cell_morphogenesis |
| GO:0001939 | 7 | female_pronucleus |
| GO:0001941 | 7 | postsynaptic_membrane_organization |
| GO:0001945 | 7 | lymph_vessel_development |
| GO:0001964 | 7 | startle_response |
| GO:0002016 | 7 | regulation_of_blood_volume_by_renin-angiotensin |
| GO:0002021 | 7 | response_to_dietary_excess |
| GO:0002116 | 7 | semaphorin_receptor_complex |
| GO:0002118 | 7 | aggressive_behavior |
| GO:0002161 | 7 | aminoacyl-tRNA_editing_activity |
| GO:0002228 | 7 | natural_killer_cell_mediated_immunity |
| GO:0002446 | 7 | neutrophil_mediated_immunity |
| GO:0002507 | 7 | tolerance_induction |
| GO:0002606 | 7 | positive_regulation_of_dendritic_cell_antigen_processing_and_presentation |
| GO:0002639 | 7 | positive_regulation_of_immunoglobulin_production |
| GO:0002679 | 7 | respiratory_burst_involved_in_defense_response |
| GO:0002739 | 7 | regulation_of_cytokine_secretion_involved_in_immune_response |
| GO:0002866 | 7 | positive_regulation_of_acute_inflammatory_response_to_antigenic_stimulus |
| GO:0002925 | 7 | positive_regulation_of_humoral_immune_response_mediated_by_circulating_immunoglobulin |
| GO:0003093 | 7 | regulation_of_glomerular_filtration |
| GO:0003094 | 7 | glomerular_filtration |
| GO:0003215 | 7 | cardiac_right_ventricle_morphogenesis |
| GO:0003307 | 7 | regulation_of_Wnt_receptor_signaling_pathway_involved_in_heart_development |
| GO:0003407 | 7 | neural_retina_development |
| GO:0003680 | 7 | AT_DNA_binding |
| GO:0003696 | 7 | satellite_DNA_binding |
| GO:0003708 | 7 | retinoic_acid_receptor_activity |
| GO:0003854 | 7 | 3-beta-hydroxy-delta5-steroid_dehydrogenase_activity |
| GO:0004030 | 7 | aldehyde_dehydrogenase_[NAD(P)+]_activity |
| GO:0004032 | 7 | alditol:NADP+_1-oxidoreductase_activity |
| GO:0004176 | 7 | ATP-dependent_peptidase_activity |
| GO:0004415 | 7 | hyalurononglucosaminidase_activity |
| GO:0004445 | 7 | inositol-polyphosphate_5-phosphatase_activity |
| GO:0004559 | 7 | alpha-mannosidase_activity |
| GO:0004568 | 7 | chitinase_activity |
| GO:0004679 | 7 | AMP-activated_protein_kinase_activity |
| GO:0004691 | 7 | cAMP-dependent_protein_kinase_activity |
| GO:0004703 | 7 | G-protein_coupled_receptor_kinase_activity |
| GO:0004767 | 7 | sphingomyelin_phosphodiesterase_activity |
| GO:0004908 | 7 | interleukin-1_receptor_activity |
| GO:0004931 | 7 | extracellular_ATP-gated_cation_channel_activity |
| GO:0004977 | 7 | melanocortin_receptor_activity |
| GO:0005000 | 7 | vasopressin_receptor_activity |
| GO:0005110 | 7 | frizzled-2_binding |
| GO:0005111 | 7 | type_2_fibroblast_growth_factor_receptor_binding |
| GO:0005113 | 7 | patched_binding |
| GO:0005114 | 7 | type_II_transforming_growth_factor_beta_receptor_binding |
| GO:0005165 | 7 | neurotrophin_receptor_binding |
| GO:0005172 | 7 | vascular_endothelial_growth_factor_receptor_binding |
| GO:0005579 | 7 | membrane_attack_complex |
| GO:0005606 | 7 | laminin-1_complex |
| GO:0005641 | 7 | nuclear_envelope_lumen |
| GO:0005655 | 7 | nucleolar_ribonuclease_P_complex |
| GO:0005662 | 7 | DNA_replication_factor_A_complex |
| GO:0005666 | 7 | DNA-directed_RNA_polymerase_III_complex |
| GO:0005672 | 7 | transcription_factor_TFIIA_complex |
| GO:0005677 | 7 | chromatin_silencing_complex |
| GO:0005683 | 7 | U7_snRNP |
| GO:0005742 | 7 | mitochondrial_outer_membrane_translocase_complex |
| GO:0005750 | 7 | mitochondrial_respiratory_chain_complex_III |
| GO:0005832 | 7 | chaperonin-containing_T-complex |
| GO:0005885 | 7 | Arp2/3_protein_complex |
| GO:0005890 | 7 | sodium:potassium-exchanging_ATPase_complex |
| GO:0005981 | 7 | regulation_of_glycogen_catabolic_process |
| GO:0005984 | 7 | disaccharide_metabolic_process |
| GO:0006002 | 7 | fructose_6-phosphate_metabolic_process |
| GO:0006030 | 7 | chitin_metabolic_process |
| GO:0006032 | 7 | chitin_catabolic_process |
| GO:0006042 | 7 | glucosamine_biosynthetic_process |
| GO:0006045 | 7 | N-acetylglucosamine_biosynthetic_process |
| GO:0006072 | 7 | glycerol-3-phosphate_metabolic_process |
| GO:0006101 | 7 | citrate_metabolic_process |
| GO:0006307 | 7 | DNA_dealkylation_involved_in_DNA_repair |
| GO:0006448 | 7 | regulation_of_translational_elongation |
| GO:0006449 | 7 | regulation_of_translational_termination |
| GO:0006477 | 7 | protein_sulfation |
| GO:0006534 | 7 | cysteine_metabolic_process |
| GO:0006538 | 7 | glutamate_catabolic_process |
| GO:0006547 | 7 | histidine_metabolic_process |
| GO:0006551 | 7 | leucine_metabolic_process |
| GO:0006561 | 7 | proline_biosynthetic_process |
| GO:0006578 | 7 | betaine_biosynthetic_process |
| GO:0006658 | 7 | phosphatidylserine_metabolic_process |
| GO:0006705 | 7 | mineralocorticoid_biosynthetic_process |
| GO:0006777 | 7 | Mo-molybdopterin_cofactor_biosynthetic_process |
| GO:0006837 | 7 | serotonin_transport |
| GO:0006863 | 7 | purine_base_transport |
| GO:0006907 | 7 | pinocytosis |
| GO:0006925 | 7 | inflammatory_cell_apoptotic_process |
| GO:0007097 | 7 | nuclear_migration |
| GO:0007184 | 7 | SMAD_protein_import_into_nucleus |
| GO:0007185 | 7 | transmembrane_receptor_protein_tyrosine_phosphatase_signaling_pathway |
| GO:0007216 | 7 | G-protein_coupled_glutamate_receptor_signaling_pathway |
| GO:0007256 | 7 | activation_of_JNKK_activity |
| GO:0007406 | 7 | negative_regulation_of_neuroblast_proliferation |
| GO:0007435 | 7 | salivary_gland_morphogenesis |
| GO:0007506 | 7 | gonadal_mesoderm_development |
| GO:0007549 | 7 | dosage_compensation |
| GO:0007638 | 7 | mechanosensory_behavior |
| GO:0008035 | 7 | high-density_lipoprotein_particle_binding |
| GO:0008046 | 7 | axon_guidance_receptor_activity |
| GO:0008053 | 7 | mitochondrial_fusion |
| GO:0008212 | 7 | mineralocorticoid_metabolic_process |
| GO:0008228 | 7 | opsonization |
| GO:0008274 | 7 | gamma-tubulin_ring_complex |
| GO:0008343 | 7 | adult_feeding_behavior |
| GO:0008356 | 7 | asymmetric_cell_division |
| GO:0008440 | 7 | inositol-1,4,5-trisphosphate_3-kinase_activity |
| GO:0008443 | 7 | phosphofructokinase_activity |
| GO:0008469 | 7 | histone-arginine_N-methyltransferase_activity |
| GO:0008494 | 7 | translation_activator_activity |
| GO:0008504 | 7 | monoamine_transmembrane_transporter_activity |
| GO:0008649 | 7 | rRNA_methyltransferase_activity |
| GO:0009075 | 7 | histidine_family_amino_acid_metabolic_process |
| GO:0009111 | 7 | vitamin_catabolic_process |
| GO:0009133 | 7 | nucleoside_diphosphate_biosynthetic_process |
| GO:0009223 | 7 | pyrimidine_deoxyribonucleotide_catabolic_process |
| GO:0009226 | 7 | nucleotide-sugar_biosynthetic_process |
| GO:0009253 | 7 | peptidoglycan_catabolic_process |
| GO:0009304 | 7 | tRNA_transcription |
| GO:0009996 | 7 | negative_regulation_of_cell_fate_specification |
| GO:0010447 | 7 | response_to_acidity |
| GO:0010528 | 7 | regulation_of_transposition |
| GO:0010529 | 7 | negative_regulation_of_transposition |
| GO:0010533 | 7 | regulation_of_activation_of_Janus_kinase_activity |
| GO:0010536 | 7 | positive_regulation_of_activation_of_Janus_kinase_activity |
| GO:0010561 | 7 | negative_regulation_of_glycoprotein_biosynthetic_process |
| GO:0010614 | 7 | negative_regulation_of_cardiac_muscle_hypertrophy |
| GO:0010642 | 7 | negative_regulation_of_platelet-derived_growth_factor_receptor_signaling_pathway |
| GO:0010759 | 7 | positive_regulation_of_macrophage_chemotaxis |
| GO:0010761 | 7 | fibroblast_migration |
| GO:0010801 | 7 | negative_regulation_of_peptidyl-threonine_phosphorylation |
| GO:0010873 | 7 | positive_regulation_of_cholesterol_esterification |
| GO:0010882 | 7 | regulation_of_cardiac_muscle_contraction_by_calcium_ion_signaling |
| GO:0010886 | 7 | positive_regulation_of_cholesterol_storage |
| GO:0014033 | 7 | neural_crest_cell_differentiation |
| GO:0014731 | 7 | spectrin-associated_cytoskeleton |
| GO:0014889 | 7 | muscle_atrophy |
| GO:0015166 | 7 | polyol_transmembrane_transporter_activity |
| GO:0015174 | 7 | basic_amino_acid_transmembrane_transporter_activity |
| GO:0015198 | 7 | oligopeptide_transporter_activity |
| GO:0015205 | 7 | nucleobase_transmembrane_transporter_activity |
| GO:0015232 | 7 | heme_transporter_activity |
| GO:0015368 | 7 | calcium:cation_antiporter_activity |
| GO:0015780 | 7 | nucleotide-sugar_transport |
| GO:0015824 | 7 | proline_transport |
| GO:0015858 | 7 | nucleoside_transport |
| GO:0016018 | 7 | cyclosporin_A_binding |
| GO:0016048 | 7 | detection_of_temperature_stimulus |
| GO:0016081 | 7 | synaptic_vesicle_docking_involved_in_exocytosis |
| GO:0016091 | 7 | prenol_biosynthetic_process |
| GO:0016255 | 7 | attachment_of_GPI_anchor_to_protein |
| GO:0016308 | 7 | 1-phosphatidylinositol-4-phosphate_5-kinase_activity |
| GO:0016327 | 7 | apicolateral_plasma_membrane |
| GO:0016461 | 7 | unconventional_myosin_complex |
| GO:0016556 | 7 | mRNA_modification |
| GO:0016593 | 7 | Cdc73/Paf1_complex |
| GO:0016615 | 7 | malate_dehydrogenase_activity |
| GO:0016670 | 7 | oxidoreductase_activity,_acting_on_a_sulfur_group_of_donors,_oxygen_as_acceptor |
| GO:0016714 | 7 | oxidoreductase_activity,_acting_on_paired_donors,_with_incorporation_or_reduction_of_molecular_oxygen,_reduced_pteridine_as_one_donor,_and_incorporation_of_one_atom_of_oxygen |
| GO:0016717 | 7 | oxidoreductase_activity,_acting_on_paired_donors,_with_oxidation_of_a_pair_of_donors_resulting_in_the_reduction_of_molecular_oxygen_to_two_molecules_of_water |
| GO:0016783 | 7 | sulfurtransferase_activity |
| GO:0016803 | 7 | ether_hydrolase_activity |
| GO:0016885 | 7 | ligase_activity,_forming_carbon-carbon_bonds |
| GO:0016889 | 7 | endodeoxyribonuclease_activity,_producing_3'-phosphomonoesters |
| GO:0017002 | 7 | activin-activated_receptor_activity |
| GO:0018023 | 7 | peptidyl-lysine_trimethylation |
| GO:0018206 | 7 | peptidyl-methionine_modification |
| GO:0019211 | 7 | phosphatase_activator_activity |
| GO:0019240 | 7 | citrulline_biosynthetic_process |
| GO:0019441 | 7 | tryptophan_catabolic_process_to_kynurenine |
| GO:0019530 | 7 | taurine_metabolic_process |
| GO:0019563 | 7 | glycerol_catabolic_process |
| GO:0019693 | 7 | ribose_phosphate_metabolic_process |
| GO:0019695 | 7 | choline_metabolic_process |
| GO:0019720 | 7 | Mo-molybdopterin_cofactor_metabolic_process |
| GO:0019798 | 7 | procollagen-proline_dioxygenase_activity |
| GO:0019886 | 7 | antigen_processing_and_presentation_of_exogenous_peptide_antigen_via_MHC_class_II |
| GO:0019957 | 7 | C-C_chemokine_binding |
| GO:0019985 | 7 | translesion_synthesis |
| GO:0021516 | 7 | dorsal_spinal_cord_development |
| GO:0021535 | 7 | cell_migration_in_hindbrain |
| GO:0021546 | 7 | rhombomere_development |
| GO:0021781 | 7 | glial_cell_fate_commitment |
| GO:0021879 | 7 | forebrain_neuron_differentiation |
| GO:0022027 | 7 | interkinetic_nuclear_migration |
| GO:0022601 | 7 | menstrual_cycle_phase |
| GO:0030130 | 7 | clathrin_coat_of_trans-Golgi_network_vesicle |
| GO:0030157 | 7 | pancreatic_juice_secretion |
| GO:0030210 | 7 | heparin_biosynthetic_process |
| GO:0030219 | 7 | megakaryocyte_differentiation |
| GO:0030235 | 7 | nitric-oxide_synthase_regulator_activity |
| GO:0030249 | 7 | guanylate_cyclase_regulator_activity |
| GO:0030252 | 7 | growth_hormone_secretion |
| GO:0030656 | 7 | regulation_of_vitamin_metabolic_process |
| GO:0030669 | 7 | clathrin-coated_endocytic_vesicle_membrane |
| GO:0030677 | 7 | ribonuclease_P_complex |
| GO:0030681 | 7 | multimeric_ribonuclease_P_complex |
| GO:0030828 | 7 | positive_regulation_of_cGMP_biosynthetic_process |
| GO:0030850 | 7 | prostate_gland_development |
| GO:0030854 | 7 | positive_regulation_of_granulocyte_differentiation |
| GO:0030867 | 7 | rough_endoplasmic_reticulum_membrane |
| GO:0030893 | 7 | meiotic_cohesin_complex |
| GO:0030915 | 7 | Smc5-Smc6_complex |
| GO:0030935 | 7 | sheet-forming_collagen |
| GO:0030955 | 7 | potassium_ion_binding |
| GO:0030976 | 7 | thiamine_pyrophosphate_binding |
| GO:0031054 | 7 | pre-miRNA_processing |
| GO:0031061 | 7 | negative_regulation_of_histone_methylation |
| GO:0031092 | 7 | platelet_alpha_granule_membrane |
| GO:0031115 | 7 | negative_regulation_of_microtubule_polymerization |
| GO:0031297 | 7 | replication_fork_processing |
| GO:0031338 | 7 | regulation_of_vesicle_fusion |
| GO:0031442 | 7 | positive_regulation_of_mRNA_3'-end_processing |
| GO:0031464 | 7 | Cul4A-RING_ubiquitin_ligase_complex |
| GO:0031507 | 7 | heterochromatin_assembly |
| GO:0031512 | 7 | motile_primary_cilium |
| GO:0031588 | 7 | AMP-activated_protein_kinase_complex |
| GO:0031659 | 7 | positive_regulation_of_cyclin-dependent_protein_kinase_activity_involved_in_G1/S |
| GO:0031664 | 7 | regulation_of_lipopolysaccharide-mediated_signaling_pathway |
| GO:0031915 | 7 | positive_regulation_of_synaptic_plasticity |
| GO:0031936 | 7 | negative_regulation_of_chromatin_silencing |
| GO:0031946 | 7 | regulation_of_glucocorticoid_biosynthetic_process |
| GO:0031957 | 7 | very_long-chain_fatty_acid-CoA_ligase_activity |
| GO:0031994 | 7 | insulin-like_growth_factor_I_binding |
| GO:0032059 | 7 | bleb |
| GO:0032210 | 7 | regulation_of_telomere_maintenance_via_telomerase |
| GO:0032236 | 7 | positive_regulation_of_calcium_ion_transport_via_store-operated_calcium_channel_activity |
| GO:0032317 | 7 | regulation_of_Rap_GTPase_activity |
| GO:0032324 | 7 | molybdopterin_cofactor_biosynthetic_process |
| GO:0032353 | 7 | negative_regulation_of_hormone_biosynthetic_process |
| GO:0032515 | 7 | negative_regulation_of_phosphoprotein_phosphatase_activity |
| GO:0032516 | 7 | positive_regulation_of_phosphoprotein_phosphatase_activity |
| GO:0032585 | 7 | multivesicular_body_membrane |
| GO:0032609 | 7 | interferon-gamma_production |
| GO:0032667 | 7 | regulation_of_interleukin-23_production |
| GO:0032817 | 7 | regulation_of_natural_killer_cell_proliferation |
| GO:0032823 | 7 | regulation_of_natural_killer_cell_differentiation |
| GO:0032825 | 7 | positive_regulation_of_natural_killer_cell_differentiation |
| GO:0032909 | 7 | regulation_of_transforming_growth_factor_beta2_production |
| GO:0032927 | 7 | positive_regulation_of_activin_receptor_signaling_pathway |
| GO:0033018 | 7 | sarcoplasmic_reticulum_lumen |
| GO:0033084 | 7 | regulation_of_immature_T_cell_proliferation_in_thymus |
| GO:0033089 | 7 | positive_regulation_of_T_cell_differentiation_in_thymus |
| GO:0033120 | 7 | positive_regulation_of_RNA_splicing |
| GO:0033127 | 7 | regulation_of_histone_phosphorylation |
| GO:0033169 | 7 | histone_H3-K9_demethylation |
| GO:0033604 | 7 | negative_regulation_of_catecholamine_secretion |
| GO:0033623 | 7 | regulation_of_integrin_activation |
| GO:0033632 | 7 | regulation_of_cell-cell_adhesion_mediated_by_integrin |
| GO:0033683 | 7 | nucleotide-excision_repair,_DNA_incision |
| GO:0033691 | 7 | sialic_acid_binding |
| GO:0033962 | 7 | cytoplasmic_mRNA_processing_body_assembly |
| GO:0034063 | 7 | stress_granule_assembly |
| GO:0034235 | 7 | GPI_anchor_binding |
| GO:0034237 | 7 | protein_kinase_A_regulatory_subunit_binding |
| GO:0034333 | 7 | adherens_junction_assembly |
| GO:0034372 | 7 | very-low-density_lipoprotein_particle_remodeling |
| GO:0034713 | 7 | type_I_transforming_growth_factor_beta_receptor_binding |
| GO:0034747 | 7 | Axin-APC-beta-catenin-GSK3B_complex |
| GO:0035058 | 7 | nonmotile_primary_cilium_assembly |
| GO:0035089 | 7 | establishment_of_apical/basal_cell_polarity |
| GO:0035371 | 7 | microtubule_plus_end |
| GO:0035381 | 7 | ATP-gated_ion_channel_activity |
| GO:0035518 | 7 | histone_H2A_monoubiquitination |
| GO:0035640 | 7 | exploration_behavior |
| GO:0035909 | 7 | aorta_morphogenesis |
| GO:0035970 | 7 | peptidyl-threonine_dephosphorylation |
| GO:0035999 | 7 | tetrahydrofolate_interconversion |
| GO:0038061 | 7 | NIK/NF-kappaB_cascade |
| GO:0038092 | 7 | nodal_signaling_pathway |
| GO:0040015 | 7 | negative_regulation_of_multicellular_organism_growth |
| GO:0040016 | 7 | embryonic_cleavage |
| GO:0042117 | 7 | monocyte_activation |
| GO:0042178 | 7 | xenobiotic_catabolic_process |
| GO:0042267 | 7 | natural_killer_cell_mediated_cytotoxicity |
| GO:0042268 | 7 | regulation_of_cytolysis |
| GO:0042363 | 7 | fat-soluble_vitamin_catabolic_process |
| GO:0042415 | 7 | norepinephrine_metabolic_process |
| GO:0042416 | 7 | dopamine_biosynthetic_process |
| GO:0042482 | 7 | positive_regulation_of_odontogenesis |
| GO:0042532 | 7 | negative_regulation_of_tyrosine_phosphorylation_of_STAT_protein |
| GO:0042622 | 7 | photoreceptor_outer_segment_membrane |
| GO:0042693 | 7 | muscle_cell_fate_commitment |
| GO:0042711 | 7 | maternal_behavior |
| GO:0042761 | 7 | very_long-chain_fatty_acid_biosynthetic_process |
| GO:0042921 | 7 | glucocorticoid_receptor_signaling_pathway |
| GO:0043083 | 7 | synaptic_cleft |
| GO:0043220 | 7 | Schmidt-Lanterman_incisure |
| GO:0043237 | 7 | laminin-1_binding |
| GO:0043248 | 7 | proteasome_assembly |
| GO:0043268 | 7 | positive_regulation_of_potassium_ion_transport |
| GO:0043297 | 7 | apical_junction_assembly |
| GO:0043380 | 7 | regulation_of_memory_T_cell_differentiation |
| GO:0043501 | 7 | skeletal_muscle_adaptation |
| GO:0043539 | 7 | protein_serine/threonine_kinase_activator_activity |
| GO:0043545 | 7 | molybdopterin_cofactor_metabolic_process |
| GO:0043589 | 7 | skin_morphogenesis |
| GO:0043596 | 7 | nuclear_replication_fork |
| GO:0043983 | 7 | histone_H4-K12_acetylation |
| GO:0044068 | 7 | modulation_by_symbiont_of_host_cellular_process |
| GO:0044295 | 7 | axonal_growth_cone |
| GO:0045006 | 7 | DNA_deamination |
| GO:0045039 | 7 | protein_import_into_mitochondrial_inner_membrane |
| GO:0045056 | 7 | transcytosis |
| GO:0045091 | 7 | regulation_of_retroviral_genome_replication |
| GO:0045116 | 7 | protein_neddylation |
| GO:0045123 | 7 | cellular_extravasation |
| GO:0045171 | 7 | intercellular_bridge |
| GO:0045178 | 7 | basal_part_of_cell |
| GO:0045179 | 7 | apical_cortex |
| GO:0045188 | 7 | regulation_of_circadian_sleep/wake_cycle,_non-REM_sleep |
| GO:0045275 | 7 | respiratory_chain_complex_III |
| GO:0045359 | 7 | positive_regulation_of_interferon-beta_biosynthetic_process |
| GO:0045586 | 7 | regulation_of_gamma-delta_T_cell_differentiation |
| GO:0045588 | 7 | positive_regulation_of_gamma-delta_T_cell_differentiation |
| GO:0045714 | 7 | regulation_of_low-density_lipoprotein_particle_receptor_biosynthetic_process |
| GO:0045719 | 7 | negative_regulation_of_glycogen_biosynthetic_process |
| GO:0045721 | 7 | negative_regulation_of_gluconeogenesis |
| GO:0045743 | 7 | positive_regulation_of_fibroblast_growth_factor_receptor_signaling_pathway |
| GO:0045836 | 7 | positive_regulation_of_meiosis |
| GO:0045842 | 7 | positive_regulation_of_mitotic_metaphase/anaphase_transition |
| GO:0045945 | 7 | positive_regulation_of_transcription_from_RNA_polymerase_III_promoter |
| GO:0045988 | 7 | negative_regulation_of_striated_muscle_contraction |
| GO:0046007 | 7 | negative_regulation_of_activated_T_cell_proliferation |
| GO:0046037 | 7 | GMP_metabolic_process |
| GO:0046146 | 7 | tetrahydrobiopterin_metabolic_process |
| GO:0046325 | 7 | negative_regulation_of_glucose_import |
| GO:0046349 | 7 | amino_sugar_biosynthetic_process |
| GO:0046485 | 7 | ether_lipid_metabolic_process |
| GO:0046500 | 7 | S-adenosylmethionine_metabolic_process |
| GO:0046668 | 7 | regulation_of_retinal_cell_programmed_cell_death |
| GO:0046950 | 7 | cellular_ketone_body_metabolic_process |
| GO:0047372 | 7 | acylglycerol_lipase_activity |
| GO:0048096 | 7 | chromatin-mediated_maintenance_of_transcription |
| GO:0048102 | 7 | autophagic_cell_death |
| GO:0048148 | 7 | behavioral_response_to_cocaine |
| GO:0048156 | 7 | tau_protein_binding |
| GO:0048256 | 7 | flap_endonuclease_activity |
| GO:0048304 | 7 | positive_regulation_of_isotype_switching_to_IgG_isotypes |
| GO:0048332 | 7 | mesoderm_morphogenesis |
| GO:0048541 | 7 | Peyer's_patch_development |
| GO:0048642 | 7 | negative_regulation_of_skeletal_muscle_tissue_development |
| GO:0048664 | 7 | neuron_fate_determination |
| GO:0048739 | 7 | cardiac_muscle_fiber_development |
| GO:0048793 | 7 | pronephros_development |
| GO:0050687 | 7 | negative_regulation_of_defense_response_to_virus |
| GO:0050733 | 7 | RS_domain_binding |
| GO:0050748 | 7 | negative_regulation_of_lipoprotein_metabolic_process |
| GO:0050786 | 7 | RAGE_receptor_binding |
| GO:0050855 | 7 | regulation_of_B_cell_receptor_signaling_pathway |
| GO:0051001 | 7 | negative_regulation_of_nitric-oxide_synthase_activity |
| GO:0051024 | 7 | positive_regulation_of_immunoglobulin_secretion |
| GO:0051189 | 7 | prosthetic_group_metabolic_process |
| GO:0051290 | 7 | protein_heterotetramerization |
| GO:0051307 | 7 | meiotic_chromosome_separation |
| GO:0051371 | 7 | muscle_alpha-actinin_binding |
| GO:0051488 | 7 | activation_of_anaphase-promoting_complex_activity |
| GO:0051567 | 7 | histone_H3-K9_methylation |
| GO:0051570 | 7 | regulation_of_histone_H3-K9_methylation |
| GO:0051584 | 7 | regulation_of_dopamine_uptake_involved_in_synaptic_transmission |
| GO:0051608 | 7 | histamine_transport |
| GO:0051642 | 7 | centrosome_localization |
| GO:0051647 | 7 | nucleus_localization |
| GO:0051775 | 7 | response_to_redox_state |
| GO:0051788 | 7 | response_to_misfolded_protein |
| GO:0051798 | 7 | positive_regulation_of_hair_follicle_development |
| GO:0051823 | 7 | regulation_of_synapse_structural_plasticity |
| GO:0051864 | 7 | histone_demethylase_activity_(H3-K36_specific) |
| GO:0051895 | 7 | negative_regulation_of_focal_adhesion_assembly |
| GO:0051940 | 7 | regulation_of_catecholamine_uptake_involved_in_synaptic_transmission |
| GO:0051974 | 7 | negative_regulation_of_telomerase_activity |
| GO:0052031 | 7 | modulation_by_symbiont_of_host_defense_response |
| GO:0052255 | 7 | modulation_by_organism_of_defense_response_of_other_organism_involved_in_symbiotic_interaction |
| GO:0052509 | 7 | positive_regulation_by_symbiont_of_host_defense_response |
| GO:0052510 | 7 | positive_regulation_by_organism_of_defense_response_of_other_organism_involved_in_symbiotic_interaction |
| GO:0052564 | 7 | response_to_immune_response_of_other_organism_involved_in_symbiotic_interaction |
| GO:0052572 | 7 | response_to_host_immune_response |
| GO:0060008 | 7 | Sertoli_cell_differentiation |
| GO:0060022 | 7 | hard_palate_development |
| GO:0060088 | 7 | auditory_receptor_cell_stereocilium_organization |
| GO:0060124 | 7 | positive_regulation_of_growth_hormone_secretion |
| GO:0060136 | 7 | embryonic_process_involved_in_female_pregnancy |
| GO:0060155 | 7 | platelet_dense_granule_organization |
| GO:0060259 | 7 | regulation_of_feeding_behavior |
| GO:0060297 | 7 | regulation_of_sarcomere_organization |
| GO:0060306 | 7 | regulation_of_membrane_repolarization |
| GO:0060405 | 7 | regulation_of_penile_erection |
| GO:0060428 | 7 | lung_epithelium_development |
| GO:0060558 | 7 | regulation_of_calcidiol_1-monooxygenase_activity |
| GO:0060664 | 7 | epithelial_cell_proliferation_involved_in_salivary_gland_morphogenesis |
| GO:0060665 | 7 | regulation_of_branching_involved_in_salivary_gland_morphogenesis_by_mesenchymal-epithelial_signaling |
| GO:0060684 | 7 | epithelial-mesenchymal_cell_signaling |
| GO:0060685 | 7 | regulation_of_prostatic_bud_formation |
| GO:0060732 | 7 | positive_regulation_of_inositol_phosphate_biosynthetic_process |
| GO:0060746 | 7 | parental_behavior |
| GO:0060770 | 7 | negative_regulation_of_epithelial_cell_proliferation_involved_in_prostate_gland_development |
| GO:0060973 | 7 | cell_migration_involved_in_heart_development |
| GO:0060986 | 7 | endocrine_hormone_secretion |
| GO:0061053 | 7 | somite_development |
| GO:0061072 | 7 | iris_morphogenesis |
| GO:0061180 | 7 | mammary_gland_epithelium_development |
| GO:0061298 | 7 | retina_vasculature_development_in_camera-type_eye |
| GO:0061383 | 7 | trabecula_morphogenesis |
| GO:0070061 | 7 | fructose_binding |
| GO:0070102 | 7 | interleukin-6-mediated_signaling_pathway |
| GO:0070170 | 7 | regulation_of_tooth_mineralization |
| GO:0070296 | 7 | sarcoplasmic_reticulum_calcium_ion_transport |
| GO:0070391 | 7 | response_to_lipoteichoic_acid |
| GO:0070419 | 7 | nonhomologous_end_joining_complex |
| GO:0070493 | 7 | thrombin_receptor_signaling_pathway |
| GO:0070509 | 7 | calcium_ion_import |
| GO:0070528 | 7 | protein_kinase_C_signaling_cascade |
| GO:0070531 | 7 | BRCA1-A_complex |
| GO:0070567 | 7 | cytidylyltransferase_activity |
| GO:0070569 | 7 | uridylyltransferase_activity |
| GO:0070775 | 7 | H3_histone_acetyltransferase_complex |
| GO:0070776 | 7 | MOZ/MORF_histone_acetyltransferase_complex |
| GO:0070828 | 7 | heterochromatin_organization |
| GO:0070933 | 7 | histone_H4_deacetylation |
| GO:0071107 | 7 | response_to_parathyroid_hormone_stimulus |
| GO:0071223 | 7 | cellular_response_to_lipoteichoic_acid |
| GO:0071243 | 7 | cellular_response_to_arsenic-containing_substance |
| GO:0071372 | 7 | cellular_response_to_follicle-stimulating_hormone_stimulus |
| GO:0071380 | 7 | cellular_response_to_prostaglandin_E_stimulus |
| GO:0071415 | 7 | cellular_response_to_purine-containing_compound |
| GO:0071480 | 7 | cellular_response_to_gamma_radiation |
| GO:0072074 | 7 | kidney_mesenchyme_development |
| GO:0072075 | 7 | metanephric_mesenchyme_development |
| GO:0072132 | 7 | mesenchyme_morphogenesis |
| GO:0072178 | 7 | nephric_duct_morphogenesis |
| GO:0072205 | 7 | metanephric_collecting_duct_development |
| GO:0072207 | 7 | metanephric_epithelium_development |
| GO:0072321 | 7 | chaperone-mediated_protein_transport |
| GO:0072350 | 7 | tricarboxylic_acid_metabolic_process |
| GO:0072357 | 7 | PTW/PP1_phosphatase_complex |
| GO:0085020 | 7 | protein_K6-linked_ubiquitination |
| GO:0086001 | 7 | regulation_of_cardiac_muscle_cell_action_potential |
| GO:0086002 | 7 | regulation_of_cardiac_muscle_cell_action_potential_involved_in_contraction |
| GO:0086036 | 7 | regulation_of_cardiac_muscle_cell_membrane_potential |
| GO:0086065 | 7 | cell_communication_involved_in_cardiac_conduction |
| GO:0090036 | 7 | regulation_of_protein_kinase_C_signaling_cascade |
| GO:0090037 | 7 | positive_regulation_of_protein_kinase_C_signaling_cascade |
| GO:0090128 | 7 | regulation_of_synapse_maturation |
| GO:0090178 | 7 | regulation_of_establishment_of_planar_polarity_involved_in_neural_tube_closure |
| GO:0090179 | 7 | planar_cell_polarity_pathway_involved_in_neural_tube_closure |
| GO:0090196 | 7 | regulation_of_chemokine_secretion |
| GO:0090205 | 7 | positive_regulation_of_cholesterol_metabolic_process |
| GO:0090224 | 7 | regulation_of_spindle_organization |
| GO:0090303 | 7 | positive_regulation_of_wound_healing |
| GO:0090330 | 7 | regulation_of_platelet_aggregation |
| GO:0097191 | 7 | extrinsic_apoptotic_signaling_pathway |
| GO:0097205 | 7 | renal_filtration |
| GO:1901020 | 7 | negative_regulation_of_calcium_ion_transmembrane_transporter_activity |
| GO:1901073 | 7 | glucosamine-containing_compound_biosynthetic_process |
| GO:2000018 | 7 | regulation_of_male_gonad_development |
| GO:2000060 | 7 | positive_regulation_of_protein_ubiquitination_involved_in_ubiquitin-dependent_protein_catabolic_process |
| GO:2000114 | 7 | regulation_of_establishment_of_cell_polarity |
| GO:2000134 | 7 | negative_regulation_of_G1/S_transition_of_mitotic_cell_cycle |
| GO:2000144 | 7 | positive_regulation_of_DNA-dependent_transcription,_initiation |
| GO:2000300 | 7 | regulation_of_synaptic_vesicle_exocytosis |
| GO:2000543 | 7 | positive_regulation_of_gastrulation |
| GO:2000573 | 7 | positive_regulation_of_DNA_biosynthetic_process |
| GO:2000794 | 7 | regulation_of_epithelial_cell_proliferation_involved_in_lung_morphogenesis |
| GO:0000014 | 6 | single-stranded_DNA_specific_endodeoxyribonuclease_activity |
| GO:0000028 | 6 | ribosomal_small_subunit_assembly |
| GO:0000089 | 6 | mitotic_metaphase |
| GO:0000125 | 6 | PCAF_complex |
| GO:0000127 | 6 | transcription_factor_TFIIIC_complex |
| GO:0000153 | 6 | cytoplasmic_ubiquitin_ligase_complex |
| GO:0000176 | 6 | nuclear_exosome_(RNase_complex) |
| GO:0000177 | 6 | cytoplasmic_exosome_(RNase_complex) |
| GO:0000244 | 6 | assembly_of_spliceosomal_tri-snRNP |
| GO:0000268 | 6 | peroxisome_targeting_sequence_binding |
| GO:0000394 | 6 | RNA_splicing,_via_endonucleolytic_cleavage_and_ligation |
| GO:0000405 | 6 | bubble_DNA_binding |
| GO:0000422 | 6 | mitochondrion_degradation |
| GO:0000710 | 6 | meiotic_mismatch_repair |
| GO:0000729 | 6 | DNA_double-strand_break_processing |
| GO:0000796 | 6 | condensin_complex |
| GO:0000800 | 6 | lateral_element |
| GO:0000805 | 6 | X_chromosome |
| GO:0001514 | 6 | selenocysteine_incorporation |
| GO:0001547 | 6 | antral_ovarian_follicle_growth |
| GO:0001561 | 6 | fatty_acid_alpha-oxidation |
| GO:0001618 | 6 | viral_receptor_activity |
| GO:0001640 | 6 | adenylate_cyclase_inhibiting_G-protein_coupled_glutamate_receptor_activity |
| GO:0001710 | 6 | mesodermal_cell_fate_commitment |
| GO:0001711 | 6 | endodermal_cell_fate_commitment |
| GO:0001727 | 6 | lipid_kinase_activity |
| GO:0001739 | 6 | sex_chromatin |
| GO:0001821 | 6 | histamine_secretion |
| GO:0001846 | 6 | opsonin_binding |
| GO:0001957 | 6 | intramembranous_ossification |
| GO:0001976 | 6 | neurological_system_process_involved_in_regulation_of_systemic_arterial_blood_pressure |
| GO:0002029 | 6 | desensitization_of_G-protein_coupled_receptor_protein_signaling_pathway |
| GO:0002067 | 6 | glandular_epithelial_cell_differentiation |
| GO:0002178 | 6 | palmitoyltransferase_complex |
| GO:0002251 | 6 | organ_or_tissue_specific_immune_response |
| GO:0002313 | 6 | mature_B_cell_differentiation_involved_in_immune_response |
| GO:0002347 | 6 | response_to_tumor_cell |
| GO:0002483 | 6 | antigen_processing_and_presentation_of_endogenous_peptide_antigen |
| GO:0002691 | 6 | regulation_of_cellular_extravasation |
| GO:0002765 | 6 | immune_response-inhibiting_signal_transduction |
| GO:0002829 | 6 | negative_regulation_of_type_2_immune_response |
| GO:0002855 | 6 | regulation_of_natural_killer_cell_mediated_immune_response_to_tumor_cell |
| GO:0002857 | 6 | positive_regulation_of_natural_killer_cell_mediated_immune_response_to_tumor_cell |
| GO:0002858 | 6 | regulation_of_natural_killer_cell_mediated_cytotoxicity_directed_against_tumor_cell_target |
| GO:0002860 | 6 | positive_regulation_of_natural_killer_cell_mediated_cytotoxicity_directed_against_tumor_cell_target |
| GO:0002885 | 6 | positive_regulation_of_hypersensitivity |
| GO:0002921 | 6 | negative_regulation_of_humoral_immune_response |
| GO:0003091 | 6 | renal_water_homeostasis |
| GO:0003128 | 6 | heart_field_specification |
| GO:0003139 | 6 | secondary_heart_field_specification |
| GO:0003159 | 6 | morphogenesis_of_an_endothelium |
| GO:0003181 | 6 | atrioventricular_valve_morphogenesis |
| GO:0003214 | 6 | cardiac_left_ventricle_morphogenesis |
| GO:0003264 | 6 | regulation_of_cardioblast_proliferation |
| GO:0003266 | 6 | regulation_of_secondary_heart_field_cardioblast_proliferation |
| GO:0003300 | 6 | cardiac_muscle_hypertrophy |
| GO:0003308 | 6 | negative_regulation_of_Wnt_receptor_signaling_pathway_involved_in_heart_development |
| GO:0003688 | 6 | DNA_replication_origin_binding |
| GO:0003747 | 6 | translation_release_factor_activity |
| GO:0003916 | 6 | DNA_topoisomerase_activity |
| GO:0003956 | 6 | NAD(P)+-protein-arginine_ADP-ribosyltransferase_activity |
| GO:0003988 | 6 | acetyl-CoA_C-acyltransferase_activity |
| GO:0003997 | 6 | acyl-CoA_oxidase_activity |
| GO:0004028 | 6 | 3-chloroallyl_aldehyde_dehydrogenase_activity |
| GO:0004035 | 6 | alkaline_phosphatase_activity |
| GO:0004046 | 6 | aminoacylase_activity |
| GO:0004047 | 6 | aminomethyltransferase_activity |
| GO:0004075 | 6 | biotin_carboxylase_activity |
| GO:0004331 | 6 | fructose-2,6-bisphosphate_2-phosphatase_activity |
| GO:0004366 | 6 | glycerol-3-phosphate_O-acyltransferase_activity |
| GO:0004439 | 6 | phosphatidylinositol-4,5-bisphosphate_5-phosphatase_activity |
| GO:0004457 | 6 | lactate_dehydrogenase_activity |
| GO:0004499 | 6 | N,N-dimethylaniline_monooxygenase_activity |
| GO:0004558 | 6 | alpha-glucosidase_activity |
| GO:0004630 | 6 | phospholipase_D_activity |
| GO:0004652 | 6 | polynucleotide_adenylyltransferase_activity |
| GO:0004661 | 6 | protein_geranylgeranyltransferase_activity |
| GO:0004704 | 6 | NF-kappaB-inducing_kinase_activity |
| GO:0004723 | 6 | calcium-dependent_protein_serine/threonine_phosphatase_activity |
| GO:0004936 | 6 | alpha-adrenergic_receptor_activity |
| GO:0004948 | 6 | calcitonin_receptor_activity |
| GO:0004957 | 6 | prostaglandin_E_receptor_activity |
| GO:0004969 | 6 | histamine_receptor_activity |
| GO:0004972 | 6 | N-methyl-D-aspartate_selective_glutamate_receptor_activity |
| GO:0005138 | 6 | interleukin-6_receptor_binding |
| GO:0005152 | 6 | interleukin-1_receptor_antagonist_activity |
| GO:0005237 | 6 | inhibitory_extracellular_ligand-gated_ion_channel_activity |
| GO:0005375 | 6 | copper_ion_transmembrane_transporter_activity |
| GO:0005499 | 6 | vitamin_D_binding |
| GO:0005587 | 6 | collagen_type_IV |
| GO:0005593 | 6 | FACIT_collagen |
| GO:0005652 | 6 | nuclear_lamina |
| GO:0005658 | 6 | alpha_DNA_polymerase:primase_complex |
| GO:0005663 | 6 | DNA_replication_factor_C_complex |
| GO:0005682 | 6 | U5_snRNP |
| GO:0005697 | 6 | telomerase_holoenzyme_complex |
| GO:0005786 | 6 | signal_recognition_particle,_endoplasmic_reticulum_targeting |
| GO:0005851 | 6 | eukaryotic_translation_initiation_factor_2B_complex |
| GO:0005853 | 6 | eukaryotic_translation_elongation_factor_1_complex |
| GO:0005869 | 6 | dynactin_complex |
| GO:0005958 | 6 | DNA-dependent_protein_kinase-DNA_ligase_4_complex |
| GO:0006068 | 6 | ethanol_catabolic_process |
| GO:0006085 | 6 | acetyl-CoA_biosynthetic_process |
| GO:0006102 | 6 | isocitrate_metabolic_process |
| GO:0006189 | 6 | 'de_novo'_IMP_biosynthetic_process |
| GO:0006287 | 6 | base-excision_repair,_gap-filling |
| GO:0006290 | 6 | pyrimidine_dimer_repair |
| GO:0006346 | 6 | methylation-dependent_chromatin_silencing |
| GO:0006388 | 6 | tRNA_splicing,_via_endonucleolytic_cleavage_and_ligation |
| GO:0006398 | 6 | histone_mRNA_3'-end_processing |
| GO:0006451 | 6 | translational_readthrough |
| GO:0006467 | 6 | protein_thiol-disulfide_exchange |
| GO:0006537 | 6 | glutamate_biosynthetic_process |
| GO:0006545 | 6 | glycine_biosynthetic_process |
| GO:0006548 | 6 | histidine_catabolic_process |
| GO:0006591 | 6 | ornithine_metabolic_process |
| GO:0006642 | 6 | triglyceride_mobilization |
| GO:0006657 | 6 | CDP-choline_pathway |
| GO:0006729 | 6 | tetrahydrobiopterin_biosynthetic_process |
| GO:0006787 | 6 | porphyrin-containing_compound_catabolic_process |
| GO:0006817 | 6 | phosphate_ion_transport |
| GO:0006828 | 6 | manganese_ion_transport |
| GO:0006862 | 6 | nucleotide_transport |
| GO:0006927 | 6 | transformed_cell_apoptotic_process |
| GO:0006975 | 6 | DNA_damage_induced_protein_phosphorylation |
| GO:0007064 | 6 | mitotic_sister_chromatid_cohesion |
| GO:0007089 | 6 | traversing_start_control_point_of_mitotic_cell_cycle |
| GO:0007098 | 6 | centrosome_cycle |
| GO:0007129 | 6 | synapsis |
| GO:0007143 | 6 | female_meiosis |
| GO:0007253 | 6 | cytoplasmic_sequestering_of_NF-kappaB |
| GO:0007262 | 6 | STAT_protein_import_into_nucleus |
| GO:0007289 | 6 | spermatid_nucleus_differentiation |
| GO:0007341 | 6 | penetration_of_zona_pellucida |
| GO:0007614 | 6 | short-term_memory |
| GO:0008079 | 6 | translation_termination_factor_activity |
| GO:0008090 | 6 | retrograde_axon_cargo_transport |
| GO:0008113 | 6 | peptide-methionine-(S)-S-oxide_reductase_activity |
| GO:0008172 | 6 | S-methyltransferase_activity |
| GO:0008179 | 6 | adenylate_cyclase_binding |
| GO:0008294 | 6 | calcium-_and_calmodulin-responsive_adenylate_cyclase_activity |
| GO:0008312 | 6 | 7S_RNA_binding |
| GO:0008503 | 6 | benzodiazepine_receptor_activity |
| GO:0008508 | 6 | bile_acid:sodium_symporter_activity |
| GO:0008599 | 6 | protein_phosphatase_type_1_regulator_activity |
| GO:0008762 | 6 | UDP-N-acetylmuramate_dehydrogenase_activity |
| GO:0009077 | 6 | histidine_family_amino_acid_catabolic_process |
| GO:0009134 | 6 | nucleoside_diphosphate_catabolic_process |
| GO:0009162 | 6 | deoxyribonucleoside_monophosphate_metabolic_process |
| GO:0009211 | 6 | pyrimidine_deoxyribonucleoside_triphosphate_metabolic_process |
| GO:0009221 | 6 | pyrimidine_deoxyribonucleotide_biosynthetic_process |
| GO:0009235 | 6 | cobalamin_metabolic_process |
| GO:0009586 | 6 | rhodopsin_mediated_phototransduction |
| GO:0009648 | 6 | photoperiodism |
| GO:0010172 | 6 | embryonic_body_morphogenesis |
| GO:0010369 | 6 | chromocenter |
| GO:0010455 | 6 | positive_regulation_of_cell_fate_commitment |
| GO:0010534 | 6 | regulation_of_activation_of_JAK2_kinase_activity |
| GO:0010535 | 6 | positive_regulation_of_activation_of_JAK2_kinase_activity |
| GO:0010592 | 6 | positive_regulation_of_lamellipodium_assembly |
| GO:0010631 | 6 | epithelial_cell_migration |
| GO:0010633 | 6 | negative_regulation_of_epithelial_cell_migration |
| GO:0010661 | 6 | positive_regulation_of_muscle_cell_apoptotic_process |
| GO:0010694 | 6 | positive_regulation_of_alkaline_phosphatase_activity |
| GO:0010715 | 6 | regulation_of_extracellular_matrix_disassembly |
| GO:0010763 | 6 | positive_regulation_of_fibroblast_migration |
| GO:0010804 | 6 | negative_regulation_of_tumor_necrosis_factor-mediated_signaling_pathway |
| GO:0010853 | 6 | cyclase_activator_activity |
| GO:0010861 | 6 | thyroid_hormone_receptor_activator_activity |
| GO:0010871 | 6 | negative_regulation_of_receptor_biosynthetic_process |
| GO:0010881 | 6 | regulation_of_cardiac_muscle_contraction_by_regulation_of_the_release_of_sequestered_calcium_ion |
| GO:0010887 | 6 | negative_regulation_of_cholesterol_storage |
| GO:0010898 | 6 | positive_regulation_of_triglyceride_catabolic_process |
| GO:0010940 | 6 | positive_regulation_of_necrotic_cell_death |
| GO:0010985 | 6 | negative_regulation_of_lipoprotein_particle_clearance |
| GO:0010988 | 6 | regulation_of_low-density_lipoprotein_particle_clearance |
| GO:0014012 | 6 | peripheral_nervous_system_axon_regeneration |
| GO:0014037 | 6 | Schwann_cell_differentiation |
| GO:0014060 | 6 | regulation_of_epinephrine_secretion |
| GO:0014820 | 6 | tonic_smooth_muscle_contraction |
| GO:0014897 | 6 | striated_muscle_hypertrophy |
| GO:0014909 | 6 | smooth_muscle_cell_migration |
| GO:0015037 | 6 | peptide_disulfide_oxidoreductase_activity |
| GO:0015095 | 6 | magnesium_ion_transmembrane_transporter_activity |
| GO:0015106 | 6 | bicarbonate_transmembrane_transporter_activity |
| GO:0015165 | 6 | pyrimidine_nucleotide_sugar_transmembrane_transporter_activity |
| GO:0015168 | 6 | glycerol_transmembrane_transporter_activity |
| GO:0015187 | 6 | glycine_transmembrane_transporter_activity |
| GO:0015279 | 6 | store-operated_calcium_channel_activity |
| GO:0015651 | 6 | quaternary_ammonium_group_transmembrane_transporter_activity |
| GO:0015696 | 6 | ammonium_transport |
| GO:0015781 | 6 | pyrimidine_nucleotide-sugar_transport |
| GO:0015791 | 6 | polyol_transport |
| GO:0015802 | 6 | basic_amino_acid_transport |
| GO:0015810 | 6 | aspartate_transport |
| GO:0015872 | 6 | dopamine_transport |
| GO:0015886 | 6 | heme_transport |
| GO:0015942 | 6 | formate_metabolic_process |
| GO:0016004 | 6 | phospholipase_activator_activity |
| GO:0016012 | 6 | sarcoglycan_complex |
| GO:0016098 | 6 | monoterpenoid_metabolic_process |
| GO:0016165 | 6 | lipoxygenase_activity |
| GO:0016188 | 6 | synaptic_vesicle_maturation |
| GO:0016198 | 6 | axon_choice_point_recognition |
| GO:0016242 | 6 | negative_regulation_of_macroautophagy |
| GO:0016264 | 6 | gap_junction_assembly |
| GO:0016406 | 6 | carnitine_O-acyltransferase_activity |
| GO:0016417 | 6 | S-acyltransferase_activity |
| GO:0016421 | 6 | CoA_carboxylase_activity |
| GO:0016503 | 6 | pheromone_receptor_activity |
| GO:0016553 | 6 | base_conversion_or_substitution_editing |
| GO:0016584 | 6 | nucleosome_positioning |
| GO:0016589 | 6 | NURF_complex |
| GO:0016778 | 6 | diphosphotransferase_activity |
| GO:0016886 | 6 | ligase_activity,_forming_phosphoric_ester_bonds |
| GO:0016973 | 6 | poly(A)+_mRNA_export_from_nucleus |
| GO:0017040 | 6 | ceramidase_activity |
| GO:0017059 | 6 | serine_C-palmitoyltransferase_complex |
| GO:0018026 | 6 | peptidyl-lysine_monomethylation |
| GO:0018879 | 6 | biphenyl_metabolic_process |
| GO:0018894 | 6 | dibenzo-p-dioxin_metabolic_process |
| GO:0019103 | 6 | pyrimidine_nucleotide_binding |
| GO:0019236 | 6 | response_to_pheromone |
| GO:0019237 | 6 | centromeric_DNA_binding |
| GO:0019302 | 6 | D-ribose_biosynthetic_process |
| GO:0019368 | 6 | fatty_acid_elongation,_unsaturated_fatty_acid |
| GO:0019374 | 6 | galactolipid_metabolic_process |
| GO:0019388 | 6 | galactose_catabolic_process |
| GO:0019534 | 6 | toxin_transporter_activity |
| GO:0019885 | 6 | antigen_processing_and_presentation_of_endogenous_peptide_antigen_via_MHC_class_I |
| GO:0019907 | 6 | cyclin-dependent_protein_kinase_activating_kinase_holoenzyme_complex |
| GO:0019908 | 6 | nuclear_cyclin-dependent_protein_kinase_holoenzyme_complex |
| GO:0020017 | 6 | flagellar_membrane |
| GO:0021587 | 6 | cerebellum_morphogenesis |
| GO:0021604 | 6 | cranial_nerve_structural_organization |
| GO:0021707 | 6 | cerebellar_granule_cell_differentiation |
| GO:0021860 | 6 | pyramidal_neuron_development |
| GO:0021873 | 6 | forebrain_neuroblast_division |
| GO:0021984 | 6 | adenohypophysis_development |
| GO:0022038 | 6 | corpus_callosum_development |
| GO:0022401 | 6 | negative_adaptation_of_signaling_pathway |
| GO:0023058 | 6 | adaptation_of_signaling_pathway |
| GO:0030020 | 6 | extracellular_matrix_structural_constituent_conferring_tensile_strength |
| GO:0030021 | 6 | extracellular_matrix_structural_constituent_conferring_compression_resistance |
| GO:0030033 | 6 | microvillus_assembly |
| GO:0030061 | 6 | mitochondrial_crista |
| GO:0030121 | 6 | AP-1_adaptor_complex |
| GO:0030132 | 6 | clathrin_coat_of_coated_pit |
| GO:0030137 | 6 | COPI-coated_vesicle |
| GO:0030174 | 6 | regulation_of_DNA-dependent_DNA_replication_initiation |
| GO:0030214 | 6 | hyaluronan_catabolic_process |
| GO:0030215 | 6 | semaphorin_receptor_binding |
| GO:0030250 | 6 | guanylate_cyclase_activator_activity |
| GO:0030274 | 6 | LIM_domain_binding |
| GO:0030300 | 6 | regulation_of_intestinal_cholesterol_absorption |
| GO:0030306 | 6 | ADP-ribosylation_factor_binding |
| GO:0030345 | 6 | structural_constituent_of_tooth_enamel |
| GO:0030346 | 6 | protein_phosphatase_2B_binding |
| GO:0030388 | 6 | fructose_1,6-bisphosphate_metabolic_process |
| GO:0030492 | 6 | hemoglobin_binding |
| GO:0030579 | 6 | ubiquitin-dependent_SMAD_protein_catabolic_process |
| GO:0030643 | 6 | cellular_phosphate_ion_homeostasis |
| GO:0030853 | 6 | negative_regulation_of_granulocyte_differentiation |
| GO:0030877 | 6 | beta-catenin_destruction_complex |
| GO:0030916 | 6 | otic_vesicle_formation |
| GO:0030957 | 6 | Tat_protein_binding |
| GO:0030992 | 6 | intraflagellar_transport_particle_B |
| GO:0031010 | 6 | ISWI_complex |
| GO:0031045 | 6 | dense_core_granule |
| GO:0031211 | 6 | endoplasmic_reticulum_palmitoyltransferase_complex |
| GO:0031264 | 6 | death-inducing_signaling_complex |
| GO:0031342 | 6 | negative_regulation_of_cell_killing |
| GO:0031392 | 6 | regulation_of_prostaglandin_biosynthetic_process |
| GO:0031462 | 6 | Cul2-RING_ubiquitin_ligase_complex |
| GO:0031616 | 6 | spindle_pole_centrosome |
| GO:0031639 | 6 | plasminogen_activation |
| GO:0031702 | 6 | type_1_angiotensin_receptor_binding |
| GO:0031726 | 6 | CCR1_chemokine_receptor_binding |
| GO:0032020 | 6 | ISG15-protein_conjugation |
| GO:0032025 | 6 | response_to_cobalt_ion |
| GO:0032036 | 6 | myosin_heavy_chain_binding |
| GO:0032045 | 6 | guanyl-nucleotide_exchange_factor_complex |
| GO:0032052 | 6 | bile_acid_binding |
| GO:0032055 | 6 | negative_regulation_of_translation_in_response_to_stress |
| GO:0032135 | 6 | DNA_insertion_or_deletion_binding |
| GO:0032222 | 6 | regulation_of_synaptic_transmission,_cholinergic |
| GO:0032277 | 6 | negative_regulation_of_gonadotropin_secretion |
| GO:0032288 | 6 | myelin_assembly |
| GO:0032313 | 6 | regulation_of_Rab_GTPase_activity |
| GO:0032344 | 6 | regulation_of_aldosterone_metabolic_process |
| GO:0032347 | 6 | regulation_of_aldosterone_biosynthetic_process |
| GO:0032364 | 6 | oxygen_homeostasis |
| GO:0032405 | 6 | MutLalpha_complex_binding |
| GO:0032506 | 6 | cytokinetic_process |
| GO:0032528 | 6 | microvillus_organization |
| GO:0032552 | 6 | deoxyribonucleotide_binding |
| GO:0032584 | 6 | growth_cone_membrane |
| GO:0032590 | 6 | dendrite_membrane |
| GO:0032682 | 6 | negative_regulation_of_chemokine_production |
| GO:0032693 | 6 | negative_regulation_of_interleukin-10_production |
| GO:0032776 | 6 | DNA_methylation_on_cytosine |
| GO:0032801 | 6 | receptor_catabolic_process |
| GO:0032809 | 6 | neuronal_cell_body_membrane |
| GO:0032819 | 6 | positive_regulation_of_natural_killer_cell_proliferation |
| GO:0032908 | 6 | regulation_of_transforming_growth_factor_beta1_production |
| GO:0032964 | 6 | collagen_biosynthetic_process |
| GO:0033004 | 6 | negative_regulation_of_mast_cell_activation |
| GO:0033015 | 6 | tetrapyrrole_catabolic_process |
| GO:0033033 | 6 | negative_regulation_of_myeloid_cell_apoptotic_process |
| GO:0033131 | 6 | regulation_of_glucokinase_activity |
| GO:0033147 | 6 | negative_regulation_of_intracellular_estrogen_receptor_signaling_pathway |
| GO:0033179 | 6 | proton-transporting_V-type_ATPase,_V0_domain |
| GO:0033539 | 6 | fatty_acid_beta-oxidation_using_acyl-CoA_dehydrogenase |
| GO:0033603 | 6 | positive_regulation_of_dopamine_secretion |
| GO:0033629 | 6 | negative_regulation_of_cell_adhesion_mediated_by_integrin |
| GO:0033689 | 6 | negative_regulation_of_osteoblast_proliferation |
| GO:0034067 | 6 | protein_localization_in_Golgi_apparatus |
| GO:0034101 | 6 | erythrocyte_homeostasis |
| GO:0034115 | 6 | negative_regulation_of_heterotypic_cell-cell_adhesion |
| GO:0034212 | 6 | peptide_N-acetyltransferase_activity |
| GO:0034236 | 6 | protein_kinase_A_catalytic_subunit_binding |
| GO:0034261 | 6 | negative_regulation_of_Ras_GTPase_activity |
| GO:0034314 | 6 | Arp2/3_complex-mediated_actin_nucleation |
| GO:0034350 | 6 | regulation_of_glial_cell_apoptotic_process |
| GO:0034351 | 6 | negative_regulation_of_glial_cell_apoptotic_process |
| GO:0034379 | 6 | very-low-density_lipoprotein_particle_assembly |
| GO:0034382 | 6 | chylomicron_remnant_clearance |
| GO:0034384 | 6 | high-density_lipoprotein_particle_clearance |
| GO:0034501 | 6 | protein_localization_to_kinetochore |
| GO:0034629 | 6 | cellular_protein_complex_localization |
| GO:0034643 | 6 | establishment_of_mitochondrion_localization,_microtubule-mediated |
| GO:0034720 | 6 | histone_H3-K4_demethylation |
| GO:0034767 | 6 | positive_regulation_of_ion_transmembrane_transport |
| GO:0034776 | 6 | response_to_histamine |
| GO:0034993 | 6 | SUN-KASH_complex |
| GO:0035014 | 6 | phosphatidylinositol_3-kinase_regulator_activity |
| GO:0035095 | 6 | behavioral_response_to_nicotine |
| GO:0035278 | 6 | negative_regulation_of_translation_involved_in_gene_silencing_by_miRNA |
| GO:0035305 | 6 | negative_regulation_of_dephosphorylation |
| GO:0035315 | 6 | hair_cell_differentiation |
| GO:0035330 | 6 | regulation_of_hippo_signaling_cascade |
| GO:0035358 | 6 | regulation_of_peroxisome_proliferator_activated_receptor_signaling_pathway |
| GO:0035418 | 6 | protein_localization_to_synapse |
| GO:0035434 | 6 | copper_ion_transmembrane_transport |
| GO:0035457 | 6 | cellular_response_to_interferon-alpha |
| GO:0035497 | 6 | cAMP_response_element_binding |
| GO:0035581 | 6 | sequestering_of_extracellular_ligand_from_receptor |
| GO:0035634 | 6 | response_to_stilbenoid |
| GO:0035673 | 6 | oligopeptide_transmembrane_transporter_activity |
| GO:0035728 | 6 | response_to_hepatocyte_growth_factor_stimulus |
| GO:0035729 | 6 | cellular_response_to_hepatocyte_growth_factor_stimulus |
| GO:0035767 | 6 | endothelial_cell_chemotaxis |
| GO:0035774 | 6 | positive_regulation_of_insulin_secretion_involved_in_cellular_response_to_glucose_stimulus |
| GO:0036005 | 6 | response_to_macrophage_colony-stimulating_factor_stimulus |
| GO:0036006 | 6 | cellular_response_to_macrophage_colony-stimulating_factor_stimulus |
| GO:0036037 | 6 | CD8-positive,_alpha-beta_T_cell_activation |
| GO:0036041 | 6 | long-chain_fatty_acid_binding |
| GO:0036056 | 6 | filtration_diaphragm |
| GO:0036057 | 6 | slit_diaphragm |
| GO:0036064 | 6 | cilium_basal_body |
| GO:0036072 | 6 | direct_ossification |
| GO:0038003 | 6 | opioid_receptor_signaling_pathway |
| GO:0038036 | 6 | sphingosine-1-phosphate_receptor_activity |
| GO:0040033 | 6 | negative_regulation_of_translation,_ncRNA-mediated |
| GO:0042153 | 6 | RPTP-like_protein_binding |
| GO:0042167 | 6 | heme_catabolic_process |
| GO:0042320 | 6 | regulation_of_circadian_sleep/wake_cycle,_REM_sleep |
| GO:0042321 | 6 | negative_regulation_of_circadian_sleep/wake_cycle,_sleep |
| GO:0042373 | 6 | vitamin_K_metabolic_process |
| GO:0042382 | 6 | paraspeckles |
| GO:0042487 | 6 | regulation_of_odontogenesis_of_dentin-containing_tooth |
| GO:0042491 | 6 | auditory_receptor_cell_differentiation |
| GO:0042538 | 6 | hyperosmotic_salinity_response |
| GO:0042587 | 6 | glycogen_granule |
| GO:0042713 | 6 | sperm_ejaculation |
| GO:0042719 | 6 | mitochondrial_intermembrane_space_protein_transporter_complex |
| GO:0042754 | 6 | negative_regulation_of_circadian_rhythm |
| GO:0042760 | 6 | very_long-chain_fatty_acid_catabolic_process |
| GO:0042789 | 6 | mRNA_transcription_from_RNA_polymerase_II_promoter |
| GO:0042791 | 6 | 5S_class_rRNA_transcription_from_RNA_polymerase_III_type_1_promoter |
| GO:0042797 | 6 | tRNA_transcription_from_RNA_polymerase_III_promoter |
| GO:0042984 | 6 | regulation_of_amyloid_precursor_protein_biosynthetic_process |
| GO:0043096 | 6 | purine_base_salvage |
| GO:0043117 | 6 | positive_regulation_of_vascular_permeability |
| GO:0043138 | 6 | 3'-5'_DNA_helicase_activity |
| GO:0043190 | 6 | ATP-binding_cassette_(ABC)_transporter_complex |
| GO:0043374 | 6 | CD8-positive,_alpha-beta_T_cell_differentiation |
| GO:0043457 | 6 | regulation_of_cellular_respiration |
| GO:0043497 | 6 | regulation_of_protein_heterodimerization_activity |
| GO:0043558 | 6 | regulation_of_translational_initiation_in_response_to_stress |
| GO:0043619 | 6 | regulation_of_transcription_from_RNA_polymerase_II_promoter_in_response_to_oxidative_stress |
| GO:0043628 | 6 | ncRNA_3'-end_processing |
| GO:0043653 | 6 | mitochondrial_fragmentation_involved_in_apoptotic_process |
| GO:0044091 | 6 | membrane_biogenesis |
| GO:0044224 | 6 | juxtaparanode_region_of_axon |
| GO:0044298 | 6 | cell_body_membrane |
| GO:0044380 | 6 | protein_localization_to_cytoskeleton |
| GO:0044453 | 6 | nuclear_membrane_part |
| GO:0044557 | 6 | relaxation_of_smooth_muscle |
| GO:0045046 | 6 | protein_import_into_peroxisome_membrane |
| GO:0045084 | 6 | positive_regulation_of_interleukin-12_biosynthetic_process |
| GO:0045136 | 6 | development_of_secondary_sexual_characteristics |
| GO:0045236 | 6 | CXCR_chemokine_receptor_binding |
| GO:0045239 | 6 | tricarboxylic_acid_cycle_enzyme_complex |
| GO:0045261 | 6 | proton-transporting_ATP_synthase_complex,_catalytic_core_F(1) |
| GO:0045334 | 6 | clathrin-coated_endocytic_vesicle |
| GO:0045343 | 6 | regulation_of_MHC_class_I_biosynthetic_process |
| GO:0045410 | 6 | positive_regulation_of_interleukin-6_biosynthetic_process |
| GO:0045542 | 6 | positive_regulation_of_cholesterol_biosynthetic_process |
| GO:0045630 | 6 | positive_regulation_of_T-helper_2_cell_differentiation |
| GO:0045634 | 6 | regulation_of_melanocyte_differentiation |
| GO:0045650 | 6 | negative_regulation_of_macrophage_differentiation |
| GO:0045683 | 6 | negative_regulation_of_epidermis_development |
| GO:0045793 | 6 | positive_regulation_of_cell_size |
| GO:0045835 | 6 | negative_regulation_of_meiosis |
| GO:0045898 | 6 | regulation_of_RNA_polymerase_II_transcriptional_preinitiation_complex_assembly |
| GO:0045924 | 6 | regulation_of_female_receptivity |
| GO:0045974 | 6 | regulation_of_translation,_ncRNA-mediated |
| GO:0045989 | 6 | positive_regulation_of_striated_muscle_contraction |
| GO:0045990 | 6 | carbon_catabolite_regulation_of_transcription |
| GO:0046031 | 6 | ADP_metabolic_process |
| GO:0046069 | 6 | cGMP_catabolic_process |
| GO:0046100 | 6 | hypoxanthine_metabolic_process |
| GO:0046149 | 6 | pigment_catabolic_process |
| GO:0046185 | 6 | aldehyde_catabolic_process |
| GO:0046390 | 6 | ribose_phosphate_biosynthetic_process |
| GO:0046473 | 6 | phosphatidic_acid_metabolic_process |
| GO:0046475 | 6 | glycerophospholipid_catabolic_process |
| GO:0046477 | 6 | glycosylceramide_catabolic_process |
| GO:0046498 | 6 | S-adenosylhomocysteine_metabolic_process |
| GO:0046512 | 6 | sphingosine_biosynthetic_process |
| GO:0046548 | 6 | retinal_rod_cell_development |
| GO:0046628 | 6 | positive_regulation_of_insulin_receptor_signaling_pathway |
| GO:0046790 | 6 | virion_binding |
| GO:0046880 | 6 | regulation_of_follicle-stimulating_hormone_secretion |
| GO:0046912 | 6 | transferase_activity,_transferring_acyl_groups,_acyl_groups_converted_into_alkyl_on_transfer |
| GO:0047429 | 6 | nucleoside-triphosphate_diphosphatase_activity |
| GO:0047497 | 6 | mitochondrion_transport_along_microtubule |
| GO:0048027 | 6 | mRNA_5'-UTR_binding |
| GO:0048149 | 6 | behavioral_response_to_ethanol |
| GO:0048251 | 6 | elastic_fiber_assembly |
| GO:0048293 | 6 | regulation_of_isotype_switching_to_IgE_isotypes |
| GO:0048406 | 6 | nerve_growth_factor_binding |
| GO:0048500 | 6 | signal_recognition_particle |
| GO:0048569 | 6 | post-embryonic_organ_development |
| GO:0048667 | 6 | cell_morphogenesis_involved_in_neuron_differentiation |
| GO:0048752 | 6 | semicircular_canal_morphogenesis |
| GO:0048845 | 6 | venous_blood_vessel_morphogenesis |
| GO:0048864 | 6 | stem_cell_development |
| GO:0050542 | 6 | icosanoid_binding |
| GO:0050543 | 6 | icosatetraenoic_acid_binding |
| GO:0050656 | 6 | 3'-phosphoadenosine_5'-phosphosulfate_binding |
| GO:0050701 | 6 | interleukin-1_secretion |
| GO:0050798 | 6 | activated_T_cell_proliferation |
| GO:0050847 | 6 | progesterone_receptor_signaling_pathway |
| GO:0050862 | 6 | positive_regulation_of_T_cell_receptor_signaling_pathway |
| GO:0050932 | 6 | regulation_of_pigment_cell_differentiation |
| GO:0050957 | 6 | equilibrioception |
| GO:0051014 | 6 | actin_filament_severing |
| GO:0051016 | 6 | barbed-end_actin_filament_capping |
| GO:0051103 | 6 | DNA_ligation_involved_in_DNA_repair |
| GO:0051182 | 6 | coenzyme_transport |
| GO:0051379 | 6 | epinephrine_binding |
| GO:0051409 | 6 | response_to_nitrosative_stress |
| GO:0051459 | 6 | regulation_of_corticotropin_secretion |
| GO:0051497 | 6 | negative_regulation_of_stress_fiber_assembly |
| GO:0051590 | 6 | positive_regulation_of_neurotransmitter_transport |
| GO:0051636 | 6 | Gram-negative_bacterial_cell_surface_binding |
| GO:0051709 | 6 | regulation_of_killing_of_cells_of_other_organism |
| GO:0051739 | 6 | ammonia_transmembrane_transporter_activity |
| GO:0051782 | 6 | negative_regulation_of_cell_division |
| GO:0051852 | 6 | disruption_by_host_of_symbiont_cells |
| GO:0051873 | 6 | killing_by_host_of_symbiont_cells |
| GO:0051901 | 6 | positive_regulation_of_mitochondrial_depolarization |
| GO:0055012 | 6 | ventricular_cardiac_muscle_cell_differentiation |
| GO:0055057 | 6 | neuroblast_division |
| GO:0055075 | 6 | potassium_ion_homeostasis |
| GO:0055098 | 6 | response_to_low-density_lipoprotein_particle_stimulus |
| GO:0060013 | 6 | righting_reflex |
| GO:0060017 | 6 | parathyroid_gland_development |
| GO:0060019 | 6 | radial_glial_cell_differentiation |
| GO:0060023 | 6 | soft_palate_development |
| GO:0060024 | 6 | rhythmic_synaptic_transmission |
| GO:0060159 | 6 | regulation_of_dopamine_receptor_signaling_pathway |
| GO:0060180 | 6 | female_mating_behavior |
| GO:0060218 | 6 | hemopoietic_stem_cell_differentiation |
| GO:0060252 | 6 | positive_regulation_of_glial_cell_proliferation |
| GO:0060261 | 6 | positive_regulation_of_transcription_initiation_from_RNA_polymerase_II_promoter |
| GO:0060323 | 6 | head_morphogenesis |
| GO:0060501 | 6 | positive_regulation_of_epithelial_cell_proliferation_involved_in_lung_morphogenesis |
| GO:0060510 | 6 | Type_II_pneumocyte_differentiation |
| GO:0060547 | 6 | negative_regulation_of_necrotic_cell_death |
| GO:0060556 | 6 | regulation_of_vitamin_D_biosynthetic_process |
| GO:0060603 | 6 | mammary_gland_duct_morphogenesis |
| GO:0060631 | 6 | regulation_of_meiosis_I |
| GO:0060632 | 6 | regulation_of_microtubule-based_movement |
| GO:0060710 | 6 | chorio-allantoic_fusion |
| GO:0060907 | 6 | positive_regulation_of_macrophage_cytokine_production |
| GO:0061154 | 6 | endothelial_tube_morphogenesis |
| GO:0061318 | 6 | renal_filtration_cell_differentiation |
| GO:0065002 | 6 | intracellular_protein_transmembrane_transport |
| GO:0070062 | 6 | extracellular_vesicular_exosome |
| GO:0070087 | 6 | chromo_shadow_domain_binding |
| GO:0070099 | 6 | regulation_of_chemokine-mediated_signaling_pathway |
| GO:0070198 | 6 | protein_localization_to_chromosome,_telomeric_region |
| GO:0070231 | 6 | T_cell_apoptotic_process |
| GO:0070245 | 6 | positive_regulation_of_thymocyte_apoptotic_process |
| GO:0070265 | 6 | necrotic_cell_death |
| GO:0070295 | 6 | renal_water_absorption |
| GO:0070365 | 6 | hepatocyte_differentiation |
| GO:0070382 | 6 | exocytic_vesicle |
| GO:0070389 | 6 | chaperone_cofactor-dependent_protein_refolding |
| GO:0070472 | 6 | regulation_of_uterine_smooth_muscle_contraction |
| GO:0070474 | 6 | positive_regulation_of_uterine_smooth_muscle_contraction |
| GO:0070486 | 6 | leukocyte_aggregation |
| GO:0070544 | 6 | histone_H3-K36_demethylation |
| GO:0070568 | 6 | guanylyltransferase_activity |
| GO:0070571 | 6 | negative_regulation_of_neuron_projection_regeneration |
| GO:0070679 | 6 | inositol_1,4,5_trisphosphate_binding |
| GO:0070761 | 6 | pre-snoRNP_complex |
| GO:0070857 | 6 | regulation_of_bile_acid_biosynthetic_process |
| GO:0070938 | 6 | contractile_ring |
| GO:0070986 | 6 | left/right_axis_specification |
| GO:0071204 | 6 | histone_pre-mRNA_3'end_processing_complex |
| GO:0071313 | 6 | cellular_response_to_caffeine |
| GO:0071360 | 6 | cellular_response_to_exogenous_dsRNA |
| GO:0071458 | 6 | integral_to_cytosolic_side_of_endoplasmic_reticulum_membrane |
| GO:0071503 | 6 | response_to_heparin |
| GO:0071732 | 6 | cellular_response_to_nitric_oxide |
| GO:0071778 | 6 | WINAC_complex |
| GO:0071806 | 6 | protein_transmembrane_transport |
| GO:0071830 | 6 | triglyceride-rich_lipoprotein_particle_clearance |
| GO:0071949 | 6 | FAD_binding |
| GO:0072001 | 6 | renal_system_development |
| GO:0072050 | 6 | S-shaped_body_morphogenesis |
| GO:0072112 | 6 | glomerular_visceral_epithelial_cell_differentiation |
| GO:0072124 | 6 | regulation_of_glomerular_mesangial_cell_proliferation |
| GO:0072161 | 6 | mesenchymal_cell_differentiation_involved_in_kidney_development |
| GO:0072298 | 6 | regulation_of_metanephric_glomerulus_development |
| GO:0072311 | 6 | glomerular_epithelial_cell_differentiation |
| GO:0072348 | 6 | sulfur_compound_transport |
| GO:0072393 | 6 | microtubule_anchoring_at_microtubule_organizing_center |
| GO:0072502 | 6 | cellular_trivalent_inorganic_anion_homeostasis |
| GO:0072678 | 6 | T_cell_migration |
| GO:0086091 | 6 | regulation_of_heart_rate_by_cardiac_conduction |
| GO:0090031 | 6 | positive_regulation_of_steroid_hormone_biosynthetic_process |
| GO:0090050 | 6 | positive_regulation_of_cell_migration_involved_in_sprouting_angiogenesis |
| GO:0090072 | 6 | positive_regulation_of_sodium_ion_transport_via_voltage-gated_sodium_channel_activity |
| GO:0090073 | 6 | positive_regulation_of_protein_homodimerization_activity |
| GO:0090129 | 6 | positive_regulation_of_synapse_maturation |
| GO:0090197 | 6 | positive_regulation_of_chemokine_secretion |
| GO:0090343 | 6 | positive_regulation_of_cell_aging |
| GO:0090381 | 6 | regulation_of_heart_induction |
| GO:0090400 | 6 | stress-induced_premature_senescence |
| GO:0097110 | 6 | scaffold_protein_binding |
| GO:1900078 | 6 | positive_regulation_of_cellular_response_to_insulin_stimulus |
| GO:2000020 | 6 | positive_regulation_of_male_gonad_development |
| GO:2000047 | 6 | regulation_of_cell-cell_adhesion_mediated_by_cadherin |
| GO:2000050 | 6 | regulation_of_non-canonical_Wnt_receptor_signaling_pathway |
| GO:2000192 | 6 | negative_regulation_of_fatty_acid_transport |
| GO:2000279 | 6 | negative_regulation_of_DNA_biosynthetic_process |
| GO:2000316 | 6 | regulation_of_T-helper_17_type_immune_response |
| GO:2000465 | 6 | regulation_of_glycogen_(starch)_synthase_activity |
| GO:2000501 | 6 | regulation_of_natural_killer_cell_chemotaxis |
| GO:2000505 | 6 | regulation_of_energy_homeostasis |
| GO:2000651 | 6 | positive_regulation_of_sodium_ion_transmembrane_transporter_activity |
| GO:2000679 | 6 | positive_regulation_of_transcription_regulatory_region_DNA_binding |
| GO:2001012 | 6 | mesenchymal_cell_differentiation_involved_in_renal_system_development |
| GO:2001212 | 6 | regulation_of_vasculogenesis |
| GO:2001243 | 6 | negative_regulation_of_intrinsic_apoptotic_signaling_pathway |
| GO:2001279 | 6 | regulation_of_unsaturated_fatty_acid_biosynthetic_process |
| GO:0000099 | 5 | sulfur_amino_acid_transmembrane_transporter_activity |
| GO:0000179 | 5 | rRNA_(adenine-N6,N6-)-dimethyltransferase_activity |
| GO:0000182 | 5 | rDNA_binding |
| GO:0000275 | 5 | mitochondrial_proton-transporting_ATP_synthase_complex,_catalytic_core_F(1) |
| GO:0000347 | 5 | THO_complex |
| GO:0000389 | 5 | nuclear_mRNA_3'-splice_site_recognition |
| GO:0000400 | 5 | four-way_junction_DNA_binding |
| GO:0000403 | 5 | Y-form_DNA_binding |
| GO:0000445 | 5 | THO_complex_part_of_transcription_export_complex |
| GO:0000730 | 5 | DNA_recombinase_assembly |
| GO:0000801 | 5 | central_element |
| GO:0000828 | 5 | inositol_hexakisphosphate_kinase_activity |
| GO:0000832 | 5 | inositol_hexakisphosphate_5-kinase_activity |
| GO:0000900 | 5 | translation_repressor_activity,_nucleic_acid_binding |
| GO:0001515 | 5 | opioid_peptide_activity |
| GO:0001517 | 5 | N-acetylglucosamine_6-O-sulfotransferase_activity |
| GO:0001537 | 5 | N-acetylgalactosamine_4-O-sulfotransferase_activity |
| GO:0001595 | 5 | angiotensin_receptor_activity |
| GO:0001661 | 5 | conditioned_taste_aversion |
| GO:0001714 | 5 | endodermal_cell_fate_specification |
| GO:0001754 | 5 | eye_photoreceptor_cell_differentiation |
| GO:0001767 | 5 | establishment_of_lymphocyte_polarity |
| GO:0001768 | 5 | establishment_of_T_cell_polarity |
| GO:0001778 | 5 | plasma_membrane_repair |
| GO:0001820 | 5 | serotonin_secretion |
| GO:0001825 | 5 | blastocyst_formation |
| GO:0001875 | 5 | lipopolysaccharide_receptor_activity |
| GO:0001880 | 5 | Mullerian_duct_regression |
| GO:0001881 | 5 | receptor_recycling |
| GO:0001893 | 5 | maternal_placenta_development |
| GO:0001911 | 5 | negative_regulation_of_leukocyte_mediated_cytotoxicity |
| GO:0001940 | 5 | male_pronucleus |
| GO:0001955 | 5 | blood_vessel_maturation |
| GO:0002066 | 5 | columnar/cuboidal_epithelial_cell_development |
| GO:0002162 | 5 | dystroglycan_binding |
| GO:0002238 | 5 | response_to_molecule_of_fungal_origin |
| GO:0002282 | 5 | microglial_cell_activation_involved_in_immune_response |
| GO:0002318 | 5 | myeloid_progenitor_cell_differentiation |
| GO:0002326 | 5 | B_cell_lineage_commitment |
| GO:0002327 | 5 | immature_B_cell_differentiation |
| GO:0002328 | 5 | pro-B_cell_differentiation |
| GO:0002329 | 5 | pre-B_cell_differentiation |
| GO:0002377 | 5 | immunoglobulin_production |
| GO:0002455 | 5 | humoral_immune_response_mediated_by_circulating_immunoglobulin |
| GO:0002475 | 5 | antigen_processing_and_presentation_via_MHC_class_Ib |
| GO:0002568 | 5 | somatic_diversification_of_T_cell_receptor_genes |
| GO:0002667 | 5 | regulation_of_T_cell_anergy |
| GO:0002678 | 5 | positive_regulation_of_chronic_inflammatory_response |
| GO:0002681 | 5 | somatic_recombination_of_T_cell_receptor_gene_segments |
| GO:0002689 | 5 | negative_regulation_of_leukocyte_chemotaxis |
| GO:0002740 | 5 | negative_regulation_of_cytokine_secretion_involved_in_immune_response |
| GO:0002767 | 5 | immune_response-inhibiting_cell_surface_receptor_signaling_pathway |
| GO:0002830 | 5 | positive_regulation_of_type_2_immune_response |
| GO:0002911 | 5 | regulation_of_lymphocyte_anergy |
| GO:0003032 | 5 | detection_of_oxygen |
| GO:0003057 | 5 | regulation_of_the_force_of_heart_contraction_by_chemical_signal |
| GO:0003100 | 5 | regulation_of_systemic_arterial_blood_pressure_by_endothelin |
| GO:0003180 | 5 | aortic_valve_morphogenesis |
| GO:0003188 | 5 | heart_valve_formation |
| GO:0003198 | 5 | epithelial_to_mesenchymal_transition_involved_in_endocardial_cushion_formation |
| GO:0003299 | 5 | muscle_hypertrophy_in_response_to_stress |
| GO:0003306 | 5 | Wnt_receptor_signaling_pathway_involved_in_heart_development |
| GO:0003351 | 5 | epithelial_cilium_movement |
| GO:0003357 | 5 | noradrenergic_neuron_differentiation |
| GO:0003417 | 5 | growth_plate_cartilage_development |
| GO:0003691 | 5 | double-stranded_telomeric_DNA_binding |
| GO:0003828 | 5 | alpha-N-acetylneuraminate_alpha-2,8-sialyltransferase_activity |
| GO:0003846 | 5 | 2-acylglycerol_O-acyltransferase_activity |
| GO:0003847 | 5 | 1-alkyl-2-acetylglycerophosphocholine_esterase_activity |
| GO:0003923 | 5 | GPI-anchor_transamidase_activity |
| GO:0004062 | 5 | aryl_sulfotransferase_activity |
| GO:0004185 | 5 | serine-type_carboxypeptidase_activity |
| GO:0004300 | 5 | enoyl-CoA_hydratase_activity |
| GO:0004301 | 5 | epoxide_hydrolase_activity |
| GO:0004311 | 5 | farnesyltranstransferase_activity |
| GO:0004396 | 5 | hexokinase_activity |
| GO:0004438 | 5 | phosphatidylinositol-3-phosphatase_activity |
| GO:0004448 | 5 | isocitrate_dehydrogenase_activity |
| GO:0004459 | 5 | L-lactate_dehydrogenase_activity |
| GO:0004514 | 5 | nicotinate-nucleotide_diphosphorylase_(carboxylating)_activity |
| GO:0004516 | 5 | nicotinate_phosphoribosyltransferase_activity |
| GO:0004528 | 5 | phosphodiesterase_I_activity |
| GO:0004596 | 5 | peptide_alpha-N-acetyltransferase_activity |
| GO:0004614 | 5 | phosphoglucomutase_activity |
| GO:0004645 | 5 | phosphorylase_activity |
| GO:0004668 | 5 | protein-arginine_deiminase_activity |
| GO:0004689 | 5 | phosphorylase_kinase_activity |
| GO:0004749 | 5 | ribose_phosphate_diphosphokinase_activity |
| GO:0004758 | 5 | serine_C-palmitoyltransferase_activity |
| GO:0004791 | 5 | thioredoxin-disulfide_reductase_activity |
| GO:0004826 | 5 | phenylalanine-tRNA_ligase_activity |
| GO:0004844 | 5 | uracil_DNA_N-glycosylase_activity |
| GO:0004865 | 5 | protein_serine/threonine_phosphatase_inhibitor_activity |
| GO:0004904 | 5 | interferon_receptor_activity |
| GO:0004945 | 5 | angiotensin_type_II_receptor_activity |
| GO:0004952 | 5 | dopamine_receptor_activity |
| GO:0004974 | 5 | leukotriene_receptor_activity |
| GO:0004994 | 5 | somatostatin_receptor_activity |
| GO:0005004 | 5 | GPI-linked_ephrin_receptor_activity |
| GO:0005007 | 5 | fibroblast_growth_factor-activated_receptor_activity |
| GO:0005078 | 5 | MAP-kinase_scaffold_activity |
| GO:0005087 | 5 | Ran_guanyl-nucleotide_exchange_factor_activity |
| GO:0005105 | 5 | type_1_fibroblast_growth_factor_receptor_binding |
| GO:0005127 | 5 | ciliary_neurotrophic_factor_receptor_binding |
| GO:0005131 | 5 | growth_hormone_receptor_binding |
| GO:0005143 | 5 | interleukin-12_receptor_binding |
| GO:0005167 | 5 | neurotrophin_TRK_receptor_binding |
| GO:0005168 | 5 | neurotrophin_TRKA_receptor_binding |
| GO:0005219 | 5 | ryanodine-sensitive_calcium-release_channel_activity |
| GO:0005345 | 5 | purine_base_transmembrane_transporter_activity |
| GO:0005384 | 5 | manganese_ion_transmembrane_transporter_activity |
| GO:0005432 | 5 | calcium:sodium_antiporter_activity |
| GO:0005497 | 5 | androgen_binding |
| GO:0005638 | 5 | lamin_filament |
| GO:0005639 | 5 | integral_to_nuclear_inner_membrane |
| GO:0005751 | 5 | mitochondrial_respiratory_chain_complex_IV |
| GO:0005766 | 5 | primary_lysosome |
| GO:0005767 | 5 | secondary_lysosome |
| GO:0005797 | 5 | Golgi_medial_cisterna |
| GO:0005828 | 5 | kinetochore_microtubule |
| GO:0005964 | 5 | phosphorylase_kinase_complex |
| GO:0006048 | 5 | UDP-N-acetylglucosamine_biosynthetic_process |
| GO:0006122 | 5 | mitochondrial_electron_transport,_ubiquinol_to_cytochrome_c |
| GO:0006123 | 5 | mitochondrial_electron_transport,_cytochrome_c_to_oxygen |
| GO:0006165 | 5 | nucleoside_diphosphate_phosphorylation |
| GO:0006166 | 5 | purine_ribonucleoside_salvage |
| GO:0006167 | 5 | AMP_biosynthetic_process |
| GO:0006177 | 5 | GMP_biosynthetic_process |
| GO:0006522 | 5 | alanine_metabolic_process |
| GO:0006526 | 5 | arginine_biosynthetic_process |
| GO:0006552 | 5 | leucine_catabolic_process |
| GO:0006572 | 5 | tyrosine_catabolic_process |
| GO:0006598 | 5 | polyamine_catabolic_process |
| GO:0006654 | 5 | phosphatidic_acid_biosynthetic_process |
| GO:0006678 | 5 | glucosylceramide_metabolic_process |
| GO:0006681 | 5 | galactosylceramide_metabolic_process |
| GO:0006685 | 5 | sphingomyelin_catabolic_process |
| GO:0006686 | 5 | sphingomyelin_biosynthetic_process |
| GO:0006701 | 5 | progesterone_biosynthetic_process |
| GO:0006824 | 5 | cobalt_ion_transport |
| GO:0006855 | 5 | drug_transmembrane_transport |
| GO:0006924 | 5 | activation-induced_cell_death_of_T_cells |
| GO:0006929 | 5 | substrate-dependent_cell_migration |
| GO:0006999 | 5 | nuclear_pore_organization |
| GO:0007000 | 5 | nucleolus_organization |
| GO:0007021 | 5 | tubulin_complex_assembly |
| GO:0007023 | 5 | post-chaperonin_tubulin_folding_pathway |
| GO:0007195 | 5 | adenylate_cyclase-inhibiting_dopamine_receptor_signaling_pathway |
| GO:0007386 | 5 | compartment_pattern_specification |
| GO:0007501 | 5 | mesodermal_cell_fate_specification |
| GO:0007598 | 5 | blood_coagulation,_extrinsic_pathway |
| GO:0008020 | 5 | G-protein_coupled_photoreceptor_activity |
| GO:0008048 | 5 | calcium_sensitive_guanylate_cyclase_activator_activity |
| GO:0008131 | 5 | primary_amine_oxidase_activity |
| GO:0008190 | 5 | eukaryotic_initiation_factor_4E_binding |
| GO:0008298 | 5 | intracellular_mRNA_localization |
| GO:0008327 | 5 | methyl-CpG_binding |
| GO:0008330 | 5 | protein_tyrosine/threonine_phosphatase_activity |
| GO:0008420 | 5 | CTD_phosphatase_activity |
| GO:0008429 | 5 | phosphatidylethanolamine_binding |
| GO:0008467 | 5 | [heparan_sulfate]-glucosamine_3-sulfotransferase_1_activity |
| GO:0008517 | 5 | folic_acid_transporter_activity |
| GO:0008526 | 5 | phosphatidylinositol_transporter_activity |
| GO:0008568 | 5 | microtubule-severing_ATPase_activity |
| GO:0008611 | 5 | ether_lipid_biosynthetic_process |
| GO:0008612 | 5 | peptidyl-lysine_modification_to_hypusine |
| GO:0008967 | 5 | phosphoglycolate_phosphatase_activity |
| GO:0009008 | 5 | DNA-methyltransferase_activity |
| GO:0009048 | 5 | dosage_compensation_by_inactivation_of_X_chromosome |
| GO:0009078 | 5 | pyruvate_family_amino_acid_metabolic_process |
| GO:0009120 | 5 | deoxyribonucleoside_metabolic_process |
| GO:0009128 | 5 | purine_nucleoside_monophosphate_catabolic_process |
| GO:0009155 | 5 | purine_deoxyribonucleotide_catabolic_process |
| GO:0009186 | 5 | deoxyribonucleoside_diphosphate_metabolic_process |
| GO:0009191 | 5 | ribonucleoside_diphosphate_catabolic_process |
| GO:0009215 | 5 | purine_deoxyribonucleoside_triphosphate_metabolic_process |
| GO:0009249 | 5 | protein_lipoylation |
| GO:0009298 | 5 | GDP-mannose_biosynthetic_process |
| GO:0009374 | 5 | biotin_binding |
| GO:0009410 | 5 | response_to_xenobiotic_stimulus |
| GO:0009414 | 5 | response_to_water_deprivation |
| GO:0009642 | 5 | response_to_light_intensity |
| GO:0009804 | 5 | coumarin_metabolic_process |
| GO:0010002 | 5 | cardioblast_differentiation |
| GO:0010138 | 5 | pyrimidine_ribonucleotide_salvage |
| GO:0010193 | 5 | response_to_ozone |
| GO:0010314 | 5 | phosphatidylinositol-5-phosphate_binding |
| GO:0010340 | 5 | carboxyl-O-methyltransferase_activity |
| GO:0010624 | 5 | regulation_of_Schwann_cell_proliferation |
| GO:0010635 | 5 | regulation_of_mitochondrial_fusion |
| GO:0010700 | 5 | negative_regulation_of_norepinephrine_secretion |
| GO:0010713 | 5 | negative_regulation_of_collagen_metabolic_process |
| GO:0010739 | 5 | positive_regulation_of_protein_kinase_A_signaling_cascade |
| GO:0010746 | 5 | regulation_of_plasma_membrane_long-chain_fatty_acid_transport |
| GO:0010766 | 5 | negative_regulation_of_sodium_ion_transport |
| GO:0010839 | 5 | negative_regulation_of_keratinocyte_proliferation |
| GO:0010890 | 5 | positive_regulation_of_sequestering_of_triglyceride |
| GO:0010901 | 5 | regulation_of_very-low-density_lipoprotein_particle_remodeling |
| GO:0014009 | 5 | glial_cell_proliferation |
| GO:0014034 | 5 | neural_crest_cell_fate_commitment |
| GO:0014050 | 5 | negative_regulation_of_glutamate_secretion |
| GO:0014052 | 5 | regulation_of_gamma-aminobutyric_acid_secretion |
| GO:0014808 | 5 | release_of_sequestered_calcium_ion_into_cytosol_by_sarcoplasmic_reticulum |
| GO:0014819 | 5 | regulation_of_skeletal_muscle_contraction |
| GO:0014824 | 5 | artery_smooth_muscle_contraction |
| GO:0014831 | 5 | gastro-intestinal_system_smooth_muscle_contraction |
| GO:0014887 | 5 | cardiac_muscle_adaptation |
| GO:0014898 | 5 | cardiac_muscle_hypertrophy_in_response_to_stress |
| GO:0015038 | 5 | glutathione_disulfide_oxidoreductase_activity |
| GO:0015057 | 5 | thrombin_receptor_activity |
| GO:0015184 | 5 | L-cystine_transmembrane_transporter_activity |
| GO:0015185 | 5 | L-gamma-aminobutyric_acid_transmembrane_transporter_activity |
| GO:0015186 | 5 | L-glutamine_transmembrane_transporter_activity |
| GO:0015193 | 5 | L-proline_transmembrane_transporter_activity |
| GO:0015252 | 5 | hydrogen_ion_channel_activity |
| GO:0015321 | 5 | sodium-dependent_phosphate_transmembrane_transporter_activity |
| GO:0015347 | 5 | sodium-independent_organic_anion_transmembrane_transporter_activity |
| GO:0015355 | 5 | secondary_active_monocarboxylate_transmembrane_transporter_activity |
| GO:0015440 | 5 | peptide-transporting_ATPase_activity |
| GO:0015793 | 5 | glycerol_transport |
| GO:0015840 | 5 | urea_transport |
| GO:0015865 | 5 | purine_nucleotide_transport |
| GO:0015868 | 5 | purine_ribonucleotide_transport |
| GO:0015925 | 5 | galactosidase_activity |
| GO:0016015 | 5 | morphogen_activity |
| GO:0016019 | 5 | peptidoglycan_receptor_activity |
| GO:0016093 | 5 | polyprenol_metabolic_process |
| GO:0016139 | 5 | glycoside_catabolic_process |
| GO:0016174 | 5 | NAD(P)H_oxidase_activity |
| GO:0016199 | 5 | axon_midline_choice_point_recognition |
| GO:0016236 | 5 | macroautophagy |
| GO:0016413 | 5 | O-acetyltransferase_activity |
| GO:0016423 | 5 | tRNA_(guanine)_methyltransferase_activity |
| GO:0016433 | 5 | rRNA_(adenine)_methyltransferase_activity |
| GO:0016454 | 5 | C-palmitoyltransferase_activity |
| GO:0016520 | 5 | growth_hormone-releasing_hormone_receptor_activity |
| GO:0016530 | 5 | metallochaperone_activity |
| GO:0016559 | 5 | peroxisome_fission |
| GO:0016715 | 5 | oxidoreductase_activity,_acting_on_paired_donors,_with_incorporation_or_reduction_of_molecular_oxygen,_reduced_ascorbate_as_one_donor,_and_incorporation_of_one_atom_of_oxygen |
| GO:0016723 | 5 | oxidoreductase_activity,_oxidizing_metal_ions,_NAD_or_NADP_as_acceptor |
| GO:0016833 | 5 | oxo-acid-lyase_activity |
| GO:0016841 | 5 | ammonia-lyase_activity |
| GO:0016847 | 5 | 1-aminocyclopropane-1-carboxylate_synthase_activity |
| GO:0016901 | 5 | oxidoreductase_activity,_acting_on_the_CH-OH_group_of_donors,_quinone_or_similar_compound_as_acceptor |
| GO:0016907 | 5 | G-protein_coupled_acetylcholine_receptor_activity |
| GO:0017014 | 5 | protein_nitrosylation |
| GO:0017056 | 5 | structural_constituent_of_nuclear_pore |
| GO:0017070 | 5 | U6_snRNA_binding |
| GO:0017081 | 5 | chloride_channel_regulator_activity |
| GO:0017089 | 5 | glycolipid_transporter_activity |
| GO:0017108 | 5 | 5'-flap_endonuclease_activity |
| GO:0017121 | 5 | phospholipid_scrambling |
| GO:0017176 | 5 | phosphatidylinositol_N-acetylglucosaminyltransferase_activity |
| GO:0017182 | 5 | peptidyl-diphthamide_metabolic_process |
| GO:0017183 | 5 | peptidyl-diphthamide_biosynthetic_process_from_peptidyl-histidine |
| GO:0018027 | 5 | peptidyl-lysine_dimethylation |
| GO:0018101 | 5 | peptidyl-citrulline_biosynthetic_process_from_peptidyl-arginine |
| GO:0018106 | 5 | peptidyl-histidine_phosphorylation |
| GO:0018119 | 5 | peptidyl-cysteine_S-nitrosylation |
| GO:0018126 | 5 | protein_hydroxylation |
| GO:0018685 | 5 | alkane_1-monooxygenase_activity |
| GO:0019060 | 5 | intracellular_transport_of_viral_proteins_in_host_cell |
| GO:0019204 | 5 | nucleotide_phosphatase_activity |
| GO:0019227 | 5 | neuronal_action_potential_propagation |
| GO:0019238 | 5 | cyclohydrolase_activity |
| GO:0019307 | 5 | mannose_biosynthetic_process |
| GO:0019348 | 5 | dolichol_metabolic_process |
| GO:0019367 | 5 | fatty_acid_elongation,_saturated_fatty_acid |
| GO:0019418 | 5 | sulfide_oxidation |
| GO:0019605 | 5 | butyrate_metabolic_process |
| GO:0019614 | 5 | catechol-containing_compound_catabolic_process |
| GO:0019626 | 5 | short-chain_fatty_acid_catabolic_process |
| GO:0019855 | 5 | calcium_channel_inhibitor_activity |
| GO:0019863 | 5 | IgE_binding |
| GO:0019896 | 5 | axon_transport_of_mitochondrion |
| GO:0019934 | 5 | cGMP-mediated_signaling |
| GO:0019987 | 5 | negative_regulation_of_anti-apoptosis |
| GO:0021517 | 5 | ventral_spinal_cord_development |
| GO:0021696 | 5 | cerebellar_cortex_morphogenesis |
| GO:0021778 | 5 | oligodendrocyte_cell_fate_specification |
| GO:0021780 | 5 | glial_cell_fate_specification |
| GO:0021797 | 5 | forebrain_anterior/posterior_pattern_specification |
| GO:0021826 | 5 | substrate-independent_telencephalic_tangential_migration |
| GO:0021830 | 5 | interneuron_migration_from_the_subpallium_to_the_cortex |
| GO:0021843 | 5 | substrate-independent_telencephalic_tangential_interneuron_migration |
| GO:0021853 | 5 | cerebral_cortex_GABAergic_interneuron_migration |
| GO:0021869 | 5 | forebrain_ventricular_zone_progenitor_cell_division |
| GO:0022417 | 5 | protein_maturation_by_protein_folding |
| GO:0022614 | 5 | membrane_to_membrane_docking |
| GO:0022824 | 5 | transmitter-gated_ion_channel_activity |
| GO:0022835 | 5 | transmitter-gated_channel_activity |
| GO:0030011 | 5 | maintenance_of_cell_polarity |
| GO:0030042 | 5 | actin_filament_depolymerization |
| GO:0030062 | 5 | mitochondrial_tricarboxylic_acid_cycle_enzyme_complex |
| GO:0030134 | 5 | ER_to_Golgi_transport_vesicle |
| GO:0030213 | 5 | hyaluronan_biosynthetic_process |
| GO:0030240 | 5 | skeletal_muscle_thin_filament_assembly |
| GO:0030259 | 5 | lipid_glycosylation |
| GO:0030262 | 5 | apoptotic_nuclear_change |
| GO:0030284 | 5 | estrogen_receptor_activity |
| GO:0030292 | 5 | protein_tyrosine_kinase_inhibitor_activity |
| GO:0030314 | 5 | junctional_membrane_complex |
| GO:0030321 | 5 | transepithelial_chloride_transport |
| GO:0030375 | 5 | thyroid_hormone_receptor_coactivator_activity |
| GO:0030581 | 5 | symbiont_intracellular_protein_transport_in_host |
| GO:0030618 | 5 | transforming_growth_factor_beta_receptor,_pathway-specific_cytoplasmic_mediator_activity |
| GO:0030859 | 5 | polarized_epithelial_cell_differentiation |
| GO:0030868 | 5 | smooth_endoplasmic_reticulum_membrane |
| GO:0030870 | 5 | Mre11_complex |
| GO:0030911 | 5 | TPR_domain_binding |
| GO:0030913 | 5 | paranodal_junction_assembly |
| GO:0030917 | 5 | midbrain-hindbrain_boundary_development |
| GO:0030970 | 5 | retrograde_protein_transport,_ER_to_cytosol |
| GO:0031013 | 5 | troponin_I_binding |
| GO:0031050 | 5 | dsRNA_fragmentation |
| GO:0031053 | 5 | primary_miRNA_processing |
| GO:0031077 | 5 | post-embryonic_camera-type_eye_development |
| GO:0031088 | 5 | platelet_dense_granule_membrane |
| GO:0031223 | 5 | auditory_behavior |
| GO:0031229 | 5 | intrinsic_to_nuclear_inner_membrane |
| GO:0031232 | 5 | extrinsic_to_external_side_of_plasma_membrane |
| GO:0031394 | 5 | positive_regulation_of_prostaglandin_biosynthetic_process |
| GO:0031557 | 5 | induction_of_programmed_cell_death_in_response_to_chemical_stimulus |
| GO:0031573 | 5 | intra-S_DNA_damage_checkpoint |
| GO:0031665 | 5 | negative_regulation_of_lipopolysaccharide-mediated_signaling_pathway |
| GO:0031685 | 5 | adenosine_receptor_binding |
| GO:0031694 | 5 | alpha-2A_adrenergic_receptor_binding |
| GO:0031730 | 5 | CCR5_chemokine_receptor_binding |
| GO:0031944 | 5 | negative_regulation_of_glucocorticoid_metabolic_process |
| GO:0031947 | 5 | negative_regulation_of_glucocorticoid_biosynthetic_process |
| GO:0032060 | 5 | bleb_assembly |
| GO:0032202 | 5 | telomere_assembly |
| GO:0032206 | 5 | positive_regulation_of_telomere_maintenance |
| GO:0032211 | 5 | negative_regulation_of_telomere_maintenance_via_telomerase |
| GO:0032239 | 5 | regulation_of_nucleobase-containing_compound_transport |
| GO:0032262 | 5 | pyrimidine_nucleotide_salvage |
| GO:0032264 | 5 | IMP_salvage |
| GO:0032278 | 5 | positive_regulation_of_gonadotropin_secretion |
| GO:0032328 | 5 | alanine_transport |
| GO:0032366 | 5 | intracellular_sterol_transport |
| GO:0032367 | 5 | intracellular_cholesterol_transport |
| GO:0032415 | 5 | regulation_of_sodium:hydrogen_antiporter_activity |
| GO:0032430 | 5 | positive_regulation_of_phospholipase_A2_activity |
| GO:0032454 | 5 | histone_demethylase_activity_(H3-K9_specific) |
| GO:0032486 | 5 | Rap_protein_signal_transduction |
| GO:0032493 | 5 | response_to_bacterial_lipoprotein |
| GO:0032543 | 5 | mitochondrial_translation |
| GO:0032554 | 5 | purine_deoxyribonucleotide_binding |
| GO:0032558 | 5 | adenyl_deoxyribonucleotide_binding |
| GO:0032713 | 5 | negative_regulation_of_interleukin-4_production |
| GO:0032717 | 5 | negative_regulation_of_interleukin-8_production |
| GO:0032725 | 5 | positive_regulation_of_granulocyte_macrophage_colony-stimulating_factor_production |
| GO:0032747 | 5 | positive_regulation_of_interleukin-23_production |
| GO:0032785 | 5 | negative_regulation_of_DNA-dependent_transcription,_elongation |
| GO:0032800 | 5 | receptor_biosynthetic_process |
| GO:0032811 | 5 | negative_regulation_of_epinephrine_secretion |
| GO:0032847 | 5 | regulation_of_cellular_pH_reduction |
| GO:0032851 | 5 | positive_regulation_of_Rab_GTPase_activity |
| GO:0032854 | 5 | positive_regulation_of_Rap_GTPase_activity |
| GO:0032875 | 5 | regulation_of_DNA_endoreduplication |
| GO:0032876 | 5 | negative_regulation_of_DNA_endoreduplication |
| GO:0032930 | 5 | positive_regulation_of_superoxide_anion_generation |
| GO:0032957 | 5 | inositol_trisphosphate_metabolic_process |
| GO:0032958 | 5 | inositol_phosphate_biosynthetic_process |
| GO:0032983 | 5 | kainate_selective_glutamate_receptor_complex |
| GO:0032988 | 5 | ribonucleoprotein_complex_disassembly |
| GO:0033085 | 5 | negative_regulation_of_T_cell_differentiation_in_thymus |
| GO:0033129 | 5 | positive_regulation_of_histone_phosphorylation |
| GO:0033130 | 5 | acetylcholine_receptor_binding |
| GO:0033133 | 5 | positive_regulation_of_glucokinase_activity |
| GO:0033152 | 5 | immunoglobulin_V(D)J_recombination |
| GO:0033153 | 5 | T_cell_receptor_V(D)J_recombination |
| GO:0033210 | 5 | leptin-mediated_signaling_pathway |
| GO:0033256 | 5 | I-kappaB/NF-kappaB_complex |
| GO:0033601 | 5 | positive_regulation_of_mammary_gland_epithelial_cell_proliferation |
| GO:0033631 | 5 | cell-cell_adhesion_mediated_by_integrin |
| GO:0033687 | 5 | osteoblast_proliferation |
| GO:0034111 | 5 | negative_regulation_of_homotypic_cell-cell_adhesion |
| GO:0034112 | 5 | positive_regulation_of_homotypic_cell-cell_adhesion |
| GO:0034135 | 5 | regulation_of_toll-like_receptor_2_signaling_pathway |
| GO:0034145 | 5 | positive_regulation_of_toll-like_receptor_4_signaling_pathway |
| GO:0034201 | 5 | response_to_oleic_acid |
| GO:0034214 | 5 | protein_hexamerization |
| GO:0034313 | 5 | diol_catabolic_process |
| GO:0034380 | 5 | high-density_lipoprotein_particle_assembly |
| GO:0034393 | 5 | positive_regulation_of_smooth_muscle_cell_apoptotic_process |
| GO:0034399 | 5 | nuclear_periphery |
| GO:0034416 | 5 | bisphosphoglycerate_phosphatase_activity |
| GO:0034442 | 5 | regulation_of_lipoprotein_oxidation |
| GO:0034452 | 5 | dynactin_binding |
| GO:0034481 | 5 | chondroitin_sulfotransferase_activity |
| GO:0034498 | 5 | early_endosome_to_Golgi_transport |
| GO:0034505 | 5 | tooth_mineralization |
| GO:0034594 | 5 | phosphatidylinositol_trisphosphate_phosphatase_activity |
| GO:0034983 | 5 | peptidyl-lysine_deacetylation |
| GO:0035068 | 5 | micro-ribonucleoprotein_complex |
| GO:0035104 | 5 | positive_regulation_of_transcription_via_sterol_regulatory_element_binding |
| GO:0035117 | 5 | embryonic_arm_morphogenesis |
| GO:0035140 | 5 | arm_morphogenesis |
| GO:0035184 | 5 | histone_threonine_kinase_activity |
| GO:0035197 | 5 | siRNA_binding |
| GO:0035238 | 5 | vitamin_A_biosynthetic_process |
| GO:0035247 | 5 | peptidyl-arginine_omega-N-methylation |
| GO:0035284 | 5 | brain_segmentation |
| GO:0035308 | 5 | negative_regulation_of_protein_dephosphorylation |
| GO:0035404 | 5 | histone-serine_phosphorylation |
| GO:0035405 | 5 | histone-threonine_phosphorylation |
| GO:0035426 | 5 | extracellular_matrix-cell_signaling |
| GO:0035456 | 5 | response_to_interferon-beta |
| GO:0035520 | 5 | monoubiquitinated_protein_deubiquitination |
| GO:0035564 | 5 | regulation_of_kidney_size |
| GO:0035624 | 5 | receptor_transactivation |
| GO:0035748 | 5 | myelin_sheath_abaxonal_region |
| GO:0035749 | 5 | myelin_sheath_adaxonal_region |
| GO:0035811 | 5 | negative_regulation_of_urine_volume |
| GO:0035855 | 5 | megakaryocyte_development |
| GO:0035860 | 5 | glial_cell-derived_neurotrophic_factor_receptor_signaling_pathway |
| GO:0035886 | 5 | vascular_smooth_muscle_cell_differentiation |
| GO:0036119 | 5 | response_to_platelet-derived_growth_factor_stimulus |
| GO:0038169 | 5 | somatostatin_receptor_signaling_pathway |
| GO:0038170 | 5 | somatostatin_signaling_pathway |
| GO:0042118 | 5 | endothelial_cell_activation |
| GO:0042158 | 5 | lipoprotein_biosynthetic_process |
| GO:0042289 | 5 | MHC_class_II_protein_binding |
| GO:0042424 | 5 | catecholamine_catabolic_process |
| GO:0042435 | 5 | indole-containing_compound_biosynthetic_process |
| GO:0042503 | 5 | tyrosine_phosphorylation_of_Stat3_protein |
| GO:0042536 | 5 | negative_regulation_of_tumor_necrosis_factor_biosynthetic_process |
| GO:0042541 | 5 | hemoglobin_biosynthetic_process |
| GO:0042582 | 5 | azurophil_granule |
| GO:0042588 | 5 | zymogen_granule |
| GO:0042608 | 5 | T_cell_receptor_binding |
| GO:0042609 | 5 | CD4_receptor_binding |
| GO:0042661 | 5 | regulation_of_mesodermal_cell_fate_specification |
| GO:0042723 | 5 | thiamine-containing_compound_metabolic_process |
| GO:0042726 | 5 | flavin-containing_compound_metabolic_process |
| GO:0042765 | 5 | GPI-anchor_transamidase_complex |
| GO:0042788 | 5 | polysomal_ribosome |
| GO:0042904 | 5 | 9-cis-retinoic_acid_biosynthetic_process |
| GO:0042905 | 5 | 9-cis-retinoic_acid_metabolic_process |
| GO:0042940 | 5 | D-amino_acid_transport |
| GO:0042979 | 5 | ornithine_decarboxylase_regulator_activity |
| GO:0043008 | 5 | ATP-dependent_protein_binding |
| GO:0043031 | 5 | negative_regulation_of_macrophage_activation |
| GO:0043049 | 5 | otic_placode_formation |
| GO:0043084 | 5 | penile_erection |
| GO:0043125 | 5 | ErbB-3_class_receptor_binding |
| GO:0043139 | 5 | 5'-3'_DNA_helicase_activity |
| GO:0043140 | 5 | ATP-dependent_3'-5'_DNA_helicase_activity |
| GO:0043217 | 5 | myelin_maintenance |
| GO:0043219 | 5 | lateral_loop |
| GO:0043301 | 5 | negative_regulation_of_leukocyte_degranulation |
| GO:0043312 | 5 | neutrophil_degranulation |
| GO:0043382 | 5 | positive_regulation_of_memory_T_cell_differentiation |
| GO:0043426 | 5 | MRF_binding |
| GO:0043546 | 5 | molybdopterin_cofactor_binding |
| GO:0043559 | 5 | insulin_binding |
| GO:0043570 | 5 | maintenance_of_DNA_repeat_elements |
| GO:0043584 | 5 | nose_development |
| GO:0043615 | 5 | astrocyte_cell_migration |
| GO:0043902 | 5 | positive_regulation_of_multi-organism_process |
| GO:0043931 | 5 | ossification_involved_in_bone_maturation |
| GO:0043951 | 5 | negative_regulation_of_cAMP-mediated_signaling |
| GO:0043985 | 5 | histone_H4-R3_methylation |
| GO:0044206 | 5 | UMP_salvage |
| GO:0044245 | 5 | polysaccharide_digestion |
| GO:0044252 | 5 | negative_regulation_of_multicellular_organismal_metabolic_process |
| GO:0044268 | 5 | multicellular_organismal_protein_metabolic_process |
| GO:0044341 | 5 | sodium-dependent_phosphate_transport |
| GO:0045008 | 5 | depyrimidination |
| GO:0045066 | 5 | regulatory_T_cell_differentiation |
| GO:0045077 | 5 | negative_regulation_of_interferon-gamma_biosynthetic_process |
| GO:0045132 | 5 | meiotic_chromosome_segregation |
| GO:0045137 | 5 | development_of_primary_sexual_characteristics |
| GO:0045254 | 5 | pyruvate_dehydrogenase_complex |
| GO:0045277 | 5 | respiratory_chain_complex_IV |
| GO:0045329 | 5 | carnitine_biosynthetic_process |
| GO:0045347 | 5 | negative_regulation_of_MHC_class_II_biosynthetic_process |
| GO:0045351 | 5 | type_I_interferon_biosynthetic_process |
| GO:0045354 | 5 | regulation_of_interferon-alpha_biosynthetic_process |
| GO:0045409 | 5 | negative_regulation_of_interleukin-6_biosynthetic_process |
| GO:0045578 | 5 | negative_regulation_of_B_cell_differentiation |
| GO:0045591 | 5 | positive_regulation_of_regulatory_T_cell_differentiation |
| GO:0045627 | 5 | positive_regulation_of_T-helper_1_cell_differentiation |
| GO:0045631 | 5 | regulation_of_mechanoreceptor_differentiation |
| GO:0045651 | 5 | positive_regulation_of_macrophage_differentiation |
| GO:0045654 | 5 | positive_regulation_of_megakaryocyte_differentiation |
| GO:0045657 | 5 | positive_regulation_of_monocyte_differentiation |
| GO:0045722 | 5 | positive_regulation_of_gluconeogenesis |
| GO:0045738 | 5 | negative_regulation_of_DNA_repair |
| GO:0045750 | 5 | positive_regulation_of_S_phase_of_mitotic_cell_cycle |
| GO:0045764 | 5 | positive_regulation_of_cellular_amino_acid_metabolic_process |
| GO:0045820 | 5 | negative_regulation_of_glycolysis |
| GO:0045837 | 5 | negative_regulation_of_membrane_potential |
| GO:0045869 | 5 | negative_regulation_of_retroviral_genome_replication |
| GO:0045906 | 5 | negative_regulation_of_vasoconstriction |
| GO:0045938 | 5 | positive_regulation_of_circadian_sleep/wake_cycle,_sleep |
| GO:0045943 | 5 | positive_regulation_of_transcription_from_RNA_polymerase_I_promoter |
| GO:0046015 | 5 | regulation_of_transcription_by_glucose |
| GO:0046101 | 5 | hypoxanthine_biosynthetic_process |
| GO:0046137 | 5 | negative_regulation_of_vitamin_metabolic_process |
| GO:0046219 | 5 | indolalkylamine_biosynthetic_process |
| GO:0046416 | 5 | D-amino_acid_metabolic_process |
| GO:0046469 | 5 | platelet_activating_factor_metabolic_process |
| GO:0046516 | 5 | hypusine_metabolic_process |
| GO:0046530 | 5 | photoreceptor_cell_differentiation |
| GO:0046532 | 5 | regulation_of_photoreceptor_cell_differentiation |
| GO:0046642 | 5 | negative_regulation_of_alpha-beta_T_cell_proliferation |
| GO:0046696 | 5 | lipopolysaccharide_receptor_complex |
| GO:0046785 | 5 | microtubule_polymerization |
| GO:0046826 | 5 | negative_regulation_of_protein_export_from_nucleus |
| GO:0046874 | 5 | quinolinate_metabolic_process |
| GO:0046881 | 5 | positive_regulation_of_follicle-stimulating_hormone_secretion |
| GO:0046951 | 5 | ketone_body_biosynthetic_process |
| GO:0046974 | 5 | histone_methyltransferase_activity_(H3-K9_specific) |
| GO:0047499 | 5 | calcium-independent_phospholipase_A2_activity |
| GO:0048012 | 5 | hepatocyte_growth_factor_receptor_signaling_pathway |
| GO:0048087 | 5 | positive_regulation_of_developmental_pigmentation |
| GO:0048240 | 5 | sperm_capacitation |
| GO:0048280 | 5 | vesicle_fusion_with_Golgi_apparatus |
| GO:0048339 | 5 | paraxial_mesoderm_development |
| GO:0048539 | 5 | bone_marrow_development |
| GO:0048563 | 5 | post-embryonic_organ_morphogenesis |
| GO:0048676 | 5 | axon_extension_involved_in_development |
| GO:0048681 | 5 | negative_regulation_of_axon_regeneration |
| GO:0048702 | 5 | embryonic_neurocranium_morphogenesis |
| GO:0048808 | 5 | male_genitalia_morphogenesis |
| GO:0048859 | 5 | formation_of_anatomical_boundary |
| GO:0050327 | 5 | testosterone_17-beta-dehydrogenase_(NAD+)_activity |
| GO:0050654 | 5 | chondroitin_sulfate_proteoglycan_metabolic_process |
| GO:0050765 | 5 | negative_regulation_of_phagocytosis |
| GO:0050917 | 5 | sensory_perception_of_umami_taste |
| GO:0050961 | 5 | detection_of_temperature_stimulus_involved_in_sensory_perception |
| GO:0050965 | 5 | detection_of_temperature_stimulus_involved_in_sensory_perception_of_pain |
| GO:0051005 | 5 | negative_regulation_of_lipoprotein_lipase_activity |
| GO:0051029 | 5 | rRNA_transport |
| GO:0051096 | 5 | positive_regulation_of_helicase_activity |
| GO:0051133 | 5 | regulation_of_NK_T_cell_activation |
| GO:0051135 | 5 | positive_regulation_of_NK_T_cell_activation |
| GO:0051255 | 5 | spindle_midzone_assembly |
| GO:0051387 | 5 | negative_regulation_of_nerve_growth_factor_receptor_signaling_pathway |
| GO:0051450 | 5 | myoblast_proliferation |
| GO:0051461 | 5 | positive_regulation_of_corticotropin_secretion |
| GO:0051534 | 5 | negative_regulation_of_NFAT_protein_import_into_nucleus |
| GO:0051546 | 5 | keratinocyte_migration |
| GO:0051547 | 5 | regulation_of_keratinocyte_migration |
| GO:0051549 | 5 | positive_regulation_of_keratinocyte_migration |
| GO:0051552 | 5 | flavone_metabolic_process |
| GO:0051560 | 5 | mitochondrial_calcium_ion_homeostasis |
| GO:0051561 | 5 | elevation_of_mitochondrial_calcium_ion_concentration |
| GO:0051581 | 5 | negative_regulation_of_neurotransmitter_uptake |
| GO:0051611 | 5 | regulation_of_serotonin_uptake |
| GO:0051612 | 5 | negative_regulation_of_serotonin_uptake |
| GO:0051637 | 5 | Gram-positive_bacterial_cell_surface_binding |
| GO:0051708 | 5 | intracellular_protein_transport_in_other_organism_involved_in_symbiotic_interaction |
| GO:0051712 | 5 | positive_regulation_of_killing_of_cells_of_other_organism |
| GO:0051891 | 5 | positive_regulation_of_cardioblast_differentiation |
| GO:0051961 | 5 | negative_regulation_of_nervous_system_development |
| GO:0051973 | 5 | positive_regulation_of_telomerase_activity |
| GO:0051998 | 5 | protein_carboxyl_O-methyltransferase_activity |
| GO:0052552 | 5 | modulation_by_organism_of_immune_response_of_other_organism_involved_in_symbiotic_interaction |
| GO:0052553 | 5 | modulation_by_symbiont_of_host_immune_response |
| GO:0052555 | 5 | positive_regulation_by_organism_of_immune_response_of_other_organism_involved_in_symbiotic_interaction |
| GO:0052556 | 5 | positive_regulation_by_symbiont_of_host_immune_response |
| GO:0052696 | 5 | flavonoid_glucuronidation |
| GO:0052697 | 5 | xenobiotic_glucuronidation |
| GO:0052723 | 5 | inositol_hexakisphosphate_1-kinase_activity |
| GO:0052724 | 5 | inositol_hexakisphosphate_3-kinase_activity |
| GO:0052743 | 5 | inositol_tetrakisphosphate_phosphatase_activity |
| GO:0052744 | 5 | phosphatidylinositol_monophosphate_phosphatase_activity |
| GO:0055103 | 5 | ligase_regulator_activity |
| GO:0055129 | 5 | L-proline_biosynthetic_process |
| GO:0060029 | 5 | convergent_extension_involved_in_organogenesis |
| GO:0060040 | 5 | retinal_bipolar_neuron_differentiation |
| GO:0060042 | 5 | retina_morphogenesis_in_camera-type_eye |
| GO:0060049 | 5 | regulation_of_protein_glycosylation |
| GO:0060054 | 5 | positive_regulation_of_epithelial_cell_proliferation_involved_in_wound_healing |
| GO:0060087 | 5 | relaxation_of_vascular_smooth_muscle |
| GO:0060137 | 5 | maternal_process_involved_in_parturition |
| GO:0060147 | 5 | regulation_of_posttranscriptional_gene_silencing |
| GO:0060157 | 5 | urinary_bladder_development |
| GO:0060164 | 5 | regulation_of_timing_of_neuron_differentiation |
| GO:0060228 | 5 | phosphatidylcholine-sterol_O-acyltransferase_activator_activity |
| GO:0060235 | 5 | lens_induction_in_camera-type_eye |
| GO:0060236 | 5 | regulation_of_mitotic_spindle_organization |
| GO:0060307 | 5 | regulation_of_ventricular_cardiomyocyte_membrane_repolarization |
| GO:0060315 | 5 | negative_regulation_of_ryanodine-sensitive_calcium-release_channel_activity |
| GO:0060371 | 5 | regulation_of_atrial_cardiomyocyte_membrane_depolarization |
| GO:0060394 | 5 | negative_regulation_of_pathway-restricted_SMAD_protein_phosphorylation |
| GO:0060406 | 5 | positive_regulation_of_penile_erection |
| GO:0060456 | 5 | positive_regulation_of_digestive_system_process |
| GO:0060463 | 5 | lung_lobe_morphogenesis |
| GO:0060572 | 5 | morphogenesis_of_an_epithelial_bud |
| GO:0060575 | 5 | intestinal_epithelial_cell_differentiation |
| GO:0060591 | 5 | chondroblast_differentiation |
| GO:0060605 | 5 | tube_lumen_cavitation |
| GO:0060662 | 5 | salivary_gland_cavitation |
| GO:0060666 | 5 | dichotomous_subdivision_of_terminal_units_involved_in_salivary_gland_branching |
| GO:0060676 | 5 | ureteric_bud_formation |
| GO:0060696 | 5 | regulation_of_phospholipid_catabolic_process |
| GO:0060712 | 5 | spongiotrophoblast_layer_development |
| GO:0060750 | 5 | epithelial_cell_proliferation_involved_in_mammary_gland_duct_elongation |
| GO:0060753 | 5 | regulation_of_mast_cell_chemotaxis |
| GO:0060754 | 5 | positive_regulation_of_mast_cell_chemotaxis |
| GO:0060789 | 5 | hair_follicle_placode_formation |
| GO:0060836 | 5 | lymphatic_endothelial_cell_differentiation |
| GO:0060964 | 5 | regulation_of_gene_silencing_by_miRNA |
| GO:0060966 | 5 | regulation_of_gene_silencing_by_RNA |
| GO:0060993 | 5 | kidney_morphogenesis |
| GO:0060996 | 5 | dendritic_spine_development |
| GO:0061000 | 5 | negative_regulation_of_dendritic_spine_development |
| GO:0061003 | 5 | positive_regulation_of_dendritic_spine_morphogenesis |
| GO:0061013 | 5 | regulation_of_mRNA_catabolic_process |
| GO:0061037 | 5 | negative_regulation_of_cartilage_development |
| GO:0061046 | 5 | regulation_of_branching_involved_in_lung_morphogenesis |
| GO:0061088 | 5 | regulation_of_sequestering_of_zinc_ion |
| GO:0061133 | 5 | endopeptidase_activator_activity |
| GO:0061312 | 5 | BMP_signaling_pathway_involved_in_heart_development |
| GO:0070044 | 5 | synaptobrevin_2-SNAP-25-syntaxin-1a_complex |
| GO:0070071 | 5 | proton-transporting_two-sector_ATPase_complex_assembly |
| GO:0070083 | 5 | clathrin_sculpted_monoamine_transport_vesicle_membrane |
| GO:0070092 | 5 | regulation_of_glucagon_secretion |
| GO:0070095 | 5 | fructose-6-phosphate_binding |
| GO:0070120 | 5 | ciliary_neurotrophic_factor-mediated_signaling_pathway |
| GO:0070127 | 5 | tRNA_aminoacylation_for_mitochondrial_protein_translation |
| GO:0070141 | 5 | response_to_UV-A |
| GO:0070215 | 5 | MDM2_binding |
| GO:0070221 | 5 | sulfide_oxidation,_using_sulfide:quinone_oxidoreductase |
| GO:0070244 | 5 | negative_regulation_of_thymocyte_apoptotic_process |
| GO:0070273 | 5 | phosphatidylinositol-4-phosphate_binding |
| GO:0070286 | 5 | axonemal_dynein_complex_assembly |
| GO:0070290 | 5 | NAPE-specific_phospholipase_D_activity |
| GO:0070307 | 5 | lens_fiber_cell_development |
| GO:0070339 | 5 | response_to_bacterial_lipopeptide |
| GO:0070344 | 5 | regulation_of_fat_cell_proliferation |
| GO:0070424 | 5 | regulation_of_nucleotide-binding_oligomerization_domain_containing_signaling_pathway |
| GO:0070530 | 5 | K63-linked_polyubiquitin_binding |
| GO:0070695 | 5 | FHF_complex |
| GO:0070700 | 5 | BMP_receptor_binding |
| GO:0070841 | 5 | inclusion_body_assembly |
| GO:0070842 | 5 | aggresome_assembly |
| GO:0070884 | 5 | regulation_of_calcineurin-NFAT_signaling_cascade |
| GO:0070918 | 5 | production_of_small_RNA_involved_in_gene_silencing_by_RNA |
| GO:0070932 | 5 | histone_H3_deacetylation |
| GO:0070934 | 5 | CRD-mediated_mRNA_stabilization |
| GO:0070937 | 5 | CRD-mediated_mRNA_stability_complex |
| GO:0070995 | 5 | NADPH_oxidation |
| GO:0071025 | 5 | RNA_surveillance |
| GO:0071071 | 5 | regulation_of_phospholipid_biosynthetic_process |
| GO:0071218 | 5 | cellular_response_to_misfolded_protein |
| GO:0071220 | 5 | cellular_response_to_bacterial_lipoprotein |
| GO:0071221 | 5 | cellular_response_to_bacterial_lipopeptide |
| GO:0071257 | 5 | cellular_response_to_electrical_stimulus |
| GO:0071280 | 5 | cellular_response_to_copper_ion |
| GO:0071318 | 5 | cellular_response_to_ATP |
| GO:0071361 | 5 | cellular_response_to_ethanol |
| GO:0071374 | 5 | cellular_response_to_parathyroid_hormone_stimulus |
| GO:0071436 | 5 | sodium_ion_export |
| GO:0071493 | 5 | cellular_response_to_UV-B |
| GO:0071504 | 5 | cellular_response_to_heparin |
| GO:0071546 | 5 | pi-body |
| GO:0071625 | 5 | vocalization_behavior |
| GO:0071637 | 5 | regulation_of_monocyte_chemotactic_protein-1_production |
| GO:0071680 | 5 | response_to_indole-3-methanol |
| GO:0071681 | 5 | cellular_response_to_indole-3-methanol |
| GO:0071694 | 5 | maintenance_of_protein_location_in_extracellular_region |
| GO:0071817 | 5 | MMXD_complex |
| GO:0072012 | 5 | glomerulus_vasculature_development |
| GO:0072104 | 5 | glomerular_capillary_formation |
| GO:0072162 | 5 | metanephric_mesenchymal_cell_differentiation |
| GO:0072201 | 5 | negative_regulation_of_mesenchymal_cell_proliferation |
| GO:0072307 | 5 | regulation_of_metanephric_nephron_tubule_epithelial_cell_differentiation |
| GO:0072338 | 5 | cellular_lactam_metabolic_process |
| GO:0072383 | 5 | plus-end-directed_vesicle_transport_along_microtubule |
| GO:0072386 | 5 | plus-end-directed_organelle_transport_along_microtubule |
| GO:0072498 | 5 | embryonic_skeletal_joint_development |
| GO:0072583 | 5 | clathrin-mediated_endocytosis |
| GO:0072669 | 5 | tRNA-splicing_ligase_complex |
| GO:0086006 | 5 | voltage-gated_sodium_channel_activity_involved_in_regulation_of_cardiac_muscle_cell_action_potential |
| GO:0086009 | 5 | membrane_repolarization |
| GO:0086011 | 5 | membrane_repolarization_involved_in_regulation_of_action_potential |
| GO:0086013 | 5 | membrane_repolarization_involved_in_regulation_of_cardiac_muscle_cell_action_potential |
| GO:0090032 | 5 | negative_regulation_of_steroid_hormone_biosynthetic_process |
| GO:0090083 | 5 | regulation_of_inclusion_body_assembly |
| GO:0090140 | 5 | regulation_of_mitochondrial_fission |
| GO:0090174 | 5 | organelle_membrane_fusion |
| GO:0090239 | 5 | regulation_of_histone_H4_acetylation |
| GO:0090280 | 5 | positive_regulation_of_calcium_ion_import |
| GO:0090286 | 5 | cytoskeletal_anchoring_at_nuclear_membrane |
| GO:0090370 | 5 | negative_regulation_of_cholesterol_efflux |
| GO:0090407 | 5 | organophosphate_biosynthetic_process |
| GO:0097104 | 5 | postsynaptic_membrane_assembly |
| GO:0097306 | 5 | cellular_response_to_alcohol |
| GO:1900006 | 5 | positive_regulation_of_dendrite_development |
| GO:1900101 | 5 | regulation_of_endoplasmic_reticulum_unfolded_protein_response |
| GO:1900407 | 5 | regulation_of_cellular_response_to_oxidative_stress |
| GO:1901031 | 5 | regulation_of_response_to_reactive_oxygen_species |
| GO:2000049 | 5 | positive_regulation_of_cell-cell_adhesion_mediated_by_cadherin |
| GO:2000064 | 5 | regulation_of_cortisol_biosynthetic_process |
| GO:2000074 | 5 | regulation_of_type_B_pancreatic_cell_development |
| GO:2000095 | 5 | regulation_of_Wnt_receptor_signaling_pathway,_planar_cell_polarity_pathway |
| GO:2000194 | 5 | regulation_of_female_gonad_development |
| GO:2000209 | 5 | regulation_of_anoikis |
| GO:2000251 | 5 | positive_regulation_of_actin_cytoskeleton_reorganization |
| GO:2000318 | 5 | positive_regulation_of_T-helper_17_type_immune_response |
| GO:2000319 | 5 | regulation_of_T-helper_17_cell_differentiation |
| GO:2000341 | 5 | regulation_of_chemokine_(C-X-C_motif)_ligand_2_production |
| GO:2000344 | 5 | positive_regulation_of_acrosome_reaction |
| GO:2000369 | 5 | regulation_of_clathrin-mediated_endocytosis |
| GO:2000503 | 5 | positive_regulation_of_natural_killer_cell_chemotaxis |
| GO:2000551 | 5 | regulation_of_T-helper_2_cell_cytokine_production |
| GO:2000641 | 5 | regulation_of_early_endosome_to_late_endosome_transport |
| GO:2000644 | 5 | regulation_of_receptor_catabolic_process |
| GO:2000648 | 5 | positive_regulation_of_stem_cell_proliferation |
| GO:2000727 | 5 | positive_regulation_of_cardiac_muscle_cell_differentiation |
| GO:2000737 | 5 | negative_regulation_of_stem_cell_differentiation |
| GO:2000739 | 5 | regulation_of_mesenchymal_stem_cell_differentiation |
| GO:2000980 | 5 | regulation_of_inner_ear_receptor_cell_differentiation |
| GO:2001016 | 5 | positive_regulation_of_skeletal_muscle_cell_differentiation |
| GO:2001026 | 5 | regulation_of_endothelial_cell_chemotaxis |
| GO:2001053 | 5 | regulation_of_mesenchymal_cell_apoptotic_process |
| GO:2001054 | 5 | negative_regulation_of_mesenchymal_cell_apoptotic_process |
| GO:2001214 | 5 | positive_regulation_of_vasculogenesis |
| GO:2001239 | 5 | regulation_of_extrinsic_apoptotic_signaling_pathway_in_absence_of_ligand |
| GO:2001280 | 5 | positive_regulation_of_unsaturated_fatty_acid_biosynthetic_process |
| GO:0000015 | 4 | phosphopyruvate_hydratase_complex |
| GO:0000019 | 4 | regulation_of_mitotic_recombination |
| GO:0000064 | 4 | L-ornithine_transmembrane_transporter_activity |
| GO:0000101 | 4 | sulfur_amino_acid_transport |
| GO:0000104 | 4 | succinate_dehydrogenase_activity |
| GO:0000137 | 4 | Golgi_cis_cisterna |
| GO:0000138 | 4 | Golgi_trans_cisterna |
| GO:0000164 | 4 | protein_phosphatase_type_1_complex |
| GO:0000212 | 4 | meiotic_spindle_organization |
| GO:0000247 | 4 | C-8_sterol_isomerase_activity |
| GO:0000255 | 4 | allantoin_metabolic_process |
| GO:0000293 | 4 | ferric-chelate_reductase_activity |
| GO:0000300 | 4 | peripheral_to_membrane_of_membrane_fraction |
| GO:0000320 | 4 | re-entry_into_mitotic_cell_cycle |
| GO:0000407 | 4 | pre-autophagosomal_structure |
| GO:0000429 | 4 | carbon_catabolite_regulation_of_transcription_from_RNA_polymerase_II_promoter |
| GO:0000436 | 4 | carbon_catabolite_activation_of_transcription_from_RNA_polymerase_II_promoter |
| GO:0000444 | 4 | MIS12/MIND_type_complex |
| GO:0000447 | 4 | endonucleolytic_cleavage_in_ITS1_to_separate_SSU-rRNA_from_5.8S_rRNA_and_LSU-rRNA_from_tricistronic_rRNA_transcript_(SSU-rRNA,_5.8S_rRNA,_LSU-rRNA) |
| GO:0000469 | 4 | cleavage_involved_in_rRNA_processing |
| GO:0000478 | 4 | endonucleolytic_cleavage_involved_in_rRNA_processing |
| GO:0000479 | 4 | endonucleolytic_cleavage_of_tricistronic_rRNA_transcript_(SSU-rRNA,_5.8S_rRNA,_LSU-rRNA) |
| GO:0000733 | 4 | DNA_strand_renaturation |
| GO:0000739 | 4 | DNA_strand_annealing_activity |
| GO:0000966 | 4 | RNA_5'-end_processing |
| GO:0001016 | 4 | RNA_polymerase_III_regulatory_region_DNA_binding |
| GO:0001520 | 4 | outer_dense_fiber |
| GO:0001601 | 4 | peptide_YY_receptor_activity |
| GO:0001652 | 4 | granular_component |
| GO:0001660 | 4 | fever_generation |
| GO:0001672 | 4 | regulation_of_chromatin_assembly_or_disassembly |
| GO:0001696 | 4 | gastric_acid_secretion |
| GO:0001730 | 4 | 2'-5'-oligoadenylate_synthetase_activity |
| GO:0001733 | 4 | galactosylceramide_sulfotransferase_activity |
| GO:0001740 | 4 | Barr_body |
| GO:0001757 | 4 | somite_specification |
| GO:0001771 | 4 | immunological_synapse_formation |
| GO:0001835 | 4 | blastocyst_hatching |
| GO:0001913 | 4 | T_cell_mediated_cytotoxicity |
| GO:0002043 | 4 | blood_vessel_endothelial_cell_proliferation_involved_in_sprouting_angiogenesis |
| GO:0002054 | 4 | nucleobase_binding |
| GO:0002069 | 4 | columnar/cuboidal_epithelial_cell_maturation |
| GO:0002097 | 4 | tRNA_wobble_base_modification |
| GO:0002098 | 4 | tRNA_wobble_uridine_modification |
| GO:0002125 | 4 | maternal_aggressive_behavior |
| GO:0002142 | 4 | stereocilia_ankle_link_complex |
| GO:0002220 | 4 | innate_immune_response_activating_cell_surface_receptor_signaling_pathway |
| GO:0002227 | 4 | innate_immune_response_in_mucosa |
| GO:0002309 | 4 | T_cell_proliferation_involved_in_immune_response |
| GO:0002381 | 4 | immunoglobulin_production_involved_in_immunoglobulin_mediated_immune_response |
| GO:0002385 | 4 | mucosal_immune_response |
| GO:0002523 | 4 | leukocyte_migration_involved_in_inflammatory_response |
| GO:0002551 | 4 | mast_cell_chemotaxis |
| GO:0002669 | 4 | positive_regulation_of_T_cell_anergy |
| GO:0002725 | 4 | negative_regulation_of_T_cell_cytokine_production |
| GO:0002913 | 4 | positive_regulation_of_lymphocyte_anergy |
| GO:0003056 | 4 | regulation_of_vascular_smooth_muscle_contraction |
| GO:0003099 | 4 | positive_regulation_of_the_force_of_heart_contraction_by_chemical_signal |
| GO:0003143 | 4 | embryonic_heart_tube_morphogenesis |
| GO:0003183 | 4 | mitral_valve_morphogenesis |
| GO:0003203 | 4 | endocardial_cushion_morphogenesis |
| GO:0003219 | 4 | cardiac_right_ventricle_formation |
| GO:0003222 | 4 | ventricular_trabecula_myocardium_morphogenesis |
| GO:0003283 | 4 | atrial_septum_development |
| GO:0003289 | 4 | atrial_septum_primum_morphogenesis |
| GO:0003334 | 4 | keratinocyte_development |
| GO:0003352 | 4 | regulation_of_cilium_movement |
| GO:0003416 | 4 | endochondral_bone_growth |
| GO:0003720 | 4 | telomerase_activity |
| GO:0003831 | 4 | beta-N-acetylglucosaminylglycopeptide_beta-1,4-galactosyltransferase_activity |
| GO:0003873 | 4 | 6-phosphofructo-2-kinase_activity |
| GO:0003886 | 4 | DNA_(cytosine-5-)-methyltransferase_activity |
| GO:0003896 | 4 | DNA_primase_activity |
| GO:0003917 | 4 | DNA_topoisomerase_type_I_activity |
| GO:0003918 | 4 | DNA_topoisomerase_(ATP-hydrolyzing)_activity |
| GO:0003945 | 4 | N-acetyllactosamine_synthase_activity |
| GO:0003960 | 4 | NADPH:quinone_reductase_activity |
| GO:0004024 | 4 | alcohol_dehydrogenase_activity,_zinc-dependent |
| GO:0004045 | 4 | aminoacyl-tRNA_hydrolase_activity |
| GO:0004064 | 4 | arylesterase_activity |
| GO:0004082 | 4 | bisphosphoglycerate_mutase_activity |
| GO:0004083 | 4 | bisphosphoglycerate_2-phosphatase_activity |
| GO:0004090 | 4 | carbonyl_reductase_(NADPH)_activity |
| GO:0004095 | 4 | carnitine_O-palmitoyltransferase_activity |
| GO:0004111 | 4 | creatine_kinase_activity |
| GO:0004127 | 4 | cytidylate_kinase_activity |
| GO:0004144 | 4 | diacylglycerol_O-acyltransferase_activity |
| GO:0004165 | 4 | dodecenoyl-CoA_delta-isomerase_activity |
| GO:0004169 | 4 | dolichyl-phosphate-mannose-protein_mannosyltransferase_activity |
| GO:0004305 | 4 | ethanolamine_kinase_activity |
| GO:0004308 | 4 | exo-alpha-sialidase_activity |
| GO:0004322 | 4 | ferroxidase_activity |
| GO:0004370 | 4 | glycerol_kinase_activity |
| GO:0004427 | 4 | inorganic_diphosphatase_activity |
| GO:0004430 | 4 | 1-phosphatidylinositol_4-kinase_activity |
| GO:0004470 | 4 | malic_enzyme_activity |
| GO:0004488 | 4 | methylenetetrahydrofolate_dehydrogenase_(NADP+)_activity |
| GO:0004515 | 4 | nicotinate-nucleotide_adenylyltransferase_activity |
| GO:0004517 | 4 | nitric-oxide_synthase_activity |
| GO:0004535 | 4 | poly(A)-specific_ribonuclease_activity |
| GO:0004563 | 4 | beta-N-acetylhexosaminidase_activity |
| GO:0004594 | 4 | pantothenate_kinase_activity |
| GO:0004616 | 4 | phosphogluconate_dehydrogenase_(decarboxylating)_activity |
| GO:0004619 | 4 | phosphoglycerate_mutase_activity |
| GO:0004634 | 4 | phosphopyruvate_hydratase_activity |
| GO:0004647 | 4 | phosphoserine_phosphatase_activity |
| GO:0004656 | 4 | procollagen-proline_4-dioxygenase_activity |
| GO:0004663 | 4 | Rab_geranylgeranyltransferase_activity |
| GO:0004677 | 4 | DNA-dependent_protein_kinase_activity |
| GO:0004694 | 4 | eukaryotic_translation_initiation_factor_2alpha_kinase_activity |
| GO:0004706 | 4 | JUN_kinase_kinase_kinase_activity |
| GO:0004711 | 4 | ribosomal_protein_S6_kinase_activity |
| GO:0004720 | 4 | protein-lysine_6-oxidase_activity |
| GO:0004735 | 4 | pyrroline-5-carboxylate_reductase_activity |
| GO:0004740 | 4 | pyruvate_dehydrogenase_(acetyl-transferring)_kinase_activity |
| GO:0004774 | 4 | succinate-CoA_ligase_activity |
| GO:0004784 | 4 | superoxide_dismutase_activity |
| GO:0004828 | 4 | serine-tRNA_ligase_activity |
| GO:0004829 | 4 | threonine-tRNA_ligase_activity |
| GO:0004849 | 4 | uridine_kinase_activity |
| GO:0004862 | 4 | cAMP-dependent_protein_kinase_inhibitor_activity |
| GO:0004897 | 4 | ciliary_neurotrophic_factor_receptor_activity |
| GO:0004958 | 4 | prostaglandin_F_receptor_activity |
| GO:0004971 | 4 | alpha-amino-3-hydroxy-5-methyl-4-isoxazole_propionate_selective_glutamate_receptor_activity |
| GO:0004980 | 4 | melanocyte-stimulating_hormone_receptor_activity |
| GO:0005006 | 4 | epidermal_growth_factor-activated_receptor_activity |
| GO:0005010 | 4 | insulin-like_growth_factor-activated_receptor_activity |
| GO:0005025 | 4 | transforming_growth_factor_beta_receptor_activity,_type_I |
| GO:0005030 | 4 | neurotrophin_receptor_activity |
| GO:0005042 | 4 | netrin_receptor_activity |
| GO:0005047 | 4 | signal_recognition_particle_binding |
| GO:0005148 | 4 | prolactin_receptor_binding |
| GO:0005220 | 4 | inositol_1,4,5-trisphosphate-sensitive_calcium-release_channel_activity |
| GO:0005315 | 4 | inorganic_phosphate_transmembrane_transporter_activity |
| GO:0005324 | 4 | long-chain_fatty_acid_transporter_activity |
| GO:0005332 | 4 | gamma-aminobutyric_acid:sodium_symporter_activity |
| GO:0005381 | 4 | iron_ion_transmembrane_transporter_activity |
| GO:0005534 | 4 | galactose_binding |
| GO:0005623 | 4 | cell |
| GO:0005736 | 4 | DNA-directed_RNA_polymerase_I_complex |
| GO:0005749 | 4 | mitochondrial_respiratory_chain_complex_II |
| GO:0005826 | 4 | actomyosin_contractile_ring |
| GO:0005827 | 4 | polar_microtubule |
| GO:0005862 | 4 | muscle_thin_filament_tropomyosin |
| GO:0005947 | 4 | mitochondrial_alpha-ketoglutarate_dehydrogenase_complex |
| GO:0005954 | 4 | calcium-_and_calmodulin-dependent_protein_kinase_complex |
| GO:0005955 | 4 | calcineurin_complex |
| GO:0005997 | 4 | xylulose_metabolic_process |
| GO:0006003 | 4 | fructose_2,6-bisphosphate_metabolic_process |
| GO:0006015 | 4 | 5-phosphoribose_1-diphosphate_biosynthetic_process |
| GO:0006021 | 4 | inositol_biosynthetic_process |
| GO:0006054 | 4 | N-acetylneuraminate_metabolic_process |
| GO:0006089 | 4 | lactate_metabolic_process |
| GO:0006104 | 4 | succinyl-CoA_metabolic_process |
| GO:0006114 | 4 | glycerol_biosynthetic_process |
| GO:0006172 | 4 | ADP_biosynthetic_process |
| GO:0006269 | 4 | DNA_replication,_synthesis_of_RNA_primer |
| GO:0006273 | 4 | lagging_strand_elongation |
| GO:0006432 | 4 | phenylalanyl-tRNA_aminoacylation |
| GO:0006434 | 4 | seryl-tRNA_aminoacylation |
| GO:0006435 | 4 | threonyl-tRNA_aminoacylation |
| GO:0006524 | 4 | alanine_catabolic_process |
| GO:0006531 | 4 | aspartate_metabolic_process |
| GO:0006533 | 4 | aspartate_catabolic_process |
| GO:0006549 | 4 | isoleucine_metabolic_process |
| GO:0006562 | 4 | proline_catabolic_process |
| GO:0006564 | 4 | L-serine_biosynthetic_process |
| GO:0006566 | 4 | threonine_metabolic_process |
| GO:0006579 | 4 | betaine_catabolic_process |
| GO:0006610 | 4 | ribosomal_protein_import_into_nucleus |
| GO:0006655 | 4 | phosphatidylglycerol_biosynthetic_process |
| GO:0006659 | 4 | phosphatidylserine_biosynthetic_process |
| GO:0006663 | 4 | platelet_activating_factor_biosynthetic_process |
| GO:0006689 | 4 | ganglioside_catabolic_process |
| GO:0006726 | 4 | eye_pigment_biosynthetic_process |
| GO:0006771 | 4 | riboflavin_metabolic_process |
| GO:0006868 | 4 | glutamine_transport |
| GO:0006883 | 4 | cellular_sodium_ion_homeostasis |
| GO:0006900 | 4 | membrane_budding |
| GO:0006923 | 4 | cleavage_of_cytoskeletal_proteins_involved_in_apoptosis |
| GO:0006930 | 4 | substrate-dependent_cell_migration,_cell_extension |
| GO:0006971 | 4 | hypotonic_response |
| GO:0007008 | 4 | outer_mitochondrial_membrane_organization |
| GO:0007019 | 4 | microtubule_depolymerization |
| GO:0007028 | 4 | cytoplasm_organization |
| GO:0007035 | 4 | vacuolar_acidification |
| GO:0007042 | 4 | lysosomal_lumen_acidification |
| GO:0007258 | 4 | JUN_phosphorylation |
| GO:0007288 | 4 | sperm_axoneme_assembly |
| GO:0007320 | 4 | insemination |
| GO:0007412 | 4 | axon_target_recognition |
| GO:0007431 | 4 | salivary_gland_development |
| GO:0007442 | 4 | hindgut_morphogenesis |
| GO:0007500 | 4 | mesodermal_cell_fate_determination |
| GO:0007621 | 4 | negative_regulation_of_female_receptivity |
| GO:0008054 | 4 | cyclin_catabolic_process |
| GO:0008061 | 4 | chitin_binding |
| GO:0008073 | 4 | ornithine_decarboxylase_inhibitor_activity |
| GO:0008097 | 4 | 5S_rRNA_binding |
| GO:0008174 | 4 | mRNA_methyltransferase_activity |
| GO:0008215 | 4 | spermine_metabolic_process |
| GO:0008216 | 4 | spermidine_metabolic_process |
| GO:0008291 | 4 | acetylcholine_metabolic_process |
| GO:0008331 | 4 | high_voltage-gated_calcium_channel_activity |
| GO:0008379 | 4 | thioredoxin_peroxidase_activity |
| GO:0008391 | 4 | arachidonic_acid_monooxygenase_activity |
| GO:0008392 | 4 | arachidonic_acid_epoxygenase_activity |
| GO:0008449 | 4 | N-acetylglucosamine-6-sulfatase_activity |
| GO:0008486 | 4 | diphosphoinositol-polyphosphate_diphosphatase_activity |
| GO:0008518 | 4 | reduced_folate_carrier_activity |
| GO:0008574 | 4 | plus-end-directed_microtubule_motor_activity |
| GO:0008588 | 4 | release_of_cytoplasmic_sequestered_NF-kappaB |
| GO:0008607 | 4 | phosphorylase_kinase_regulator_activity |
| GO:0008617 | 4 | guanosine_metabolic_process |
| GO:0008628 | 4 | induction_of_apoptosis_by_hormones |
| GO:0008745 | 4 | N-acetylmuramoyl-L-alanine_amidase_activity |
| GO:0008821 | 4 | crossover_junction_endodeoxyribonuclease_activity |
| GO:0008853 | 4 | exodeoxyribonuclease_III_activity |
| GO:0009080 | 4 | pyruvate_family_amino_acid_catabolic_process |
| GO:0009136 | 4 | purine_nucleoside_diphosphate_biosynthetic_process |
| GO:0009158 | 4 | ribonucleoside_monophosphate_catabolic_process |
| GO:0009169 | 4 | purine_ribonucleoside_monophosphate_catabolic_process |
| GO:0009176 | 4 | pyrimidine_deoxyribonucleoside_monophosphate_metabolic_process |
| GO:0009180 | 4 | purine_ribonucleoside_diphosphate_biosynthetic_process |
| GO:0009188 | 4 | ribonucleoside_diphosphate_biosynthetic_process |
| GO:0009202 | 4 | deoxyribonucleoside_triphosphate_biosynthetic_process |
| GO:0009301 | 4 | snRNA_transcription |
| GO:0009313 | 4 | oligosaccharide_catabolic_process |
| GO:0009445 | 4 | putrescine_metabolic_process |
| GO:0009448 | 4 | gamma-aminobutyric_acid_metabolic_process |
| GO:0009624 | 4 | response_to_nematode |
| GO:0009726 | 4 | detection_of_endogenous_stimulus |
| GO:0009753 | 4 | response_to_jasmonic_acid_stimulus |
| GO:0009756 | 4 | carbohydrate_mediated_signaling |
| GO:0009922 | 4 | fatty_acid_elongase_activity |
| GO:0010107 | 4 | potassium_ion_import |
| GO:0010452 | 4 | histone_H3-K36_methylation |
| GO:0010458 | 4 | exit_from_mitosis |
| GO:0010459 | 4 | negative_regulation_of_heart_rate |
| GO:0010519 | 4 | negative_regulation_of_phospholipase_activity |
| GO:0010520 | 4 | regulation_of_reciprocal_meiotic_recombination |
| GO:0010572 | 4 | positive_regulation_of_platelet_activation |
| GO:0010626 | 4 | negative_regulation_of_Schwann_cell_proliferation |
| GO:0010637 | 4 | negative_regulation_of_mitochondrial_fusion |
| GO:0010748 | 4 | negative_regulation_of_plasma_membrane_long-chain_fatty_acid_transport |
| GO:0010756 | 4 | positive_regulation_of_plasminogen_activation |
| GO:0010757 | 4 | negative_regulation_of_plasminogen_activation |
| GO:0010815 | 4 | bradykinin_catabolic_process |
| GO:0010818 | 4 | T_cell_chemotaxis |
| GO:0010891 | 4 | negative_regulation_of_sequestering_of_triglyceride |
| GO:0010915 | 4 | regulation_of_very-low-density_lipoprotein_particle_clearance |
| GO:0010916 | 4 | negative_regulation_of_very-low-density_lipoprotein_particle_clearance |
| GO:0010936 | 4 | negative_regulation_of_macrophage_cytokine_production |
| GO:0010957 | 4 | negative_regulation_of_vitamin_D_biosynthetic_process |
| GO:0014054 | 4 | positive_regulation_of_gamma-aminobutyric_acid_secretion |
| GO:0014732 | 4 | skeletal_muscle_atrophy |
| GO:0014805 | 4 | smooth_muscle_adaptation |
| GO:0014826 | 4 | vein_smooth_muscle_contraction |
| GO:0014848 | 4 | urinary_tract_smooth_muscle_contraction |
| GO:0014874 | 4 | response_to_stimulus_involved_in_regulation_of_muscle_adaptation |
| GO:0014891 | 4 | striated_muscle_atrophy |
| GO:0015014 | 4 | heparan_sulfate_proteoglycan_biosynthetic_process,_polysaccharide_chain_biosynthetic_process |
| GO:0015016 | 4 | [heparan_sulfate]-glucosamine_N-sulfotransferase_activity |
| GO:0015065 | 4 | uridine_nucleotide_receptor_activity |
| GO:0015180 | 4 | L-alanine_transmembrane_transporter_activity |
| GO:0015183 | 4 | L-aspartate_transmembrane_transporter_activity |
| GO:0015204 | 4 | urea_transmembrane_transporter_activity |
| GO:0015220 | 4 | choline_transmembrane_transporter_activity |
| GO:0015254 | 4 | glycerol_channel_activity |
| GO:0015379 | 4 | potassium:chloride_symporter_activity |
| GO:0015421 | 4 | oligopeptide-transporting_ATPase_activity |
| GO:0015467 | 4 | G-protein_activated_inward_rectifier_potassium_channel_activity |
| GO:0015670 | 4 | carbon_dioxide_transport |
| GO:0015691 | 4 | cadmium_ion_transport |
| GO:0015705 | 4 | iodide_transport |
| GO:0015712 | 4 | hexose_phosphate_transport |
| GO:0015722 | 4 | canalicular_bile_acid_transport |
| GO:0015760 | 4 | glucose-6-phosphate_transport |
| GO:0015801 | 4 | aromatic_amino_acid_transport |
| GO:0015811 | 4 | L-cystine_transport |
| GO:0015853 | 4 | adenine_transport |
| GO:0015867 | 4 | ATP_transport |
| GO:0015871 | 4 | choline_transport |
| GO:0015889 | 4 | cobalamin_transport |
| GO:0016082 | 4 | synaptic_vesicle_priming |
| GO:0016160 | 4 | amylase_activity |
| GO:0016286 | 4 | small_conductance_calcium-activated_potassium_channel_activity |
| GO:0016361 | 4 | activin_receptor_activity,_type_I |
| GO:0016416 | 4 | O-palmitoyltransferase_activity |
| GO:0016441 | 4 | posttranscriptional_gene_silencing |
| GO:0016453 | 4 | C-acetyltransferase_activity |
| GO:0016528 | 4 | sarcoplasm |
| GO:0016531 | 4 | copper_chaperone_activity |
| GO:0016557 | 4 | peroxisome_membrane_biogenesis |
| GO:0016602 | 4 | CCAAT-binding_factor_complex |
| GO:0016635 | 4 | oxidoreductase_activity,_acting_on_the_CH-CH_group_of_donors,_quinone_or_related_compound_as_acceptor |
| GO:0016661 | 4 | oxidoreductase_activity,_acting_on_other_nitrogenous_compounds_as_donors |
| GO:0016721 | 4 | oxidoreductase_activity,_acting_on_superoxide_radicals_as_acceptor |
| GO:0016724 | 4 | oxidoreductase_activity,_oxidizing_metal_ions,_oxygen_as_acceptor |
| GO:0016744 | 4 | transferase_activity,_transferring_aldehyde_or_ketonic_groups |
| GO:0016822 | 4 | hydrolase_activity,_acting_on_acid_carbon-carbon_bonds |
| GO:0016823 | 4 | hydrolase_activity,_acting_on_acid_carbon-carbon_bonds,_in_ketonic_substances |
| GO:0016838 | 4 | carbon-oxygen_lyase_activity,_acting_on_phosphates |
| GO:0016842 | 4 | amidine-lyase_activity |
| GO:0016882 | 4 | cyclo-ligase_activity |
| GO:0016900 | 4 | oxidoreductase_activity,_acting_on_the_CH-OH_group_of_donors,_disulfide_as_acceptor |
| GO:0016933 | 4 | extracellular-glycine-gated_ion_channel_activity |
| GO:0016934 | 4 | extracellular-glycine-gated_chloride_channel_activity |
| GO:0016936 | 4 | galactoside_binding |
| GO:0016942 | 4 | insulin-like_growth_factor_binding_protein_complex |
| GO:0016972 | 4 | thiol_oxidase_activity |
| GO:0016997 | 4 | alpha-sialidase_activity |
| GO:0017004 | 4 | cytochrome_complex_assembly |
| GO:0017071 | 4 | intracellular_cyclic_nucleotide_activated_cation_channel_complex |
| GO:0017112 | 4 | Rab_guanyl-nucleotide_exchange_factor_activity |
| GO:0017128 | 4 | phospholipid_scramblase_activity |
| GO:0017150 | 4 | tRNA_dihydrouridine_synthase_activity |
| GO:0017160 | 4 | Ral_GTPase_binding |
| GO:0018095 | 4 | protein_polyglutamylation |
| GO:0018199 | 4 | peptidyl-glutamine_modification |
| GO:0018344 | 4 | protein_geranylgeranylation |
| GO:0018963 | 4 | phthalate_metabolic_process |
| GO:0019054 | 4 | modulation_by_virus_of_host_cellular_process |
| GO:0019087 | 4 | transformation_of_host_cell_by_virus |
| GO:0019136 | 4 | deoxynucleoside_kinase_activity |
| GO:0019144 | 4 | ADP-sugar_diphosphatase_activity |
| GO:0019215 | 4 | intermediate_filament_binding |
| GO:0019323 | 4 | pentose_catabolic_process |
| GO:0019373 | 4 | epoxygenase_P450_pathway |
| GO:0019401 | 4 | alditol_biosynthetic_process |
| GO:0019474 | 4 | L-lysine_catabolic_process_to_acetyl-CoA |
| GO:0019477 | 4 | L-lysine_catabolic_process |
| GO:0019478 | 4 | D-amino_acid_catabolic_process |
| GO:0019482 | 4 | beta-alanine_metabolic_process |
| GO:0019511 | 4 | peptidyl-proline_hydroxylation |
| GO:0019556 | 4 | histidine_catabolic_process_to_glutamate_and_formamide |
| GO:0019557 | 4 | histidine_catabolic_process_to_glutamate_and_formate |
| GO:0019763 | 4 | immunoglobulin_receptor_activity |
| GO:0019788 | 4 | NEDD8_ligase_activity |
| GO:0019911 | 4 | structural_constituent_of_myelin_sheath |
| GO:0019966 | 4 | interleukin-1_binding |
| GO:0021520 | 4 | spinal_cord_motor_neuron_cell_fate_specification |
| GO:0021555 | 4 | midbrain-hindbrain_boundary_morphogenesis |
| GO:0021559 | 4 | trigeminal_nerve_development |
| GO:0021612 | 4 | facial_nerve_structural_organization |
| GO:0021631 | 4 | optic_nerve_morphogenesis |
| GO:0021819 | 4 | layer_formation_in_cerebral_cortex |
| GO:0021861 | 4 | forebrain_radial_glial_cell_differentiation |
| GO:0021871 | 4 | forebrain_regionalization |
| GO:0021877 | 4 | forebrain_neuron_fate_commitment |
| GO:0021913 | 4 | regulation_of_transcription_from_RNA_polymerase_II_promoter_involved_in_ventral_spinal_cord_interneuron_specification |
| GO:0021936 | 4 | regulation_of_cerebellar_granule_cell_precursor_proliferation |
| GO:0021938 | 4 | smoothened_signaling_pathway_involved_in_regulation_of_cerebellar_granule_cell_precursor_cell_proliferation |
| GO:0021978 | 4 | telencephalon_regionalization |
| GO:0022009 | 4 | central_nervous_system_vasculogenesis |
| GO:0022028 | 4 | tangential_migration_from_the_subventricular_zone_to_the_olfactory_bulb |
| GO:0022626 | 4 | cytosolic_ribosome |
| GO:0022821 | 4 | potassium_ion_antiporter_activity |
| GO:0022858 | 4 | alanine_transmembrane_transporter_activity |
| GO:0030091 | 4 | protein_repair |
| GO:0030151 | 4 | molybdenum_ion_binding |
| GO:0030160 | 4 | GKAP/Homer_scaffold_activity |
| GO:0030172 | 4 | troponin_C_binding |
| GO:0030197 | 4 | extracellular_matrix_constituent,_lubricant_activity |
| GO:0030285 | 4 | integral_to_synaptic_vesicle_membrane |
| GO:0030297 | 4 | transmembrane_receptor_protein_tyrosine_kinase_activator_activity |
| GO:0030320 | 4 | cellular_monovalent_inorganic_anion_homeostasis |
| GO:0030490 | 4 | maturation_of_SSU-rRNA |
| GO:0030540 | 4 | female_genitalia_development |
| GO:0030575 | 4 | nuclear_body_organization |
| GO:0030644 | 4 | cellular_chloride_ion_homeostasis |
| GO:0030686 | 4 | 90S_preribosome |
| GO:0030718 | 4 | germ-line_stem_cell_maintenance |
| GO:0030836 | 4 | positive_regulation_of_actin_filament_depolymerization |
| GO:0030891 | 4 | VCB_complex |
| GO:0030904 | 4 | retromer_complex |
| GO:0030948 | 4 | negative_regulation_of_vascular_endothelial_growth_factor_receptor_signaling_pathway |
| GO:0030952 | 4 | establishment_or_maintenance_of_cytoskeleton_polarity |
| GO:0031014 | 4 | troponin_T_binding |
| GO:0031033 | 4 | myosin_filament_organization |
| GO:0031034 | 4 | myosin_filament_assembly |
| GO:0031262 | 4 | Ndc80_complex |
| GO:0031265 | 4 | CD95_death-inducing_signaling_complex |
| GO:0031268 | 4 | pseudopodium_organization |
| GO:0031282 | 4 | regulation_of_guanylate_cyclase_activity |
| GO:0031313 | 4 | extrinsic_to_endosome_membrane |
| GO:0031340 | 4 | positive_regulation_of_vesicle_fusion |
| GO:0031402 | 4 | sodium_ion_binding |
| GO:0031404 | 4 | chloride_ion_binding |
| GO:0031427 | 4 | response_to_methotrexate |
| GO:0031428 | 4 | box_C/D_snoRNP_complex |
| GO:0031465 | 4 | Cul4B-RING_ubiquitin_ligase_complex |
| GO:0031466 | 4 | Cul5-RING_ubiquitin_ligase_complex |
| GO:0031536 | 4 | positive_regulation_of_exit_from_mitosis |
| GO:0031545 | 4 | peptidyl-proline_4-dioxygenase_activity |
| GO:0031558 | 4 | induction_of_apoptosis_in_response_to_chemical_stimulus |
| GO:0031649 | 4 | heat_generation |
| GO:0031821 | 4 | G-protein_coupled_serotonin_receptor_binding |
| GO:0031848 | 4 | protection_from_non-homologous_end_joining_at_telomere |
| GO:0031931 | 4 | TORC1_complex |
| GO:0031932 | 4 | TORC2_complex |
| GO:0032009 | 4 | early_phagosome |
| GO:0032010 | 4 | phagolysosome |
| GO:0032021 | 4 | NELF_complex |
| GO:0032048 | 4 | cardiolipin_metabolic_process |
| GO:0032049 | 4 | cardiolipin_biosynthetic_process |
| GO:0032057 | 4 | negative_regulation_of_translational_initiation_in_response_to_stress |
| GO:0032070 | 4 | regulation_of_deoxyribonuclease_activity |
| GO:0032133 | 4 | chromosome_passenger_complex |
| GO:0032137 | 4 | guanine/thymine_mispair_binding |
| GO:0032138 | 4 | single_base_insertion_or_deletion_binding |
| GO:0032190 | 4 | acrosin_binding |
| GO:0032224 | 4 | positive_regulation_of_synaptic_transmission,_cholinergic |
| GO:0032226 | 4 | positive_regulation_of_synaptic_transmission,_dopaminergic |
| GO:0032227 | 4 | negative_regulation_of_synaptic_transmission,_dopaminergic |
| GO:0032237 | 4 | activation_of_store-operated_calcium_channel_activity |
| GO:0032329 | 4 | serine_transport |
| GO:0032345 | 4 | negative_regulation_of_aldosterone_metabolic_process |
| GO:0032348 | 4 | negative_regulation_of_aldosterone_biosynthetic_process |
| GO:0032396 | 4 | inhibitory_MHC_class_I_receptor_activity |
| GO:0032400 | 4 | melanosome_localization |
| GO:0032418 | 4 | lysosome_localization |
| GO:0032421 | 4 | stereocilium_bundle |
| GO:0032431 | 4 | activation_of_phospholipase_A2_activity |
| GO:0032455 | 4 | nerve_growth_factor_processing |
| GO:0032471 | 4 | reduction_of_endoplasmic_reticulum_calcium_ion_concentration |
| GO:0032488 | 4 | Cdc42_protein_signal_transduction |
| GO:0032536 | 4 | regulation_of_cell_projection_size |
| GO:0032549 | 4 | ribonucleoside_binding |
| GO:0032564 | 4 | dATP_binding |
| GO:0032571 | 4 | response_to_vitamin_K |
| GO:0032606 | 4 | type_I_interferon_production |
| GO:0032661 | 4 | regulation_of_interleukin-18_production |
| GO:0032714 | 4 | negative_regulation_of_interleukin-5_production |
| GO:0032736 | 4 | positive_regulation_of_interleukin-13_production |
| GO:0032763 | 4 | regulation_of_mast_cell_cytokine_production |
| GO:0032777 | 4 | Piccolo_NuA4_histone_acetyltransferase_complex |
| GO:0032780 | 4 | negative_regulation_of_ATPase_activity |
| GO:0032792 | 4 | negative_regulation_of_CREB_transcription_factor_activity |
| GO:0032795 | 4 | heterotrimeric_G-protein_binding |
| GO:0032803 | 4 | regulation_of_low-density_lipoprotein_particle_receptor_catabolic_process |
| GO:0032808 | 4 | lacrimal_gland_development |
| GO:0032849 | 4 | positive_regulation_of_cellular_pH_reduction |
| GO:0032914 | 4 | positive_regulation_of_transforming_growth_factor_beta1_production |
| GO:0032933 | 4 | SREBP-mediated_signaling_pathway |
| GO:0032960 | 4 | regulation_of_inositol_trisphosphate_biosynthetic_process |
| GO:0032966 | 4 | negative_regulation_of_collagen_biosynthetic_process |
| GO:0033025 | 4 | regulation_of_mast_cell_apoptotic_process |
| GO:0033087 | 4 | negative_regulation_of_immature_T_cell_proliferation |
| GO:0033088 | 4 | negative_regulation_of_immature_T_cell_proliferation_in_thymus |
| GO:0033091 | 4 | positive_regulation_of_immature_T_cell_proliferation |
| GO:0033192 | 4 | calmodulin-dependent_protein_phosphatase_activity |
| GO:0033206 | 4 | cytokinesis_after_meiosis |
| GO:0033211 | 4 | adiponectin-mediated_signaling_pathway |
| GO:0033265 | 4 | choline_binding |
| GO:0033270 | 4 | paranode_region_of_axon |
| GO:0033299 | 4 | secretion_of_lysosomal_enzymes |
| GO:0033503 | 4 | HULC_complex |
| GO:0033553 | 4 | rDNA_heterochromatin |
| GO:0033600 | 4 | negative_regulation_of_mammary_gland_epithelial_cell_proliferation |
| GO:0033625 | 4 | positive_regulation_of_integrin_activation |
| GO:0033634 | 4 | positive_regulation_of_cell-cell_adhesion_mediated_by_integrin |
| GO:0033643 | 4 | host_cell_part |
| GO:0033646 | 4 | host_intracellular_part |
| GO:0033695 | 4 | oxidoreductase_activity,_acting_on_CH_or_CH2_groups,_quinone_or_similar_compound_as_acceptor |
| GO:0033765 | 4 | steroid_dehydrogenase_activity,_acting_on_the_CH-CH_group_of_donors |
| GO:0033860 | 4 | regulation_of_NAD(P)H_oxidase_activity |
| GO:0034056 | 4 | estrogen_response_element_binding |
| GO:0034086 | 4 | maintenance_of_sister_chromatid_cohesion |
| GO:0034088 | 4 | maintenance_of_mitotic_sister_chromatid_cohesion |
| GO:0034098 | 4 | Cdc48p-Npl4p-Ufd1p_AAA_ATPase_complex |
| GO:0034116 | 4 | positive_regulation_of_heterotypic_cell-cell_adhesion |
| GO:0034139 | 4 | regulation_of_toll-like_receptor_3_signaling_pathway |
| GO:0034144 | 4 | negative_regulation_of_toll-like_receptor_4_signaling_pathway |
| GO:0034190 | 4 | apolipoprotein_receptor_binding |
| GO:0034227 | 4 | tRNA_thio-modification |
| GO:0034363 | 4 | intermediate-density_lipoprotein_particle |
| GO:0034389 | 4 | lipid_particle_organization |
| GO:0034392 | 4 | negative_regulation_of_smooth_muscle_cell_apoptotic_process |
| GO:0034443 | 4 | negative_regulation_of_lipoprotein_oxidation |
| GO:0034454 | 4 | microtubule_anchoring_at_centrosome |
| GO:0034617 | 4 | tetrahydrobiopterin_binding |
| GO:0034618 | 4 | arginine_binding |
| GO:0034625 | 4 | fatty_acid_elongation,_monounsaturated_fatty_acid |
| GO:0034626 | 4 | fatty_acid_elongation,_polyunsaturated_fatty_acid |
| GO:0034648 | 4 | histone_demethylase_activity_(H3-dimethyl-K4_specific) |
| GO:0034755 | 4 | iron_ion_transmembrane_transport |
| GO:0034770 | 4 | histone_H4-K20_methylation |
| GO:0034875 | 4 | caffeine_oxidase_activity |
| GO:0035033 | 4 | histone_deacetylase_regulator_activity |
| GO:0035081 | 4 | induction_of_programmed_cell_death_by_hormones |
| GO:0035084 | 4 | flagellar_axoneme_assembly |
| GO:0035092 | 4 | sperm_chromatin_condensation |
| GO:0035188 | 4 | hatching |
| GO:0035189 | 4 | Rb-E2F_complex |
| GO:0035194 | 4 | posttranscriptional_gene_silencing_by_RNA |
| GO:0035242 | 4 | protein-arginine_omega-N_asymmetric_methyltransferase_activity |
| GO:0035252 | 4 | UDP-xylosyltransferase_activity |
| GO:0035256 | 4 | G-protein_coupled_glutamate_receptor_binding |
| GO:0035372 | 4 | protein_localization_to_microtubule |
| GO:0035458 | 4 | cellular_response_to_interferon-beta |
| GO:0035519 | 4 | protein_K29-linked_ubiquitination |
| GO:0035584 | 4 | calcium-mediated_signaling_using_intracellular_calcium_source |
| GO:0035588 | 4 | G-protein_coupled_purinergic_receptor_signaling_pathway |
| GO:0035608 | 4 | protein_deglutamylation |
| GO:0035625 | 4 | epidermal_growth_factor-activated_receptor_transactivation_by_G-protein_coupled_receptor_signaling_pathway |
| GO:0035641 | 4 | locomotory_exploration_behavior |
| GO:0035751 | 4 | regulation_of_lysosomal_lumen_pH |
| GO:0035791 | 4 | platelet-derived_growth_factor_receptor-beta_signaling_pathway |
| GO:0035799 | 4 | ureter_maturation |
| GO:0035814 | 4 | negative_regulation_of_renal_sodium_excretion |
| GO:0035864 | 4 | response_to_potassium_ion |
| GO:0035865 | 4 | cellular_response_to_potassium_ion |
| GO:0035988 | 4 | chondrocyte_proliferation |
| GO:0036020 | 4 | endolysosome_membrane |
| GO:0036021 | 4 | endolysosome_lumen |
| GO:0036120 | 4 | cellular_response_to_platelet-derived_growth_factor_stimulus |
| GO:0039528 | 4 | cytoplasmic_pattern_recognition_receptor_signaling_pathway_in_response_to_virus |
| GO:0039531 | 4 | regulation_of_viral-induced_cytoplasmic_pattern_recognition_receptor_signaling_pathway |
| GO:0039535 | 4 | regulation_of_RIG-I_signaling_pathway |
| GO:0042048 | 4 | olfactory_behavior |
| GO:0042092 | 4 | type_2_immune_response |
| GO:0042105 | 4 | alpha-beta_T_cell_receptor_complex |
| GO:0042159 | 4 | lipoprotein_catabolic_process |
| GO:0042219 | 4 | cellular_modified_amino_acid_catabolic_process |
| GO:0042249 | 4 | establishment_of_planar_polarity_of_embryonic_epithelium |
| GO:0042285 | 4 | xylosyltransferase_activity |
| GO:0042296 | 4 | ISG15_ligase_activity |
| GO:0042360 | 4 | vitamin_E_metabolic_process |
| GO:0042396 | 4 | phosphagen_biosynthetic_process |
| GO:0042405 | 4 | nuclear_inclusion_body |
| GO:0042420 | 4 | dopamine_catabolic_process |
| GO:0042421 | 4 | norepinephrine_biosynthetic_process |
| GO:0042441 | 4 | eye_pigment_metabolic_process |
| GO:0042518 | 4 | negative_regulation_of_tyrosine_phosphorylation_of_Stat3_protein |
| GO:0042519 | 4 | regulation_of_tyrosine_phosphorylation_of_Stat4_protein |
| GO:0042520 | 4 | positive_regulation_of_tyrosine_phosphorylation_of_Stat4_protein |
| GO:0042577 | 4 | lipid_phosphatase_activity |
| GO:0042584 | 4 | chromaffin_granule_membrane |
| GO:0042732 | 4 | D-xylose_metabolic_process |
| GO:0042759 | 4 | long-chain_fatty_acid_biosynthetic_process |
| GO:0042762 | 4 | regulation_of_sulfur_metabolic_process |
| GO:0042773 | 4 | ATP_synthesis_coupled_electron_transport |
| GO:0042806 | 4 | fucose_binding |
| GO:0042825 | 4 | TAP_complex |
| GO:0042851 | 4 | L-alanine_metabolic_process |
| GO:0042853 | 4 | L-alanine_catabolic_process |
| GO:0042866 | 4 | pyruvate_biosynthetic_process |
| GO:0042887 | 4 | amide_transmembrane_transporter_activity |
| GO:0042910 | 4 | xenobiotic_transporter_activity |
| GO:0042976 | 4 | activation_of_Janus_kinase_activity |
| GO:0043024 | 4 | ribosomal_small_subunit_binding |
| GO:0043045 | 4 | DNA_methylation_involved_in_embryo_development |
| GO:0043064 | 4 | flagellum_organization |
| GO:0043103 | 4 | hypoxanthine_salvage |
| GO:0043137 | 4 | DNA_replication,_removal_of_RNA_primer |
| GO:0043142 | 4 | single-stranded_DNA-dependent_ATPase_activity |
| GO:0043153 | 4 | entrainment_of_circadian_clock_by_photoperiod |
| GO:0043162 | 4 | ubiquitin-dependent_protein_catabolic_process_via_the_multivesicular_body_sorting_pathway |
| GO:0043184 | 4 | vascular_endothelial_growth_factor_receptor_2_binding |
| GO:0043196 | 4 | varicosity |
| GO:0043201 | 4 | response_to_leucine |
| GO:0043218 | 4 | compact_myelin |
| GO:0043247 | 4 | telomere_maintenance_in_response_to_DNA_damage |
| GO:0043313 | 4 | regulation_of_neutrophil_degranulation |
| GO:0043324 | 4 | pigment_metabolic_process_involved_in_developmental_pigmentation |
| GO:0043353 | 4 | enucleate_erythrocyte_differentiation |
| GO:0043474 | 4 | pigment_metabolic_process_involved_in_pigmentation |
| GO:0043495 | 4 | protein_anchor |
| GO:0043515 | 4 | kinetochore_binding |
| GO:0043950 | 4 | positive_regulation_of_cAMP-mediated_signaling |
| GO:0044183 | 4 | protein_binding_involved_in_protein_folding |
| GO:0044217 | 4 | other_organism_part |
| GO:0044241 | 4 | lipid_digestion |
| GO:0044254 | 4 | multicellular_organismal_protein_catabolic_process |
| GO:0044266 | 4 | multicellular_organismal_macromolecule_catabolic_process |
| GO:0044314 | 4 | protein_K27-linked_ubiquitination |
| GO:0044332 | 4 | Wnt_receptor_signaling_pathway_involved_in_dorsal/ventral_axis_specification |
| GO:0044539 | 4 | long-chain_fatty_acid_import |
| GO:0045040 | 4 | protein_import_into_mitochondrial_outer_membrane |
| GO:0045085 | 4 | negative_regulation_of_interleukin-2_biosynthetic_process |
| GO:0045113 | 4 | regulation_of_integrin_biosynthetic_process |
| GO:0045134 | 4 | uridine-diphosphatase_activity |
| GO:0045159 | 4 | myosin_II_binding |
| GO:0045198 | 4 | establishment_of_epithelial_cell_apical/basal_polarity |
| GO:0045240 | 4 | dihydrolipoyl_dehydrogenase_complex |
| GO:0045257 | 4 | succinate_dehydrogenase_complex_(ubiquinone) |
| GO:0045281 | 4 | succinate_dehydrogenase_complex |
| GO:0045283 | 4 | fumarate_reductase_complex |
| GO:0045322 | 4 | unmethylated_CpG_binding |
| GO:0045345 | 4 | positive_regulation_of_MHC_class_I_biosynthetic_process |
| GO:0045356 | 4 | positive_regulation_of_interferon-alpha_biosynthetic_process |
| GO:0045402 | 4 | regulation_of_interleukin-4_biosynthetic_process |
| GO:0045415 | 4 | negative_regulation_of_interleukin-8_biosynthetic_process |
| GO:0045423 | 4 | regulation_of_granulocyte_macrophage_colony-stimulating_factor_biosynthetic_process |
| GO:0045475 | 4 | locomotor_rhythm |
| GO:0045499 | 4 | chemorepellent_activity |
| GO:0045545 | 4 | syndecan_binding |
| GO:0045569 | 4 | TRAIL_binding |
| GO:0045602 | 4 | negative_regulation_of_endothelial_cell_differentiation |
| GO:0045605 | 4 | negative_regulation_of_epidermal_cell_differentiation |
| GO:0045607 | 4 | regulation_of_auditory_receptor_cell_differentiation |
| GO:0045617 | 4 | negative_regulation_of_keratinocyte_differentiation |
| GO:0045636 | 4 | positive_regulation_of_melanocyte_differentiation |
| GO:0045716 | 4 | positive_regulation_of_low-density_lipoprotein_particle_receptor_biosynthetic_process |
| GO:0045916 | 4 | negative_regulation_of_complement_activation |
| GO:0045919 | 4 | positive_regulation_of_cytolysis |
| GO:0045955 | 4 | negative_regulation_of_calcium_ion-dependent_exocytosis |
| GO:0045991 | 4 | carbon_catabolite_activation_of_transcription |
| GO:0046102 | 4 | inosine_metabolic_process |
| GO:0046322 | 4 | negative_regulation_of_fatty_acid_oxidation |
| GO:0046323 | 4 | glucose_import |
| GO:0046351 | 4 | disaccharide_biosynthetic_process |
| GO:0046391 | 4 | 5-phosphoribose_1-diphosphate_metabolic_process |
| GO:0046398 | 4 | UDP-glucuronate_metabolic_process |
| GO:0046440 | 4 | L-lysine_metabolic_process |
| GO:0046449 | 4 | creatinine_metabolic_process |
| GO:0046471 | 4 | phosphatidylglycerol_metabolic_process |
| GO:0046523 | 4 | S-methyl-5-thioribose-1-phosphate_isomerase_activity |
| GO:0046533 | 4 | negative_regulation_of_photoreceptor_cell_differentiation |
| GO:0046540 | 4 | U4/U6_x_U5_tri-snRNP_complex |
| GO:0046543 | 4 | development_of_secondary_female_sexual_characteristics |
| GO:0046545 | 4 | development_of_primary_female_sexual_characteristics |
| GO:0046683 | 4 | response_to_organophosphorus |
| GO:0046684 | 4 | response_to_pyrethroid |
| GO:0046796 | 4 | viral_genome_transport_in_host_cell |
| GO:0046813 | 4 | virion_attachment,_binding_of_host_cell_surface_receptor |
| GO:0046831 | 4 | regulation_of_RNA_export_from_nucleus |
| GO:0046877 | 4 | regulation_of_saliva_secretion |
| GO:0046882 | 4 | negative_regulation_of_follicle-stimulating_hormone_secretion |
| GO:0046923 | 4 | ER_retention_sequence_binding |
| GO:0046931 | 4 | pore_complex_assembly |
| GO:0046934 | 4 | phosphatidylinositol-4,5-bisphosphate_3-kinase_activity |
| GO:0046935 | 4 | 1-phosphatidylinositol-3-kinase_regulator_activity |
| GO:0046952 | 4 | ketone_body_catabolic_process |
| GO:0046977 | 4 | TAP_binding |
| GO:0046978 | 4 | TAP1_binding |
| GO:0046979 | 4 | TAP2_binding |
| GO:0047023 | 4 | androsterone_dehydrogenase_activity |
| GO:0047057 | 4 | vitamin-K-epoxide_reductase_(warfarin-sensitive)_activity |
| GO:0047115 | 4 | trans-1,2-dihydrobenzene-1,2-diol_dehydrogenase_activity |
| GO:0047184 | 4 | 1-acylglycerophosphocholine_O-acyltransferase_activity |
| GO:0047238 | 4 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan_4-beta-N-acetylgalactosaminyltransferase_activity |
| GO:0047631 | 4 | ADP-ribose_diphosphatase_activity |
| GO:0047961 | 4 | glycine_N-acyltransferase_activity |
| GO:0048026 | 4 | positive_regulation_of_nuclear_mRNA_splicing,_via_spliceosome |
| GO:0048263 | 4 | determination_of_dorsal_identity |
| GO:0048312 | 4 | intracellular_distribution_of_mitochondria |
| GO:0048333 | 4 | mesodermal_cell_differentiation |
| GO:0048343 | 4 | paraxial_mesodermal_cell_fate_commitment |
| GO:0048386 | 4 | positive_regulation_of_retinoic_acid_receptor_signaling_pathway |
| GO:0048495 | 4 | Roundabout_binding |
| GO:0048496 | 4 | maintenance_of_organ_identity |
| GO:0048548 | 4 | regulation_of_pinocytosis |
| GO:0048552 | 4 | regulation_of_metalloenzyme_activity |
| GO:0048554 | 4 | positive_regulation_of_metalloenzyme_activity |
| GO:0048671 | 4 | negative_regulation_of_collateral_sprouting |
| GO:0048672 | 4 | positive_regulation_of_collateral_sprouting |
| GO:0048680 | 4 | positive_regulation_of_axon_regeneration |
| GO:0048733 | 4 | sebaceous_gland_development |
| GO:0048755 | 4 | branching_morphogenesis_of_a_nerve |
| GO:0048799 | 4 | organ_maturation |
| GO:0048861 | 4 | leukemia_inhibitory_factor_signaling_pathway |
| GO:0050051 | 4 | leukotriene-B4_20-monooxygenase_activity |
| GO:0050501 | 4 | hyaluronan_synthase_activity |
| GO:0050510 | 4 | N-acetylgalactosaminyl-proteoglycan_3-beta-glucuronosyltransferase_activity |
| GO:0050544 | 4 | arachidonic_acid_binding |
| GO:0050650 | 4 | chondroitin_sulfate_proteoglycan_biosynthetic_process |
| GO:0050689 | 4 | negative_regulation_of_defense_response_to_virus_by_host |
| GO:0050693 | 4 | LBD_domain_binding |
| GO:0050694 | 4 | galactose_3-O-sulfotransferase_activity |
| GO:0050702 | 4 | interleukin-1_beta_secretion |
| GO:0050803 | 4 | regulation_of_synapse_structure_and_activity |
| GO:0050815 | 4 | phosphoserine_binding |
| GO:0050849 | 4 | negative_regulation_of_calcium-mediated_signaling |
| GO:0050897 | 4 | cobalt_ion_binding |
| GO:0050916 | 4 | sensory_perception_of_sweet_taste |
| GO:0050923 | 4 | regulation_of_negative_chemotaxis |
| GO:0050942 | 4 | positive_regulation_of_pigment_cell_differentiation |
| GO:0050966 | 4 | detection_of_mechanical_stimulus_involved_in_sensory_perception_of_pain |
| GO:0051026 | 4 | chiasma_assembly |
| GO:0051045 | 4 | negative_regulation_of_membrane_protein_ectodomain_proteolysis |
| GO:0051136 | 4 | regulation_of_NK_T_cell_differentiation |
| GO:0051138 | 4 | positive_regulation_of_NK_T_cell_differentiation |
| GO:0051152 | 4 | positive_regulation_of_smooth_muscle_cell_differentiation |
| GO:0051156 | 4 | glucose_6-phosphate_metabolic_process |
| GO:0051177 | 4 | meiotic_sister_chromatid_cohesion |
| GO:0051185 | 4 | coenzyme_transporter_activity |
| GO:0051208 | 4 | sequestering_of_calcium_ion |
| GO:0051256 | 4 | spindle_midzone_assembly_involved_in_mitosis |
| GO:0051299 | 4 | centrosome_separation |
| GO:0051325 | 4 | interphase |
| GO:0051380 | 4 | norepinephrine_binding |
| GO:0051425 | 4 | PTB_domain_binding |
| GO:0051434 | 4 | BH3_domain_binding |
| GO:0051503 | 4 | adenine_nucleotide_transport |
| GO:0051573 | 4 | negative_regulation_of_histone_H3-K9_methylation |
| GO:0051575 | 4 | 5'-deoxyribose-5-phosphate_lyase_activity |
| GO:0051599 | 4 | response_to_hydrostatic_pressure |
| GO:0051665 | 4 | membrane_raft_localization |
| GO:0051731 | 4 | polynucleotide_5'-hydroxyl-kinase_activity |
| GO:0051875 | 4 | pigment_granule_localization |
| GO:0051919 | 4 | positive_regulation_of_fibrinolysis |
| GO:0051932 | 4 | synaptic_transmission,_GABAergic |
| GO:0051957 | 4 | positive_regulation_of_amino_acid_transport |
| GO:0051964 | 4 | negative_regulation_of_synapse_assembly |
| GO:0052033 | 4 | pathogen-associated_molecular_pattern_dependent_induction_by_symbiont_of_host_innate_immune_response |
| GO:0052166 | 4 | positive_regulation_by_symbiont_of_host_innate_immune_response |
| GO:0052167 | 4 | modulation_by_symbiont_of_host_innate_immune_response |
| GO:0052169 | 4 | pathogen-associated_molecular_pattern_dependent_modulation_by_symbiont_of_host_innate_immune_response |
| GO:0052257 | 4 | pathogen-associated_molecular_pattern_dependent_induction_by_organism_of_innate_immune_response_of_other_organism_involved_in_symbiotic_interaction |
| GO:0052305 | 4 | positive_regulation_by_organism_of_innate_immune_response_in_other_organism_involved_in_symbiotic_interaction |
| GO:0052306 | 4 | modulation_by_organism_of_innate_immune_response_in_other_organism_involved_in_symbiotic_interaction |
| GO:0052308 | 4 | pathogen-associated_molecular_pattern_dependent_modulation_by_organism_of_innate_immune_response_in_other_organism_involved_in_symbiotic_interaction |
| GO:0052650 | 4 | NADP-retinol_dehydrogenase_activity |
| GO:0052658 | 4 | inositol-1,4,5-trisphosphate_5-phosphatase_activity |
| GO:0052794 | 4 | exo-alpha-(2->3)-sialidase_activity |
| GO:0052795 | 4 | exo-alpha-(2->6)-sialidase_activity |
| GO:0052796 | 4 | exo-alpha-(2->8)-sialidase_activity |
| GO:0052813 | 4 | phosphatidylinositol_bisphosphate_kinase_activity |
| GO:0052840 | 4 | inositol_diphosphate_tetrakisphosphate_diphosphatase_activity |
| GO:0052841 | 4 | inositol_bisdiphosphate_tetrakisphosphate_diphosphatase_activity |
| GO:0052842 | 4 | inositol_diphosphate_pentakisphosphate_diphosphatase_activity |
| GO:0052843 | 4 | inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate_diphosphatase_activity |
| GO:0052844 | 4 | inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate_diphosphatase_activity |
| GO:0052845 | 4 | inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate_diphosphatase_activity |
| GO:0052846 | 4 | inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate_1-diphosphatase_activity |
| GO:0052847 | 4 | inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate_5-diphosphatase_activity |
| GO:0052848 | 4 | inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate_5-diphosphatase_activity |
| GO:0055005 | 4 | ventricular_cardiac_myofibril_development |
| GO:0055009 | 4 | atrial_cardiac_muscle_tissue_morphogenesis |
| GO:0055064 | 4 | chloride_ion_homeostasis |
| GO:0055083 | 4 | monovalent_inorganic_anion_homeostasis |
| GO:0055106 | 4 | ubiquitin-protein_ligase_regulator_activity |
| GO:0055119 | 4 | relaxation_of_cardiac_muscle |
| GO:0060011 | 4 | Sertoli_cell_proliferation |
| GO:0060018 | 4 | astrocyte_fate_commitment |
| GO:0060044 | 4 | negative_regulation_of_cardiac_muscle_cell_proliferation |
| GO:0060187 | 4 | cell_pole |
| GO:0060203 | 4 | clathrin_sculpted_glutamate_transport_vesicle_membrane |
| GO:0060214 | 4 | endocardium_formation |
| GO:0060267 | 4 | positive_regulation_of_respiratory_burst |
| GO:0060285 | 4 | ciliary_cell_motility |
| GO:0060287 | 4 | epithelial_cilium_movement_involved_in_determination_of_left/right_asymmetry |
| GO:0060298 | 4 | positive_regulation_of_sarcomere_organization |
| GO:0060312 | 4 | regulation_of_blood_vessel_remodeling |
| GO:0060316 | 4 | positive_regulation_of_ryanodine-sensitive_calcium-release_channel_activity |
| GO:0060318 | 4 | definitive_erythrocyte_differentiation |
| GO:0060331 | 4 | negative_regulation_of_response_to_interferon-gamma |
| GO:0060332 | 4 | positive_regulation_of_response_to_interferon-gamma |
| GO:0060335 | 4 | positive_regulation_of_interferon-gamma-mediated_signaling_pathway |
| GO:0060336 | 4 | negative_regulation_of_interferon-gamma-mediated_signaling_pathway |
| GO:0060340 | 4 | positive_regulation_of_type_I_interferon-mediated_signaling_pathway |
| GO:0060364 | 4 | frontal_suture_morphogenesis |
| GO:0060398 | 4 | regulation_of_growth_hormone_receptor_signaling_pathway |
| GO:0060421 | 4 | positive_regulation_of_heart_growth |
| GO:0060430 | 4 | lung_saccule_development |
| GO:0060453 | 4 | regulation_of_gastric_acid_secretion |
| GO:0060523 | 4 | prostate_epithelial_cord_elongation |
| GO:0060528 | 4 | secretory_columnal_luminar_epithelial_cell_differentiation_involved_in_prostate_glandular_acinus_development |
| GO:0060535 | 4 | trachea_cartilage_morphogenesis |
| GO:0060544 | 4 | regulation_of_necroptosis |
| GO:0060564 | 4 | negative_regulation_of_mitotic_anaphase-promoting_complex_activity |
| GO:0060586 | 4 | multicellular_organismal_iron_ion_homeostasis |
| GO:0060596 | 4 | mammary_placode_formation |
| GO:0060628 | 4 | regulation_of_ER_to_Golgi_vesicle-mediated_transport |
| GO:0060677 | 4 | ureteric_bud_elongation |
| GO:0060686 | 4 | negative_regulation_of_prostatic_bud_formation |
| GO:0060708 | 4 | spongiotrophoblast_differentiation |
| GO:0060737 | 4 | prostate_gland_morphogenetic_growth |
| GO:0060743 | 4 | epithelial_cell_maturation_involved_in_prostate_gland_development |
| GO:0060744 | 4 | mammary_gland_branching_involved_in_thelarche |
| GO:0060745 | 4 | mammary_gland_branching_involved_in_pregnancy |
| GO:0060762 | 4 | regulation_of_branching_involved_in_mammary_gland_duct_morphogenesis |
| GO:0060763 | 4 | mammary_duct_terminal_end_bud_growth |
| GO:0060841 | 4 | venous_blood_vessel_development |
| GO:0060896 | 4 | neural_plate_pattern_specification |
| GO:0060911 | 4 | cardiac_cell_fate_commitment |
| GO:0060913 | 4 | cardiac_cell_fate_determination |
| GO:0060923 | 4 | cardiac_muscle_cell_fate_commitment |
| GO:0060977 | 4 | coronary_vasculature_morphogenesis |
| GO:0061002 | 4 | negative_regulation_of_dendritic_spine_morphogenesis |
| GO:0061014 | 4 | positive_regulation_of_mRNA_catabolic_process |
| GO:0061028 | 4 | establishment_of_endothelial_barrier |
| GO:0061030 | 4 | epithelial_cell_differentiation_involved_in_mammary_gland_alveolus_development |
| GO:0061038 | 4 | uterus_morphogenesis |
| GO:0061043 | 4 | regulation_of_vascular_wound_healing |
| GO:0061045 | 4 | negative_regulation_of_wound_healing |
| GO:0061183 | 4 | regulation_of_dermatome_development |
| GO:0061299 | 4 | retina_vasculature_morphogenesis_in_camera-type_eye |
| GO:0061316 | 4 | canonical_Wnt_receptor_signaling_pathway_involved_in_heart_development |
| GO:0061384 | 4 | heart_trabecula_morphogenesis |
| GO:0061418 | 4 | regulation_of_transcription_from_RNA_polymerase_II_promoter_in_response_to_hypoxia |
| GO:0065001 | 4 | specification_of_axis_polarity |
| GO:0070050 | 4 | neuron_homeostasis |
| GO:0070053 | 4 | thrombospondin_receptor_activity |
| GO:0070097 | 4 | delta-catenin_binding |
| GO:0070100 | 4 | negative_regulation_of_chemokine-mediated_signaling_pathway |
| GO:0070255 | 4 | regulation_of_mucus_secretion |
| GO:0070305 | 4 | response_to_cGMP |
| GO:0070309 | 4 | lens_fiber_cell_morphogenesis |
| GO:0070316 | 4 | regulation_of_G0_to_G1_transition |
| GO:0070324 | 4 | thyroid_hormone_binding |
| GO:0070326 | 4 | very-low-density_lipoprotein_particle_receptor_binding |
| GO:0070431 | 4 | nucleotide-binding_oligomerization_domain_containing_2_signaling_pathway |
| GO:0070546 | 4 | L-phenylalanine_aminotransferase_activity |
| GO:0070552 | 4 | BRISC_complex |
| GO:0070562 | 4 | regulation_of_vitamin_D_receptor_signaling_pathway |
| GO:0070572 | 4 | positive_regulation_of_neuron_projection_regeneration |
| GO:0070574 | 4 | cadmium_ion_transmembrane_transport |
| GO:0070579 | 4 | methylcytosine_dioxygenase_activity |
| GO:0070669 | 4 | response_to_interleukin-2 |
| GO:0070671 | 4 | response_to_interleukin-12 |
| GO:0070697 | 4 | activin_receptor_binding |
| GO:0070728 | 4 | leucine_binding |
| GO:0070815 | 4 | peptidyl-lysine_5-dioxygenase_activity |
| GO:0070852 | 4 | cell_body_fiber |
| GO:0070876 | 4 | SOSS_complex |
| GO:0070942 | 4 | neutrophil_mediated_cytotoxicity |
| GO:0070943 | 4 | neutrophil_mediated_killing_of_symbiont_cell |
| GO:0071027 | 4 | nuclear_RNA_surveillance |
| GO:0071028 | 4 | nuclear_mRNA_surveillance |
| GO:0071072 | 4 | negative_regulation_of_phospholipid_biosynthetic_process |
| GO:0071141 | 4 | SMAD_protein_complex |
| GO:0071208 | 4 | histone_pre-mRNA_DCP_binding |
| GO:0071286 | 4 | cellular_response_to_magnesium_ion |
| GO:0071287 | 4 | cellular_response_to_manganese_ion |
| GO:0071316 | 4 | cellular_response_to_nicotine |
| GO:0071340 | 4 | skeletal_muscle_acetylcholine-gated_channel_clustering |
| GO:0071395 | 4 | cellular_response_to_jasmonic_acid_stimulus |
| GO:0071460 | 4 | cellular_response_to_cell-matrix_adhesion |
| GO:0071462 | 4 | cellular_response_to_water |
| GO:0071464 | 4 | cellular_response_to_hydrostatic_pressure |
| GO:0071468 | 4 | cellular_response_to_acidity |
| GO:0071474 | 4 | cellular_hyperosmotic_response |
| GO:0071476 | 4 | cellular_hypotonic_response |
| GO:0071547 | 4 | piP-body |
| GO:0071553 | 4 | G-protein_coupled_pyrimidinergic_nucleotide_receptor_activity |
| GO:0071569 | 4 | protein_ufmylation |
| GO:0071599 | 4 | otic_vesicle_development |
| GO:0071635 | 4 | negative_regulation_of_transforming_growth_factor_beta_production |
| GO:0071679 | 4 | commissural_neuron_axon_guidance |
| GO:0071684 | 4 | organism_emergence_from_protective_structure |
| GO:0071711 | 4 | basement_membrane_organization |
| GO:0071723 | 4 | lipopeptide_binding |
| GO:0071786 | 4 | endoplasmic_reticulum_tubular_network_organization |
| GO:0071801 | 4 | regulation_of_podosome_assembly |
| GO:0071875 | 4 | adrenergic_receptor_signaling_pathway |
| GO:0071985 | 4 | multivesicular_body_sorting_pathway |
| GO:0072011 | 4 | glomerular_endothelium_development |
| GO:0072021 | 4 | ascending_thin_limb_development |
| GO:0072025 | 4 | distal_convoluted_tubule_development |
| GO:0072049 | 4 | comma-shaped_body_morphogenesis |
| GO:0072070 | 4 | loop_of_Henle_development |
| GO:0072089 | 4 | stem_cell_proliferation |
| GO:0072095 | 4 | regulation_of_branch_elongation_involved_in_ureteric_bud_branching |
| GO:0072106 | 4 | regulation_of_ureteric_bud_formation |
| GO:0072107 | 4 | positive_regulation_of_ureteric_bud_formation |
| GO:0072108 | 4 | positive_regulation_of_mesenchymal_to_epithelial_transition_involved_in_metanephros_morphogenesis |
| GO:0072143 | 4 | mesangial_cell_development |
| GO:0072170 | 4 | metanephric_tubule_development |
| GO:0072171 | 4 | mesonephric_tubule_morphogenesis |
| GO:0072173 | 4 | metanephric_tubule_morphogenesis |
| GO:0072176 | 4 | nephric_duct_development |
| GO:0072177 | 4 | mesonephric_duct_development |
| GO:0072179 | 4 | nephric_duct_formation |
| GO:0072180 | 4 | mesonephric_duct_morphogenesis |
| GO:0072199 | 4 | regulation_of_mesenchymal_cell_proliferation_involved_in_ureter_development |
| GO:0072218 | 4 | metanephric_ascending_thin_limb_development |
| GO:0072221 | 4 | metanephric_distal_convoluted_tubule_development |
| GO:0072282 | 4 | metanephric_nephron_tubule_morphogenesis |
| GO:0072284 | 4 | metanephric_S-shaped_body_morphogenesis |
| GO:0072289 | 4 | metanephric_nephron_tubule_formation |
| GO:0072367 | 4 | regulation_of_lipid_transport_by_regulation_of_transcription_from_RNA_polymerase_II_promoter |
| GO:0072487 | 4 | MSL_complex |
| GO:0072526 | 4 | pyridine-containing_compound_catabolic_process |
| GO:0072578 | 4 | neurotransmitter-gated_ion_channel_clustering |
| GO:0072584 | 4 | caveolin-mediated_endocytosis |
| GO:0072698 | 4 | protein_localization_to_microtubule_cytoskeleton |
| GO:0080030 | 4 | methyl_indole-3-acetate_esterase_activity |
| GO:0080031 | 4 | methyl_salicylate_esterase_activity |
| GO:0080032 | 4 | methyl_jasmonate_esterase_activity |
| GO:0086005 | 4 | regulation_of_ventricular_cardiac_muscle_cell_action_potential |
| GO:0090005 | 4 | negative_regulation_of_establishment_of_protein_localization_in_plasma_membrane |
| GO:0090116 | 4 | C-5_methylation_of_cytosine |
| GO:0090141 | 4 | positive_regulation_of_mitochondrial_fission |
| GO:0090170 | 4 | regulation_of_Golgi_inheritance |
| GO:0090185 | 4 | negative_regulation_of_kidney_development |
| GO:0090191 | 4 | negative_regulation_of_branching_involved_in_ureteric_bud_morphogenesis |
| GO:0090193 | 4 | positive_regulation_of_glomerulus_development |
| GO:0090209 | 4 | negative_regulation_of_triglyceride_metabolic_process |
| GO:0090232 | 4 | positive_regulation_of_spindle_checkpoint |
| GO:0090266 | 4 | regulation_of_mitotic_cell_cycle_spindle_assembly_checkpoint |
| GO:0090273 | 4 | regulation_of_somatostatin_secretion |
| GO:0090308 | 4 | regulation_of_methylation-dependent_chromatin_silencing |
| GO:0097029 | 4 | mature_dendritic_cell_differentiation |
| GO:0097056 | 4 | selenocysteinyl-tRNA(Sec)_biosynthetic_process |
| GO:0097084 | 4 | vascular_smooth_muscle_cell_development |
| GO:0097119 | 4 | postsynaptic_density_protein_95_clustering |
| GO:0097136 | 4 | Bcl-2_family_protein_complex |
| GO:0097178 | 4 | ruffle_assembly |
| GO:1900103 | 4 | positive_regulation_of_endoplasmic_reticulum_unfolded_protein_response |
| GO:1900107 | 4 | regulation_of_nodal_signaling_pathway |
| GO:1900120 | 4 | regulation_of_receptor_binding |
| GO:1900125 | 4 | regulation_of_hyaluronan_biosynthetic_process |
| GO:1900145 | 4 | regulation_of_nodal_signaling_pathway_involved_in_determination_of_left/right_asymmetry |
| GO:1900619 | 4 | acetate_ester_metabolic_process |
| GO:2000015 | 4 | regulation_of_determination_of_dorsal_identity |
| GO:2000043 | 4 | regulation_of_cardiac_cell_fate_specification |
| GO:2000052 | 4 | positive_regulation_of_non-canonical_Wnt_receptor_signaling_pathway |
| GO:2000053 | 4 | regulation_of_Wnt_receptor_signaling_pathway_involved_in_dorsal/ventral_axis_specification |
| GO:2000065 | 4 | negative_regulation_of_cortisol_biosynthetic_process |
| GO:2000105 | 4 | positive_regulation_of_DNA-dependent_DNA_replication |
| GO:2000110 | 4 | negative_regulation_of_macrophage_apoptotic_process |
| GO:2000111 | 4 | positive_regulation_of_macrophage_apoptotic_process |
| GO:2000121 | 4 | regulation_of_removal_of_superoxide_radicals |
| GO:2000136 | 4 | regulation_of_cell_proliferation_involved_in_heart_morphogenesis |
| GO:2000171 | 4 | negative_regulation_of_dendrite_development |
| GO:2000188 | 4 | regulation_of_cholesterol_homeostasis |
| GO:2000258 | 4 | negative_regulation_of_protein_activation_cascade |
| GO:2000328 | 4 | regulation_of_T-helper_17_cell_lineage_commitment |
| GO:2000330 | 4 | positive_regulation_of_T-helper_17_cell_lineage_commitment |
| GO:2000345 | 4 | regulation_of_hepatocyte_proliferation |
| GO:2000353 | 4 | positive_regulation_of_endothelial_cell_apoptotic_process |
| GO:2000482 | 4 | regulation_of_interleukin-8_secretion |
| GO:2000484 | 4 | positive_regulation_of_interleukin-8_secretion |
| GO:2000589 | 4 | regulation_of_metanephric_mesenchymal_cell_migration |
| GO:2000668 | 4 | regulation_of_dendritic_cell_apoptotic_process |
| GO:2000774 | 4 | positive_regulation_of_cellular_senescence |
| GO:2000778 | 4 | positive_regulation_of_interleukin-6_secretion |
| GO:2000780 | 4 | negative_regulation_of_double-strand_break_repair |
| GO:2000810 | 4 | regulation_of_tight_junction_assembly |
| GO:2000831 | 4 | regulation_of_steroid_hormone_secretion |
| GO:2001023 | 4 | regulation_of_response_to_drug |
| GO:2001185 | 4 | regulation_of_CD8-positive,_alpha-beta_T_cell_activation |
| GO:2001198 | 4 | regulation_of_dendritic_cell_differentiation |
| GO:2001222 | 4 | regulation_of_neuron_migration |
| GO:0000026 | 3 | alpha-1,2-mannosyltransferase_activity |
| GO:0000033 | 3 | alpha-1,3-mannosyltransferase_activity |
| GO:0000046 | 3 | autophagic_vacuole_fusion |
| GO:0000103 | 3 | sulfate_assimilation |
| GO:0000120 | 3 | RNA_polymerase_I_transcription_factor_complex |
| GO:0000124 | 3 | SAGA_complex |
| GO:0000214 | 3 | tRNA-intron_endonuclease_complex |
| GO:0000253 | 3 | 3-keto_sterol_reductase_activity |
| GO:0000309 | 3 | nicotinamide-nucleotide_adenylyltransferase_activity |
| GO:0000318 | 3 | protein-methionine-R-oxide_reductase_activity |
| GO:0000395 | 3 | nuclear_mRNA_5'-splice_site_recognition |
| GO:0000404 | 3 | loop_DNA_binding |
| GO:0000430 | 3 | regulation_of_transcription_from_RNA_polymerase_II_promoter_by_glucose |
| GO:0000432 | 3 | positive_regulation_of_transcription_from_RNA_polymerase_II_promoter_by_glucose |
| GO:0000462 | 3 | maturation_of_SSU-rRNA_from_tricistronic_rRNA_transcript_(SSU-rRNA,_5.8S_rRNA,_LSU-rRNA) |
| GO:0000463 | 3 | maturation_of_LSU-rRNA_from_tricistronic_rRNA_transcript_(SSU-rRNA,_5.8S_rRNA,_LSU-rRNA) |
| GO:0000470 | 3 | maturation_of_LSU-rRNA |
| GO:0000491 | 3 | small_nucleolar_ribonucleoprotein_complex_assembly |
| GO:0000492 | 3 | box_C/D_snoRNP_assembly |
| GO:0000788 | 3 | nuclear_nucleosome |
| GO:0000942 | 3 | condensed_nuclear_chromosome_outer_kinetochore |
| GO:0001030 | 3 | RNA_polymerase_III_type_1_promoter_DNA_binding |
| GO:0001031 | 3 | RNA_polymerase_III_type_2_promoter_DNA_binding |
| GO:0001032 | 3 | RNA_polymerase_III_type_3_promoter_DNA_binding |
| GO:0001302 | 3 | replicative_cell_aging |
| GO:0001306 | 3 | age-dependent_response_to_oxidative_stress |
| GO:0001543 | 3 | ovarian_follicle_rupture |
| GO:0001554 | 3 | luteolysis |
| GO:0001560 | 3 | regulation_of_cell_growth_by_extracellular_stimulus |
| GO:0001591 | 3 | dopamine_receptor_activity,_coupled_via_Gi/Go |
| GO:0001602 | 3 | pancreatic_polypeptide_receptor_activity |
| GO:0001632 | 3 | leukotriene_B4_receptor_activity |
| GO:0001665 | 3 | alpha-N-acetylgalactosaminide_alpha-2,6-sialyltransferase_activity |
| GO:0001674 | 3 | female_germ_cell_nucleus |
| GO:0001738 | 3 | morphogenesis_of_a_polarized_epithelium |
| GO:0001743 | 3 | optic_placode_formation |
| GO:0001766 | 3 | membrane_raft_polarization |
| GO:0001780 | 3 | neutrophil_homeostasis |
| GO:0001781 | 3 | neutrophil_apoptotic_process |
| GO:0001783 | 3 | B_cell_apoptotic_process |
| GO:0001796 | 3 | regulation_of_type_IIa_hypersensitivity |
| GO:0001798 | 3 | positive_regulation_of_type_IIa_hypersensitivity |
| GO:0001807 | 3 | regulation_of_type_IV_hypersensitivity |
| GO:0001826 | 3 | inner_cell_mass_cell_differentiation |
| GO:0001832 | 3 | blastocyst_growth |
| GO:0001839 | 3 | neural_plate_morphogenesis |
| GO:0001842 | 3 | neural_fold_formation |
| GO:0001847 | 3 | opsonin_receptor_activity |
| GO:0001849 | 3 | complement_component_C1q_binding |
| GO:0001883 | 3 | purine_nucleoside_binding |
| GO:0001897 | 3 | cytolysis_by_symbiont_of_host_cells |
| GO:0001907 | 3 | killing_by_symbiont_of_host_cells |
| GO:0001915 | 3 | negative_regulation_of_T_cell_mediated_cytotoxicity |
| GO:0001922 | 3 | B-1_B_cell_homeostasis |
| GO:0001950 | 3 | plasma_membrane_enriched_fraction |
| GO:0001956 | 3 | positive_regulation_of_neurotransmitter_secretion |
| GO:0001973 | 3 | adenosine_receptor_signaling_pathway |
| GO:0002005 | 3 | angiotensin_catabolic_process_in_blood |
| GO:0002018 | 3 | renin-angiotensin_regulation_of_aldosterone_production |
| GO:0002019 | 3 | regulation_of_renal_output_by_angiotensin |
| GO:0002023 | 3 | reduction_of_food_intake_in_response_to_dietary_excess |
| GO:0002025 | 3 | vasodilation_by_norepinephrine-epinephrine_involved_in_regulation_of_systemic_arterial_blood_pressure |
| GO:0002032 | 3 | desensitization_of_G-protein_coupled_receptor_protein_signaling_pathway_by_arrestin |
| GO:0002034 | 3 | regulation_of_blood_vessel_size_by_renin-angiotensin |
| GO:0002060 | 3 | purine_base_binding |
| GO:0002175 | 3 | protein_localization_to_paranode_region_of_axon |
| GO:0002193 | 3 | MAML1-RBP-Jkappa-_ICN1_complex |
| GO:0002248 | 3 | connective_tissue_replacement_involved_in_inflammatory_response_wound_healing |
| GO:0002295 | 3 | T-helper_cell_lineage_commitment |
| GO:0002323 | 3 | natural_killer_cell_activation_involved_in_immune_response |
| GO:0002331 | 3 | pre-B_cell_allelic_exclusion |
| GO:0002361 | 3 | CD4-positive,_CD25-positive,_alpha-beta_regulatory_T_cell_differentiation |
| GO:0002369 | 3 | T_cell_cytokine_production |
| GO:0002430 | 3 | complement_receptor_mediated_signaling_pathway |
| GO:0002442 | 3 | serotonin_secretion_involved_in_inflammatory_response |
| GO:0002513 | 3 | tolerance_induction_to_self_antigen |
| GO:0002554 | 3 | serotonin_secretion_by_platelet |
| GO:0002636 | 3 | positive_regulation_of_germinal_center_formation |
| GO:0002677 | 3 | negative_regulation_of_chronic_inflammatory_response |
| GO:0002693 | 3 | positive_regulation_of_cellular_extravasation |
| GO:0002716 | 3 | negative_regulation_of_natural_killer_cell_mediated_immunity |
| GO:0002774 | 3 | Fc_receptor_mediated_inhibitory_signaling_pathway |
| GO:0002826 | 3 | negative_regulation_of_T-helper_1_type_immune_response |
| GO:0002865 | 3 | negative_regulation_of_acute_inflammatory_response_to_antigenic_stimulus |
| GO:0002874 | 3 | regulation_of_chronic_inflammatory_response_to_antigenic_stimulus |
| GO:0002888 | 3 | positive_regulation_of_myeloid_leukocyte_mediated_immunity |
| GO:0002892 | 3 | regulation_of_type_II_hypersensitivity |
| GO:0002894 | 3 | positive_regulation_of_type_II_hypersensitivity |
| GO:0002905 | 3 | regulation_of_mature_B_cell_apoptotic_process |
| GO:0002906 | 3 | negative_regulation_of_mature_B_cell_apoptotic_process |
| GO:0003072 | 3 | renal_control_of_peripheral_vascular_resistance_involved_in_regulation_of_systemic_arterial_blood_pressure |
| GO:0003095 | 3 | pressure_natriuresis |
| GO:0003096 | 3 | renal_sodium_ion_transport |
| GO:0003104 | 3 | positive_regulation_of_glomerular_filtration |
| GO:0003105 | 3 | negative_regulation_of_glomerular_filtration |
| GO:0003127 | 3 | detection_of_nodal_flow |
| GO:0003130 | 3 | BMP_signaling_pathway_involved_in_heart_induction |
| GO:0003136 | 3 | negative_regulation_of_heart_induction_by_canonical_Wnt_receptor_signaling_pathway |
| GO:0003140 | 3 | determination_of_left/right_asymmetry_in_lateral_mesoderm |
| GO:0003158 | 3 | endothelium_development |
| GO:0003166 | 3 | bundle_of_His_development |
| GO:0003170 | 3 | heart_valve_development |
| GO:0003229 | 3 | ventricular_cardiac_muscle_tissue_development |
| GO:0003253 | 3 | cardiac_neural_crest_cell_migration_involved_in_outflow_tract_morphogenesis |
| GO:0003256 | 3 | regulation_of_transcription_from_RNA_polymerase_II_promoter_involved_in_myocardial_precursor_cell_differentiation |
| GO:0003257 | 3 | positive_regulation_of_transcription_from_RNA_polymerase_II_promoter_involved_in_myocardial_precursor_cell_differentiation |
| GO:0003309 | 3 | type_B_pancreatic_cell_differentiation |
| GO:0003340 | 3 | negative_regulation_of_mesenchymal_to_epithelial_transition_involved_in_metanephros_morphogenesis |
| GO:0003406 | 3 | retinal_pigment_epithelium_development |
| GO:0003413 | 3 | chondrocyte_differentiation_involved_in_endochondral_bone_morphogenesis |
| GO:0003726 | 3 | double-stranded_RNA_adenosine_deaminase_activity |
| GO:0003827 | 3 | alpha-1,3-mannosylglycoprotein_2-beta-N-acetylglucosaminyltransferase_activity |
| GO:0003829 | 3 | beta-1,3-galactosyl-O-glycosyl-glycoprotein_beta-1,6-N-acetylglucosaminyltransferase_activity |
| GO:0003836 | 3 | beta-galactoside_(CMP)_alpha-2,3-sialyltransferase_activity |
| GO:0003865 | 3 | 3-oxo-5-alpha-steroid_4-dehydrogenase_activity |
| GO:0003872 | 3 | 6-phosphofructokinase_activity |
| GO:0003876 | 3 | AMP_deaminase_activity |
| GO:0003909 | 3 | DNA_ligase_activity |
| GO:0003910 | 3 | DNA_ligase_(ATP)_activity |
| GO:0003941 | 3 | L-serine_ammonia-lyase_activity |
| GO:0003951 | 3 | NAD+_kinase_activity |
| GO:0003953 | 3 | NAD+_nucleosidase_activity |
| GO:0003980 | 3 | UDP-glucose:glycoprotein_glucosyltransferase_activity |
| GO:0003985 | 3 | acetyl-CoA_C-acetyltransferase_activity |
| GO:0003986 | 3 | acetyl-CoA_hydrolase_activity |
| GO:0003987 | 3 | acetate-CoA_ligase_activity |
| GO:0003989 | 3 | acetyl-CoA_carboxylase_activity |
| GO:0003994 | 3 | aconitate_hydratase_activity |
| GO:0004013 | 3 | adenosylhomocysteinase_activity |
| GO:0004060 | 3 | arylamine_N-acetyltransferase_activity |
| GO:0004104 | 3 | cholinesterase_activity |
| GO:0004117 | 3 | calmodulin-dependent_cyclic-nucleotide_phosphodiesterase_activity |
| GO:0004118 | 3 | cGMP-stimulated_cyclic-nucleotide_phosphodiesterase_activity |
| GO:0004142 | 3 | diacylglycerol_cholinephosphotransferase_activity |
| GO:0004157 | 3 | dihydropyrimidinase_activity |
| GO:0004167 | 3 | dopachrome_isomerase_activity |
| GO:0004307 | 3 | ethanolaminephosphotransferase_activity |
| GO:0004329 | 3 | formate-tetrahydrofolate_ligase_activity |
| GO:0004332 | 3 | fructose-bisphosphate_aldolase_activity |
| GO:0004346 | 3 | glucose-6-phosphatase_activity |
| GO:0004351 | 3 | glutamate_decarboxylase_activity |
| GO:0004367 | 3 | glycerol-3-phosphate_dehydrogenase_[NAD+]_activity |
| GO:0004416 | 3 | hydroxyacylglutathione_hydrolase_activity |
| GO:0004449 | 3 | isocitrate_dehydrogenase_(NAD+)_activity |
| GO:0004461 | 3 | lactose_synthase_activity |
| GO:0004464 | 3 | leukotriene-C4_synthase_activity |
| GO:0004465 | 3 | lipoprotein_lipase_activity |
| GO:0004466 | 3 | long-chain-acyl-CoA_dehydrogenase_activity |
| GO:0004476 | 3 | mannose-6-phosphate_isomerase_activity |
| GO:0004477 | 3 | methenyltetrahydrofolate_cyclohydrolase_activity |
| GO:0004500 | 3 | dopamine_beta-monooxygenase_activity |
| GO:0004505 | 3 | phenylalanine_4-monooxygenase_activity |
| GO:0004525 | 3 | ribonuclease_III_activity |
| GO:0004556 | 3 | alpha-amylase_activity |
| GO:0004560 | 3 | alpha-L-fucosidase_activity |
| GO:0004565 | 3 | beta-galactosidase_activity |
| GO:0004566 | 3 | beta-glucuronidase_activity |
| GO:0004582 | 3 | dolichyl-phosphate_beta-D-mannosyltransferase_activity |
| GO:0004591 | 3 | oxoglutarate_dehydrogenase_(succinyl-transferring)_activity |
| GO:0004660 | 3 | protein_farnesyltransferase_activity |
| GO:0004687 | 3 | myosin_light_chain_kinase_activity |
| GO:0004705 | 3 | JUN_kinase_activity |
| GO:0004719 | 3 | protein-L-isoaspartate_(D-aspartate)_O-methyltransferase_activity |
| GO:0004738 | 3 | pyruvate_dehydrogenase_activity |
| GO:0004739 | 3 | pyruvate_dehydrogenase_(acetyl-transferring)_activity |
| GO:0004743 | 3 | pyruvate_kinase_activity |
| GO:0004748 | 3 | ribonucleoside-diphosphate_reductase_activity,_thioredoxin_disulfide_as_acceptor |
| GO:0004765 | 3 | shikimate_kinase_activity |
| GO:0004768 | 3 | stearoyl-CoA_9-desaturase_activity |
| GO:0004769 | 3 | steroid_delta-isomerase_activity |
| GO:0004775 | 3 | succinate-CoA_ligase_(ADP-forming)_activity |
| GO:0004792 | 3 | thiosulfate_sulfurtransferase_activity |
| GO:0004800 | 3 | thyroxine_5'-deiodinase_activity |
| GO:0004802 | 3 | transketolase_activity |
| GO:0004803 | 3 | transposase_activity |
| GO:0004813 | 3 | alanine-tRNA_ligase_activity |
| GO:0004814 | 3 | arginine-tRNA_ligase_activity |
| GO:0004815 | 3 | aspartate-tRNA_ligase_activity |
| GO:0004821 | 3 | histidine-tRNA_ligase_activity |
| GO:0004832 | 3 | valine-tRNA_ligase_activity |
| GO:0004833 | 3 | tryptophan_2,3-dioxygenase_activity |
| GO:0004873 | 3 | asialoglycoprotein_receptor_activity |
| GO:0004875 | 3 | complement_receptor_activity |
| GO:0004882 | 3 | androgen_receptor_activity |
| GO:0004886 | 3 | 9-cis_retinoic_acid_receptor_activity |
| GO:0004911 | 3 | interleukin-2_receptor_activity |
| GO:0004924 | 3 | oncostatin-M_receptor_activity |
| GO:0004937 | 3 | alpha1-adrenergic_receptor_activity |
| GO:0004938 | 3 | alpha2-adrenergic_receptor_activity |
| GO:0004939 | 3 | beta-adrenergic_receptor_activity |
| GO:0004942 | 3 | anaphylatoxin_receptor_activity |
| GO:0004946 | 3 | bombesin_receptor_activity |
| GO:0004949 | 3 | cannabinoid_receptor_activity |
| GO:0004965 | 3 | G-protein_coupled_GABA_receptor_activity |
| GO:0004966 | 3 | galanin_receptor_activity |
| GO:0004967 | 3 | glucagon_receptor_activity |
| GO:0004992 | 3 | platelet_activating_factor_receptor_activity |
| GO:0004995 | 3 | tachykinin_receptor_activity |
| GO:0004999 | 3 | vasoactive_intestinal_polypeptide_receptor_activity |
| GO:0005017 | 3 | platelet-derived_growth_factor-activated_receptor_activity |
| GO:0005049 | 3 | nuclear_export_signal_receptor_activity |
| GO:0005094 | 3 | Rho_GDP-dissociation_inhibitor_activity |
| GO:0005134 | 3 | interleukin-2_receptor_binding |
| GO:0005173 | 3 | stem_cell_factor_receptor_binding |
| GO:0005176 | 3 | ErbB-2_class_receptor_binding |
| GO:0005222 | 3 | intracellular_cAMP_activated_cation_channel_activity |
| GO:0005229 | 3 | intracellular_calcium_activated_chloride_channel_activity |
| GO:0005250 | 3 | A-type_(transient_outward)_potassium_channel_activity |
| GO:0005294 | 3 | neutral_L-amino_acid_secondary_active_transmembrane_transporter_activity |
| GO:0005329 | 3 | dopamine_transmembrane_transporter_activity |
| GO:0005350 | 3 | pyrimidine_base_transmembrane_transporter_activity |
| GO:0005415 | 3 | nucleoside:sodium_symporter_activity |
| GO:0005549 | 3 | odorant_binding |
| GO:0005588 | 3 | collagen_type_V |
| GO:0005589 | 3 | collagen_type_VI |
| GO:0005594 | 3 | collagen_type_IX |
| GO:0005610 | 3 | laminin-5_complex |
| GO:0005686 | 3 | U2_snRNP |
| GO:0005726 | 3 | perichromatin_fibrils |
| GO:0005787 | 3 | signal_peptidase_complex |
| GO:0005850 | 3 | eukaryotic_translation_initiation_factor_2_complex |
| GO:0005863 | 3 | striated_muscle_myosin_thick_filament |
| GO:0005865 | 3 | striated_muscle_thin_filament |
| GO:0005896 | 3 | interleukin-6_receptor_complex |
| GO:0005899 | 3 | insulin_receptor_complex |
| GO:0005900 | 3 | oncostatin-M_receptor_complex |
| GO:0005927 | 3 | muscle_tendon_junction |
| GO:0005945 | 3 | 6-phosphofructokinase_complex |
| GO:0005956 | 3 | protein_kinase_CK2_complex |
| GO:0005991 | 3 | trehalose_metabolic_process |
| GO:0006011 | 3 | UDP-glucose_metabolic_process |
| GO:0006065 | 3 | UDP-glucuronate_biosynthetic_process |
| GO:0006086 | 3 | acetyl-CoA_biosynthetic_process_from_pyruvate |
| GO:0006106 | 3 | fumarate_metabolic_process |
| GO:0006145 | 3 | purine_base_catabolic_process |
| GO:0006154 | 3 | adenosine_catabolic_process |
| GO:0006168 | 3 | adenine_salvage |
| GO:0006207 | 3 | 'de_novo'_pyrimidine_base_biosynthetic_process |
| GO:0006210 | 3 | thymine_catabolic_process |
| GO:0006235 | 3 | dTTP_biosynthetic_process |
| GO:0006295 | 3 | nucleotide-excision_repair,_DNA_incision,_3'-to_lesion |
| GO:0006296 | 3 | nucleotide-excision_repair,_DNA_incision,_5'-to_lesion |
| GO:0006344 | 3 | maintenance_of_chromatin_silencing |
| GO:0006384 | 3 | transcription_initiation_from_RNA_polymerase_III_promoter |
| GO:0006391 | 3 | transcription_initiation_from_mitochondrial_promoter |
| GO:0006419 | 3 | alanyl-tRNA_aminoacylation |
| GO:0006420 | 3 | arginyl-tRNA_aminoacylation |
| GO:0006421 | 3 | asparaginyl-tRNA_aminoacylation |
| GO:0006427 | 3 | histidyl-tRNA_aminoacylation |
| GO:0006438 | 3 | valyl-tRNA_aminoacylation |
| GO:0006452 | 3 | translational_frameshifting |
| GO:0006498 | 3 | N-terminal_protein_lipidation |
| GO:0006528 | 3 | asparagine_metabolic_process |
| GO:0006543 | 3 | glutamine_catabolic_process |
| GO:0006556 | 3 | S-adenosylmethionine_biosynthetic_process |
| GO:0006565 | 3 | L-serine_catabolic_process |
| GO:0006574 | 3 | valine_catabolic_process |
| GO:0006667 | 3 | sphinganine_metabolic_process |
| GO:0006772 | 3 | thiamine_metabolic_process |
| GO:0006922 | 3 | cleavage_of_lamin |
| GO:0006926 | 3 | virus-infected_cell_apoptotic_process |
| GO:0006931 | 3 | substrate-dependent_cell_migration,_cell_attachment_to_substrate |
| GO:0006933 | 3 | negative_regulation_of_cell_adhesion_involved_in_substrate-bound_cell_migration |
| GO:0006982 | 3 | response_to_lipid_hydroperoxide |
| GO:0006991 | 3 | response_to_sterol_depletion |
| GO:0007113 | 3 | endomitotic_cell_cycle |
| GO:0007132 | 3 | meiotic_metaphase_I |
| GO:0007144 | 3 | female_meiosis_I |
| GO:0007181 | 3 | transforming_growth_factor_beta_receptor_complex_assembly |
| GO:0007182 | 3 | common-partner_SMAD_protein_phosphorylation |
| GO:0007207 | 3 | phospholipase_C-activating_G-protein_coupled_acetylcholine_receptor_signaling_pathway |
| GO:0007221 | 3 | positive_regulation_of_transcription_of_Notch_receptor_target |
| GO:0007228 | 3 | positive_regulation_of_hh_target_transcription_factor_activity |
| GO:0007343 | 3 | egg_activation |
| GO:0007352 | 3 | zygotic_specification_of_dorsal/ventral_axis |
| GO:0007403 | 3 | glial_cell_fate_determination |
| GO:0007418 | 3 | ventral_midline_development |
| GO:0007521 | 3 | muscle_cell_fate_determination |
| GO:0007525 | 3 | somatic_muscle_development |
| GO:0007538 | 3 | primary_sex_determination |
| GO:0007618 | 3 | mating |
| GO:0008107 | 3 | galactoside_2-alpha-L-fucosyltransferase_activity |
| GO:0008109 | 3 | N-acetyllactosaminide_beta-1,6-N-acetylglucosaminyltransferase_activity |
| GO:0008142 | 3 | oxysterol_binding |
| GO:0008184 | 3 | glycogen_phosphorylase_activity |
| GO:0008192 | 3 | RNA_guanylyltransferase_activity |
| GO:0008241 | 3 | peptidyl-dipeptidase_activity |
| GO:0008282 | 3 | ATP-sensitive_potassium_channel_complex |
| GO:0008295 | 3 | spermidine_biosynthetic_process |
| GO:0008296 | 3 | 3'-5'-exodeoxyribonuclease_activity |
| GO:0008300 | 3 | isoprenoid_catabolic_process |
| GO:0008332 | 3 | low_voltage-gated_calcium_channel_activity |
| GO:0008384 | 3 | IkappaB_kinase_activity |
| GO:0008401 | 3 | retinoic_acid_4-hydroxylase_activity |
| GO:0008422 | 3 | beta-glucosidase_activity |
| GO:0008426 | 3 | protein_kinase_C_inhibitor_activity |
| GO:0008431 | 3 | vitamin_E_binding |
| GO:0008453 | 3 | alanine-glyoxylate_transaminase_activity |
| GO:0008454 | 3 | alpha-1,3-mannosylglycoprotein_4-beta-N-acetylglucosaminyltransferase_activity |
| GO:0008474 | 3 | palmitoyl-(protein)_hydrolase_activity |
| GO:0008475 | 3 | procollagen-lysine_5-dioxygenase_activity |
| GO:0008502 | 3 | melatonin_receptor_activity |
| GO:0008510 | 3 | sodium:bicarbonate_symporter_activity |
| GO:0008519 | 3 | ammonium_transmembrane_transporter_activity |
| GO:0008523 | 3 | sodium-dependent_multivitamin_transmembrane_transporter_activity |
| GO:0008537 | 3 | proteasome_activator_complex |
| GO:0008540 | 3 | proteasome_regulatory_particle,_base_subcomplex |
| GO:0008559 | 3 | xenobiotic-transporting_ATPase_activity |
| GO:0008670 | 3 | 2,4-dienoyl-CoA_reductase_(NADPH)_activity |
| GO:0008898 | 3 | homocysteine_S-methyltransferase_activity |
| GO:0008900 | 3 | hydrogen:potassium-exchanging_ATPase_activity |
| GO:0008934 | 3 | inositol_monophosphate_1-phosphatase_activity |
| GO:0008969 | 3 | phosphohistidine_phosphatase_activity |
| GO:0008970 | 3 | phosphatidylcholine_1-acylhydrolase_activity |
| GO:0009051 | 3 | pentose-phosphate_shunt,_oxidative_branch |
| GO:0009052 | 3 | pentose-phosphate_shunt,_non-oxidative_branch |
| GO:0009106 | 3 | lipoate_metabolic_process |
| GO:0009137 | 3 | purine_nucleoside_diphosphate_catabolic_process |
| GO:0009138 | 3 | pyrimidine_nucleoside_diphosphate_metabolic_process |
| GO:0009157 | 3 | deoxyribonucleoside_monophosphate_biosynthetic_process |
| GO:0009159 | 3 | deoxyribonucleoside_monophosphate_catabolic_process |
| GO:0009177 | 3 | pyrimidine_deoxyribonucleoside_monophosphate_biosynthetic_process |
| GO:0009181 | 3 | purine_ribonucleoside_diphosphate_catabolic_process |
| GO:0009189 | 3 | deoxyribonucleoside_diphosphate_biosynthetic_process |
| GO:0009212 | 3 | pyrimidine_deoxyribonucleoside_triphosphate_biosynthetic_process |
| GO:0009236 | 3 | cobalamin_biosynthetic_process |
| GO:0009240 | 3 | isopentenyl_diphosphate_biosynthetic_process |
| GO:0009279 | 3 | cell_outer_membrane |
| GO:0009296 | 3 | flagellum_assembly |
| GO:0009331 | 3 | glycerol-3-phosphate_dehydrogenase_complex |
| GO:0009436 | 3 | glyoxylate_catabolic_process |
| GO:0009597 | 3 | detection_of_virus |
| GO:0009602 | 3 | detection_of_symbiont |
| GO:0009608 | 3 | response_to_symbiont |
| GO:0009637 | 3 | response_to_blue_light |
| GO:0009730 | 3 | detection_of_carbohydrate_stimulus |
| GO:0009732 | 3 | detection_of_hexose_stimulus |
| GO:0009786 | 3 | regulation_of_asymmetric_cell_division |
| GO:0009822 | 3 | alkaloid_catabolic_process |
| GO:0009826 | 3 | unidimensional_cell_growth |
| GO:0009957 | 3 | epidermal_cell_fate_specification |
| GO:0009997 | 3 | negative_regulation_of_cardioblast_cell_fate_specification |
| GO:0010040 | 3 | response_to_iron(II)_ion |
| GO:0010041 | 3 | response_to_iron(III)_ion |
| GO:0010044 | 3 | response_to_aluminum_ion |
| GO:0010133 | 3 | proline_catabolic_process_to_glutamate |
| GO:0010159 | 3 | specification_of_organ_position |
| GO:0010216 | 3 | maintenance_of_DNA_methylation |
| GO:0010248 | 3 | establishment_or_maintenance_of_transmembrane_electrochemical_gradient |
| GO:0010424 | 3 | DNA_methylation_on_cytosine_within_a_CG_sequence |
| GO:0010457 | 3 | centriole-centriole_cohesion |
| GO:0010511 | 3 | regulation_of_phosphatidylinositol_biosynthetic_process |
| GO:0010521 | 3 | telomerase_inhibitor_activity |
| GO:0010523 | 3 | negative_regulation_of_calcium_ion_transport_into_cytosol |
| GO:0010586 | 3 | miRNA_metabolic_process |
| GO:0010587 | 3 | miRNA_catabolic_process |
| GO:0010621 | 3 | negative_regulation_of_transcription_by_transcription_factor_localization |
| GO:0010657 | 3 | muscle_cell_apoptotic_process |
| GO:0010693 | 3 | negative_regulation_of_alkaline_phosphatase_activity |
| GO:0010701 | 3 | positive_regulation_of_norepinephrine_secretion |
| GO:0010716 | 3 | negative_regulation_of_extracellular_matrix_disassembly |
| GO:0010724 | 3 | regulation_of_definitive_erythrocyte_differentiation |
| GO:0010742 | 3 | macrophage_derived_foam_cell_differentiation |
| GO:0010764 | 3 | negative_regulation_of_fibroblast_migration |
| GO:0010792 | 3 | DNA_double-strand_break_processing_involved_in_repair_via_single-strand_annealing |
| GO:0010807 | 3 | regulation_of_synaptic_vesicle_priming |
| GO:0010826 | 3 | negative_regulation_of_centrosome_duplication |
| GO:0010831 | 3 | positive_regulation_of_myotube_differentiation |
| GO:0010834 | 3 | telomere_maintenance_via_telomere_shortening |
| GO:0010838 | 3 | positive_regulation_of_keratinocyte_proliferation |
| GO:0010899 | 3 | regulation_of_phosphatidylcholine_catabolic_process |
| GO:0010903 | 3 | negative_regulation_of_very-low-density_lipoprotein_particle_remodeling |
| GO:0010911 | 3 | regulation_of_isomerase_activity |
| GO:0010912 | 3 | positive_regulation_of_isomerase_activity |
| GO:0010956 | 3 | negative_regulation_of_calcidiol_1-monooxygenase_activity |
| GO:0010966 | 3 | regulation_of_phosphate_transport |
| GO:0010979 | 3 | regulation_of_vitamin_D_24-hydroxylase_activity |
| GO:0010980 | 3 | positive_regulation_of_vitamin_D_24-hydroxylase_activity |
| GO:0010982 | 3 | regulation_of_high-density_lipoprotein_particle_clearance |
| GO:0010986 | 3 | positive_regulation_of_lipoprotein_particle_clearance |
| GO:0010989 | 3 | negative_regulation_of_low-density_lipoprotein_particle_clearance |
| GO:0010990 | 3 | regulation_of_SMAD_protein_complex_assembly |
| GO:0010991 | 3 | negative_regulation_of_SMAD_protein_complex_assembly |
| GO:0010997 | 3 | anaphase-promoting_complex_binding |
| GO:0014038 | 3 | regulation_of_Schwann_cell_differentiation |
| GO:0014041 | 3 | regulation_of_neuron_maturation |
| GO:0014044 | 3 | Schwann_cell_development |
| GO:0014056 | 3 | regulation_of_acetylcholine_secretion |
| GO:0014071 | 3 | response_to_cycloalkane |
| GO:0014707 | 3 | branchiomeric_skeletal_muscle_development |
| GO:0014738 | 3 | regulation_of_muscle_hyperplasia |
| GO:0014745 | 3 | negative_regulation_of_muscle_adaptation |
| GO:0014816 | 3 | satellite_cell_differentiation |
| GO:0014832 | 3 | urinary_bladder_smooth_muscle_contraction |
| GO:0014834 | 3 | satellite_cell_maintenance_involved_in_skeletal_muscle_regeneration |
| GO:0014846 | 3 | esophagus_smooth_muscle_contraction |
| GO:0015015 | 3 | heparan_sulfate_proteoglycan_biosynthetic_process,_enzymatic_modification |
| GO:0015018 | 3 | galactosylgalactosylxylosylprotein_3-beta-glucuronosyltransferase_activity |
| GO:0015137 | 3 | citrate_transmembrane_transporter_activity |
| GO:0015142 | 3 | tricarboxylic_acid_transmembrane_transporter_activity |
| GO:0015181 | 3 | arginine_transmembrane_transporter_activity |
| GO:0015194 | 3 | L-serine_transmembrane_transporter_activity |
| GO:0015211 | 3 | purine_nucleoside_transmembrane_transporter_activity |
| GO:0015215 | 3 | nucleotide_transmembrane_transporter_activity |
| GO:0015222 | 3 | serotonin_transmembrane_transporter_activity |
| GO:0015226 | 3 | carnitine_transporter_activity |
| GO:0015272 | 3 | ATP-activated_inward_rectifier_potassium_channel_activity |
| GO:0015556 | 3 | C4-dicarboxylate_transmembrane_transporter_activity |
| GO:0015562 | 3 | efflux_transmembrane_transporter_activity |
| GO:0015803 | 3 | branched-chain_aliphatic_amino_acid_transport |
| GO:0015808 | 3 | L-alanine_transport |
| GO:0015812 | 3 | gamma-aminobutyric_acid_transport |
| GO:0015816 | 3 | glycine_transport |
| GO:0015820 | 3 | leucine_transport |
| GO:0015825 | 3 | L-serine_transport |
| GO:0015842 | 3 | synaptic_vesicle_amine_transport |
| GO:0015860 | 3 | purine_nucleoside_transport |
| GO:0015864 | 3 | pyrimidine_nucleoside_transport |
| GO:0015866 | 3 | ADP_transport |
| GO:0015917 | 3 | aminophospholipid_transport |
| GO:0015928 | 3 | fucosidase_activity |
| GO:0015956 | 3 | bis(5'-nucleosidyl)_oligophosphate_metabolic_process |
| GO:0015959 | 3 | diadenosine_polyphosphate_metabolic_process |
| GO:0016056 | 3 | rhodopsin_mediated_signaling_pathway |
| GO:0016080 | 3 | synaptic_vesicle_targeting |
| GO:0016094 | 3 | polyprenol_biosynthetic_process |
| GO:0016149 | 3 | translation_release_factor_activity,_codon_specific |
| GO:0016151 | 3 | nickel_cation_binding |
| GO:0016155 | 3 | formyltetrahydrofolate_dehydrogenase_activity |
| GO:0016176 | 3 | superoxide-generating_NADPH_oxidase_activator_activity |
| GO:0016212 | 3 | kynurenine-oxoglutarate_transaminase_activity |
| GO:0016215 | 3 | acyl-CoA_desaturase_activity |
| GO:0016233 | 3 | telomere_capping |
| GO:0016309 | 3 | 1-phosphatidylinositol-5-phosphate_4-kinase_activity |
| GO:0016312 | 3 | inositol_bisphosphate_phosphatase_activity |
| GO:0016362 | 3 | activin_receptor_activity,_type_II |
| GO:0016480 | 3 | negative_regulation_of_transcription_from_RNA_polymerase_III_promoter |
| GO:0016482 | 3 | cytoplasmic_transport |
| GO:0016508 | 3 | long-chain-enoyl-CoA_hydratase_activity |
| GO:0016533 | 3 | cyclin-dependent_protein_kinase_5_holoenzyme_complex |
| GO:0016561 | 3 | protein_import_into_peroxisome_matrix,_translocation |
| GO:0016576 | 3 | histone_dephosphorylation |
| GO:0016600 | 3 | flotillin_complex |
| GO:0016619 | 3 | malate_dehydrogenase_(oxaloacetate-decarboxylating)_activity |
| GO:0016623 | 3 | oxidoreductase_activity,_acting_on_the_aldehyde_or_oxo_group_of_donors,_oxygen_as_acceptor |
| GO:0016647 | 3 | oxidoreductase_activity,_acting_on_the_CH-NH_group_of_donors,_oxygen_as_acceptor |
| GO:0016672 | 3 | oxidoreductase_activity,_acting_on_a_sulfur_group_of_donors,_quinone_or_similar_compound_as_acceptor |
| GO:0016716 | 3 | oxidoreductase_activity,_acting_on_paired_donors,_with_incorporation_or_reduction_of_molecular_oxygen,_another_compound_as_one_donor,_and_incorporation_of_one_atom_of_oxygen |
| GO:0016728 | 3 | oxidoreductase_activity,_acting_on_CH_or_CH2_groups,_disulfide_as_acceptor |
| GO:0016748 | 3 | succinyltransferase_activity |
| GO:0016774 | 3 | phosphotransferase_activity,_carboxyl_group_as_acceptor |
| GO:0016802 | 3 | trialkylsulfonium_hydrolase_activity |
| GO:0016909 | 3 | SAP_kinase_activity |
| GO:0016926 | 3 | protein_desumoylation |
| GO:0016929 | 3 | SUMO-specific_protease_activity |
| GO:0016939 | 3 | kinesin_II_complex |
| GO:0016941 | 3 | natriuretic_peptide_receptor_activity |
| GO:0017060 | 3 | 3-galactosyl-N-acetylglucosaminide_4-alpha-L-fucosyltransferase_activity |
| GO:0017123 | 3 | Ral_GTPase_activator_activity |
| GO:0017151 | 3 | DEAD/H-box_RNA_helicase_binding |
| GO:0017159 | 3 | pantetheine_hydrolase_activity |
| GO:0017162 | 3 | aryl_hydrocarbon_receptor_binding |
| GO:0018076 | 3 | N-terminal_peptidyl-lysine_acetylation |
| GO:0018094 | 3 | protein_polyglycylation |
| GO:0018125 | 3 | peptidyl-cysteine_methylation |
| GO:0018158 | 3 | protein_oxidation |
| GO:0018230 | 3 | peptidyl-L-cysteine_S-palmitoylation |
| GO:0018231 | 3 | peptidyl-S-diacylglycerol-L-cysteine_biosynthetic_process_from_peptidyl-cysteine |
| GO:0018242 | 3 | protein_O-linked_glycosylation_via_serine |
| GO:0018243 | 3 | protein_O-linked_glycosylation_via_threonine |
| GO:0018343 | 3 | protein_farnesylation |
| GO:0018377 | 3 | protein_myristoylation |
| GO:0018401 | 3 | peptidyl-proline_hydroxylation_to_4-hydroxy-L-proline |
| GO:0018636 | 3 | phenanthrene_9,10-monooxygenase_activity |
| GO:0019049 | 3 | evasion_of_host_defenses_by_virus |
| GO:0019064 | 3 | viral_envelope_fusion_with_host_membrane |
| GO:0019166 | 3 | trans-2-enoyl-CoA_reductase_(NADPH)_activity |
| GO:0019254 | 3 | carnitine_metabolic_process,_CoA-linked |
| GO:0019276 | 3 | UDP-N-acetylgalactosamine_metabolic_process |
| GO:0019287 | 3 | isopentenyl_diphosphate_biosynthetic_process,_mevalonate_pathway |
| GO:0019344 | 3 | cysteine_biosynthetic_process |
| GO:0019408 | 3 | dolichol_biosynthetic_process |
| GO:0019471 | 3 | 4-hydroxyproline_metabolic_process |
| GO:0019531 | 3 | oxalate_transmembrane_transporter_activity |
| GO:0019532 | 3 | oxalate_transport |
| GO:0019682 | 3 | glyceraldehyde-3-phosphate_metabolic_process |
| GO:0019718 | 3 | rough_microsome |
| GO:0019767 | 3 | IgE_receptor_activity |
| GO:0019797 | 3 | procollagen-proline_3-dioxygenase_activity |
| GO:0019805 | 3 | quinolinate_biosynthetic_process |
| GO:0019814 | 3 | immunoglobulin_complex |
| GO:0019815 | 3 | B_cell_receptor_complex |
| GO:0019826 | 3 | oxygen_sensor_activity |
| GO:0019834 | 3 | phospholipase_A2_inhibitor_activity |
| GO:0019836 | 3 | hemolysis_by_symbiont_of_host_erythrocytes |
| GO:0019856 | 3 | pyrimidine_base_biosynthetic_process |
| GO:0019859 | 3 | thymine_metabolic_process |
| GO:0019862 | 3 | IgA_binding |
| GO:0019870 | 3 | potassium_channel_inhibitor_activity |
| GO:0019871 | 3 | sodium_channel_inhibitor_activity |
| GO:0019919 | 3 | peptidyl-arginine_methylation,_to_asymmetrical-dimethyl_arginine |
| GO:0019958 | 3 | C-X-C_chemokine_binding |
| GO:0019959 | 3 | interleukin-8_binding |
| GO:0019976 | 3 | interleukin-2_binding |
| GO:0021508 | 3 | floor_plate_formation |
| GO:0021513 | 3 | spinal_cord_dorsal/ventral_patterning |
| GO:0021530 | 3 | spinal_cord_oligodendrocyte_cell_fate_specification |
| GO:0021554 | 3 | optic_nerve_development |
| GO:0021570 | 3 | rhombomere_4_development |
| GO:0021571 | 3 | rhombomere_5_development |
| GO:0021575 | 3 | hindbrain_morphogenesis |
| GO:0021592 | 3 | fourth_ventricle_development |
| GO:0021603 | 3 | cranial_nerve_formation |
| GO:0021678 | 3 | third_ventricle_development |
| GO:0021695 | 3 | cerebellar_cortex_development |
| GO:0021754 | 3 | facial_nucleus_development |
| GO:0021796 | 3 | cerebral_cortex_regionalization |
| GO:0021798 | 3 | forebrain_dorsal/ventral_pattern_formation |
| GO:0021800 | 3 | cerebral_cortex_tangential_migration |
| GO:0021834 | 3 | chemorepulsion_involved_in_embryonic_olfactory_bulb_interneuron_precursor_migration |
| GO:0021889 | 3 | olfactory_bulb_interneuron_differentiation |
| GO:0021891 | 3 | olfactory_bulb_interneuron_development |
| GO:0021910 | 3 | smoothened_signaling_pathway_involved_in_ventral_spinal_cord_patterning |
| GO:0021912 | 3 | regulation_of_transcription_from_RNA_polymerase_II_promoter_involved_in_spinal_cord_motor_neuron_fate_specification |
| GO:0021940 | 3 | positive_regulation_of_cerebellar_granule_cell_precursor_proliferation |
| GO:0021960 | 3 | anterior_commissure_morphogenesis |
| GO:0021979 | 3 | hypothalamus_cell_differentiation |
| GO:0022007 | 3 | convergent_extension_involved_in_neural_plate_elongation |
| GO:0022889 | 3 | serine_transmembrane_transporter_activity |
| GO:0023041 | 3 | neuronal_signal_transduction |
| GO:0030008 | 3 | TRAPP_complex |
| GO:0030023 | 3 | extracellular_matrix_constituent_conferring_elasticity |
| GO:0030035 | 3 | microspike_assembly |
| GO:0030185 | 3 | nitric_oxide_transport |
| GO:0030186 | 3 | melatonin_metabolic_process |
| GO:0030229 | 3 | very-low-density_lipoprotein_particle_receptor_activity |
| GO:0030241 | 3 | skeletal_muscle_myosin_thick_filament_assembly |
| GO:0030263 | 3 | apoptotic_chromosome_condensation |
| GO:0030272 | 3 | 5-formyltetrahydrofolate_cyclo-ligase_activity |
| GO:0030309 | 3 | poly-N-acetyllactosamine_metabolic_process |
| GO:0030323 | 3 | respiratory_tube_development |
| GO:0030343 | 3 | vitamin_D3_25-hydroxylase_activity |
| GO:0030350 | 3 | iron-responsive_element_binding |
| GO:0030354 | 3 | melanin-concentrating_hormone_activity |
| GO:0030395 | 3 | lactose_binding |
| GO:0030422 | 3 | production_of_siRNA_involved_in_RNA_interference |
| GO:0030423 | 3 | targeting_of_mRNA_for_destruction_involved_in_RNA_interference |
| GO:0030478 | 3 | actin_cap |
| GO:0030485 | 3 | smooth_muscle_contractile_fiber |
| GO:0030505 | 3 | inorganic_diphosphate_transport |
| GO:0030538 | 3 | embryonic_genitalia_morphogenesis |
| GO:0030573 | 3 | bile_acid_catabolic_process |
| GO:0030578 | 3 | PML_body_organization |
| GO:0030613 | 3 | oxidoreductase_activity,_acting_on_phosphorus_or_arsenic_in_donors |
| GO:0030614 | 3 | oxidoreductase_activity,_acting_on_phosphorus_or_arsenic_in_donors,_disulfide_as_acceptor |
| GO:0030688 | 3 | preribosome,_small_subunit_precursor |
| GO:0030730 | 3 | sequestering_of_triglyceride |
| GO:0030881 | 3 | beta-2-microglobulin_binding |
| GO:0030895 | 3 | apolipoprotein_B_mRNA_editing_enzyme_complex |
| GO:0030899 | 3 | calcium-dependent_ATPase_activity |
| GO:0030910 | 3 | olfactory_placode_formation |
| GO:0030951 | 3 | establishment_or_maintenance_of_microtubule_cytoskeleton_polarity |
| GO:0030953 | 3 | astral_microtubule_organization |
| GO:0030956 | 3 | glutamyl-tRNA(Gln)_amidotransferase_complex |
| GO:0030984 | 3 | kininogen_binding |
| GO:0031005 | 3 | filamin_binding |
| GO:0031022 | 3 | nuclear_migration_along_microfilament |
| GO:0031094 | 3 | platelet_dense_tubular_network |
| GO:0031117 | 3 | positive_regulation_of_microtubule_depolymerization |
| GO:0031125 | 3 | rRNA_3'-end_processing |
| GO:0031127 | 3 | alpha-(1,2)-fucosyltransferase_activity |
| GO:0031133 | 3 | regulation_of_axon_diameter |
| GO:0031177 | 3 | phosphopantetheine_binding |
| GO:0031205 | 3 | endoplasmic_reticulum_Sec_complex |
| GO:0031269 | 3 | pseudopodium_assembly |
| GO:0031284 | 3 | positive_regulation_of_guanylate_cyclase_activity |
| GO:0031314 | 3 | extrinsic_to_mitochondrial_inner_membrane |
| GO:0031489 | 3 | myosin_V_binding |
| GO:0031501 | 3 | mannosyltransferase_complex |
| GO:0031544 | 3 | peptidyl-proline_3-dioxygenase_activity |
| GO:0031580 | 3 | membrane_raft_distribution |
| GO:0031584 | 3 | activation_of_phospholipase_D_activity |
| GO:0031585 | 3 | regulation_of_inositol_1,4,5-trisphosphate-sensitive_calcium-release_channel_activity |
| GO:0031618 | 3 | nuclear_centromeric_heterochromatin |
| GO:0031628 | 3 | opioid_receptor_binding |
| GO:0031642 | 3 | negative_regulation_of_myelination |
| GO:0031643 | 3 | positive_regulation_of_myelination |
| GO:0031681 | 3 | G-protein_beta-subunit_binding |
| GO:0031735 | 3 | CCR10_chemokine_receptor_binding |
| GO:0031779 | 3 | melanocortin_receptor_binding |
| GO:0031781 | 3 | type_3_melanocortin_receptor_binding |
| GO:0031782 | 3 | type_4_melanocortin_receptor_binding |
| GO:0031838 | 3 | haptoglobin-hemoglobin_complex |
| GO:0031849 | 3 | olfactory_receptor_binding |
| GO:0031905 | 3 | early_endosome_lumen |
| GO:0031933 | 3 | telomeric_heterochromatin |
| GO:0031981 | 3 | nuclear_lumen |
| GO:0031987 | 3 | locomotion_involved_in_locomotory_behavior |
| GO:0031995 | 3 | insulin-like_growth_factor_II_binding |
| GO:0032002 | 3 | interleukin-28_receptor_complex |
| GO:0032066 | 3 | nucleolus_to_nucleoplasm_transport |
| GO:0032071 | 3 | regulation_of_endodeoxyribonuclease_activity |
| GO:0032097 | 3 | positive_regulation_of_response_to_food |
| GO:0032100 | 3 | positive_regulation_of_appetite |
| GO:0032139 | 3 | dinucleotide_insertion_or_deletion_binding |
| GO:0032142 | 3 | single_guanine_insertion_binding |
| GO:0032183 | 3 | SUMO_binding |
| GO:0032203 | 3 | telomere_formation_via_telomerase |
| GO:0032212 | 3 | positive_regulation_of_telomere_maintenance_via_telomerase |
| GO:0032217 | 3 | riboflavin_transporter_activity |
| GO:0032218 | 3 | riboflavin_transport |
| GO:0032274 | 3 | gonadotropin_secretion |
| GO:0032287 | 3 | peripheral_nervous_system_myelin_maintenance |
| GO:0032299 | 3 | ribonuclease_H2_complex |
| GO:0032315 | 3 | regulation_of_Ral_GTPase_activity |
| GO:0032341 | 3 | aldosterone_metabolic_process |
| GO:0032342 | 3 | aldosterone_biosynthetic_process |
| GO:0032356 | 3 | oxidized_DNA_binding |
| GO:0032357 | 3 | oxidized_purine_DNA_binding |
| GO:0032417 | 3 | positive_regulation_of_sodium:hydrogen_antiporter_activity |
| GO:0032422 | 3 | purine-rich_negative_regulatory_element_binding |
| GO:0032426 | 3 | stereocilium_bundle_tip |
| GO:0032444 | 3 | activin_responsive_factor_complex |
| GO:0032449 | 3 | CBM_complex |
| GO:0032450 | 3 | maltose_alpha-glucosidase_activity |
| GO:0032460 | 3 | negative_regulation_of_protein_oligomerization |
| GO:0032463 | 3 | negative_regulation_of_protein_homooligomerization |
| GO:0032497 | 3 | detection_of_lipopolysaccharide |
| GO:0032509 | 3 | endosome_transport_via_multivesicular_body_sorting_pathway |
| GO:0032512 | 3 | regulation_of_protein_phosphatase_type_2B_activity |
| GO:0032527 | 3 | protein_exit_from_endoplasmic_reticulum |
| GO:0032530 | 3 | regulation_of_microvillus_organization |
| GO:0032557 | 3 | pyrimidine_ribonucleotide_binding |
| GO:0032591 | 3 | dendritic_spine_membrane |
| GO:0032593 | 3 | insulin-responsive_compartment |
| GO:0032608 | 3 | interferon-beta_production |
| GO:0032650 | 3 | regulation_of_interleukin-1_alpha_production |
| GO:0032696 | 3 | negative_regulation_of_interleukin-13_production |
| GO:0032730 | 3 | positive_regulation_of_interleukin-1_alpha_production |
| GO:0032754 | 3 | positive_regulation_of_interleukin-5_production |
| GO:0032790 | 3 | ribosome_disassembly |
| GO:0032797 | 3 | SMN_complex |
| GO:0032852 | 3 | positive_regulation_of_Ral_GTPase_activity |
| GO:0032859 | 3 | activation_of_Ral_GTPase_activity |
| GO:0032864 | 3 | activation_of_Cdc42_GTPase_activity |
| GO:0032910 | 3 | regulation_of_transforming_growth_factor_beta3_production |
| GO:0032962 | 3 | positive_regulation_of_inositol_trisphosphate_biosynthetic_process |
| GO:0032971 | 3 | regulation_of_muscle_filament_sliding |
| GO:0033007 | 3 | negative_regulation_of_mast_cell_activation_involved_in_immune_response |
| GO:0033026 | 3 | negative_regulation_of_mast_cell_apoptotic_process |
| GO:0033028 | 3 | myeloid_cell_apoptotic_process |
| GO:0033034 | 3 | positive_regulation_of_myeloid_cell_apoptotic_process |
| GO:0033038 | 3 | bitter_taste_receptor_activity |
| GO:0033059 | 3 | cellular_pigmentation |
| GO:0033076 | 3 | isoquinoline_alkaloid_metabolic_process |
| GO:0033092 | 3 | positive_regulation_of_immature_T_cell_proliferation_in_thymus |
| GO:0033150 | 3 | cytoskeletal_calyx |
| GO:0033159 | 3 | negative_regulation_of_protein_import_into_nucleus,_translocation |
| GO:0033185 | 3 | dolichol-phosphate-mannose_synthase_complex |
| GO:0033186 | 3 | CAF-1_complex |
| GO:0033188 | 3 | sphingomyelin_synthase_activity |
| GO:0033239 | 3 | negative_regulation_of_cellular_amine_metabolic_process |
| GO:0033269 | 3 | internode_region_of_axon |
| GO:0033300 | 3 | dehydroascorbic_acid_transporter_activity |
| GO:0033314 | 3 | mitotic_cell_cycle_DNA_replication_checkpoint |
| GO:0033504 | 3 | floor_plate_development |
| GO:0033512 | 3 | L-lysine_catabolic_process_to_acetyl-CoA_via_saccharopine |
| GO:0033596 | 3 | TSC1-TSC2_complex |
| GO:0033655 | 3 | host_cell_cytoplasm_part |
| GO:0033829 | 3 | O-fucosylpeptide_3-beta-N-acetylglucosaminyltransferase_activity |
| GO:0034014 | 3 | response_to_triglyceride |
| GO:0034046 | 3 | poly(G)_RNA_binding |
| GO:0034047 | 3 | regulation_of_protein_phosphatase_type_2A_activity |
| GO:0034113 | 3 | heterotypic_cell-cell_adhesion |
| GO:0034124 | 3 | regulation_of_MyD88-dependent_toll-like_receptor_signaling_pathway |
| GO:0034136 | 3 | negative_regulation_of_toll-like_receptor_2_signaling_pathway |
| GO:0034141 | 3 | positive_regulation_of_toll-like_receptor_3_signaling_pathway |
| GO:0034186 | 3 | apolipoprotein_A-I_binding |
| GO:0034205 | 3 | beta-amyloid_formation |
| GO:0034219 | 3 | carbohydrate_transmembrane_transport |
| GO:0034244 | 3 | negative_regulation_of_transcription_elongation_from_RNA_polymerase_II_promoter |
| GO:0034259 | 3 | negative_regulation_of_Rho_GTPase_activity |
| GO:0034287 | 3 | detection_of_monosaccharide_stimulus |
| GO:0034344 | 3 | regulation_of_type_III_interferon_production |
| GO:0034371 | 3 | chylomicron_remodeling |
| GO:0034421 | 3 | post-translational_protein_acetylation |
| GO:0034433 | 3 | steroid_esterification |
| GO:0034434 | 3 | sterol_esterification |
| GO:0034435 | 3 | cholesterol_esterification |
| GO:0034444 | 3 | regulation_of_plasma_lipoprotein_particle_oxidation |
| GO:0034445 | 3 | negative_regulation_of_plasma_lipoprotein_particle_oxidation |
| GO:0034447 | 3 | very-low-density_lipoprotein_particle_clearance |
| GO:0034584 | 3 | piRNA_binding |
| GO:0034650 | 3 | cortisol_metabolic_process |
| GO:0034651 | 3 | cortisol_biosynthetic_process |
| GO:0034653 | 3 | retinoic_acid_catabolic_process |
| GO:0034663 | 3 | endoplasmic_reticulum_chaperone_complex |
| GO:0034673 | 3 | inhibin-betaglycan-ActRII_complex |
| GO:0034678 | 3 | alpha8-beta1_integrin_complex |
| GO:0034711 | 3 | inhibin_binding |
| GO:0034723 | 3 | DNA_replication-dependent_nucleosome_organization |
| GO:0034742 | 3 | APC-Axin-1-beta-catenin_complex |
| GO:0034766 | 3 | negative_regulation_of_ion_transmembrane_transport |
| GO:0034987 | 3 | immunoglobulin_receptor_binding |
| GO:0035005 | 3 | 1-phosphatidylinositol-4-phosphate_3-kinase_activity |
| GO:0035021 | 3 | negative_regulation_of_Rac_protein_signal_transduction |
| GO:0035022 | 3 | positive_regulation_of_Rac_protein_signal_transduction |
| GO:0035054 | 3 | embryonic_heart_tube_anterior/posterior_pattern_specification |
| GO:0035083 | 3 | cilium_axoneme_assembly |
| GO:0035174 | 3 | histone_serine_kinase_activity |
| GO:0035195 | 3 | gene_silencing_by_miRNA |
| GO:0035196 | 3 | production_of_miRNAs_involved_in_gene_silencing_by_miRNA |
| GO:0035234 | 3 | germ_cell_programmed_cell_death |
| GO:0035241 | 3 | protein-arginine_omega-N_monomethyltransferase_activity |
| GO:0035268 | 3 | protein_mannosylation |
| GO:0035269 | 3 | protein_O-linked_mannosylation |
| GO:0035331 | 3 | negative_regulation_of_hippo_signaling_cascade |
| GO:0035356 | 3 | cellular_triglyceride_homeostasis |
| GO:0035360 | 3 | positive_regulation_of_peroxisome_proliferator_activated_receptor_signaling_pathway |
| GO:0035376 | 3 | sterol_import |
| GO:0035382 | 3 | sterol_transmembrane_transport |
| GO:0035385 | 3 | Roundabout_signaling_pathway |
| GO:0035441 | 3 | cell_migration_involved_in_vasculogenesis |
| GO:0035459 | 3 | cargo_loading_into_vesicle |
| GO:0035461 | 3 | vitamin_transmembrane_transport |
| GO:0035502 | 3 | metanephric_ureteric_bud_development |
| GO:0035507 | 3 | regulation_of_myosin-light-chain-phosphatase_activity |
| GO:0035511 | 3 | oxidative_DNA_demethylation |
| GO:0035514 | 3 | DNA_demethylase_activity |
| GO:0035529 | 3 | NADH_pyrophosphatase_activity |
| GO:0035582 | 3 | sequestering_of_BMP_in_extracellular_matrix |
| GO:0035583 | 3 | sequestering_of_TGFbeta_in_extracellular_matrix |
| GO:0035609 | 3 | C-terminal_protein_deglutamylation |
| GO:0035610 | 3 | protein_side_chain_deglutamylation |
| GO:0035630 | 3 | bone_mineralization_involved_in_bone_maturation |
| GO:0035694 | 3 | mitochondrial_protein_catabolic_process |
| GO:0035722 | 3 | interleukin-12-mediated_signaling_pathway |
| GO:0035747 | 3 | natural_killer_cell_chemotaxis |
| GO:0035754 | 3 | B_cell_chemotaxis |
| GO:0035771 | 3 | interleukin-4-mediated_signaling_pathway |
| GO:0035793 | 3 | positive_regulation_of_metanephric_mesenchymal_cell_migration_by_platelet-derived_growth_factor_receptor-beta_signaling_pathway |
| GO:0035803 | 3 | egg_coat_formation |
| GO:0035845 | 3 | photoreceptor_cell_outer_segment_organization |
| GO:0035927 | 3 | RNA_import_into_mitochondrion |
| GO:0035928 | 3 | rRNA_import_into_mitochondrion |
| GO:0035929 | 3 | steroid_hormone_secretion |
| GO:0035939 | 3 | microsatellite_binding |
| GO:0035962 | 3 | response_to_interleukin-13 |
| GO:0035985 | 3 | senescence-associated_heterochromatin_focus |
| GO:0035986 | 3 | senescence-associated_heterochromatin_focus_assembly |
| GO:0035993 | 3 | deltoid_tuberosity_development |
| GO:0036003 | 3 | positive_regulation_of_transcription_from_RNA_polymerase_II_promoter_in_response_to_stress |
| GO:0036015 | 3 | response_to_interleukin-3 |
| GO:0036016 | 3 | cellular_response_to_interleukin-3 |
| GO:0036017 | 3 | response_to_erythropoietin |
| GO:0036018 | 3 | cellular_response_to_erythropoietin |
| GO:0036022 | 3 | limb_joint_morphogenesis |
| GO:0036023 | 3 | embryonic_skeletal_limb_joint_morphogenesis |
| GO:0036100 | 3 | leukotriene_catabolic_process |
| GO:0036101 | 3 | leukotriene_B4_catabolic_process |
| GO:0036102 | 3 | leukotriene_B4_metabolic_process |
| GO:0036122 | 3 | BMP_binding |
| GO:0036137 | 3 | kynurenine_aminotransferase_activity |
| GO:0038026 | 3 | reelin-mediated_signaling_pathway |
| GO:0038028 | 3 | insulin_receptor_signaling_pathway_via_phosphatidylinositol_3-kinase_cascade |
| GO:0038030 | 3 | non-canonical_Wnt_receptor_signaling_pathway_via_MAPK_cascade |
| GO:0038085 | 3 | vascular_endothelial_growth_factor_binding |
| GO:0038086 | 3 | VEGF-activated_platelet-derived_growth_factor_receptor_signaling_pathway |
| GO:0038091 | 3 | positive_regulation_of_cell_proliferation_by_VEGF-activated_platelet_derived_growth_factor_receptor_signaling_pathway |
| GO:0038093 | 3 | Fc_receptor_signaling_pathway |
| GO:0038108 | 3 | negative_regulation_of_appetite_by_leptin-mediated_signaling_pathway |
| GO:0038110 | 3 | interleukin-2-mediated_signaling_pathway |
| GO:0038165 | 3 | oncostatin-M-mediated_signaling_pathway |
| GO:0038171 | 3 | cannabinoid_signaling_pathway |
| GO:0040009 | 3 | regulation_of_growth_rate |
| GO:0042045 | 3 | epithelial_fluid_transport |
| GO:0042063 | 3 | gliogenesis |
| GO:0042094 | 3 | interleukin-2_biosynthetic_process |
| GO:0042126 | 3 | nitrate_metabolic_process |
| GO:0042138 | 3 | meiotic_DNA_double-strand_break_formation |
| GO:0042175 | 3 | nuclear_outer_membrane-endoplasmic_reticulum_membrane_network |
| GO:0042256 | 3 | mature_ribosome_assembly |
| GO:0042271 | 3 | susceptibility_to_natural_killer_cell_mediated_cytotoxicity |
| GO:0042272 | 3 | nuclear_RNA_export_factor_complex |
| GO:0042297 | 3 | vocal_learning |
| GO:0042339 | 3 | keratan_sulfate_metabolic_process |
| GO:0042369 | 3 | vitamin_D_catabolic_process |
| GO:0042371 | 3 | vitamin_K_biosynthetic_process |
| GO:0042407 | 3 | cristae_formation |
| GO:0042414 | 3 | epinephrine_metabolic_process |
| GO:0042427 | 3 | serotonin_biosynthetic_process |
| GO:0042471 | 3 | ear_morphogenesis |
| GO:0042494 | 3 | detection_of_bacterial_lipoprotein |
| GO:0042500 | 3 | aspartic_endopeptidase_activity,_intramembrane_cleaving |
| GO:0042506 | 3 | tyrosine_phosphorylation_of_Stat5_protein |
| GO:0042524 | 3 | negative_regulation_of_tyrosine_phosphorylation_of_Stat5_protein |
| GO:0042629 | 3 | mast_cell_granule |
| GO:0042633 | 3 | hair_cycle |
| GO:0042662 | 3 | negative_regulation_of_mesodermal_cell_fate_specification |
| GO:0042663 | 3 | regulation_of_endodermal_cell_fate_specification |
| GO:0042668 | 3 | auditory_receptor_cell_fate_determination |
| GO:0042686 | 3 | regulation_of_cardioblast_cell_fate_specification |
| GO:0042699 | 3 | follicle-stimulating_hormone_signaling_pathway |
| GO:0042731 | 3 | PH_domain_binding |
| GO:0042775 | 3 | mitochondrial_ATP_synthesis_coupled_electron_transport |
| GO:0042799 | 3 | histone_methyltransferase_activity_(H4-K20_specific) |
| GO:0042816 | 3 | vitamin_B6_metabolic_process |
| GO:0042819 | 3 | vitamin_B6_biosynthetic_process |
| GO:0042895 | 3 | antibiotic_transporter_activity |
| GO:0042977 | 3 | activation_of_JAK2_kinase_activity |
| GO:0042985 | 3 | negative_regulation_of_amyloid_precursor_protein_biosynthetic_process |
| GO:0042996 | 3 | regulation_of_Golgi_to_plasma_membrane_protein_transport |
| GO:0043047 | 3 | single-stranded_telomeric_DNA_binding |
| GO:0043141 | 3 | ATP-dependent_5'-3'_DNA_helicase_activity |
| GO:0043159 | 3 | acrosomal_matrix |
| GO:0043203 | 3 | axon_hillock |
| GO:0043259 | 3 | laminin-10_complex |
| GO:0043260 | 3 | laminin-11_complex |
| GO:0043276 | 3 | anoikis |
| GO:0043293 | 3 | apoptosome |
| GO:0043320 | 3 | natural_killer_cell_degranulation |
| GO:0043398 | 3 | HLH_domain_binding |
| GO:0043416 | 3 | regulation_of_skeletal_muscle_tissue_regeneration |
| GO:0043451 | 3 | alkene_catabolic_process |
| GO:0043461 | 3 | proton-transporting_ATP_synthase_complex_assembly |
| GO:0043476 | 3 | pigment_accumulation |
| GO:0043490 | 3 | malate-aspartate_shuttle |
| GO:0043533 | 3 | inositol_1,3,4,5_tetrakisphosphate_binding |
| GO:0043578 | 3 | nuclear_matrix_organization |
| GO:0043585 | 3 | nose_morphogenesis |
| GO:0043625 | 3 | delta_DNA_polymerase_complex |
| GO:0043633 | 3 | polyadenylation-dependent_RNA_catabolic_process |
| GO:0043650 | 3 | dicarboxylic_acid_biosynthetic_process |
| GO:0043654 | 3 | recognition_of_apoptotic_cell |
| GO:0043734 | 3 | DNA-N1-methyladenine_dioxygenase_activity |
| GO:0043932 | 3 | ossification_involved_in_bone_remodeling |
| GO:0044004 | 3 | disruption_by_symbiont_of_host_cells |
| GO:0044020 | 3 | histone_methyltransferase_activity_(H4-R3_specific) |
| GO:0044026 | 3 | DNA_hypermethylation |
| GO:0044027 | 3 | hypermethylation_of_CpG_island |
| GO:0044154 | 3 | histone_H3-K14_acetylation |
| GO:0044179 | 3 | hemolysis_in_other_organism |
| GO:0044205 | 3 | 'de_novo'_UMP_biosynthetic_process |
| GO:0044208 | 3 | 'de_novo'_AMP_biosynthetic_process |
| GO:0044211 | 3 | CTP_salvage |
| GO:0044240 | 3 | multicellular_organismal_lipid_catabolic_process |
| GO:0044292 | 3 | dendrite_terminus |
| GO:0044299 | 3 | C-fiber |
| GO:0044323 | 3 | retinoic_acid-responsive_element_binding |
| GO:0044340 | 3 | canonical_Wnt_receptor_signaling_pathway_involved_in_regulation_of_cell_proliferation |
| GO:0044413 | 3 | avoidance_of_host_defenses |
| GO:0044415 | 3 | evasion_or_tolerance_of_host_defenses |
| GO:0045007 | 3 | depurination |
| GO:0045048 | 3 | protein_insertion_into_ER_membrane |
| GO:0045063 | 3 | T-helper_1_cell_differentiation |
| GO:0045074 | 3 | regulation_of_interleukin-10_biosynthetic_process |
| GO:0045079 | 3 | negative_regulation_of_chemokine_biosynthetic_process |
| GO:0045098 | 3 | type_III_intermediate_filament |
| GO:0045143 | 3 | homologous_chromosome_segregation |
| GO:0045162 | 3 | clustering_of_voltage-gated_sodium_channels |
| GO:0045163 | 3 | clustering_of_voltage-gated_potassium_channels |
| GO:0045213 | 3 | neurotransmitter_receptor_metabolic_process |
| GO:0045218 | 3 | zonula_adherens_maintenance |
| GO:0045298 | 3 | tubulin_complex |
| GO:0045323 | 3 | interleukin-1_receptor_complex |
| GO:0045338 | 3 | farnesyl_diphosphate_metabolic_process |
| GO:0045404 | 3 | positive_regulation_of_interleukin-4_biosynthetic_process |
| GO:0045425 | 3 | positive_regulation_of_granulocyte_macrophage_colony-stimulating_factor_biosynthetic_process |
| GO:0045541 | 3 | negative_regulation_of_cholesterol_biosynthetic_process |
| GO:0045590 | 3 | negative_regulation_of_regulatory_T_cell_differentiation |
| GO:0045626 | 3 | negative_regulation_of_T-helper_1_cell_differentiation |
| GO:0045629 | 3 | negative_regulation_of_T-helper_2_cell_differentiation |
| GO:0045632 | 3 | negative_regulation_of_mechanoreceptor_differentiation |
| GO:0045653 | 3 | negative_regulation_of_megakaryocyte_differentiation |
| GO:0045656 | 3 | negative_regulation_of_monocyte_differentiation |
| GO:0045715 | 3 | negative_regulation_of_low-density_lipoprotein_particle_receptor_biosynthetic_process |
| GO:0045726 | 3 | positive_regulation_of_integrin_biosynthetic_process |
| GO:0045759 | 3 | negative_regulation_of_action_potential |
| GO:0045829 | 3 | negative_regulation_of_isotype_switching |
| GO:0045896 | 3 | regulation_of_transcription_during_mitosis |
| GO:0045897 | 3 | positive_regulation_of_transcription_during_mitosis |
| GO:0045899 | 3 | positive_regulation_of_RNA_polymerase_II_transcriptional_preinitiation_complex_assembly |
| GO:0045901 | 3 | positive_regulation_of_translational_elongation |
| GO:0045905 | 3 | positive_regulation_of_translational_termination |
| GO:0045908 | 3 | negative_regulation_of_vasodilation |
| GO:0045918 | 3 | negative_regulation_of_cytolysis |
| GO:0045953 | 3 | negative_regulation_of_natural_killer_cell_mediated_cytotoxicity |
| GO:0046005 | 3 | positive_regulation_of_circadian_sleep/wake_cycle,_REM_sleep |
| GO:0046010 | 3 | positive_regulation_of_circadian_sleep/wake_cycle,_non-REM_sleep |
| GO:0046016 | 3 | positive_regulation_of_transcription_by_glucose |
| GO:0046021 | 3 | regulation_of_transcription_from_RNA_polymerase_II_promoter,_mitotic |
| GO:0046022 | 3 | positive_regulation_of_transcription_from_RNA_polymerase_II_promoter_during_mitosis |
| GO:0046060 | 3 | dATP_metabolic_process |
| GO:0046075 | 3 | dTTP_metabolic_process |
| GO:0046078 | 3 | dUMP_metabolic_process |
| GO:0046080 | 3 | dUTP_metabolic_process |
| GO:0046083 | 3 | adenine_metabolic_process |
| GO:0046084 | 3 | adenine_biosynthetic_process |
| GO:0046098 | 3 | guanine_metabolic_process |
| GO:0046103 | 3 | inosine_biosynthetic_process |
| GO:0046121 | 3 | deoxyribonucleoside_catabolic_process |
| GO:0046125 | 3 | pyrimidine_deoxyribonucleoside_metabolic_process |
| GO:0046399 | 3 | glucuronate_biosynthetic_process |
| GO:0046404 | 3 | ATP-dependent_polydeoxyribonucleotide_5'-hydroxyl-kinase_activity |
| GO:0046476 | 3 | glycosylceramide_biosynthetic_process |
| GO:0046490 | 3 | isopentenyl_diphosphate_metabolic_process |
| GO:0046499 | 3 | S-adenosylmethioninamine_metabolic_process |
| GO:0046541 | 3 | saliva_secretion |
| GO:0046544 | 3 | development_of_secondary_male_sexual_characteristics |
| GO:0046582 | 3 | Rap_GTPase_activator_activity |
| GO:0046602 | 3 | regulation_of_mitotic_centrosome_separation |
| GO:0046633 | 3 | alpha-beta_T_cell_proliferation |
| GO:0046654 | 3 | tetrahydrofolate_biosynthetic_process |
| GO:0046703 | 3 | natural_killer_cell_lectin-like_receptor_binding |
| GO:0046848 | 3 | hydroxyapatite_binding |
| GO:0046878 | 3 | positive_regulation_of_saliva_secretion |
| GO:0046898 | 3 | response_to_cycloheximide |
| GO:0046958 | 3 | nonassociative_learning |
| GO:0046968 | 3 | peptide_antigen_transport |
| GO:0046975 | 3 | histone_methyltransferase_activity_(H3-K36_specific) |
| GO:0046976 | 3 | histone_methyltransferase_activity_(H3-K27_specific) |
| GO:0046980 | 3 | tapasin_binding |
| GO:0046984 | 3 | regulation_of_hemoglobin_biosynthetic_process |
| GO:0047045 | 3 | testosterone_17-beta-dehydrogenase_(NADP+)_activity |
| GO:0047086 | 3 | ketosteroid_monooxygenase_activity |
| GO:0047374 | 3 | methylumbelliferyl-acetate_deacetylase_activity |
| GO:0047484 | 3 | regulation_of_response_to_osmotic_stress |
| GO:0047718 | 3 | indanol_dehydrogenase_activity |
| GO:0047756 | 3 | chondroitin_4-sulfotransferase_activity |
| GO:0047894 | 3 | flavonol_3-sulfotransferase_activity |
| GO:0048003 | 3 | antigen_processing_and_presentation_of_lipid_antigen_via_MHC_class_Ib |
| GO:0048014 | 3 | Tie_receptor_signaling_pathway |
| GO:0048030 | 3 | disaccharide_binding |
| GO:0048039 | 3 | ubiquinone_binding |
| GO:0048143 | 3 | astrocyte_activation |
| GO:0048144 | 3 | fibroblast_proliferation |
| GO:0048170 | 3 | positive_regulation_of_long-term_neuronal_synaptic_plasticity |
| GO:0048179 | 3 | activin_receptor_complex |
| GO:0048273 | 3 | mitogen-activated_protein_kinase_p38_binding |
| GO:0048294 | 3 | negative_regulation_of_isotype_switching_to_IgE_isotypes |
| GO:0048295 | 3 | positive_regulation_of_isotype_switching_to_IgE_isotypes |
| GO:0048305 | 3 | immunoglobulin_secretion |
| GO:0048341 | 3 | paraxial_mesoderm_formation |
| GO:0048388 | 3 | endosomal_lumen_acidification |
| GO:0048478 | 3 | replication_fork_protection |
| GO:0048515 | 3 | spermatid_differentiation |
| GO:0048619 | 3 | embryonic_hindgut_morphogenesis |
| GO:0048625 | 3 | myoblast_cell_fate_commitment |
| GO:0048630 | 3 | skeletal_muscle_tissue_growth |
| GO:0048631 | 3 | regulation_of_skeletal_muscle_tissue_growth |
| GO:0048659 | 3 | smooth_muscle_cell_proliferation |
| GO:0048668 | 3 | collateral_sprouting |
| GO:0048769 | 3 | sarcomerogenesis |
| GO:0048791 | 3 | calcium_ion-dependent_exocytosis_of_neurotransmitter |
| GO:0048807 | 3 | female_genitalia_morphogenesis |
| GO:0048818 | 3 | positive_regulation_of_hair_follicle_maturation |
| GO:0048819 | 3 | regulation_of_hair_follicle_maturation |
| GO:0048880 | 3 | sensory_system_development |
| GO:0050213 | 3 | progesterone_5-alpha-reductase_activity |
| GO:0050220 | 3 | prostaglandin-E_synthase_activity |
| GO:0050254 | 3 | rhodopsin_kinase_activity |
| GO:0050265 | 3 | RNA_uridylyltransferase_activity |
| GO:0050294 | 3 | steroid_sulfotransferase_activity |
| GO:0050309 | 3 | sugar-terminal-phosphatase_activity |
| GO:0050432 | 3 | catecholamine_secretion |
| GO:0050508 | 3 | glucuronosyl-N-acetylglucosaminyl-proteoglycan_4-alpha-N-acetylglucosaminyltransferase_activity |
| GO:0050567 | 3 | glutaminyl-tRNA_synthase_(glutamine-hydrolyzing)_activity |
| GO:0050655 | 3 | dermatan_sulfate_proteoglycan_metabolic_process |
| GO:0050682 | 3 | AF-2_domain_binding |
| GO:0050698 | 3 | proteoglycan_sulfotransferase_activity |
| GO:0050711 | 3 | negative_regulation_of_interleukin-1_secretion |
| GO:0050779 | 3 | RNA_destabilization |
| GO:0050861 | 3 | positive_regulation_of_B_cell_receptor_signaling_pathway |
| GO:0050893 | 3 | sensory_processing |
| GO:0050904 | 3 | diapedesis |
| GO:0050951 | 3 | sensory_perception_of_temperature_stimulus |
| GO:0050955 | 3 | thermoception |
| GO:0051013 | 3 | microtubule_severing |
| GO:0051030 | 3 | snRNA_transport |
| GO:0051088 | 3 | PMA-inducible_membrane_protein_ectodomain_proteolysis |
| GO:0051097 | 3 | negative_regulation_of_helicase_activity |
| GO:0051140 | 3 | regulation_of_NK_T_cell_proliferation |
| GO:0051142 | 3 | positive_regulation_of_NK_T_cell_proliferation |
| GO:0051176 | 3 | positive_regulation_of_sulfur_metabolic_process |
| GO:0051286 | 3 | cell_tip |
| GO:0051305 | 3 | chromosome_movement_towards_spindle_pole |
| GO:0051321 | 3 | meiotic_cell_cycle |
| GO:0051329 | 3 | interphase_of_mitotic_cell_cycle |
| GO:0051429 | 3 | corticotropin-releasing_hormone_receptor_binding |
| GO:0051431 | 3 | corticotropin-releasing_hormone_receptor_2_binding |
| GO:0051451 | 3 | myoblast_migration |
| GO:0051525 | 3 | NFAT_protein_binding |
| GO:0051533 | 3 | positive_regulation_of_NFAT_protein_import_into_nucleus |
| GO:0051574 | 3 | positive_regulation_of_histone_H3-K9_methylation |
| GO:0051582 | 3 | positive_regulation_of_neurotransmitter_uptake |
| GO:0051583 | 3 | dopamine_uptake_involved_in_synaptic_transmission |
| GO:0051586 | 3 | positive_regulation_of_dopamine_uptake_involved_in_synaptic_transmission |
| GO:0051594 | 3 | detection_of_glucose |
| GO:0051645 | 3 | Golgi_localization |
| GO:0051653 | 3 | spindle_localization |
| GO:0051683 | 3 | establishment_of_Golgi_localization |
| GO:0051715 | 3 | cytolysis_in_other_organism |
| GO:0051733 | 3 | polydeoxyribonucleotide_kinase_activity |
| GO:0051734 | 3 | ATP-dependent_polynucleotide_kinase_activity |
| GO:0051771 | 3 | negative_regulation_of_nitric-oxide_synthase_biosynthetic_process |
| GO:0051791 | 3 | medium-chain_fatty_acid_metabolic_process |
| GO:0051801 | 3 | cytolysis_in_other_organism_involved_in_symbiotic_interaction |
| GO:0051825 | 3 | adhesion_to_other_organism_involved_in_symbiotic_interaction |
| GO:0051832 | 3 | avoidance_of_defenses_of_other_organism_involved_in_symbiotic_interaction |
| GO:0051834 | 3 | evasion_or_tolerance_of_defenses_of_other_organism_involved_in_symbiotic_interaction |
| GO:0051835 | 3 | positive_regulation_of_synapse_structural_plasticity |
| GO:0051856 | 3 | adhesion_to_symbiont |
| GO:0051877 | 3 | pigment_granule_aggregation_in_cell_center |
| GO:0051880 | 3 | G-quadruplex_DNA_binding |
| GO:0051933 | 3 | amino_acid_uptake_involved_in_synaptic_transmission |
| GO:0051934 | 3 | catecholamine_uptake_involved_in_synaptic_transmission |
| GO:0051935 | 3 | glutamate_uptake_involved_in_synaptic_transmission |
| GO:0051944 | 3 | positive_regulation_of_catecholamine_uptake_involved_in_synaptic_transmission |
| GO:0051956 | 3 | negative_regulation_of_amino_acid_transport |
| GO:0052025 | 3 | modification_by_symbiont_of_host_cell_membrane |
| GO:0052043 | 3 | modification_by_symbiont_of_host_cellular_component |
| GO:0052111 | 3 | modification_by_symbiont_of_host_structure |
| GO:0052185 | 3 | modification_of_structure_of_other_organism_involved_in_symbiotic_interaction |
| GO:0052188 | 3 | modification_of_cellular_component_in_other_organism_involved_in_symbiotic_interaction |
| GO:0052251 | 3 | induction_by_organism_of_defense_response_of_other_organism_involved_in_symbiotic_interaction |
| GO:0052331 | 3 | hemolysis_in_other_organism_involved_in_symbiotic_interaction |
| GO:0052332 | 3 | modification_by_organism_of_cell_membrane_in_other_organism_involved_in_symbiotic_interaction |
| GO:0052659 | 3 | inositol-1,3,4,5-tetrakisphosphate_5-phosphatase_activity |
| GO:0052832 | 3 | inositol_monophosphate_3-phosphatase_activity |
| GO:0052833 | 3 | inositol_monophosphate_4-phosphatase_activity |
| GO:0052834 | 3 | inositol_monophosphate_phosphatase_activity |
| GO:0052869 | 3 | arachidonic_acid_omega-hydroxylase_activity |
| GO:0055014 | 3 | atrial_cardiac_muscle_cell_development |
| GO:0055056 | 3 | D-glucose_transmembrane_transporter_activity |
| GO:0055077 | 3 | gap_junction_hemi-channel_activity |
| GO:0055087 | 3 | Ski_complex |
| GO:0055099 | 3 | response_to_high_density_lipoprotein_particle_stimulus |
| GO:0055118 | 3 | negative_regulation_of_cardiac_muscle_contraction |
| GO:0060014 | 3 | granulosa_cell_differentiation |
| GO:0060027 | 3 | convergent_extension_involved_in_gastrulation |
| GO:0060053 | 3 | neurofilament_cytoskeleton |
| GO:0060061 | 3 | Spemann_organizer_formation |
| GO:0060066 | 3 | oviduct_development |
| GO:0060074 | 3 | synapse_maturation |
| GO:0060075 | 3 | regulation_of_resting_membrane_potential |
| GO:0060084 | 3 | synaptic_transmission_involved_in_micturition |
| GO:0060099 | 3 | regulation_of_phagocytosis,_engulfment |
| GO:0060100 | 3 | positive_regulation_of_phagocytosis,_engulfment |
| GO:0060117 | 3 | auditory_receptor_cell_development |
| GO:0060126 | 3 | somatotropin_secreting_cell_differentiation |
| GO:0060129 | 3 | thyroid-stimulating_hormone-secreting_cell_differentiation |
| GO:0060148 | 3 | positive_regulation_of_posttranscriptional_gene_silencing |
| GO:0060154 | 3 | cellular_process_regulating_host_cell_cycle_in_response_to_virus |
| GO:0060161 | 3 | positive_regulation_of_dopamine_receptor_signaling_pathway |
| GO:0060166 | 3 | olfactory_pit_development |
| GO:0060177 | 3 | regulation_of_angiotensin_metabolic_process |
| GO:0060179 | 3 | male_mating_behavior |
| GO:0060197 | 3 | cloacal_septation |
| GO:0060201 | 3 | clathrin_sculpted_acetylcholine_transport_vesicle_membrane |
| GO:0060215 | 3 | primitive_hemopoiesis |
| GO:0060219 | 3 | camera-type_eye_photoreceptor_cell_differentiation |
| GO:0060230 | 3 | lipoprotein_lipase_activator_activity |
| GO:0060242 | 3 | contact_inhibition |
| GO:0060264 | 3 | regulation_of_respiratory_burst_involved_in_inflammatory_response |
| GO:0060278 | 3 | regulation_of_ovulation |
| GO:0060281 | 3 | regulation_of_oocyte_development |
| GO:0060286 | 3 | flagellar_cell_motility |
| GO:0060290 | 3 | transdifferentiation |
| GO:0060295 | 3 | regulation_of_cilium_movement_involved_in_cell_motility |
| GO:0060296 | 3 | regulation_of_cilium_beat_frequency_involved_in_ciliary_motility |
| GO:0060359 | 3 | response_to_ammonium_ion |
| GO:0060368 | 3 | regulation_of_Fc_receptor_mediated_stimulatory_signaling_pathway |
| GO:0060373 | 3 | regulation_of_ventricular_cardiomyocyte_membrane_depolarization |
| GO:0060375 | 3 | regulation_of_mast_cell_differentiation |
| GO:0060438 | 3 | trachea_development |
| GO:0060448 | 3 | dichotomous_subdivision_of_terminal_units_involved_in_lung_branching |
| GO:0060449 | 3 | bud_elongation_involved_in_lung_branching |
| GO:0060452 | 3 | positive_regulation_of_cardiac_muscle_contraction |
| GO:0060455 | 3 | negative_regulation_of_gastric_acid_secretion |
| GO:0060478 | 3 | acrosomal_vesicle_exocytosis |
| GO:0060486 | 3 | Clara_cell_differentiation |
| GO:0060492 | 3 | lung_induction |
| GO:0060509 | 3 | Type_I_pneumocyte_differentiation |
| GO:0060513 | 3 | prostatic_bud_formation |
| GO:0060541 | 3 | respiratory_system_development |
| GO:0060545 | 3 | positive_regulation_of_necroptosis |
| GO:0060553 | 3 | induction_of_necroptosis |
| GO:0060555 | 3 | induction_of_necroptosis_by_extracellular_signals |
| GO:0060559 | 3 | positive_regulation_of_calcidiol_1-monooxygenase_activity |
| GO:0060563 | 3 | neuroepithelial_cell_differentiation |
| GO:0060574 | 3 | intestinal_epithelial_cell_maturation |
| GO:0060576 | 3 | intestinal_epithelial_cell_development |
| GO:0060584 | 3 | regulation_of_prostaglandin-endoperoxide_synthase_activity |
| GO:0060585 | 3 | positive_regulation_of_prostaglandin-endoperoxide_synthase_activity |
| GO:0060592 | 3 | mammary_gland_formation |
| GO:0060613 | 3 | fat_pad_development |
| GO:0060620 | 3 | regulation_of_cholesterol_import |
| GO:0060675 | 3 | ureteric_bud_morphogenesis |
| GO:0060700 | 3 | regulation_of_ribonuclease_activity |
| GO:0060741 | 3 | prostate_gland_stromal_morphogenesis |
| GO:0060748 | 3 | tertiary_branching_involved_in_mammary_gland_duct_morphogenesis |
| GO:0060831 | 3 | smoothened_signaling_pathway_involved_in_dorsal/ventral_neural_tube_patterning |
| GO:0060897 | 3 | neural_plate_regionalization |
| GO:0060916 | 3 | mesenchymal_cell_proliferation_involved_in_lung_development |
| GO:0060947 | 3 | cardiac_vascular_smooth_muscle_cell_differentiation |
| GO:0060971 | 3 | embryonic_heart_tube_left/right_pattern_formation |
| GO:0061009 | 3 | common_bile_duct_development |
| GO:0061010 | 3 | gall_bladder_development |
| GO:0061042 | 3 | vascular_wound_healing |
| GO:0061082 | 3 | myeloid_leukocyte_cytokine_production |
| GO:0061083 | 3 | regulation_of_protein_refolding |
| GO:0061084 | 3 | negative_regulation_of_protein_refolding |
| GO:0061101 | 3 | neuroendocrine_cell_differentiation |
| GO:0061153 | 3 | trachea_gland_development |
| GO:0061184 | 3 | positive_regulation_of_dermatome_development |
| GO:0061198 | 3 | fungiform_papilla_formation |
| GO:0061205 | 3 | paramesonephric_duct_development |
| GO:0061304 | 3 | retinal_blood_vessel_morphogenesis |
| GO:0061308 | 3 | cardiac_neural_crest_cell_development_involved_in_heart_development |
| GO:0061364 | 3 | apoptotic_process_involved_in_luteolysis |
| GO:0061370 | 3 | testosterone_biosynthetic_process |
| GO:0061371 | 3 | determination_of_heart_left/right_asymmetry |
| GO:0070006 | 3 | metalloaminopeptidase_activity |
| GO:0070022 | 3 | transforming_growth_factor_beta_receptor_homodimeric_complex |
| GO:0070032 | 3 | synaptobrevin_2-SNAP-25-syntaxin-1a-complexin_I_complex |
| GO:0070052 | 3 | collagen_V_binding |
| GO:0070166 | 3 | enamel_mineralization |
| GO:0070172 | 3 | positive_regulation_of_tooth_mineralization |
| GO:0070188 | 3 | Stn1-Ten1_complex |
| GO:0070212 | 3 | protein_poly-ADP-ribosylation |
| GO:0070318 | 3 | positive_regulation_of_G0_to_G1_transition |
| GO:0070340 | 3 | detection_of_bacterial_lipopeptide |
| GO:0070346 | 3 | positive_regulation_of_fat_cell_proliferation |
| GO:0070366 | 3 | regulation_of_hepatocyte_differentiation |
| GO:0070375 | 3 | ERK5_cascade |
| GO:0070383 | 3 | DNA_cytosine_deamination |
| GO:0070487 | 3 | monocyte_aggregation |
| GO:0070508 | 3 | cholesterol_import |
| GO:0070545 | 3 | PeBoW_complex |
| GO:0070548 | 3 | L-glutamine_aminotransferase_activity |
| GO:0070576 | 3 | vitamin_D_24-hydroxylase_activity |
| GO:0070602 | 3 | regulation_of_centromeric_sister_chromatid_cohesion |
| GO:0070634 | 3 | transepithelial_ammonium_transport |
| GO:0070643 | 3 | vitamin_D_25-hydroxylase_activity |
| GO:0070644 | 3 | vitamin_D_response_element_binding |
| GO:0070653 | 3 | high-density_lipoprotein_particle_receptor_binding |
| GO:0070666 | 3 | regulation_of_mast_cell_proliferation |
| GO:0070668 | 3 | positive_regulation_of_mast_cell_proliferation |
| GO:0070681 | 3 | glutaminyl-tRNAGln_biosynthesis_via_transamidation |
| GO:0070682 | 3 | proteasome_regulatory_particle_assembly |
| GO:0070699 | 3 | type_II_activin_receptor_binding |
| GO:0070735 | 3 | protein-glycine_ligase_activity |
| GO:0070777 | 3 | D-aspartate_transport |
| GO:0070779 | 3 | D-aspartate_import |
| GO:0070820 | 3 | tertiary_granule |
| GO:0070861 | 3 | regulation_of_protein_exit_from_endoplasmic_reticulum |
| GO:0070886 | 3 | positive_regulation_of_calcineurin-NFAT_signaling_cascade |
| GO:0070891 | 3 | lipoteichoic_acid_binding |
| GO:0070897 | 3 | DNA-dependent_transcriptional_preinitiation_complex_assembly |
| GO:0070914 | 3 | UV-damage_excision_repair |
| GO:0070915 | 3 | lysophosphatidic_acid_receptor_activity |
| GO:0070940 | 3 | dephosphorylation_of_RNA_polymerase_II_C-terminal_domain |
| GO:0070944 | 3 | neutrophil_mediated_killing_of_bacterium |
| GO:0070946 | 3 | neutrophil_mediated_killing_of_gram-positive_bacterium |
| GO:0070947 | 3 | neutrophil_mediated_killing_of_fungus |
| GO:0070969 | 3 | ULK1-ATG13-FIP200_complex |
| GO:0070972 | 3 | protein_localization_in_endoplasmic_reticulum |
| GO:0070996 | 3 | type_1_melanocortin_receptor_binding |
| GO:0071001 | 3 | U4/U6_snRNP |
| GO:0071034 | 3 | CUT_catabolic_process |
| GO:0071043 | 3 | CUT_metabolic_process |
| GO:0071062 | 3 | alphav-beta3_integrin-vitronectin_complex |
| GO:0071168 | 3 | protein_localization_to_chromatin |
| GO:0071224 | 3 | cellular_response_to_peptidoglycan |
| GO:0071288 | 3 | cellular_response_to_mercury_ion |
| GO:0071315 | 3 | cellular_response_to_morphine |
| GO:0071317 | 3 | cellular_response_to_isoquinoline_alkaloid |
| GO:0071321 | 3 | cellular_response_to_cGMP |
| GO:0071336 | 3 | regulation_of_hair_follicle_cell_proliferation |
| GO:0071349 | 3 | cellular_response_to_interleukin-12 |
| GO:0071400 | 3 | cellular_response_to_oleic_acid |
| GO:0071402 | 3 | cellular_response_to_lipoprotein_particle_stimulus |
| GO:0071420 | 3 | cellular_response_to_histamine |
| GO:0071421 | 3 | manganese_ion_transmembrane_transport |
| GO:0071435 | 3 | potassium_ion_export |
| GO:0071437 | 3 | invadopodium |
| GO:0071438 | 3 | invadopodium_membrane |
| GO:0071447 | 3 | cellular_response_to_hydroperoxide |
| GO:0071459 | 3 | protein_localization_to_chromosome,_centromeric_region |
| GO:0071472 | 3 | cellular_response_to_salt_stress |
| GO:0071481 | 3 | cellular_response_to_X-ray |
| GO:0071499 | 3 | cellular_response_to_laminar_fluid_shear_stress |
| GO:0071532 | 3 | ankyrin_repeat_binding |
| GO:0071603 | 3 | endothelial_cell-cell_adhesion |
| GO:0071639 | 3 | positive_regulation_of_monocyte_chemotactic_protein-1_production |
| GO:0071664 | 3 | catenin-TCF7L2_complex |
| GO:0071671 | 3 | regulation_of_smooth_muscle_cell_chemotaxis |
| GO:0071674 | 3 | mononuclear_cell_migration |
| GO:0071688 | 3 | striated_muscle_myosin_thick_filament_assembly |
| GO:0071797 | 3 | LUBAC_complex |
| GO:0071803 | 3 | positive_regulation_of_podosome_assembly |
| GO:0071816 | 3 | tail-anchored_membrane_protein_insertion_into_ER_membrane |
| GO:0071818 | 3 | BAT3_complex |
| GO:0071821 | 3 | FANCM-MHF_complex |
| GO:0071863 | 3 | regulation_of_cell_proliferation_in_bone_marrow |
| GO:0071864 | 3 | positive_regulation_of_cell_proliferation_in_bone_marrow |
| GO:0071871 | 3 | response_to_epinephrine_stimulus |
| GO:0071872 | 3 | cellular_response_to_epinephrine_stimulus |
| GO:0071877 | 3 | regulation_of_adrenergic_receptor_signaling_pathway |
| GO:0071883 | 3 | activation_of_MAPK_activity_by_adrenergic_receptor_signaling_pathway |
| GO:0071922 | 3 | regulation_of_cohesin_localization_to_chromatin |
| GO:0071942 | 3 | XPC_complex |
| GO:0071986 | 3 | Ragulator_complex |
| GO:0072014 | 3 | proximal_tubule_development |
| GO:0072017 | 3 | distal_tubule_development |
| GO:0072034 | 3 | renal_vesicle_induction |
| GO:0072039 | 3 | regulation_of_mesenchymal_stem_cell_apoptotic_process_involved_in_nephron_morphogenesis |
| GO:0072040 | 3 | negative_regulation_of_mesenchymal_stem_cell_apoptotic_process_involved_in_nephron_morphogenesis |
| GO:0072081 | 3 | specification_of_nephron_tubule_identity |
| GO:0072102 | 3 | glomerulus_morphogenesis |
| GO:0072110 | 3 | glomerular_mesangial_cell_proliferation |
| GO:0072125 | 3 | negative_regulation_of_glomerular_mesangial_cell_proliferation |
| GO:0072131 | 3 | kidney_mesenchyme_morphogenesis |
| GO:0072133 | 3 | metanephric_mesenchyme_morphogenesis |
| GO:0072135 | 3 | kidney_mesenchymal_cell_proliferation |
| GO:0072136 | 3 | metanephric_mesenchymal_cell_proliferation_involved_in_metanephros_development |
| GO:0072138 | 3 | mesenchymal_cell_proliferation_involved_in_ureteric_bud_development |
| GO:0072144 | 3 | glomerular_mesangial_cell_development |
| GO:0072160 | 3 | nephron_tubule_epithelial_cell_differentiation |
| GO:0072172 | 3 | mesonephric_tubule_formation |
| GO:0072183 | 3 | negative_regulation_of_nephron_tubule_epithelial_cell_differentiation |
| GO:0072193 | 3 | ureter_smooth_muscle_cell_differentiation |
| GO:0072216 | 3 | positive_regulation_of_metanephros_development |
| GO:0072234 | 3 | metanephric_nephron_tubule_development |
| GO:0072243 | 3 | metanephric_nephron_epithelium_development |
| GO:0072277 | 3 | metanephric_glomerular_capillary_formation |
| GO:0072300 | 3 | positive_regulation_of_metanephric_glomerulus_development |
| GO:0072379 | 3 | ER_membrane_insertion_complex |
| GO:0072385 | 3 | minus-end-directed_organelle_transport_along_microtubule |
| GO:0072488 | 3 | ammonium_transmembrane_transport |
| GO:0072520 | 3 | seminiferous_tubule_development |
| GO:0072534 | 3 | perineuronal_net |
| GO:0072537 | 3 | fibroblast_activation |
| GO:0072542 | 3 | protein_phosphatase_activator_activity |
| GO:0072553 | 3 | terminal_button_organization |
| GO:0072592 | 3 | oxygen_metabolic_process |
| GO:0072643 | 3 | interferon-gamma_secretion |
| GO:0080084 | 3 | 5S_rDNA_binding |
| GO:0080130 | 3 | L-phenylalanine:2-oxoglutarate_aminotransferase_activity |
| GO:0080182 | 3 | histone_H3-K4_trimethylation |
| GO:0086070 | 3 | SA_node_cardiomyocyte_to_atrial_cardiomyocyte_communication |
| GO:0090027 | 3 | negative_regulation_of_monocyte_chemotaxis |
| GO:0090045 | 3 | positive_regulation_of_deacetylase_activity |
| GO:0090051 | 3 | negative_regulation_of_cell_migration_involved_in_sprouting_angiogenesis |
| GO:0090077 | 3 | foam_cell_differentiation |
| GO:0090081 | 3 | regulation_of_heart_induction_by_regulation_of_canonical_Wnt_receptor_signaling_pathway |
| GO:0090084 | 3 | negative_regulation_of_inclusion_body_assembly |
| GO:0090085 | 3 | regulation_of_protein_deubiquitination |
| GO:0090130 | 3 | tissue_migration |
| GO:0090153 | 3 | regulation_of_sphingolipid_biosynthetic_process |
| GO:0090161 | 3 | Golgi_ribbon_formation |
| GO:0090186 | 3 | regulation_of_pancreatic_juice_secretion |
| GO:0090188 | 3 | negative_regulation_of_pancreatic_juice_secretion |
| GO:0090206 | 3 | negative_regulation_of_cholesterol_metabolic_process |
| GO:0090230 | 3 | regulation_of_centromere_complex_assembly |
| GO:0090237 | 3 | regulation_of_arachidonic_acid_secretion |
| GO:0090240 | 3 | positive_regulation_of_histone_H4_acetylation |
| GO:0090244 | 3 | Wnt_receptor_signaling_pathway_involved_in_somitogenesis |
| GO:0090267 | 3 | positive_regulation_of_mitotic_cell_cycle_spindle_assembly_checkpoint |
| GO:0090289 | 3 | regulation_of_osteoclast_proliferation |
| GO:0090292 | 3 | nuclear_matrix_anchoring_at_nuclear_membrane |
| GO:0090310 | 3 | negative_regulation_of_methylation-dependent_chromatin_silencing |
| GO:0090331 | 3 | negative_regulation_of_platelet_aggregation |
| GO:0090394 | 3 | negative_regulation_of_excitatory_postsynaptic_membrane_potential |
| GO:0090403 | 3 | oxidative_stress-induced_premature_senescence |
| GO:0097011 | 3 | cellular_response_to_granulocyte_macrophage_colony-stimulating_factor_stimulus |
| GO:0097012 | 3 | response_to_granulocyte_macrophage_colony-stimulating_factor_stimulus |
| GO:0097021 | 3 | lymphocyte_migration_into_lymphoid_organs |
| GO:0097038 | 3 | perinuclear_endoplasmic_reticulum |
| GO:0097039 | 3 | protein_linear_polyubiquitination |
| GO:0097058 | 3 | CRLF-CLCF1_complex |
| GO:0097070 | 3 | ductus_arteriosus_closure |
| GO:0097091 | 3 | synaptic_vesicle_clustering |
| GO:0097108 | 3 | hedgehog_family_protein_binding |
| GO:0097113 | 3 | alpha-amino-3-hydroxy-5-methyl-4-isoxazole_propionate_receptor_clustering |
| GO:0097116 | 3 | gephyrin_clustering |
| GO:0097120 | 3 | receptor_localization_to_synapse |
| GO:0097149 | 3 | centralspindlin_complex |
| GO:0097194 | 3 | execution_phase_of_apoptosis |
| GO:0097210 | 3 | response_to_gonadotropin-releasing_hormone |
| GO:0097211 | 3 | cellular_response_to_gonadotropin-releasing_hormone |
| GO:1900037 | 3 | regulation_of_cellular_response_to_hypoxia |
| GO:1900063 | 3 | regulation_of_peroxisome_organization |
| GO:1900104 | 3 | regulation_of_hyaluranon_cable_assembly |
| GO:1900106 | 3 | positive_regulation_of_hyaluranon_cable_assembly |
| GO:1900121 | 3 | negative_regulation_of_receptor_binding |
| GO:1900127 | 3 | positive_regulation_of_hyaluronan_biosynthetic_process |
| GO:1900175 | 3 | regulation_of_nodal_signaling_pathway_involved_in_determination_of_lateral_mesoderm_left/right_asymmetry |
| GO:1900238 | 3 | regulation_of_metanephric_mesenchymal_cell_migration_by_platelet-derived_growth_factor_receptor-beta_signaling_pathway |
| GO:1900424 | 3 | regulation_of_defense_response_to_bacterium |
| GO:1900755 | 3 | branched-chain_amino-acid_anion_transport |
| GO:1901099 | 3 | negative_regulation_of_signal_transduction_in_absence_of_ligand |
| GO:2000009 | 3 | negative_regulation_of_protein_localization_at_cell_surface |
| GO:2000010 | 3 | positive_regulation_of_protein_localization_at_cell_surface |
| GO:2000017 | 3 | positive_regulation_of_determination_of_dorsal_identity |
| GO:2000044 | 3 | negative_regulation_of_cardiac_cell_fate_specification |
| GO:2000054 | 3 | negative_regulation_of_Wnt_receptor_signaling_pathway_involved_in_dorsal/ventral_axis_specification |
| GO:2000059 | 3 | negative_regulation_of_protein_ubiquitination_involved_in_ubiquitin-dependent_protein_catabolic_process |
| GO:2000078 | 3 | positive_regulation_of_type_B_pancreatic_cell_development |
| GO:2000096 | 3 | positive_regulation_of_Wnt_receptor_signaling_pathway,_planar_cell_polarity_pathway |
| GO:2000097 | 3 | regulation_of_smooth_muscle_cell-matrix_adhesion |
| GO:2000143 | 3 | negative_regulation_of_DNA-dependent_transcription,_initiation |
| GO:2000189 | 3 | positive_regulation_of_cholesterol_homeostasis |
| GO:2000195 | 3 | negative_regulation_of_female_gonad_development |
| GO:2000269 | 3 | regulation_of_fibroblast_apoptotic_process |
| GO:2000270 | 3 | negative_regulation_of_fibroblast_apoptotic_process |
| GO:2000286 | 3 | receptor_internalization_involved_in_canonical_Wnt_receptor_signaling_pathway |
| GO:2000303 | 3 | regulation_of_ceramide_biosynthetic_process |
| GO:2000343 | 3 | positive_regulation_of_chemokine_(C-X-C_motif)_ligand_2_production |
| GO:2000347 | 3 | positive_regulation_of_hepatocyte_proliferation |
| GO:2000359 | 3 | regulation_of_binding_of_sperm_to_zona_pellucida |
| GO:2000360 | 3 | negative_regulation_of_binding_of_sperm_to_zona_pellucida |
| GO:2000364 | 3 | regulation_of_STAT_protein_import_into_nucleus |
| GO:2000366 | 3 | positive_regulation_of_STAT_protein_import_into_nucleus |
| GO:2000402 | 3 | negative_regulation_of_lymphocyte_migration |
| GO:2000425 | 3 | regulation_of_apoptotic_cell_clearance |
| GO:2000466 | 3 | negative_regulation_of_glycogen_(starch)_synthase_activity |
| GO:2000467 | 3 | positive_regulation_of_glycogen_(starch)_synthase_activity |
| GO:2000508 | 3 | regulation_of_dendritic_cell_chemotaxis |
| GO:2000510 | 3 | positive_regulation_of_dendritic_cell_chemotaxis |
| GO:2000547 | 3 | regulation_of_dendritic_cell_dendrite_assembly |
| GO:2000553 | 3 | positive_regulation_of_T-helper_2_cell_cytokine_production |
| GO:2000591 | 3 | positive_regulation_of_metanephric_mesenchymal_cell_migration |
| GO:2000609 | 3 | regulation_of_thyroid_hormone_generation |
| GO:2000611 | 3 | positive_regulation_of_thyroid_hormone_generation |
| GO:2000637 | 3 | positive_regulation_of_gene_silencing_by_miRNA |
| GO:2000659 | 3 | regulation_of_interleukin-1-mediated_signaling_pathway |
| GO:2000662 | 3 | regulation_of_interleukin-5_secretion |
| GO:2000665 | 3 | regulation_of_interleukin-13_secretion |
| GO:2000669 | 3 | negative_regulation_of_dendritic_cell_apoptotic_process |
| GO:2000697 | 3 | negative_regulation_of_epithelial_cell_differentiation_involved_in_kidney_development |
| GO:2000698 | 3 | positive_regulation_of_epithelial_cell_differentiation_involved_in_kidney_development |
| GO:2000721 | 3 | positive_regulation_of_transcription_from_RNA_polymerase_II_promoter_involved_in_smooth_muscle_cell_differentiation |
| GO:2000729 | 3 | positive_regulation_of_mesenchymal_cell_proliferation_involved_in_ureter_development |
| GO:2000738 | 3 | positive_regulation_of_stem_cell_differentiation |
| GO:2000741 | 3 | positive_regulation_of_mesenchymal_stem_cell_differentiation |
| GO:2000768 | 3 | positive_regulation_of_nephron_tubule_epithelial_cell_differentiation |
| GO:2000773 | 3 | negative_regulation_of_cellular_senescence |
| GO:2000822 | 3 | regulation_of_behavioral_fear_response |
| GO:2000833 | 3 | positive_regulation_of_steroid_hormone_secretion |
| GO:2000909 | 3 | regulation_of_sterol_import |
| GO:2000969 | 3 | positive_regulation_of_alpha-amino-3-hydroxy-5-methyl-4-isoxazole_propionate_selective_glutamate_receptor_activity |
| GO:2000973 | 3 | regulation_of_pro-B_cell_differentiation |
| GO:2000974 | 3 | negative_regulation_of_pro-B_cell_differentiation |
| GO:2000981 | 3 | negative_regulation_of_inner_ear_receptor_cell_differentiation |
| GO:2001013 | 3 | epithelial_cell_proliferation_involved_in_renal_tubule_morphogenesis |
| GO:2001028 | 3 | positive_regulation_of_endothelial_cell_chemotaxis |
| GO:2001038 | 3 | regulation_of_cellular_response_to_drug |
| GO:2001070 | 3 | starch_binding |
| GO:2001179 | 3 | regulation_of_interleukin-10_secretion |
| GO:2001240 | 3 | negative_regulation_of_extrinsic_apoptotic_signaling_pathway_in_absence_of_ligand |
| GO:0000010 | 2 | trans-hexaprenyltranstransferase_activity |
| GO:0000022 | 2 | mitotic_spindle_elongation |
| GO:0000059 | 2 | protein_import_into_nucleus,_docking |
| GO:0000066 | 2 | mitochondrial_ornithine_transport |
| GO:0000072 | 2 | M_phase_specific_microtubule_process |
| GO:0000088 | 2 | mitotic_prophase |
| GO:0000105 | 2 | histidine_biosynthetic_process |
| GO:0000114 | 2 | regulation_of_transcription_involved_in_G1_phase_of_mitotic_cell_cycle |
| GO:0000115 | 2 | regulation_of_transcription_involved_in_S_phase_of_mitotic_cell_cycle |
| GO:0000117 | 2 | regulation_of_transcription_involved_in_G2/M-phase_of_mitotic_cell_cycle |
| GO:0000154 | 2 | rRNA_modification |
| GO:0000156 | 2 | two-component_response_regulator_activity |
| GO:0000172 | 2 | ribonuclease_MRP_complex |
| GO:0000189 | 2 | MAPK_import_into_nucleus |
| GO:0000213 | 2 | tRNA-intron_endonuclease_activity |
| GO:0000221 | 2 | vacuolar_proton-transporting_V-type_ATPase,_V1_domain |
| GO:0000248 | 2 | C-5_sterol_desaturase_activity |
| GO:0000290 | 2 | deadenylation-dependent_decapping_of_nuclear-transcribed_mRNA |
| GO:0000295 | 2 | adenine_nucleotide_transmembrane_transporter_activity |
| GO:0000308 | 2 | cytoplasmic_cyclin-dependent_protein_kinase_holoenzyme_complex |
| GO:0000340 | 2 | RNA_7-methylguanosine_cap_binding |
| GO:0000354 | 2 | cis_assembly_of_pre-catalytic_spliceosome |
| GO:0000406 | 2 | double-strand/single-strand_DNA_junction_binding |
| GO:0000414 | 2 | regulation_of_histone_H3-K36_methylation |
| GO:0000441 | 2 | SSL2-core_TFIIH_complex |
| GO:0000461 | 2 | endonucleolytic_cleavage_to_generate_mature_3'-end_of_SSU-rRNA_from_(SSU-rRNA,_5.8S_rRNA,_LSU-rRNA) |
| GO:0000466 | 2 | maturation_of_5.8S_rRNA_from_tricistronic_rRNA_transcript_(SSU-rRNA,_5.8S_rRNA,_LSU-rRNA) |
| GO:0000472 | 2 | endonucleolytic_cleavage_to_generate_mature_5'-end_of_SSU-rRNA_from_(SSU-rRNA,_5.8S_rRNA,_LSU-rRNA) |
| GO:0000480 | 2 | endonucleolytic_cleavage_in_5'-ETS_of_tricistronic_rRNA_transcript_(SSU-rRNA,_5.8S_rRNA,_LSU-rRNA) |
| GO:0000700 | 2 | mismatch_base_pair_DNA_N-glycosylase_activity |
| GO:0000715 | 2 | nucleotide-excision_repair,_DNA_damage_recognition |
| GO:0000774 | 2 | adenyl-nucleotide_exchange_factor_activity |
| GO:0000802 | 2 | transverse_filament |
| GO:0000806 | 2 | Y_chromosome |
| GO:0000815 | 2 | ESCRT_III_complex |
| GO:0000827 | 2 | inositol-1,3,4,5,6-pentakisphosphate_kinase_activity |
| GO:0000917 | 2 | barrier_septum_assembly |
| GO:0000939 | 2 | condensed_chromosome_inner_kinetochore |
| GO:0000957 | 2 | mitochondrial_RNA_catabolic_process |
| GO:0000958 | 2 | mitochondrial_mRNA_catabolic_process |
| GO:0000960 | 2 | regulation_of_mitochondrial_RNA_catabolic_process |
| GO:0000962 | 2 | positive_regulation_of_mitochondrial_RNA_catabolic_process |
| GO:0000963 | 2 | mitochondrial_RNA_processing |
| GO:0000965 | 2 | mitochondrial_RNA_3'-end_processing |
| GO:0000967 | 2 | rRNA_5'-end_processing |
| GO:0001025 | 2 | RNA_polymerase_III_transcription_factor_binding |
| GO:0001134 | 2 | transcription_factor_recruiting_transcription_factor_activity |
| GO:0001156 | 2 | TFIIIC-class_transcription_factor_binding |
| GO:0001300 | 2 | chronological_cell_aging |
| GO:0001301 | 2 | progressive_alteration_of_chromatin_involved_in_cell_aging |
| GO:0001304 | 2 | progressive_alteration_of_chromatin_involved_in_replicative_cell_aging |
| GO:0001309 | 2 | age-dependent_telomere_shortening |
| GO:0001507 | 2 | acetylcholine_catabolic_process_in_synaptic_cleft |
| GO:0001546 | 2 | preantral_ovarian_follicle_growth |
| GO:0001550 | 2 | ovarian_cumulus_expansion |
| GO:0001555 | 2 | oocyte_growth |
| GO:0001581 | 2 | detection_of_chemical_stimulus_involved_in_sensory_perception_of_sour_taste |
| GO:0001582 | 2 | detection_of_chemical_stimulus_involved_in_sensory_perception_of_sweet_taste |
| GO:0001588 | 2 | dopamine_receptor_activity,_coupled_via_Gs |
| GO:0001605 | 2 | adrenomedullin_receptor_activity |
| GO:0001607 | 2 | neuromedin_U_receptor_activity |
| GO:0001635 | 2 | calcitonin_gene-related_polypeptide_receptor_activity |
| GO:0001641 | 2 | group_II_metabotropic_glutamate_receptor_activity |
| GO:0001642 | 2 | group_III_metabotropic_glutamate_receptor_activity |
| GO:0001650 | 2 | fibrillar_center |
| GO:0001692 | 2 | histamine_metabolic_process |
| GO:0001694 | 2 | histamine_biosynthetic_process |
| GO:0001705 | 2 | ectoderm_formation |
| GO:0001729 | 2 | ceramide_kinase_activity |
| GO:0001732 | 2 | formation_of_translation_initiation_complex |
| GO:0001765 | 2 | membrane_raft_assembly |
| GO:0001785 | 2 | prostaglandin_J_receptor_activity |
| GO:0001803 | 2 | regulation_of_type_III_hypersensitivity |
| GO:0001805 | 2 | positive_regulation_of_type_III_hypersensitivity |
| GO:0001808 | 2 | negative_regulation_of_type_IV_hypersensitivity |
| GO:0001810 | 2 | regulation_of_type_I_hypersensitivity |
| GO:0001812 | 2 | positive_regulation_of_type_I_hypersensitivity |
| GO:0001831 | 2 | trophectodermal_cellular_morphogenesis |
| GO:0001845 | 2 | phagolysosome_assembly |
| GO:0001865 | 2 | NK_T_cell_differentiation |
| GO:0001866 | 2 | NK_T_cell_proliferation |
| GO:0001868 | 2 | regulation_of_complement_activation,_lectin_pathway |
| GO:0001869 | 2 | negative_regulation_of_complement_activation,_lectin_pathway |
| GO:0001872 | 2 | (1->3)-beta-D-glucan_binding |
| GO:0001887 | 2 | selenium_compound_metabolic_process |
| GO:0001888 | 2 | glucuronyl-galactosyl-proteoglycan_4-alpha-N-acetylglucosaminyltransferase_activity |
| GO:0001920 | 2 | negative_regulation_of_receptor_recycling |
| GO:0001994 | 2 | norepinephrine-epinephrine_vasoconstriction_involved_in_regulation_of_systemic_arterial_blood_pressure |
| GO:0001996 | 2 | positive_regulation_of_heart_rate_by_epinephrine-norepinephrine |
| GO:0002002 | 2 | regulation_of_angiotensin_levels_in_blood |
| GO:0002003 | 2 | angiotensin_maturation |
| GO:0002017 | 2 | regulation_of_blood_volume_by_renal_aldosterone |
| GO:0002024 | 2 | diet_induced_thermogenesis |
| GO:0002035 | 2 | brain_renin-angiotensin_system |
| GO:0002036 | 2 | regulation_of_L-glutamate_transport |
| GO:0002046 | 2 | opsin_binding |
| GO:0002051 | 2 | osteoblast_fate_commitment |
| GO:0002072 | 2 | optic_cup_morphogenesis_involved_in_camera-type_eye_development |
| GO:0002074 | 2 | extraocular_skeletal_muscle_development |
| GO:0002079 | 2 | inner_acrosomal_membrane |
| GO:0002081 | 2 | outer_acrosomal_membrane |
| GO:0002082 | 2 | regulation_of_oxidative_phosphorylation |
| GO:0002086 | 2 | diaphragm_contraction |
| GO:0002091 | 2 | negative_regulation_of_receptor_internalization |
| GO:0002115 | 2 | store-operated_calcium_entry |
| GO:0002133 | 2 | polycystin_complex |
| GO:0002134 | 2 | UTP_binding |
| GO:0002138 | 2 | retinoic_acid_biosynthetic_process |
| GO:0002151 | 2 | G-quadruplex_RNA_binding |
| GO:0002176 | 2 | male_germ_cell_proliferation |
| GO:0002177 | 2 | manchette |
| GO:0002223 | 2 | stimulatory_C-type_lectin_receptor_signaling_pathway |
| GO:0002246 | 2 | wound_healing_involved_in_inflammatory_response |
| GO:0002266 | 2 | follicular_dendritic_cell_activation |
| GO:0002268 | 2 | follicular_dendritic_cell_differentiation |
| GO:0002278 | 2 | eosinophil_activation_involved_in_immune_response |
| GO:0002291 | 2 | T_cell_activation_via_T_cell_receptor_contact_with_antigen_bound_to_MHC_molecule_on_antigen_presenting_cell |
| GO:0002296 | 2 | T-helper_1_cell_lineage_commitment |
| GO:0002302 | 2 | CD8-positive,_alpha-beta_T_cell_differentiation_involved_in_immune_response |
| GO:0002315 | 2 | marginal_zone_B_cell_differentiation |
| GO:0002339 | 2 | B_cell_selection |
| GO:0002357 | 2 | defense_response_to_tumor_cell |
| GO:0002378 | 2 | immunoglobulin_biosynthetic_process |
| GO:0002384 | 2 | hepatic_immune_response |
| GO:0002408 | 2 | myeloid_dendritic_cell_chemotaxis |
| GO:0002418 | 2 | immune_response_to_tumor_cell |
| GO:0002428 | 2 | antigen_processing_and_presentation_of_peptide_antigen_via_MHC_class_Ib |
| GO:0002431 | 2 | Fc_receptor_mediated_stimulatory_signaling_pathway |
| GO:0002438 | 2 | acute_inflammatory_response_to_antigenic_stimulus |
| GO:0002439 | 2 | chronic_inflammatory_response_to_antigenic_stimulus |
| GO:0002441 | 2 | histamine_secretion_involved_in_inflammatory_response |
| GO:0002477 | 2 | antigen_processing_and_presentation_of_exogenous_peptide_antigen_via_MHC_class_Ib |
| GO:0002481 | 2 | antigen_processing_and_presentation_of_exogenous_protein_antigen_via_MHC_class_Ib,_TAP-dependent |
| GO:0002522 | 2 | leukocyte_migration_involved_in_immune_response |
| GO:0002528 | 2 | regulation_of_vascular_permeability_involved_in_acute_inflammatory_response |
| GO:0002537 | 2 | nitric_oxide_production_involved_in_inflammatory_response |
| GO:0002538 | 2 | arachidonic_acid_metabolite_production_involved_in_inflammatory_response |
| GO:0002540 | 2 | leukotriene_production_involved_in_inflammatory_response |
| GO:0002542 | 2 | Factor_XII_activation |
| GO:0002543 | 2 | activation_of_blood_coagulation_via_clotting_cascade |
| GO:0002553 | 2 | histamine_secretion_by_mast_cell |
| GO:0002578 | 2 | negative_regulation_of_antigen_processing_and_presentation |
| GO:0002635 | 2 | negative_regulation_of_germinal_center_formation |
| GO:0002661 | 2 | regulation_of_B_cell_tolerance_induction |
| GO:0002663 | 2 | positive_regulation_of_B_cell_tolerance_induction |
| GO:0002692 | 2 | negative_regulation_of_cellular_extravasation |
| GO:0002713 | 2 | negative_regulation_of_B_cell_mediated_immunity |
| GO:0002727 | 2 | regulation_of_natural_killer_cell_cytokine_production |
| GO:0002729 | 2 | positive_regulation_of_natural_killer_cell_cytokine_production |
| GO:0002741 | 2 | positive_regulation_of_cytokine_secretion_involved_in_immune_response |
| GO:0002752 | 2 | cell_surface_pattern_recognition_receptor_signaling_pathway |
| GO:0002759 | 2 | regulation_of_antimicrobial_humoral_response |
| GO:0002784 | 2 | regulation_of_antimicrobial_peptide_production |
| GO:0002786 | 2 | regulation_of_antibacterial_peptide_production |
| GO:0002805 | 2 | regulation_of_antimicrobial_peptide_biosynthetic_process |
| GO:0002807 | 2 | positive_regulation_of_antimicrobial_peptide_biosynthetic_process |
| GO:0002808 | 2 | regulation_of_antibacterial_peptide_biosynthetic_process |
| GO:0002816 | 2 | regulation_of_biosynthetic_process_of_antibacterial_peptides_active_against_Gram-positive_bacteria |
| GO:0002876 | 2 | positive_regulation_of_chronic_inflammatory_response_to_antigenic_stimulus |
| GO:0002884 | 2 | negative_regulation_of_hypersensitivity |
| GO:0002890 | 2 | negative_regulation_of_immunoglobulin_mediated_immune_response |
| GO:0002924 | 2 | negative_regulation_of_humoral_immune_response_mediated_by_circulating_immunoglobulin |
| GO:0002934 | 2 | desmosome_organization |
| GO:0003010 | 2 | voluntary_skeletal_muscle_contraction |
| GO:0003011 | 2 | involuntary_skeletal_muscle_contraction |
| GO:0003051 | 2 | angiotensin-mediated_drinking_behavior |
| GO:0003062 | 2 | regulation_of_heart_rate_by_chemical_signal |
| GO:0003138 | 2 | primary_heart_field_specification |
| GO:0003149 | 2 | membranous_septum_morphogenesis |
| GO:0003157 | 2 | endocardium_development |
| GO:0003185 | 2 | sinoatrial_valve_morphogenesis |
| GO:0003195 | 2 | tricuspid_valve_formation |
| GO:0003199 | 2 | endocardial_cushion_to_mesenchymal_transition_involved_in_heart_valve_formation |
| GO:0003218 | 2 | cardiac_left_ventricle_formation |
| GO:0003221 | 2 | right_ventricular_cardiac_muscle_tissue_morphogenesis |
| GO:0003272 | 2 | endocardial_cushion_formation |
| GO:0003278 | 2 | apoptotic_process_involved_in_heart_morphogenesis |
| GO:0003290 | 2 | atrial_septum_secundum_morphogenesis |
| GO:0003310 | 2 | pancreatic_A_cell_differentiation |
| GO:0003326 | 2 | pancreatic_A_cell_fate_commitment |
| GO:0003329 | 2 | pancreatic_PP_cell_fate_commitment |
| GO:0003338 | 2 | metanephros_morphogenesis |
| GO:0003344 | 2 | pericardium_morphogenesis |
| GO:0003348 | 2 | cardiac_endothelial_cell_differentiation |
| GO:0003350 | 2 | pulmonary_myocardium_development |
| GO:0003360 | 2 | brainstem_development |
| GO:0003365 | 2 | establishment_of_cell_polarity_involved_in_ameboidal_cell_migration |
| GO:0003408 | 2 | optic_cup_formation_involved_in_camera-type_eye_development |
| GO:0003419 | 2 | growth_plate_cartilage_chondrocyte_proliferation |
| GO:0003420 | 2 | regulation_of_growth_plate_cartilage_chondrocyte_proliferation |
| GO:0003681 | 2 | bent_DNA_binding |
| GO:0003689 | 2 | DNA_clamp_loader_activity |
| GO:0003826 | 2 | alpha-ketoacid_dehydrogenase_activity |
| GO:0003835 | 2 | beta-galactoside_alpha-2,6-sialyltransferase_activity |
| GO:0003839 | 2 | gamma-glutamylcyclotransferase_activity |
| GO:0003845 | 2 | 11-beta-hydroxysteroid_dehydrogenase_[NAD(P)]_activity |
| GO:0003858 | 2 | 3-hydroxybutyrate_dehydrogenase_activity |
| GO:0003863 | 2 | 3-methyl-2-oxobutanoate_dehydrogenase_(2-methylpropanoyl-transferring)_activity |
| GO:0003868 | 2 | 4-hydroxyphenylpyruvate_dioxygenase_activity |
| GO:0003870 | 2 | 5-aminolevulinate_synthase_activity |
| GO:0003875 | 2 | ADP-ribosylarginine_hydrolase_activity |
| GO:0003878 | 2 | ATP_citrate_synthase_activity |
| GO:0003880 | 2 | protein_C-terminal_carboxyl_O-methyltransferase_activity |
| GO:0003883 | 2 | CTP_synthase_activity |
| GO:0003884 | 2 | D-amino-acid_oxidase_activity |
| GO:0003904 | 2 | deoxyribodipyrimidine_photo-lyase_activity |
| GO:0003905 | 2 | alkylbase_DNA_N-glycosylase_activity |
| GO:0003912 | 2 | DNA_nucleotidylexotransferase_activity |
| GO:0003913 | 2 | DNA_photolyase_activity |
| GO:0003914 | 2 | DNA_(6-4)_photolyase_activity |
| GO:0003920 | 2 | GMP_reductase_activity |
| GO:0003938 | 2 | IMP_dehydrogenase_activity |
| GO:0003943 | 2 | N-acetylgalactosamine-4-sulfatase_activity |
| GO:0003948 | 2 | N4-(beta-N-acetylglucosaminyl)-L-asparaginase_activity |
| GO:0003955 | 2 | NAD(P)H_dehydrogenase_(quinone)_activity |
| GO:0003963 | 2 | RNA-3'-phosphate_cyclase_activity |
| GO:0003973 | 2 | (S)-2-hydroxy-acid_oxidase_activity |
| GO:0003990 | 2 | acetylcholinesterase_activity |
| GO:0003998 | 2 | acylphosphatase_activity |
| GO:0004008 | 2 | copper-exporting_ATPase_activity |
| GO:0004019 | 2 | adenylosuccinate_synthase_activity |
| GO:0004020 | 2 | adenylylsulfate_kinase_activity |
| GO:0004021 | 2 | L-alanine:2-oxoglutarate_aminotransferase_activity |
| GO:0004027 | 2 | alcohol_sulfotransferase_activity |
| GO:0004031 | 2 | aldehyde_oxidase_activity |
| GO:0004040 | 2 | amidase_activity |
| GO:0004051 | 2 | arachidonate_5-lipoxygenase_activity |
| GO:0004052 | 2 | arachidonate_12-lipoxygenase_activity |
| GO:0004053 | 2 | arginase_activity |
| GO:0004056 | 2 | argininosuccinate_lyase_activity |
| GO:0004063 | 2 | aryldialkylphosphatase_activity |
| GO:0004066 | 2 | asparagine_synthase_(glutamine-hydrolyzing)_activity |
| GO:0004067 | 2 | asparaginase_activity |
| GO:0004069 | 2 | L-aspartate:2-oxoglutarate_aminotransferase_activity |
| GO:0004074 | 2 | biliverdin_reductase_activity |
| GO:0004084 | 2 | branched-chain-amino-acid_transaminase_activity |
| GO:0004103 | 2 | choline_kinase_activity |
| GO:0004105 | 2 | choline-phosphate_cytidylyltransferase_activity |
| GO:0004109 | 2 | coproporphyrinogen_oxidase_activity |
| GO:0004119 | 2 | cGMP-inhibited_cyclic-nucleotide_phosphodiesterase_activity |
| GO:0004140 | 2 | dephospho-CoA_kinase_activity |
| GO:0004145 | 2 | diamine_N-acetyltransferase_activity |
| GO:0004146 | 2 | dihydrofolate_reductase_activity |
| GO:0004152 | 2 | dihydroorotate_dehydrogenase_activity |
| GO:0004158 | 2 | dihydroorotate_oxidase_activity |
| GO:0004161 | 2 | dimethylallyltranstransferase_activity |
| GO:0004174 | 2 | electron-transferring-flavoprotein_dehydrogenase_activity |
| GO:0004314 | 2 | [acyl-carrier-protein]_S-malonyltransferase_activity |
| GO:0004315 | 2 | 3-oxoacyl-[acyl-carrier-protein]_synthase_activity |
| GO:0004320 | 2 | oleoyl-[acyl-carrier-protein]_hydrolase_activity |
| GO:0004321 | 2 | fatty-acyl-CoA_synthase_activity |
| GO:0004334 | 2 | fumarylacetoacetase_activity |
| GO:0004335 | 2 | galactokinase_activity |
| GO:0004337 | 2 | geranyltranstransferase_activity |
| GO:0004342 | 2 | glucosamine-6-phosphate_deaminase_activity |
| GO:0004345 | 2 | glucose-6-phosphate_dehydrogenase_activity |
| GO:0004348 | 2 | glucosylceramidase_activity |
| GO:0004352 | 2 | glutamate_dehydrogenase_(NAD+)_activity |
| GO:0004353 | 2 | glutamate_dehydrogenase_[NAD(P)+]_activity |
| GO:0004356 | 2 | glutamate-ammonia_ligase_activity |
| GO:0004357 | 2 | glutamate-cysteine_ligase_activity |
| GO:0004359 | 2 | glutaminase_activity |
| GO:0004360 | 2 | glutamine-fructose-6-phosphate_transaminase_(isomerizing)_activity |
| GO:0004365 | 2 | glyceraldehyde-3-phosphate_dehydrogenase_(NAD+)_(phosphorylating)_activity |
| GO:0004368 | 2 | glycerol-3-phosphate_dehydrogenase_activity |
| GO:0004372 | 2 | glycine_hydroxymethyltransferase_activity |
| GO:0004373 | 2 | glycogen_(starch)_synthase_activity |
| GO:0004376 | 2 | glycolipid_mannosyltransferase_activity |
| GO:0004379 | 2 | glycylpeptide_N-tetradecanoyltransferase_activity |
| GO:0004392 | 2 | heme_oxygenase_(decyclizing)_activity |
| GO:0004419 | 2 | hydroxymethylglutaryl-CoA_lyase_activity |
| GO:0004421 | 2 | hydroxymethylglutaryl-CoA_synthase_activity |
| GO:0004422 | 2 | hypoxanthine_phosphoribosyltransferase_activity |
| GO:0004447 | 2 | iodide_peroxidase_activity |
| GO:0004450 | 2 | isocitrate_dehydrogenase_(NADP+)_activity |
| GO:0004452 | 2 | isopentenyl-diphosphate_delta-isomerase_activity |
| GO:0004468 | 2 | lysine_N-acetyltransferase_activity |
| GO:0004473 | 2 | malate_dehydrogenase_(oxaloacetate-decarboxylating)_(NADP+)_activity |
| GO:0004475 | 2 | mannose-1-phosphate_guanylyltransferase_activity |
| GO:0004478 | 2 | methionine_adenosyltransferase_activity |
| GO:0004485 | 2 | methylcrotonoyl-CoA_carboxylase_activity |
| GO:0004487 | 2 | methylenetetrahydrofolate_dehydrogenase_(NAD+)_activity |
| GO:0004507 | 2 | steroid_11-beta-monooxygenase_activity |
| GO:0004509 | 2 | steroid_21-monooxygenase_activity |
| GO:0004510 | 2 | tryptophan_5-monooxygenase_activity |
| GO:0004531 | 2 | deoxyribonuclease_II_activity |
| GO:0004557 | 2 | alpha-galactosidase_activity |
| GO:0004569 | 2 | glycoprotein_endo-alpha-1,2-mannosidase_activity |
| GO:0004572 | 2 | mannosyl-oligosaccharide_1,3-1,6-alpha-mannosidase_activity |
| GO:0004605 | 2 | phosphatidate_cytidylyltransferase_activity |
| GO:0004609 | 2 | phosphatidylserine_decarboxylase_activity |
| GO:0004611 | 2 | phosphoenolpyruvate_carboxykinase_activity |
| GO:0004613 | 2 | phosphoenolpyruvate_carboxykinase_(GTP)_activity |
| GO:0004615 | 2 | phosphomannomutase_activity |
| GO:0004618 | 2 | phosphoglycerate_kinase_activity |
| GO:0004649 | 2 | poly(ADP-ribose)_glycohydrolase_activity |
| GO:0004657 | 2 | proline_dehydrogenase_activity |
| GO:0004658 | 2 | propionyl-CoA_carboxylase_activity |
| GO:0004662 | 2 | CAAX-protein_geranylgeranyltransferase_activity |
| GO:0004666 | 2 | prostaglandin-endoperoxide_synthase_activity |
| GO:0004667 | 2 | prostaglandin-D_synthase_activity |
| GO:0004692 | 2 | cGMP-dependent_protein_kinase_activity |
| GO:0004699 | 2 | calcium-independent_protein_kinase_C_activity |
| GO:0004724 | 2 | magnesium-dependent_protein_serine/threonine_phosphatase_activity |
| GO:0004727 | 2 | prenylated_protein_tyrosine_phosphatase_activity |
| GO:0004741 | 2 | [pyruvate_dehydrogenase_(lipoamide)]_phosphatase_activity |
| GO:0004756 | 2 | selenide,_water_dikinase_activity |
| GO:0004766 | 2 | spermidine_synthase_activity |
| GO:0004771 | 2 | sterol_esterase_activity |
| GO:0004772 | 2 | sterol_O-acyltransferase_activity |
| GO:0004776 | 2 | succinate-CoA_ligase_(GDP-forming)_activity |
| GO:0004779 | 2 | sulfate_adenylyltransferase_activity |
| GO:0004781 | 2 | sulfate_adenylyltransferase_(ATP)_activity |
| GO:0004793 | 2 | threonine_aldolase_activity |
| GO:0004794 | 2 | L-threonine_ammonia-lyase_activity |
| GO:0004795 | 2 | threonine_synthase_activity |
| GO:0004797 | 2 | thymidine_kinase_activity |
| GO:0004798 | 2 | thymidylate_kinase_activity |
| GO:0004799 | 2 | thymidylate_synthase_activity |
| GO:0004809 | 2 | tRNA_(guanine-N2-)-methyltransferase_activity |
| GO:0004816 | 2 | asparagine-tRNA_ligase_activity |
| GO:0004817 | 2 | cysteine-tRNA_ligase_activity |
| GO:0004818 | 2 | glutamate-tRNA_ligase_activity |
| GO:0004820 | 2 | glycine-tRNA_ligase_activity |
| GO:0004822 | 2 | isoleucine-tRNA_ligase_activity |
| GO:0004823 | 2 | leucine-tRNA_ligase_activity |
| GO:0004825 | 2 | methionine-tRNA_ligase_activity |
| GO:0004827 | 2 | proline-tRNA_ligase_activity |
| GO:0004830 | 2 | tryptophan-tRNA_ligase_activity |
| GO:0004831 | 2 | tyrosine-tRNA_ligase_activity |
| GO:0004839 | 2 | ubiquitin_activating_enzyme_activity |
| GO:0004850 | 2 | uridine_phosphorylase_activity |
| GO:0004852 | 2 | uroporphyrinogen-III_synthase_activity |
| GO:0004854 | 2 | xanthine_dehydrogenase_activity |
| GO:0004905 | 2 | type_I_interferon_receptor_activity |
| GO:0004906 | 2 | interferon-gamma_receptor_activity |
| GO:0004909 | 2 | interleukin-1,_Type_I,_activating_receptor_activity |
| GO:0004910 | 2 | interleukin-1,_Type_II,_blocking_receptor_activity |
| GO:0004912 | 2 | interleukin-3_receptor_activity |
| GO:0004913 | 2 | interleukin-4_receptor_activity |
| GO:0004914 | 2 | interleukin-5_receptor_activity |
| GO:0004915 | 2 | interleukin-6_receptor_activity |
| GO:0004917 | 2 | interleukin-7_receptor_activity |
| GO:0004918 | 2 | interleukin-8_receptor_activity |
| GO:0004920 | 2 | interleukin-10_receptor_activity |
| GO:0004923 | 2 | leukemia_inhibitory_factor_receptor_activity |
| GO:0004944 | 2 | C5a_anaphylatoxin_receptor_activity |
| GO:0004947 | 2 | bradykinin_receptor_activity |
| GO:0004951 | 2 | cholecystokinin_receptor_activity |
| GO:0004956 | 2 | prostaglandin_D_receptor_activity |
| GO:0004962 | 2 | endothelin_receptor_activity |
| GO:0004968 | 2 | gonadotropin-releasing_hormone_receptor_activity |
| GO:0004991 | 2 | parathyroid_hormone_receptor_activity |
| GO:0004996 | 2 | thyroid-stimulating_hormone_receptor_activity |
| GO:0004998 | 2 | transferrin_receptor_activity |
| GO:0005011 | 2 | macrophage_colony-stimulating_factor_receptor_activity |
| GO:0005019 | 2 | platelet-derived_growth_factor_beta-receptor_activity |
| GO:0005026 | 2 | transforming_growth_factor_beta_receptor_activity,_type_II |
| GO:0005046 | 2 | KDEL_sequence_binding |
| GO:0005052 | 2 | peroxisome_matrix_targeting_signal-1_binding |
| GO:0005053 | 2 | peroxisome_matrix_targeting_signal-2_binding |
| GO:0005093 | 2 | Rab_GDP-dissociation_inhibitor_activity |
| GO:0005119 | 2 | smoothened_binding |
| GO:0005124 | 2 | scavenger_receptor_binding |
| GO:0005133 | 2 | interferon-gamma_receptor_binding |
| GO:0005137 | 2 | interleukin-5_receptor_binding |
| GO:0005146 | 2 | leukemia_inhibitory_factor_receptor_binding |
| GO:0005151 | 2 | interleukin-1,_Type_II_receptor_binding |
| GO:0005153 | 2 | interleukin-8_receptor_binding |
| GO:0005157 | 2 | macrophage_colony-stimulating_factor_receptor_binding |
| GO:0005174 | 2 | CD40_receptor_binding |
| GO:0005185 | 2 | neurohypophyseal_hormone_activity |
| GO:0005223 | 2 | intracellular_cGMP_activated_cation_channel_activity |
| GO:0005232 | 2 | serotonin-activated_cation-selective_channel_activity |
| GO:0005277 | 2 | acetylcholine_transmembrane_transporter_activity |
| GO:0005280 | 2 | hydrogen:amino_acid_symporter_activity |
| GO:0005290 | 2 | L-histidine_transmembrane_transporter_activity |
| GO:0005314 | 2 | high-affinity_glutamate_transmembrane_transporter_activity |
| GO:0005333 | 2 | norepinephrine_transmembrane_transporter_activity |
| GO:0005347 | 2 | ATP_transmembrane_transporter_activity |
| GO:0005412 | 2 | glucose:sodium_symporter_activity |
| GO:0005427 | 2 | proton-dependent_oligopeptide_secondary_active_transmembrane_transporter_activity |
| GO:0005459 | 2 | UDP-galactose_transmembrane_transporter_activity |
| GO:0005462 | 2 | UDP-N-acetylglucosamine_transmembrane_transporter_activity |
| GO:0005471 | 2 | ATP:ADP_antiporter_activity |
| GO:0005502 | 2 | 11-cis_retinal_binding |
| GO:0005584 | 2 | collagen_type_I |
| GO:0005592 | 2 | collagen_type_XI |
| GO:0005602 | 2 | complement_component_C1_complex |
| GO:0005607 | 2 | laminin-2_complex |
| GO:0005608 | 2 | laminin-3_complex |
| GO:0005642 | 2 | annulate_lamellae |
| GO:0005674 | 2 | transcription_factor_TFIIF_complex |
| GO:0005684 | 2 | U2-type_spliceosomal_complex |
| GO:0005687 | 2 | U4_snRNP |
| GO:0005688 | 2 | U6_snRNP |
| GO:0005712 | 2 | chiasma |
| GO:0005724 | 2 | nuclear_telomeric_heterochromatin |
| GO:0005760 | 2 | gamma_DNA_polymerase_complex |
| GO:0005784 | 2 | Sec61_translocon_complex |
| GO:0005785 | 2 | signal_recognition_particle_receptor_complex |
| GO:0005873 | 2 | plus-end_kinesin_complex |
| GO:0005960 | 2 | glycine_cleavage_complex |
| GO:0005968 | 2 | Rab-protein_geranylgeranyltransferase_complex |
| GO:0005988 | 2 | lactose_metabolic_process |
| GO:0005989 | 2 | lactose_biosynthetic_process |
| GO:0005992 | 2 | trehalose_biosynthetic_process |
| GO:0005999 | 2 | xylulose_biosynthetic_process |
| GO:0006001 | 2 | fructose_catabolic_process |
| GO:0006005 | 2 | L-fucose_biosynthetic_process |
| GO:0006043 | 2 | glucosamine_catabolic_process |
| GO:0006059 | 2 | hexitol_metabolic_process |
| GO:0006083 | 2 | acetate_metabolic_process |
| GO:0006097 | 2 | glyoxylate_cycle |
| GO:0006116 | 2 | NADH_oxidation |
| GO:0006178 | 2 | guanine_salvage |
| GO:0006196 | 2 | AMP_catabolic_process |
| GO:0006212 | 2 | uracil_catabolic_process |
| GO:0006231 | 2 | dTMP_biosynthetic_process |
| GO:0006233 | 2 | dTDP_biosynthetic_process |
| GO:0006288 | 2 | base-excision_repair,_DNA_ligation |
| GO:0006311 | 2 | meiotic_gene_conversion |
| GO:0006313 | 2 | transposition,_DNA-mediated |
| GO:0006335 | 2 | DNA_replication-dependent_nucleosome_assembly |
| GO:0006348 | 2 | chromatin_silencing_at_telomere |
| GO:0006382 | 2 | adenosine_to_inosine_editing |
| GO:0006407 | 2 | rRNA_export_from_nucleus |
| GO:0006408 | 2 | snRNA_export_from_nucleus |
| GO:0006422 | 2 | aspartyl-tRNA_aminoacylation |
| GO:0006423 | 2 | cysteinyl-tRNA_aminoacylation |
| GO:0006424 | 2 | glutamyl-tRNA_aminoacylation |
| GO:0006426 | 2 | glycyl-tRNA_aminoacylation |
| GO:0006428 | 2 | isoleucyl-tRNA_aminoacylation |
| GO:0006429 | 2 | leucyl-tRNA_aminoacylation |
| GO:0006431 | 2 | methionyl-tRNA_aminoacylation |
| GO:0006433 | 2 | prolyl-tRNA_aminoacylation |
| GO:0006436 | 2 | tryptophanyl-tRNA_aminoacylation |
| GO:0006437 | 2 | tyrosyl-tRNA_aminoacylation |
| GO:0006447 | 2 | regulation_of_translational_initiation_by_iron |
| GO:0006450 | 2 | regulation_of_translational_fidelity |
| GO:0006478 | 2 | peptidyl-tyrosine_sulfation |
| GO:0006481 | 2 | C-terminal_protein_methylation |
| GO:0006489 | 2 | dolichyl_diphosphate_biosynthetic_process |
| GO:0006499 | 2 | N-terminal_protein_myristoylation |
| GO:0006517 | 2 | protein_deglycosylation |
| GO:0006529 | 2 | asparagine_biosynthetic_process |
| GO:0006532 | 2 | aspartate_biosynthetic_process |
| GO:0006540 | 2 | glutamate_decarboxylation_to_succinate |
| GO:0006542 | 2 | glutamine_biosynthetic_process |
| GO:0006581 | 2 | acetylcholine_catabolic_process |
| GO:0006583 | 2 | melanin_biosynthetic_process_from_tyrosine |
| GO:0006597 | 2 | spermine_biosynthetic_process |
| GO:0006601 | 2 | creatine_biosynthetic_process |
| GO:0006603 | 2 | phosphocreatine_metabolic_process |
| GO:0006627 | 2 | protein_processing_involved_in_protein_targeting_to_mitochondrion |
| GO:0006649 | 2 | phospholipid_transfer_to_membrane |
| GO:0006668 | 2 | sphinganine-1-phosphate_metabolic_process |
| GO:0006680 | 2 | glucosylceramide_catabolic_process |
| GO:0006682 | 2 | galactosylceramide_biosynthetic_process |
| GO:0006696 | 2 | ergosterol_biosynthetic_process |
| GO:0006710 | 2 | androgen_catabolic_process |
| GO:0006714 | 2 | sesquiterpenoid_metabolic_process |
| GO:0006738 | 2 | nicotinamide_riboside_catabolic_process |
| GO:0006742 | 2 | NADP_catabolic_process |
| GO:0006768 | 2 | biotin_metabolic_process |
| GO:0006769 | 2 | nicotinamide_metabolic_process |
| GO:0006784 | 2 | heme_a_biosynthetic_process |
| GO:0006788 | 2 | heme_oxidation |
| GO:0006808 | 2 | regulation_of_nitrogen_utilization |
| GO:0006851 | 2 | mitochondrial_calcium_ion_transport |
| GO:0006880 | 2 | intracellular_sequestering_of_iron_ion |
| GO:0006963 | 2 | positive_regulation_of_antibacterial_peptide_biosynthetic_process |
| GO:0006965 | 2 | positive_regulation_of_biosynthetic_process_of_antibacterial_peptides_active_against_Gram-positive_bacteria |
| GO:0006990 | 2 | positive_regulation_of_transcription_from_RNA_polymerase_II_promoter_involved_in_unfolded_protein_response |
| GO:0007066 | 2 | female_meiosis_sister_chromatid_cohesion |
| GO:0007079 | 2 | mitotic_chromosome_movement_towards_spindle_pole |
| GO:0007092 | 2 | activation_of_mitotic_anaphase-promoting_complex_activity |
| GO:0007100 | 2 | mitotic_centrosome_separation |
| GO:0007109 | 2 | cytokinesis,_completion_of_separation |
| GO:0007161 | 2 | calcium-independent_cell-matrix_adhesion |
| GO:0007196 | 2 | adenylate_cyclase-inhibiting_G-protein_coupled_glutamate_receptor_signaling_pathway |
| GO:0007197 | 2 | adenylate_cyclase-inhibiting_G-protein_coupled_acetylcholine_receptor_signaling_pathway |
| GO:0007206 | 2 | phospholipase_C-activating_G-protein_coupled_glutamate_receptor_signaling_pathway |
| GO:0007290 | 2 | spermatid_nucleus_elongation |
| GO:0007356 | 2 | thorax_and_anterior_abdomen_determination |
| GO:0007402 | 2 | ganglion_mother_cell_fate_determination |
| GO:0007493 | 2 | endodermal_cell_fate_determination |
| GO:0007497 | 2 | posterior_midgut_development |
| GO:0007509 | 2 | mesoderm_migration_involved_in_gastrulation |
| GO:0007518 | 2 | myoblast_cell_fate_determination |
| GO:0007529 | 2 | establishment_of_synaptic_specificity_at_neuromuscular_junction |
| GO:0007542 | 2 | primary_sex_determination,_germ-line |
| GO:0007620 | 2 | copulation |
| GO:0008043 | 2 | intracellular_ferritin_complex |
| GO:0008065 | 2 | establishment_of_blood-nerve_barrier |
| GO:0008074 | 2 | guanylate_cyclase_complex,_soluble |
| GO:0008078 | 2 | mesodermal_cell_migration |
| GO:0008108 | 2 | UDP-glucose:hexose-1-phosphate_uridylyltransferase_activity |
| GO:0008119 | 2 | thiopurine_S-methyltransferase_activity |
| GO:0008124 | 2 | 4-alpha-hydroxytetrahydrobiopterin_dehydratase_activity |
| GO:0008147 | 2 | structural_constituent_of_bone |
| GO:0008160 | 2 | protein_tyrosine_phosphatase_activator_activity |
| GO:0008176 | 2 | tRNA_(guanine-N7-)-methyltransferase_activity |
| GO:0008177 | 2 | succinate_dehydrogenase_(ubiquinone)_activity |
| GO:0008193 | 2 | tRNA_guanylyltransferase_activity |
| GO:0008204 | 2 | ergosterol_metabolic_process |
| GO:0008240 | 2 | tripeptidyl-peptidase_activity |
| GO:0008260 | 2 | 3-oxoacid_CoA-transferase_activity |
| GO:0008269 | 2 | JAK_pathway_signal_transduction_adaptor_activity |
| GO:0008273 | 2 | calcium,_potassium:sodium_antiporter_activity |
| GO:0008281 | 2 | sulfonylurea_receptor_activity |
| GO:0008292 | 2 | acetylcholine_biosynthetic_process |
| GO:0008297 | 2 | single-stranded_DNA_specific_exodeoxyribonuclease_activity |
| GO:0008309 | 2 | double-stranded_DNA_specific_exodeoxyribonuclease_activity |
| GO:0008321 | 2 | Ral_guanyl-nucleotide_exchange_factor_activity |
| GO:0008355 | 2 | olfactory_learning |
| GO:0008386 | 2 | cholesterol_monooxygenase_(side-chain-cleaving)_activity |
| GO:0008396 | 2 | oxysterol_7-alpha-hydroxylase_activity |
| GO:0008410 | 2 | CoA-transferase_activity |
| GO:0008428 | 2 | ribonuclease_inhibitor_activity |
| GO:0008441 | 2 | 3'(2'),5'-bisphosphate_nucleotidase_activity |
| GO:0008459 | 2 | chondroitin_6-sulfotransferase_activity |
| GO:0008466 | 2 | glycogenin_glucosyltransferase_activity |
| GO:0008476 | 2 | protein-tyrosine_sulfotransferase_activity |
| GO:0008479 | 2 | queuine_tRNA-ribosyltransferase_activity |
| GO:0008481 | 2 | sphinganine_kinase_activity |
| GO:0008489 | 2 | UDP-galactose:glucosylceramide_beta-1,4-galactosyltransferase_activity |
| GO:0008511 | 2 | sodium:potassium:chloride_symporter_activity |
| GO:0008513 | 2 | secondary_active_organic_cation_transmembrane_transporter_activity |
| GO:0008520 | 2 | L-ascorbate:sodium_symporter_activity |
| GO:0008525 | 2 | phosphatidylcholine_transporter_activity |
| GO:0008545 | 2 | JUN_kinase_kinase_activity |
| GO:0008582 | 2 | regulation_of_synaptic_growth_at_neuromuscular_junction |
| GO:0008594 | 2 | photoreceptor_cell_morphogenesis |
| GO:0008614 | 2 | pyridoxine_metabolic_process |
| GO:0008615 | 2 | pyridoxine_biosynthetic_process |
| GO:0008616 | 2 | queuosine_biosynthetic_process |
| GO:0008618 | 2 | 7-methylguanosine_metabolic_process |
| GO:0008623 | 2 | chromatin_accessibility_complex |
| GO:0008665 | 2 | 2'-phosphotransferase_activity |
| GO:0008732 | 2 | L-allo-threonine_aldolase_activity |
| GO:0008746 | 2 | NAD(P)+_transhydrogenase_activity |
| GO:0008753 | 2 | NADPH_dehydrogenase_(quinone)_activity |
| GO:0008781 | 2 | N-acylneuraminate_cytidylyltransferase_activity |
| GO:0008905 | 2 | mannose-phosphate_guanylyltransferase_activity |
| GO:0008907 | 2 | integrase_activity |
| GO:0008940 | 2 | nitrate_reductase_activity |
| GO:0008942 | 2 | nitrite_reductase_[NAD(P)H]_activity |
| GO:0009076 | 2 | histidine_family_amino_acid_biosynthetic_process |
| GO:0009082 | 2 | branched_chain_family_amino_acid_biosynthetic_process |
| GO:0009088 | 2 | threonine_biosynthetic_process |
| GO:0009092 | 2 | homoserine_metabolic_process |
| GO:0009093 | 2 | cysteine_catabolic_process |
| GO:0009107 | 2 | lipoate_biosynthetic_process |
| GO:0009139 | 2 | pyrimidine_nucleoside_diphosphate_biosynthetic_process |
| GO:0009153 | 2 | purine_deoxyribonucleotide_biosynthetic_process |
| GO:0009196 | 2 | pyrimidine_deoxyribonucleoside_diphosphate_metabolic_process |
| GO:0009197 | 2 | pyrimidine_deoxyribonucleoside_diphosphate_biosynthetic_process |
| GO:0009204 | 2 | deoxyribonucleoside_triphosphate_catabolic_process |
| GO:0009217 | 2 | purine_deoxyribonucleoside_triphosphate_catabolic_process |
| GO:0009256 | 2 | 10-formyltetrahydrofolate_metabolic_process |
| GO:0009258 | 2 | 10-formyltetrahydrofolate_catabolic_process |
| GO:0009292 | 2 | genetic_transfer |
| GO:0009294 | 2 | DNA_mediated_transformation |
| GO:0009346 | 2 | citrate_lyase_complex |
| GO:0009378 | 2 | four-way_junction_helicase_activity |
| GO:0009384 | 2 | N-acylmannosamine_kinase_activity |
| GO:0009397 | 2 | folic_acid-containing_compound_catabolic_process |
| GO:0009405 | 2 | pathogenesis |
| GO:0009439 | 2 | cyanate_metabolic_process |
| GO:0009440 | 2 | cyanate_catabolic_process |
| GO:0009441 | 2 | glycolate_metabolic_process |
| GO:0009446 | 2 | putrescine_biosynthetic_process |
| GO:0009447 | 2 | putrescine_catabolic_process |
| GO:0009450 | 2 | gamma-aminobutyric_acid_catabolic_process |
| GO:0009590 | 2 | detection_of_gravity |
| GO:0009785 | 2 | blue_light_signaling_pathway |
| GO:0009882 | 2 | blue_light_photoreceptor_activity |
| GO:0009912 | 2 | auditory_receptor_cell_fate_commitment |
| GO:0009992 | 2 | cellular_water_homeostasis |
| GO:0010025 | 2 | wax_biosynthetic_process |
| GO:0010045 | 2 | response_to_nickel_cation |
| GO:0010046 | 2 | response_to_mycotoxin |
| GO:0010070 | 2 | zygote_asymmetric_cell_division |
| GO:0010085 | 2 | polarity_specification_of_proximal/distal_axis |
| GO:0010157 | 2 | response_to_chlorate |
| GO:0010164 | 2 | response_to_cesium_ion |
| GO:0010166 | 2 | wax_metabolic_process |
| GO:0010430 | 2 | fatty_acid_omega-oxidation |
| GO:0010465 | 2 | nerve_growth_factor_receptor_activity |
| GO:0010484 | 2 | H3_histone_acetyltransferase_activity |
| GO:0010512 | 2 | negative_regulation_of_phosphatidylinositol_biosynthetic_process |
| GO:0010578 | 2 | regulation_of_adenylate_cyclase_activity_involved_in_G-protein_coupled_receptor_signaling_pathway |
| GO:0010579 | 2 | positive_regulation_of_adenylate_cyclase_activity_involved_in_G-protein_coupled_receptor_signaling_pathway |
| GO:0010593 | 2 | negative_regulation_of_lamellipodium_assembly |
| GO:0010641 | 2 | positive_regulation_of_platelet-derived_growth_factor_receptor_signaling_pathway |
| GO:0010658 | 2 | striated_muscle_cell_apoptotic_process |
| GO:0010659 | 2 | cardiac_muscle_cell_apoptotic_process |
| GO:0010698 | 2 | acetyltransferase_activator_activity |
| GO:0010727 | 2 | negative_regulation_of_hydrogen_peroxide_metabolic_process |
| GO:0010735 | 2 | positive_regulation_of_transcription_via_serum_response_element_binding |
| GO:0010736 | 2 | serum_response_element_binding |
| GO:0010737 | 2 | protein_kinase_A_signaling_cascade |
| GO:0010767 | 2 | regulation_of_transcription_from_RNA_polymerase_II_promoter_in_response_to_UV-induced_DNA_damage |
| GO:0010813 | 2 | neuropeptide_catabolic_process |
| GO:0010835 | 2 | regulation_of_protein_ADP-ribosylation |
| GO:0010840 | 2 | regulation_of_circadian_sleep/wake_cycle,_wakefulness |
| GO:0010841 | 2 | positive_regulation_of_circadian_sleep/wake_cycle,_wakefulness |
| GO:0010845 | 2 | positive_regulation_of_reciprocal_meiotic_recombination |
| GO:0010857 | 2 | calcium-dependent_protein_kinase_activity |
| GO:0010868 | 2 | negative_regulation_of_triglyceride_biosynthetic_process |
| GO:0010897 | 2 | negative_regulation_of_triglyceride_catabolic_process |
| GO:0010902 | 2 | positive_regulation_of_very-low-density_lipoprotein_particle_remodeling |
| GO:0010908 | 2 | regulation_of_heparan_sulfate_proteoglycan_biosynthetic_process |
| GO:0010909 | 2 | positive_regulation_of_heparan_sulfate_proteoglycan_biosynthetic_process |
| GO:0010949 | 2 | negative_regulation_of_intestinal_phytosterol_absorption |
| GO:0010958 | 2 | regulation_of_amino_acid_import |
| GO:0010960 | 2 | magnesium_ion_homeostasis |
| GO:0010963 | 2 | regulation_of_L-arginine_import |
| GO:0010971 | 2 | positive_regulation_of_G2/M_transition_of_mitotic_cell_cycle |
| GO:0010983 | 2 | positive_regulation_of_high-density_lipoprotein_particle_clearance |
| GO:0010994 | 2 | free_ubiquitin_chain_polymerization |
| GO:0010996 | 2 | response_to_auditory_stimulus |
| GO:0014010 | 2 | Schwann_cell_proliferation |
| GO:0014028 | 2 | notochord_formation |
| GO:0014040 | 2 | positive_regulation_of_Schwann_cell_differentiation |
| GO:0014043 | 2 | negative_regulation_of_neuron_maturation |
| GO:0014051 | 2 | gamma-aminobutyric_acid_secretion |
| GO:0014062 | 2 | regulation_of_serotonin_secretion |
| GO:0014063 | 2 | negative_regulation_of_serotonin_secretion |
| GO:0014719 | 2 | satellite_cell_activation |
| GO:0014721 | 2 | twitch_skeletal_muscle_contraction |
| GO:0014722 | 2 | regulation_of_skeletal_muscle_contraction_by_calcium_ion_signaling |
| GO:0014724 | 2 | regulation_of_twitch_skeletal_muscle_contraction |
| GO:0014733 | 2 | regulation_of_skeletal_muscle_adaptation |
| GO:0014740 | 2 | negative_regulation_of_muscle_hyperplasia |
| GO:0014802 | 2 | terminal_cisterna |
| GO:0014806 | 2 | smooth_muscle_hyperplasia |
| GO:0014827 | 2 | intestine_smooth_muscle_contraction |
| GO:0014854 | 2 | response_to_inactivity |
| GO:0014870 | 2 | response_to_muscle_inactivity |
| GO:0014877 | 2 | response_to_muscle_inactivity_involved_in_regulation_of_muscle_adaptation |
| GO:0014894 | 2 | response_to_denervation_involved_in_regulation_of_muscle_adaptation |
| GO:0014908 | 2 | myotube_differentiation_involved_in_skeletal_muscle_regeneration |
| GO:0014916 | 2 | regulation_of_lung_blood_pressure |
| GO:0015039 | 2 | NADPH-adrenodoxin_reductase_activity |
| GO:0015056 | 2 | corticotrophin-releasing_factor_receptor_activity |
| GO:0015093 | 2 | ferrous_iron_transmembrane_transporter_activity |
| GO:0015105 | 2 | arsenite_transmembrane_transporter_activity |
| GO:0015111 | 2 | iodide_transmembrane_transporter_activity |
| GO:0015140 | 2 | malate_transmembrane_transporter_activity |
| GO:0015141 | 2 | succinate_transmembrane_transporter_activity |
| GO:0015143 | 2 | urate_transmembrane_transporter_activity |
| GO:0015189 | 2 | L-lysine_transmembrane_transporter_activity |
| GO:0015207 | 2 | adenine_transmembrane_transporter_activity |
| GO:0015216 | 2 | purine_nucleotide_transmembrane_transporter_activity |
| GO:0015217 | 2 | ADP_transmembrane_transporter_activity |
| GO:0015228 | 2 | coenzyme_A_transmembrane_transporter_activity |
| GO:0015229 | 2 | L-ascorbic_acid_transporter_activity |
| GO:0015234 | 2 | thiamine_transmembrane_transporter_activity |
| GO:0015247 | 2 | aminophospholipid_transporter_activity |
| GO:0015265 | 2 | urea_channel_activity |
| GO:0015266 | 2 | protein_channel_activity |
| GO:0015322 | 2 | secondary_active_oligopeptide_transmembrane_transporter_activity |
| GO:0015333 | 2 | peptide:hydrogen_symporter_activity |
| GO:0015349 | 2 | thyroid_hormone_transmembrane_transporter_activity |
| GO:0015375 | 2 | glycine:sodium_symporter_activity |
| GO:0015382 | 2 | sodium:sulfate_symporter_activity |
| GO:0015386 | 2 | potassium:hydrogen_antiporter_activity |
| GO:0015389 | 2 | pyrimidine-_and_adenine-specific:sodium_symporter_activity |
| GO:0015403 | 2 | thiamine_uptake_transmembrane_transporter_activity |
| GO:0015433 | 2 | peptide_antigen-transporting_ATPase_activity |
| GO:0015501 | 2 | glutamate:sodium_symporter_activity |
| GO:0015563 | 2 | uptake_transmembrane_transporter_activity |
| GO:0015677 | 2 | copper_ion_import |
| GO:0015680 | 2 | intracellular_copper_ion_transport |
| GO:0015688 | 2 | iron_chelate_transport |
| GO:0015742 | 2 | alpha-ketoglutarate_transport |
| GO:0015747 | 2 | urate_transport |
| GO:0015755 | 2 | fructose_transport |
| GO:0015774 | 2 | polysaccharide_transport |
| GO:0015785 | 2 | UDP-galactose_transport |
| GO:0015788 | 2 | UDP-N-acetylglucosamine_transport |
| GO:0015809 | 2 | arginine_transport |
| GO:0015822 | 2 | ornithine_transport |
| GO:0015827 | 2 | tryptophan_transport |
| GO:0015855 | 2 | pyrimidine_base_transport |
| GO:0015862 | 2 | uridine_transport |
| GO:0015870 | 2 | acetylcholine_transport |
| GO:0015878 | 2 | biotin_transport |
| GO:0015880 | 2 | coenzyme_A_transport |
| GO:0015882 | 2 | L-ascorbic_acid_transport |
| GO:0015887 | 2 | pantothenate_transmembrane_transport |
| GO:0015891 | 2 | siderophore_transport |
| GO:0015911 | 2 | plasma_membrane_long-chain_fatty_acid_transport |
| GO:0015920 | 2 | lipopolysaccharide_transport |
| GO:0015958 | 2 | bis(5'-nucleosidyl)_oligophosphate_catabolic_process |
| GO:0015961 | 2 | diadenosine_polyphosphate_catabolic_process |
| GO:0015993 | 2 | molecular_hydrogen_transport |
| GO:0016013 | 2 | syntrophin_complex |
| GO:0016035 | 2 | zeta_DNA_polymerase_complex |
| GO:0016046 | 2 | detection_of_fungus |
| GO:0016074 | 2 | snoRNA_metabolic_process |
| GO:0016107 | 2 | sesquiterpenoid_catabolic_process |
| GO:0016114 | 2 | terpenoid_biosynthetic_process |
| GO:0016115 | 2 | terpenoid_catabolic_process |
| GO:0016128 | 2 | phytosteroid_metabolic_process |
| GO:0016129 | 2 | phytosteroid_biosynthetic_process |
| GO:0016138 | 2 | glycoside_biosynthetic_process |
| GO:0016167 | 2 | glial_cell-derived_neurotrophic_factor_receptor_activity |
| GO:0016206 | 2 | catechol_O-methyltransferase_activity |
| GO:0016211 | 2 | ammonia_ligase_activity |
| GO:0016262 | 2 | protein_N-acetylglucosaminyltransferase_activity |
| GO:0016295 | 2 | myristoyl-[acyl-carrier-protein]_hydrolase_activity |
| GO:0016296 | 2 | palmitoyl-[acyl-carrier-protein]_hydrolase_activity |
| GO:0016297 | 2 | acyl-[acyl-carrier-protein]_hydrolase_activity |
| GO:0016300 | 2 | tRNA_(uracil)_methyltransferase_activity |
| GO:0016314 | 2 | phosphatidylinositol-3,4,5-trisphosphate_3-phosphatase_activity |
| GO:0016316 | 2 | phosphatidylinositol-3,4-bisphosphate_4-phosphatase_activity |
| GO:0016320 | 2 | endoplasmic_reticulum_membrane_fusion |
| GO:0016340 | 2 | calcium-dependent_cell-matrix_adhesion |
| GO:0016401 | 2 | palmitoyl-CoA_oxidase_activity |
| GO:0016402 | 2 | pristanoyl-CoA_oxidase_activity |
| GO:0016403 | 2 | dimethylargininase_activity |
| GO:0016404 | 2 | 15-hydroxyprostaglandin_dehydrogenase_(NAD+)_activity |
| GO:0016415 | 2 | octanoyltransferase_activity |
| GO:0016418 | 2 | S-acetyltransferase_activity |
| GO:0016419 | 2 | S-malonyltransferase_activity |
| GO:0016420 | 2 | malonyltransferase_activity |
| GO:0016422 | 2 | mRNA_(2'-O-methyladenosine-N6-)-methyltransferase_activity |
| GO:0016426 | 2 | tRNA_(adenine)_methyltransferase_activity |
| GO:0016429 | 2 | tRNA_(adenine-N1-)-methyltransferase_activity |
| GO:0016476 | 2 | regulation_of_embryonic_cell_shape |
| GO:0016487 | 2 | farnesol_metabolic_process |
| GO:0016488 | 2 | farnesol_catabolic_process |
| GO:0016492 | 2 | G-protein_coupled_neurotensin_receptor_activity |
| GO:0016499 | 2 | orexin_receptor_activity |
| GO:0016507 | 2 | mitochondrial_fatty_acid_beta-oxidation_multienzyme_complex |
| GO:0016509 | 2 | long-chain-3-hydroxyacyl-CoA_dehydrogenase_activity |
| GO:0016524 | 2 | latrotoxin_receptor_activity |
| GO:0016532 | 2 | superoxide_dismutase_copper_chaperone_activity |
| GO:0016534 | 2 | cyclin-dependent_protein_kinase_5_activator_activity |
| GO:0016539 | 2 | intein-mediated_protein_splicing |
| GO:0016554 | 2 | cytidine_to_uridine_editing |
| GO:0016603 | 2 | glutaminyl-peptide_cyclotransferase_activity |
| GO:0016608 | 2 | growth_hormone-releasing_hormone_activity |
| GO:0016639 | 2 | oxidoreductase_activity,_acting_on_the_CH-NH2_group_of_donors,_NAD_or_NADP_as_acceptor |
| GO:0016649 | 2 | oxidoreductase_activity,_acting_on_the_CH-NH_group_of_donors,_quinone_or_similar_compound_as_acceptor |
| GO:0016652 | 2 | oxidoreductase_activity,_acting_on_NADH_or_NADPH,_NAD_or_NADP_as_acceptor |
| GO:0016657 | 2 | oxidoreductase_activity,_acting_on_NADH_or_NADPH,_nitrogenous_group_as_acceptor |
| GO:0016726 | 2 | oxidoreductase_activity,_acting_on_CH_or_CH2_groups,_NAD_or_NADP_as_acceptor |
| GO:0016743 | 2 | carboxyl-_or_carbamoyltransferase_activity |
| GO:0016749 | 2 | N-succinyltransferase_activity |
| GO:0016781 | 2 | phosphotransferase_activity,_paired_acceptors |
| GO:0016843 | 2 | amine-lyase_activity |
| GO:0016844 | 2 | strictosidine_synthase_activity |
| GO:0016855 | 2 | racemase_and_epimerase_activity,_acting_on_amino_acids_and_derivatives |
| GO:0016880 | 2 | acid-ammonia_(or_amide)_ligase_activity |
| GO:0016890 | 2 | site-specific_endodeoxyribonuclease_activity,_specific_for_altered_base |
| GO:0016899 | 2 | oxidoreductase_activity,_acting_on_the_CH-OH_group_of_donors,_oxygen_as_acceptor |
| GO:0017034 | 2 | Rap_guanyl-nucleotide_exchange_factor_activity |
| GO:0017042 | 2 | glycosylceramidase_activity |
| GO:0017050 | 2 | D-erythro-sphingosine_kinase_activity |
| GO:0017055 | 2 | negative_regulation_of_RNA_polymerase_II_transcriptional_preinitiation_complex_assembly |
| GO:0017057 | 2 | 6-phosphogluconolactonase_activity |
| GO:0017062 | 2 | respiratory_chain_complex_III_assembly |
| GO:0017077 | 2 | oxidative_phosphorylation_uncoupler_activity |
| GO:0017099 | 2 | very-long-chain-acyl-CoA_dehydrogenase_activity |
| GO:0017101 | 2 | aminoacyl-tRNA_synthetase_multienzyme_complex |
| GO:0017109 | 2 | glutamate-cysteine_ligase_complex |
| GO:0017116 | 2 | single-stranded_DNA-dependent_ATP-dependent_DNA_helicase_activity |
| GO:0017129 | 2 | triglyceride_binding |
| GO:0017143 | 2 | insecticide_metabolic_process |
| GO:0017186 | 2 | peptidyl-pyroglutamic_acid_biosynthetic_process,_using_glutaminyl-peptide_cyclotransferase |
| GO:0018153 | 2 | isopeptide_cross-linking_via_N6-(L-isoglutamyl)-L-lysine |
| GO:0018197 | 2 | peptidyl-aspartic_acid_modification |
| GO:0018262 | 2 | isopeptide_cross-linking |
| GO:0018277 | 2 | protein_deamination |
| GO:0018282 | 2 | metal_incorporation_into_metallo-sulfur_cluster |
| GO:0018283 | 2 | iron_incorporation_into_metallo-sulfur_cluster |
| GO:0018352 | 2 | protein-pyridoxal-5-phosphate_linkage |
| GO:0018675 | 2 | (S)-limonene_6-monooxygenase_activity |
| GO:0018676 | 2 | (S)-limonene_7-monooxygenase_activity |
| GO:0018874 | 2 | benzoate_metabolic_process |
| GO:0018992 | 2 | germ-line_sex_determination |
| GO:0018993 | 2 | somatic_sex_determination |
| GO:0019008 | 2 | molybdopterin_synthase_complex |
| GO:0019043 | 2 | establishment_of_viral_latency |
| GO:0019046 | 2 | reactivation_of_latent_virus |
| GO:0019062 | 2 | viral_attachment_to_host_cell |
| GO:0019067 | 2 | viral_assembly,_maturation,_egress,_and_release |
| GO:0019076 | 2 | release_of_virus_from_host |
| GO:0019086 | 2 | late_viral_mRNA_transcription |
| GO:0019100 | 2 | male_germ-line_sex_determination |
| GO:0019107 | 2 | myristoyltransferase_activity |
| GO:0019113 | 2 | limonene_monooxygenase_activity |
| GO:0019230 | 2 | proprioception |
| GO:0019255 | 2 | glucose_1-phosphate_metabolic_process |
| GO:0019264 | 2 | glycine_biosynthetic_process_from_serine |
| GO:0019265 | 2 | glycine_biosynthetic_process,_by_transamination_of_glyoxylate |
| GO:0019285 | 2 | glycine_betaine_biosynthetic_process_from_choline |
| GO:0019303 | 2 | D-ribose_catabolic_process |
| GO:0019346 | 2 | transsulfuration |
| GO:0019364 | 2 | pyridine_nucleotide_catabolic_process |
| GO:0019371 | 2 | cyclooxygenase_pathway |
| GO:0019372 | 2 | lipoxygenase_pathway |
| GO:0019375 | 2 | galactolipid_biosynthetic_process |
| GO:0019376 | 2 | galactolipid_catabolic_process |
| GO:0019442 | 2 | tryptophan_catabolic_process_to_acetyl-CoA |
| GO:0019448 | 2 | L-cysteine_catabolic_process |
| GO:0019483 | 2 | beta-alanine_biosynthetic_process |
| GO:0019541 | 2 | propionate_metabolic_process |
| GO:0019550 | 2 | glutamate_catabolic_process_to_aspartate |
| GO:0019551 | 2 | glutamate_catabolic_process_to_2-oxoglutarate |
| GO:0019566 | 2 | arabinose_metabolic_process |
| GO:0019692 | 2 | deoxyribose_phosphate_metabolic_process |
| GO:0019706 | 2 | protein-cysteine_S-palmitoleyltransferase_activity |
| GO:0019707 | 2 | protein-cysteine_S-acyltransferase_activity |
| GO:0019730 | 2 | antimicrobial_humoral_response |
| GO:0019747 | 2 | regulation_of_isoprenoid_metabolic_process |
| GO:0019777 | 2 | Atg12_ligase_activity |
| GO:0019781 | 2 | NEDD8_activating_enzyme_activity |
| GO:0019784 | 2 | NEDD8-specific_protease_activity |
| GO:0019853 | 2 | L-ascorbic_acid_biosynthetic_process |
| GO:0019858 | 2 | cytosine_metabolic_process |
| GO:0019860 | 2 | uracil_metabolic_process |
| GO:0019918 | 2 | peptidyl-arginine_methylation,_to_symmetrical-dimethyl_arginine |
| GO:0019948 | 2 | SUMO_activating_enzyme_activity |
| GO:0019961 | 2 | interferon_binding |
| GO:0019972 | 2 | interleukin-12_binding |
| GO:0019981 | 2 | interleukin-6_binding |
| GO:0020003 | 2 | symbiont-containing_vacuole |
| GO:0020028 | 2 | hemoglobin_import |
| GO:0021506 | 2 | anterior_neuropore_closure |
| GO:0021512 | 2 | spinal_cord_anterior/posterior_patterning |
| GO:0021514 | 2 | ventral_spinal_cord_interneuron_differentiation |
| GO:0021521 | 2 | ventral_spinal_cord_interneuron_specification |
| GO:0021523 | 2 | somatic_motor_neuron_differentiation |
| GO:0021524 | 2 | visceral_motor_neuron_differentiation |
| GO:0021532 | 2 | neural_tube_patterning |
| GO:0021551 | 2 | central_nervous_system_morphogenesis |
| GO:0021553 | 2 | olfactory_nerve_development |
| GO:0021557 | 2 | oculomotor_nerve_development |
| GO:0021558 | 2 | trochlear_nerve_development |
| GO:0021562 | 2 | vestibulocochlear_nerve_development |
| GO:0021568 | 2 | rhombomere_2_development |
| GO:0021569 | 2 | rhombomere_3_development |
| GO:0021610 | 2 | facial_nerve_morphogenesis |
| GO:0021615 | 2 | glossopharyngeal_nerve_morphogenesis |
| GO:0021636 | 2 | trigeminal_nerve_morphogenesis |
| GO:0021644 | 2 | vagus_nerve_morphogenesis |
| GO:0021681 | 2 | cerebellar_granular_layer_development |
| GO:0021747 | 2 | cochlear_nucleus_development |
| GO:0021750 | 2 | vestibular_nucleus_development |
| GO:0021757 | 2 | caudate_nucleus_development |
| GO:0021758 | 2 | putamen_development |
| GO:0021759 | 2 | globus_pallidus_development |
| GO:0021761 | 2 | limbic_system_development |
| GO:0021773 | 2 | striatal_medium_spiny_neuron_differentiation |
| GO:0021775 | 2 | smoothened_signaling_pathway_involved_in_ventral_spinal_cord_interneuron_specification |
| GO:0021776 | 2 | smoothened_signaling_pathway_involved_in_spinal_cord_motor_neuron_cell_fate_specification |
| GO:0021784 | 2 | postganglionic_parasympathetic_nervous_system_development |
| GO:0021801 | 2 | cerebral_cortex_radial_glia_guided_migration |
| GO:0021815 | 2 | modulation_of_microtubule_cytoskeleton_involved_in_cerebral_cortex_radial_glia_guided_migration |
| GO:0021816 | 2 | extension_of_a_leading_process_involved_in_cell_motility_in_cerebral_cortex_radial_glia_guided_migration |
| GO:0021836 | 2 | chemorepulsion_involved_in_postnatal_olfactory_bulb_interneuron_migration |
| GO:0021847 | 2 | ventricular_zone_neuroblast_division |
| GO:0021849 | 2 | neuroblast_division_in_subventricular_zone |
| GO:0021874 | 2 | Wnt_receptor_signaling_pathway_involved_in_forebrain_neuroblast_division |
| GO:0021881 | 2 | Wnt_receptor_signaling_pathway_involved_in_forebrain_neuron_fate_commitment |
| GO:0021882 | 2 | regulation_of_transcription_from_RNA_polymerase_II_promoter_involved_in_forebrain_neuron_fate_commitment |
| GO:0021892 | 2 | cerebral_cortex_GABAergic_interneuron_differentiation |
| GO:0021893 | 2 | cerebral_cortex_GABAergic_interneuron_fate_commitment |
| GO:0021903 | 2 | rostrocaudal_neural_tube_patterning |
| GO:0021932 | 2 | hindbrain_radial_glia_guided_cell_migration |
| GO:0021934 | 2 | hindbrain_tangential_cell_migration |
| GO:0021937 | 2 | cerebellar_Purkinje_cell-granule_cell_precursor_cell_signaling_involved_in_regulation_of_granule_cell_precursor_cell_proliferation |
| GO:0021943 | 2 | formation_of_radial_glial_scaffolds |
| GO:0021965 | 2 | spinal_cord_ventral_commissure_morphogenesis |
| GO:0021985 | 2 | neurohypophysis_development |
| GO:0021986 | 2 | habenula_development |
| GO:0021999 | 2 | neural_plate_anterior/posterior_regionalization |
| GO:0022897 | 2 | proton-dependent_peptide_secondary_active_transmembrane_transporter_activity |
| GO:0023035 | 2 | CD40_signaling_pathway |
| GO:0030014 | 2 | CCR4-NOT_complex |
| GO:0030050 | 2 | vesicle_transport_along_actin_filament |
| GO:0030060 | 2 | L-malate_dehydrogenase_activity |
| GO:0030070 | 2 | insulin_processing |
| GO:0030109 | 2 | HLA-B_specific_inhibitory_MHC_class_I_receptor_activity |
| GO:0030112 | 2 | glycocalyx |
| GO:0030144 | 2 | alpha-1,6-mannosylglycoprotein_6-beta-N-acetylglucosaminyltransferase_activity |
| GO:0030150 | 2 | protein_import_into_mitochondrial_matrix |
| GO:0030156 | 2 | benzodiazepine_receptor_binding |
| GO:0030158 | 2 | protein_xylosyltransferase_activity |
| GO:0030167 | 2 | proteoglycan_catabolic_process |
| GO:0030171 | 2 | voltage-gated_proton_channel_activity |
| GO:0030187 | 2 | melatonin_biosynthetic_process |
| GO:0030200 | 2 | heparan_sulfate_proteoglycan_catabolic_process |
| GO:0030205 | 2 | dermatan_sulfate_metabolic_process |
| GO:0030208 | 2 | dermatan_sulfate_biosynthetic_process |
| GO:0030221 | 2 | basophil_differentiation |
| GO:0030222 | 2 | eosinophil_differentiation |
| GO:0030223 | 2 | neutrophil_differentiation |
| GO:0030237 | 2 | female_sex_determination |
| GO:0030264 | 2 | nuclear_fragmentation_involved_in_apoptotic_nuclear_change |
| GO:0030289 | 2 | protein_phosphatase_4_complex |
| GO:0030294 | 2 | receptor_signaling_protein_tyrosine_kinase_inhibitor_activity |
| GO:0030305 | 2 | heparanase_activity |
| GO:0030311 | 2 | poly-N-acetyllactosamine_biosynthetic_process |
| GO:0030312 | 2 | external_encapsulating_structure |
| GO:0030327 | 2 | prenylated_protein_catabolic_process |
| GO:0030328 | 2 | prenylcysteine_catabolic_process |
| GO:0030329 | 2 | prenylcysteine_metabolic_process |
| GO:0030337 | 2 | DNA_polymerase_processivity_factor_activity |
| GO:0030362 | 2 | protein_phosphatase_type_4_regulator_activity |
| GO:0030368 | 2 | interleukin-17_receptor_activity |
| GO:0030382 | 2 | sperm_mitochondrion_organization |
| GO:0030389 | 2 | fructosamine_metabolic_process |
| GO:0030397 | 2 | membrane_disassembly |
| GO:0030479 | 2 | actin_cortical_patch |
| GO:0030488 | 2 | tRNA_methylation |
| GO:0030548 | 2 | acetylcholine_receptor_regulator_activity |
| GO:0030611 | 2 | arsenate_reductase_activity |
| GO:0030617 | 2 | transforming_growth_factor_beta_receptor,_inhibitory_cytoplasmic_mediator_activity |
| GO:0030628 | 2 | pre-mRNA_3'-splice_site_binding |
| GO:0030663 | 2 | COPI_coated_vesicle_membrane |
| GO:0030687 | 2 | preribosome,_large_subunit_precursor |
| GO:0030689 | 2 | Noc_complex |
| GO:0030704 | 2 | vitelline_membrane_formation |
| GO:0030719 | 2 | P_granule_organization |
| GO:0030849 | 2 | autosome |
| GO:0030860 | 2 | regulation_of_polarized_epithelial_cell_differentiation |
| GO:0030862 | 2 | positive_regulation_of_polarized_epithelial_cell_differentiation |
| GO:0030882 | 2 | lipid_antigen_binding |
| GO:0030884 | 2 | exogenous_lipid_antigen_binding |
| GO:0030885 | 2 | regulation_of_myeloid_dendritic_cell_activation |
| GO:0030892 | 2 | mitotic_cohesin_complex |
| GO:0030908 | 2 | protein_splicing |
| GO:0030942 | 2 | endoplasmic_reticulum_signal_peptide_binding |
| GO:0030967 | 2 | ER-nuclear_sterol_response_pathway |
| GO:0030977 | 2 | taurine_binding |
| GO:0030981 | 2 | cortical_microtubule_cytoskeleton |
| GO:0030997 | 2 | regulation_of_centriole-centriole_cohesion |
| GO:0031064 | 2 | negative_regulation_of_histone_deacetylation |
| GO:0031074 | 2 | nucleocytoplasmic_shuttling_complex |
| GO:0031104 | 2 | dendrite_regeneration |
| GO:0031119 | 2 | tRNA_pseudouridine_synthesis |
| GO:0031120 | 2 | snRNA_pseudouridine_synthesis |
| GO:0031134 | 2 | sister_chromatid_biorientation |
| GO:0031213 | 2 | RSF_complex |
| GO:0031247 | 2 | actin_rod_assembly |
| GO:0031296 | 2 | B_cell_costimulation |
| GO:0031298 | 2 | replication_fork_protection_complex |
| GO:0031315 | 2 | extrinsic_to_mitochondrial_outer_membrane |
| GO:0031335 | 2 | regulation_of_sulfur_amino_acid_metabolic_process |
| GO:0031372 | 2 | UBC13-MMS2_complex |
| GO:0031386 | 2 | protein_tag |
| GO:0031405 | 2 | lipoic_acid_binding |
| GO:0031436 | 2 | BRCA1-BARD1_complex |
| GO:0031444 | 2 | slow-twitch_skeletal_muscle_fiber_contraction |
| GO:0031446 | 2 | regulation_of_fast-twitch_skeletal_muscle_fiber_contraction |
| GO:0031448 | 2 | positive_regulation_of_fast-twitch_skeletal_muscle_fiber_contraction |
| GO:0031455 | 2 | glycine_betaine_metabolic_process |
| GO:0031456 | 2 | glycine_betaine_biosynthetic_process |
| GO:0031492 | 2 | nucleosomal_DNA_binding |
| GO:0031508 | 2 | centromeric_heterochromatin_assembly |
| GO:0031515 | 2 | tRNA_(m1A)_methyltransferase_complex |
| GO:0031523 | 2 | Myb_complex |
| GO:0031597 | 2 | cytosolic_proteasome_complex |
| GO:0031627 | 2 | telomeric_loop_formation |
| GO:0031630 | 2 | regulation_of_synaptic_vesicle_fusion_to_presynaptic_membrane |
| GO:0031673 | 2 | H_zone |
| GO:0031686 | 2 | A1_adenosine_receptor_binding |
| GO:0031696 | 2 | alpha-2C_adrenergic_receptor_binding |
| GO:0031698 | 2 | beta-2_adrenergic_receptor_binding |
| GO:0031711 | 2 | bradykinin_receptor_binding |
| GO:0031716 | 2 | calcitonin_receptor_binding |
| GO:0031720 | 2 | haptoglobin_binding |
| GO:0031727 | 2 | CCR2_chemokine_receptor_binding |
| GO:0031732 | 2 | CCR7_chemokine_receptor_binding |
| GO:0031752 | 2 | D5_dopamine_receptor_binding |
| GO:0031753 | 2 | endothelial_differentiation_G-protein_coupled_receptor_binding |
| GO:0031755 | 2 | Edg-2_lysophosphatidic_acid_receptor_binding |
| GO:0031780 | 2 | corticotropin_hormone_receptor_binding |
| GO:0031783 | 2 | type_5_melanocortin_receptor_binding |
| GO:0031798 | 2 | type_1_metabotropic_glutamate_receptor_binding |
| GO:0031852 | 2 | mu-type_opioid_receptor_binding |
| GO:0031862 | 2 | prostanoid_receptor_binding |
| GO:0031893 | 2 | vasopressin_receptor_binding |
| GO:0031894 | 2 | V1A_vasopressin_receptor_binding |
| GO:0031914 | 2 | negative_regulation_of_synaptic_plasticity |
| GO:0031937 | 2 | positive_regulation_of_chromatin_silencing |
| GO:0031989 | 2 | bombesin_receptor_signaling_pathway |
| GO:0031999 | 2 | negative_regulation_of_fatty_acid_beta-oxidation |
| GO:0032027 | 2 | myosin_light_chain_binding |
| GO:0032038 | 2 | myosin_II_heavy_chain_binding |
| GO:0032044 | 2 | DSIF_complex |
| GO:0032074 | 2 | negative_regulation_of_nuclease_activity |
| GO:0032089 | 2 | NACHT_domain_binding |
| GO:0032090 | 2 | Pyrin_domain_binding |
| GO:0032116 | 2 | SMC_loading_complex |
| GO:0032119 | 2 | sequestering_of_zinc_ion |
| GO:0032143 | 2 | single_thymine_insertion_binding |
| GO:0032181 | 2 | dinucleotide_repeat_insertion_binding |
| GO:0032196 | 2 | transposition |
| GO:0032213 | 2 | regulation_of_telomere_maintenance_via_semi-conservative_replication |
| GO:0032214 | 2 | negative_regulation_of_telomere_maintenance_via_semi-conservative_replication |
| GO:0032235 | 2 | negative_regulation_of_calcium_ion_transport_via_store-operated_calcium_channel_activity |
| GO:0032241 | 2 | positive_regulation_of_nucleobase-containing_compound_transport |
| GO:0032252 | 2 | secretory_granule_localization |
| GO:0032263 | 2 | GMP_salvage |
| GO:0032275 | 2 | luteinizing_hormone_secretion |
| GO:0032289 | 2 | central_nervous_system_myelin_formation |
| GO:0032298 | 2 | positive_regulation_of_DNA-dependent_DNA_replication_initiation |
| GO:0032301 | 2 | MutSalpha_complex |
| GO:0032302 | 2 | MutSbeta_complex |
| GO:0032304 | 2 | negative_regulation_of_icosanoid_secretion |
| GO:0032311 | 2 | angiogenin-PRI_complex |
| GO:0032346 | 2 | positive_regulation_of_aldosterone_metabolic_process |
| GO:0032349 | 2 | positive_regulation_of_aldosterone_biosynthetic_process |
| GO:0032390 | 2 | MutLbeta_complex |
| GO:0032416 | 2 | negative_regulation_of_sodium:hydrogen_antiporter_activity |
| GO:0032423 | 2 | regulation_of_mismatch_repair |
| GO:0032433 | 2 | filopodium_tip |
| GO:0032437 | 2 | cuticular_plate |
| GO:0032510 | 2 | endosome_to_lysosome_transport_via_multivesicular_body_sorting_pathway |
| GO:0032513 | 2 | negative_regulation_of_protein_phosphatase_type_2B_activity |
| GO:0032534 | 2 | regulation_of_microvillus_assembly |
| GO:0032594 | 2 | protein_transport_within_lipid_bilayer |
| GO:0032602 | 2 | chemokine_production |
| GO:0032633 | 2 | interleukin-4_production |
| GO:0032672 | 2 | regulation_of_interleukin-3_production |
| GO:0032707 | 2 | negative_regulation_of_interleukin-23_production |
| GO:0032741 | 2 | positive_regulation_of_interleukin-18_production |
| GO:0032764 | 2 | negative_regulation_of_mast_cell_cytokine_production |
| GO:0032765 | 2 | positive_regulation_of_mast_cell_cytokine_production |
| GO:0032767 | 2 | copper-dependent_protein_binding |
| GO:0032796 | 2 | uropod_organization |
| GO:0032798 | 2 | Swi5-Sfr1_complex |
| GO:0032799 | 2 | low-density_lipoprotein_receptor_particle_metabolic_process |
| GO:0032806 | 2 | carboxy-terminal_domain_protein_kinase_complex |
| GO:0032807 | 2 | DNA_ligase_IV_complex |
| GO:0032810 | 2 | sterol_response_element_binding |
| GO:0032829 | 2 | regulation_of_CD4-positive,_CD25-positive,_alpha-beta_regulatory_T_cell_differentiation |
| GO:0032831 | 2 | positive_regulation_of_CD4-positive,_CD25-positive,_alpha-beta_regulatory_T_cell_differentiation |
| GO:0032841 | 2 | calcitonin_binding |
| GO:0032861 | 2 | activation_of_Rap_GTPase_activity |
| GO:0032898 | 2 | neurotrophin_production |
| GO:0032899 | 2 | regulation_of_neurotrophin_production |
| GO:0032900 | 2 | negative_regulation_of_neurotrophin_production |
| GO:0032902 | 2 | nerve_growth_factor_production |
| GO:0032911 | 2 | negative_regulation_of_transforming_growth_factor_beta1_production |
| GO:0032915 | 2 | positive_regulation_of_transforming_growth_factor_beta2_production |
| GO:0032916 | 2 | positive_regulation_of_transforming_growth_factor_beta3_production |
| GO:0032929 | 2 | negative_regulation_of_superoxide_anion_generation |
| GO:0032937 | 2 | SREBP-SCAP-Insig_complex |
| GO:0032959 | 2 | inositol_trisphosphate_biosynthetic_process |
| GO:0032997 | 2 | Fc_receptor_complex |
| GO:0032998 | 2 | Fc-epsilon_receptor_I_complex |
| GO:0033010 | 2 | paranodal_junction |
| GO:0033040 | 2 | sour_taste_receptor_activity |
| GO:0033041 | 2 | sweet_taste_receptor_activity |
| GO:0033045 | 2 | regulation_of_sister_chromatid_segregation |
| GO:0033047 | 2 | regulation_of_mitotic_sister_chromatid_segregation |
| GO:0033058 | 2 | directional_locomotion |
| GO:0033061 | 2 | DNA_recombinase_mediator_complex |
| GO:0033093 | 2 | Weibel-Palade_body |
| GO:0033128 | 2 | negative_regulation_of_histone_phosphorylation |
| GO:0033132 | 2 | negative_regulation_of_glucokinase_activity |
| GO:0033136 | 2 | serine_phosphorylation_of_STAT3_protein |
| GO:0033139 | 2 | regulation_of_peptidyl-serine_phosphorylation_of_STAT_protein |
| GO:0033141 | 2 | positive_regulation_of_peptidyl-serine_phosphorylation_of_STAT_protein |
| GO:0033170 | 2 | protein-DNA_loading_ATPase_activity |
| GO:0033257 | 2 | Bcl3/NF-kappaB2_complex |
| GO:0033260 | 2 | DNA_replication_involved_in_S_phase |
| GO:0033262 | 2 | regulation_of_DNA_replication_involved_in_S_phase |
| GO:0033278 | 2 | cell_proliferation_in_midbrain |
| GO:0033292 | 2 | T-tubule_organization |
| GO:0033306 | 2 | phytol_metabolic_process |
| GO:0033313 | 2 | meiotic_cell_cycle_checkpoint |
| GO:0033326 | 2 | cerebrospinal_fluid_secretion |
| GO:0033364 | 2 | mast_cell_secretory_granule_organization |
| GO:0033383 | 2 | geranyl_diphosphate_metabolic_process |
| GO:0033384 | 2 | geranyl_diphosphate_biosynthetic_process |
| GO:0033484 | 2 | nitric_oxide_homeostasis |
| GO:0033552 | 2 | response_to_vitamin_B3 |
| GO:0033557 | 2 | Slx1-Slx4_complex |
| GO:0033561 | 2 | regulation_of_water_loss_via_skin |
| GO:0033563 | 2 | dorsal/ventral_axon_guidance |
| GO:0033566 | 2 | gamma-tubulin_complex_localization |
| GO:0033578 | 2 | protein_glycosylation_in_Golgi |
| GO:0033602 | 2 | negative_regulation_of_dopamine_secretion |
| GO:0033615 | 2 | mitochondrial_proton-transporting_ATP_synthase_complex_assembly |
| GO:0033617 | 2 | mitochondrial_respiratory_chain_complex_IV_assembly |
| GO:0033647 | 2 | host_intracellular_organelle |
| GO:0033648 | 2 | host_intracellular_membrane-bounded_organelle |
| GO:0033677 | 2 | DNA/RNA_helicase_activity |
| GO:0033743 | 2 | peptide-methionine_(R)-S-oxide_reductase_activity |
| GO:0033754 | 2 | indoleamine_2,3-dioxygenase_activity |
| GO:0033842 | 2 | N-acetyl-beta-glucosaminyl-glycoprotein_4-beta-N-acetylgalactosaminyltransferase_activity |
| GO:0033857 | 2 | diphosphoinositol-pentakisphosphate_kinase_activity |
| GO:0033861 | 2 | negative_regulation_of_NAD(P)H_oxidase_activity |
| GO:0033864 | 2 | positive_regulation_of_NAD(P)H_oxidase_activity |
| GO:0033866 | 2 | nucleoside_bisphosphate_biosynthetic_process |
| GO:0033872 | 2 | [heparan_sulfate]-glucosamine_3-sulfotransferase_3_activity |
| GO:0033883 | 2 | pyridoxal_phosphatase_activity |
| GO:0033897 | 2 | ribonuclease_T2_activity |
| GO:0033906 | 2 | hyaluronoglucuronidase_activity |
| GO:0034030 | 2 | ribonucleoside_bisphosphate_biosynthetic_process |
| GO:0034033 | 2 | purine_nucleoside_bisphosphate_biosynthetic_process |
| GO:0034036 | 2 | purine_ribonucleoside_bisphosphate_biosynthetic_process |
| GO:0034048 | 2 | negative_regulation_of_protein_phosphatase_type_2A_activity |
| GO:0034059 | 2 | response_to_anoxia |
| GO:0034091 | 2 | regulation_of_maintenance_of_sister_chromatid_cohesion |
| GO:0034093 | 2 | positive_regulation_of_maintenance_of_sister_chromatid_cohesion |
| GO:0034137 | 2 | positive_regulation_of_toll-like_receptor_2_signaling_pathway |
| GO:0034140 | 2 | negative_regulation_of_toll-like_receptor_3_signaling_pathway |
| GO:0034154 | 2 | toll-like_receptor_7_signaling_pathway |
| GO:0034162 | 2 | toll-like_receptor_9_signaling_pathway |
| GO:0034163 | 2 | regulation_of_toll-like_receptor_9_signaling_pathway |
| GO:0034182 | 2 | regulation_of_maintenance_of_mitotic_sister_chromatid_cohesion |
| GO:0034184 | 2 | positive_regulation_of_maintenance_of_mitotic_sister_chromatid_cohesion |
| GO:0034224 | 2 | cellular_response_to_zinc_ion_starvation |
| GO:0034239 | 2 | regulation_of_macrophage_fusion |
| GO:0034241 | 2 | positive_regulation_of_macrophage_fusion |
| GO:0034248 | 2 | regulation_of_cellular_amide_metabolic_process |
| GO:0034349 | 2 | glial_cell_apoptotic_process |
| GO:0034418 | 2 | urate_biosynthetic_process |
| GO:0034436 | 2 | glycoprotein_transport |
| GO:0034437 | 2 | glycoprotein_transporter_activity |
| GO:0034465 | 2 | response_to_carbon_monoxide |
| GO:0034471 | 2 | ncRNA_5'-end_processing |
| GO:0034485 | 2 | phosphatidylinositol-3,4,5-trisphosphate_5-phosphatase_activity |
| GO:0034499 | 2 | late_endosome_to_Golgi_transport |
| GO:0034509 | 2 | centromeric_core_chromatin_assembly |
| GO:0034551 | 2 | mitochondrial_respiratory_chain_complex_III_assembly |
| GO:0034596 | 2 | phosphatidylinositol_phosphate_4-phosphatase_activity |
| GO:0034597 | 2 | phosphatidylinositol-4,5-bisphosphate_4-phosphatase_activity |
| GO:0034635 | 2 | glutathione_transport |
| GO:0034638 | 2 | phosphatidylcholine_catabolic_process |
| GO:0034649 | 2 | histone_demethylase_activity_(H3-monomethyl-K4_specific) |
| GO:0034679 | 2 | alpha9-beta1_integrin_complex |
| GO:0034701 | 2 | tripeptidase_activity |
| GO:0034736 | 2 | cholesterol_O-acyltransferase_activity |
| GO:0034750 | 2 | Scrib-APC-beta-catenin_complex |
| GO:0034773 | 2 | histone_H4-K20_trimethylation |
| GO:0034970 | 2 | histone_H3-R2_methylation |
| GO:0034982 | 2 | mitochondrial_protein_processing |
| GO:0034988 | 2 | Fc-gamma_receptor_I_complex_binding |
| GO:0035090 | 2 | maintenance_of_apical/basal_cell_polarity |
| GO:0035093 | 2 | spermatogenesis,_exchange_of_chromosomal_proteins |
| GO:0035101 | 2 | FACT_complex |
| GO:0035106 | 2 | operant_conditioning |
| GO:0035166 | 2 | post-embryonic_hemopoiesis |
| GO:0035243 | 2 | protein-arginine_omega-N_symmetric_methyltransferase_activity |
| GO:0035261 | 2 | external_genitalia_morphogenesis |
| GO:0035276 | 2 | ethanol_binding |
| GO:0035279 | 2 | mRNA_cleavage_involved_in_gene_silencing_by_miRNA |
| GO:0035312 | 2 | 5'-3'_exodeoxyribonuclease_activity |
| GO:0035325 | 2 | Toll-like_receptor_binding |
| GO:0035339 | 2 | SPOTS_complex |
| GO:0035349 | 2 | coenzyme_A_transmembrane_transport |
| GO:0035354 | 2 | Toll-like_receptor_1-Toll-like_receptor_2_protein_complex |
| GO:0035355 | 2 | Toll-like_receptor_2-Toll-like_receptor_6_protein_complex |
| GO:0035357 | 2 | peroxisome_proliferator_activated_receptor_signaling_pathway |
| GO:0035359 | 2 | negative_regulation_of_peroxisome_proliferator_activated_receptor_signaling_pathway |
| GO:0035370 | 2 | UBC13-UEV1A_complex |
| GO:0035374 | 2 | chondroitin_sulfate_binding |
| GO:0035402 | 2 | histone_kinase_activity_(H3-T11_specific) |
| GO:0035403 | 2 | histone_kinase_activity_(H3-T6_specific) |
| GO:0035407 | 2 | histone_H3-T11_phosphorylation |
| GO:0035408 | 2 | histone_H3-T6_phosphorylation |
| GO:0035419 | 2 | activation_of_MAPK_activity_involved_in_innate_immune_response |
| GO:0035425 | 2 | autocrine_signaling |
| GO:0035470 | 2 | positive_regulation_of_vascular_wound_healing |
| GO:0035473 | 2 | lipase_binding |
| GO:0035494 | 2 | SNARE_complex_disassembly |
| GO:0035517 | 2 | PR-DUB_complex |
| GO:0035521 | 2 | monoubiquitinated_histone_deubiquitination |
| GO:0035522 | 2 | monoubiquitinated_histone_H2A_deubiquitination |
| GO:0035524 | 2 | proline_transmembrane_transport |
| GO:0035552 | 2 | oxidative_single-stranded_DNA_demethylation |
| GO:0035561 | 2 | regulation_of_chromatin_binding |
| GO:0035565 | 2 | regulation_of_pronephros_size |
| GO:0035566 | 2 | regulation_of_metanephros_size |
| GO:0035574 | 2 | histone_H4-K20_demethylation |
| GO:0035575 | 2 | histone_demethylase_activity_(H4-K20_specific) |
| GO:0035578 | 2 | azurophil_granule_lumen |
| GO:0035607 | 2 | fibroblast_growth_factor_receptor_signaling_pathway_involved_in_orbitofrontal_cortex_development |
| GO:0035612 | 2 | AP-2_adaptor_complex_binding |
| GO:0035617 | 2 | stress_granule_disassembly |
| GO:0035671 | 2 | enone_reductase_activity |
| GO:0035701 | 2 | hematopoietic_stem_cell_migration |
| GO:0035717 | 2 | chemokine_(C-C_motif)_ligand_7_binding |
| GO:0035726 | 2 | common_myeloid_progenitor_cell_proliferation |
| GO:0035759 | 2 | mesangial_cell-matrix_adhesion |
| GO:0035802 | 2 | adrenal_cortex_formation |
| GO:0035822 | 2 | gene_conversion |
| GO:0035854 | 2 | eosinophil_fate_commitment |
| GO:0035880 | 2 | embryonic_nail_plate_morphogenesis |
| GO:0035881 | 2 | amacrine_cell_differentiation |
| GO:0035887 | 2 | aortic_smooth_muscle_cell_differentiation |
| GO:0035904 | 2 | aorta_development |
| GO:0035912 | 2 | dorsal_aorta_morphogenesis |
| GO:0035983 | 2 | response_to_trichostatin_A |
| GO:0035984 | 2 | cellular_response_to_trichostatin_A |
| GO:0035990 | 2 | tendon_cell_differentiation |
| GO:0036093 | 2 | germ_cell_proliferation |
| GO:0036112 | 2 | medium-chain_fatty-acyl-CoA_metabolic_process |
| GO:0036117 | 2 | hyaluranon_cable |
| GO:0036125 | 2 | fatty_acid_beta-oxidation_multienzyme_complex |
| GO:0036132 | 2 | 13-prostaglandin_reductase_activity |
| GO:0036215 | 2 | response_to_stem_cell_factor_stimulus |
| GO:0036216 | 2 | cellular_response_to_stem_cell_factor_stimulus |
| GO:0038001 | 2 | paracrine_signaling |
| GO:0038007 | 2 | netrin-activated_signaling_pathway |
| GO:0038025 | 2 | reelin_receptor_activity |
| GO:0038031 | 2 | non-canonical_Wnt_receptor_signaling_pathway_via_JNK_cascade |
| GO:0038043 | 2 | interleukin-5-mediated_signaling_pathway |
| GO:0038062 | 2 | protein_tyrosine_kinase_collagen_receptor_activity |
| GO:0038063 | 2 | collagen-activated_tyrosine_kinase_receptor_signaling_pathway |
| GO:0038064 | 2 | collagen_receptor_activity |
| GO:0038065 | 2 | collagen-activated_signaling_pathway |
| GO:0038083 | 2 | peptidyl-tyrosine_autophosphorylation |
| GO:0038107 | 2 | nodal_signaling_pathway_involved_in_determination_of_left/right_asymmetry |
| GO:0038109 | 2 | Kit_signaling_pathway |
| GO:0038111 | 2 | interleukin-7-mediated_signaling_pathway |
| GO:0038112 | 2 | interleukin-8-mediated_signaling_pathway |
| GO:0038145 | 2 | macrophage_colony-stimulating_factor_signaling_pathway |
| GO:0038155 | 2 | interleukin-23-mediated_signaling_pathway |
| GO:0038156 | 2 | interleukin-3-mediated_signaling_pathway |
| GO:0038166 | 2 | angiotensin-mediated_signaling_pathway |
| GO:0039003 | 2 | pronephric_field_specification |
| GO:0039530 | 2 | MDA-5_signaling_pathway |
| GO:0039533 | 2 | regulation_of_MDA-5_signaling_pathway |
| GO:0040031 | 2 | snRNA_modification |
| GO:0040038 | 2 | polar_body_extrusion_after_meiotic_divisions |
| GO:0042019 | 2 | interleukin-23_binding |
| GO:0042020 | 2 | interleukin-23_receptor_activity |
| GO:0042030 | 2 | ATPase_inhibitor_activity |
| GO:0042073 | 2 | intraflagellar_transport |
| GO:0042078 | 2 | germ-line_stem_cell_division |
| GO:0042083 | 2 | 5,10-methylenetetrahydrofolate-dependent_methyltransferase_activity |
| GO:0042095 | 2 | interferon-gamma_biosynthetic_process |
| GO:0042109 | 2 | lymphotoxin_A_biosynthetic_process |
| GO:0042132 | 2 | fructose_1,6-bisphosphate_1-phosphatase_activity |
| GO:0042214 | 2 | terpene_metabolic_process |
| GO:0042226 | 2 | interleukin-6_biosynthetic_process |
| GO:0042245 | 2 | RNA_repair |
| GO:0042262 | 2 | DNA_protection |
| GO:0042264 | 2 | peptidyl-aspartic_acid_hydroxylation |
| GO:0042322 | 2 | negative_regulation_of_circadian_sleep/wake_cycle,_REM_sleep |
| GO:0042323 | 2 | negative_regulation_of_circadian_sleep/wake_cycle,_non-REM_sleep |
| GO:0042328 | 2 | heparan_sulfate_N-acetylglucosaminyltransferase_activity |
| GO:0042335 | 2 | cuticle_development |
| GO:0042350 | 2 | GDP-L-fucose_biosynthetic_process |
| GO:0042351 | 2 | 'de_novo'_GDP-L-fucose_biosynthetic_process |
| GO:0042353 | 2 | fucose_biosynthetic_process |
| GO:0042392 | 2 | sphingosine-1-phosphate_phosphatase_activity |
| GO:0042412 | 2 | taurine_biosynthetic_process |
| GO:0042418 | 2 | epinephrine_biosynthetic_process |
| GO:0042450 | 2 | arginine_biosynthetic_process_via_ornithine |
| GO:0042483 | 2 | negative_regulation_of_odontogenesis |
| GO:0042488 | 2 | positive_regulation_of_odontogenesis_of_dentin-containing_tooth |
| GO:0042495 | 2 | detection_of_triacyl_bacterial_lipopeptide |
| GO:0042496 | 2 | detection_of_diacyl_bacterial_lipopeptide |
| GO:0042498 | 2 | diacyl_lipopeptide_binding |
| GO:0042501 | 2 | serine_phosphorylation_of_STAT_protein |
| GO:0042508 | 2 | tyrosine_phosphorylation_of_Stat1_protein |
| GO:0042560 | 2 | pteridine-containing_compound_catabolic_process |
| GO:0042567 | 2 | insulin-like_growth_factor_ternary_complex |
| GO:0042583 | 2 | chromaffin_granule |
| GO:0042610 | 2 | CD8_receptor_binding |
| GO:0042631 | 2 | cellular_response_to_water_deprivation |
| GO:0042637 | 2 | catagen |
| GO:0042701 | 2 | progesterone_secretion |
| GO:0042706 | 2 | eye_photoreceptor_cell_fate_commitment |
| GO:0042727 | 2 | flavin-containing_compound_biosynthetic_process |
| GO:0042745 | 2 | circadian_sleep/wake_cycle |
| GO:0042758 | 2 | long-chain_fatty_acid_catabolic_process |
| GO:0042780 | 2 | tRNA_3'-end_processing |
| GO:0042822 | 2 | pyridoxal_phosphate_metabolic_process |
| GO:0042823 | 2 | pyridoxal_phosphate_biosynthetic_process |
| GO:0042824 | 2 | MHC_class_I_peptide_loading_complex |
| GO:0042835 | 2 | BRE_binding |
| GO:0042891 | 2 | antibiotic_transport |
| GO:0042903 | 2 | tubulin_deacetylase_activity |
| GO:0042939 | 2 | tripeptide_transport |
| GO:0042942 | 2 | D-serine_transport |
| GO:0042954 | 2 | lipoprotein_transporter_activity |
| GO:0042980 | 2 | cystic_fibrosis_transmembrane_conductance_regulator_binding |
| GO:0042986 | 2 | positive_regulation_of_amyloid_precursor_protein_biosynthetic_process |
| GO:0042997 | 2 | negative_regulation_of_Golgi_to_plasma_membrane_protein_transport |
| GO:0043006 | 2 | activation_of_phospholipase_A2_activity_by_calcium-mediated_signaling |
| GO:0043023 | 2 | ribosomal_large_subunit_binding |
| GO:0043120 | 2 | tumor_necrosis_factor_binding |
| GO:0043132 | 2 | NAD_transport |
| GO:0043134 | 2 | regulation_of_hindgut_contraction |
| GO:0043146 | 2 | spindle_stabilization |
| GO:0043148 | 2 | mitotic_spindle_stabilization |
| GO:0043163 | 2 | cell_envelope_organization |
| GO:0043179 | 2 | rhythmic_excitation |
| GO:0043183 | 2 | vascular_endothelial_growth_factor_receptor_1_binding |
| GO:0043185 | 2 | vascular_endothelial_growth_factor_receptor_3_binding |
| GO:0043199 | 2 | sulfate_binding |
| GO:0043262 | 2 | adenosine-diphosphatase_activity |
| GO:0043305 | 2 | negative_regulation_of_mast_cell_degranulation |
| GO:0043307 | 2 | eosinophil_activation |
| GO:0043308 | 2 | eosinophil_degranulation |
| GO:0043309 | 2 | regulation_of_eosinophil_degranulation |
| GO:0043314 | 2 | negative_regulation_of_neutrophil_degranulation |
| GO:0043366 | 2 | beta_selection |
| GO:0043369 | 2 | CD4-positive_or_CD8-positive,_alpha-beta_T_cell_lineage_commitment |
| GO:0043397 | 2 | regulation_of_corticotropin-releasing_hormone_secretion |
| GO:0043415 | 2 | positive_regulation_of_skeletal_muscle_tissue_regeneration |
| GO:0043423 | 2 | 3-phosphoinositide-dependent_protein_kinase_binding |
| GO:0043435 | 2 | response_to_corticotropin-releasing_hormone_stimulus |
| GO:0043482 | 2 | cellular_pigment_accumulation |
| GO:0043503 | 2 | skeletal_muscle_fiber_adaptation |
| GO:0043511 | 2 | inhibin_complex |
| GO:0043512 | 2 | inhibin_A_complex |
| GO:0043514 | 2 | interleukin-12_complex |
| GO:0043527 | 2 | tRNA_methyltransferase_complex |
| GO:0043532 | 2 | angiostatin_binding |
| GO:0043564 | 2 | Ku70:Ku80_complex |
| GO:0043610 | 2 | regulation_of_carbohydrate_utilization |
| GO:0043634 | 2 | polyadenylation-dependent_ncRNA_catabolic_process |
| GO:0043649 | 2 | dicarboxylic_acid_catabolic_process |
| GO:0043682 | 2 | copper-transporting_ATPase_activity |
| GO:0043987 | 2 | histone_H3-S10_phosphorylation |
| GO:0043988 | 2 | histone_H3-S28_phosphorylation |
| GO:0044028 | 2 | DNA_hypomethylation |
| GO:0044029 | 2 | hypomethylation_of_CpG_island |
| GO:0044088 | 2 | regulation_of_vacuole_organization |
| GO:0044107 | 2 | cellular_alcohol_metabolic_process |
| GO:0044108 | 2 | cellular_alcohol_biosynthetic_process |
| GO:0044194 | 2 | cytolytic_granule |
| GO:0044209 | 2 | AMP_salvage |
| GO:0044210 | 2 | 'de_novo'_CTP_biosynthetic_process |
| GO:0044294 | 2 | dendritic_growth_cone |
| GO:0044300 | 2 | cerebellar_mossy_fiber |
| GO:0044324 | 2 | regulation_of_transcription_involved_in_anterior/posterior_axis_specification |
| GO:0044334 | 2 | canonical_Wnt_receptor_signaling_pathway_involved_in_positive_regulation_of_epithelial_to_mesenchymal_transition |
| GO:0044335 | 2 | canonical_Wnt_receptor_signaling_pathway_involved_in_neural_crest_cell_differentiation |
| GO:0044336 | 2 | canonical_Wnt_receptor_signaling_pathway_involved_in_negative_regulation_of_apoptotic_process |
| GO:0044338 | 2 | canonical_Wnt_receptor_signaling_pathway_involved_in_mesenchymal_stem_cell_differentiation |
| GO:0044339 | 2 | canonical_Wnt_receptor_signaling_pathway_involved_in_osteoblast_differentiation |
| GO:0044351 | 2 | macropinocytosis |
| GO:0044416 | 2 | induction_by_symbiont_of_host_defense_response |
| GO:0044458 | 2 | motile_cilium_assembly |
| GO:0045004 | 2 | DNA_replication_proofreading |
| GO:0045013 | 2 | carbon_catabolite_repression_of_transcription |
| GO:0045014 | 2 | negative_regulation_of_transcription_by_glucose |
| GO:0045023 | 2 | G0_to_G1_transition |
| GO:0045025 | 2 | mitochondrial_degradosome |
| GO:0045029 | 2 | UDP-activated_nucleotide_receptor_activity |
| GO:0045030 | 2 | UTP-activated_nucleotide_receptor_activity |
| GO:0045031 | 2 | ATP-activated_nucleotide_receptor_activity |
| GO:0045064 | 2 | T-helper_2_cell_differentiation |
| GO:0045082 | 2 | positive_regulation_of_interleukin-10_biosynthetic_process |
| GO:0045090 | 2 | retroviral_genome_replication |
| GO:0045110 | 2 | intermediate_filament_bundle_assembly |
| GO:0045155 | 2 | electron_transporter,_transferring_electrons_from_CoQH2-cytochrome_c_reductase_complex_and_cytochrome_c_oxidase_complex_activity |
| GO:0045174 | 2 | glutathione_dehydrogenase_(ascorbate)_activity |
| GO:0045180 | 2 | basal_cortex |
| GO:0045229 | 2 | external_encapsulating_structure_organization |
| GO:0045252 | 2 | oxoglutarate_dehydrogenase_complex |
| GO:0045292 | 2 | nuclear_mRNA_cis_splicing,_via_spliceosome |
| GO:0045337 | 2 | farnesyl_diphosphate_biosynthetic_process |
| GO:0045399 | 2 | regulation_of_interleukin-3_biosynthetic_process |
| GO:0045401 | 2 | positive_regulation_of_interleukin-3_biosynthetic_process |
| GO:0045472 | 2 | response_to_ether |
| GO:0045505 | 2 | dynein_intermediate_chain_binding |
| GO:0045509 | 2 | interleukin-27_receptor_activity |
| GO:0045519 | 2 | interleukin-23_receptor_binding |
| GO:0045523 | 2 | interleukin-27_receptor_binding |
| GO:0045550 | 2 | geranylgeranyl_reductase_activity |
| GO:0045608 | 2 | negative_regulation_of_auditory_receptor_cell_differentiation |
| GO:0045658 | 2 | regulation_of_neutrophil_differentiation |
| GO:0045794 | 2 | negative_regulation_of_cell_volume |
| GO:0045796 | 2 | negative_regulation_of_intestinal_cholesterol_absorption |
| GO:0045818 | 2 | negative_regulation_of_glycogen_catabolic_process |
| GO:0045819 | 2 | positive_regulation_of_glycogen_catabolic_process |
| GO:0045852 | 2 | pH_elevation |
| GO:0045875 | 2 | negative_regulation_of_sister_chromatid_cohesion |
| GO:0045876 | 2 | positive_regulation_of_sister_chromatid_cohesion |
| GO:0045900 | 2 | negative_regulation_of_translational_elongation |
| GO:0045915 | 2 | positive_regulation_of_catecholamine_metabolic_process |
| GO:0045950 | 2 | negative_regulation_of_mitotic_recombination |
| GO:0045964 | 2 | positive_regulation_of_dopamine_metabolic_process |
| GO:0045992 | 2 | negative_regulation_of_embryonic_development |
| GO:0046013 | 2 | regulation_of_T_cell_homeostatic_proliferation |
| GO:0046038 | 2 | GMP_catabolic_process |
| GO:0046041 | 2 | ITP_metabolic_process |
| GO:0046070 | 2 | dGTP_metabolic_process |
| GO:0046072 | 2 | dTDP_metabolic_process |
| GO:0046073 | 2 | dTMP_metabolic_process |
| GO:0046099 | 2 | guanine_biosynthetic_process |
| GO:0046104 | 2 | thymidine_metabolic_process |
| GO:0046108 | 2 | uridine_metabolic_process |
| GO:0046110 | 2 | xanthine_metabolic_process |
| GO:0046114 | 2 | guanosine_biosynthetic_process |
| GO:0046116 | 2 | queuosine_metabolic_process |
| GO:0046118 | 2 | 7-methylguanosine_biosynthetic_process |
| GO:0046122 | 2 | purine_deoxyribonucleoside_metabolic_process |
| GO:0046136 | 2 | positive_regulation_of_vitamin_metabolic_process |
| GO:0046160 | 2 | heme_a_metabolic_process |
| GO:0046168 | 2 | glycerol-3-phosphate_catabolic_process |
| GO:0046184 | 2 | aldehyde_biosynthetic_process |
| GO:0046292 | 2 | formaldehyde_metabolic_process |
| GO:0046314 | 2 | phosphocreatine_biosynthetic_process |
| GO:0046340 | 2 | diacylglycerol_catabolic_process |
| GO:0046368 | 2 | GDP-L-fucose_metabolic_process |
| GO:0046373 | 2 | L-arabinose_metabolic_process |
| GO:0046386 | 2 | deoxyribose_phosphate_catabolic_process |
| GO:0046403 | 2 | polynucleotide_3'-phosphatase_activity |
| GO:0046439 | 2 | L-cysteine_metabolic_process |
| GO:0046465 | 2 | dolichyl_diphosphate_metabolic_process |
| GO:0046495 | 2 | nicotinamide_riboside_metabolic_process |
| GO:0046502 | 2 | uroporphyrinogen_III_metabolic_process |
| GO:0046511 | 2 | sphinganine_biosynthetic_process |
| GO:0046552 | 2 | photoreceptor_cell_fate_commitment |
| GO:0046556 | 2 | alpha-N-arabinofuranosidase_activity |
| GO:0046586 | 2 | regulation_of_calcium-dependent_cell-cell_adhesion |
| GO:0046592 | 2 | polyamine_oxidase_activity |
| GO:0046596 | 2 | regulation_of_viral_entry_into_host_cell |
| GO:0046600 | 2 | negative_regulation_of_centriole_replication |
| GO:0046606 | 2 | negative_regulation_of_centrosome_cycle |
| GO:0046607 | 2 | positive_regulation_of_centrosome_cycle |
| GO:0046619 | 2 | optic_placode_formation_involved_in_camera-type_eye_formation |
| GO:0046629 | 2 | gamma-delta_T_cell_activation |
| GO:0046661 | 2 | male_sex_differentiation |
| GO:0046666 | 2 | retinal_cell_programmed_cell_death |
| GO:0046670 | 2 | positive_regulation_of_retinal_cell_programmed_cell_death |
| GO:0046671 | 2 | negative_regulation_of_retinal_cell_programmed_cell_death |
| GO:0046687 | 2 | response_to_chromate |
| GO:0046691 | 2 | intracellular_canaliculus |
| GO:0046730 | 2 | induction_of_host_immune_response_by_virus |
| GO:0046732 | 2 | active_induction_of_host_immune_response_by_virus |
| GO:0046753 | 2 | non-lytic_viral_release |
| GO:0046755 | 2 | non-lytic_virus_budding |
| GO:0046778 | 2 | modification_by_virus_of_host_mRNA_processing |
| GO:0046789 | 2 | host_cell_surface_receptor_binding |
| GO:0046794 | 2 | virion_transport |
| GO:0046795 | 2 | intracellular_virion_transport |
| GO:0046812 | 2 | host_cell_surface_binding |
| GO:0046835 | 2 | carbohydrate_phosphorylation |
| GO:0046836 | 2 | glycolipid_transport |
| GO:0046857 | 2 | oxidoreductase_activity,_acting_on_other_nitrogenous_compounds_as_donors,_with_NAD_or_NADP_as_acceptor |
| GO:0046884 | 2 | follicle-stimulating_hormone_secretion |
| GO:0046899 | 2 | nucleoside_triphosphate_adenylate_kinase_activity |
| GO:0046922 | 2 | peptide-O-fucosyltransferase_activity |
| GO:0046929 | 2 | negative_regulation_of_neurotransmitter_secretion |
| GO:0046940 | 2 | nucleoside_monophosphate_phosphorylation |
| GO:0046960 | 2 | sensitization |
| GO:0046967 | 2 | cytosol_to_ER_transport |
| GO:0046985 | 2 | positive_regulation_of_hemoglobin_biosynthetic_process |
| GO:0046997 | 2 | oxidoreductase_activity,_acting_on_the_CH-NH_group_of_donors,_flavin_as_acceptor |
| GO:0047006 | 2 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one_dehydrogenase_activity |
| GO:0047026 | 2 | androsterone_dehydrogenase_(A-specific)_activity |
| GO:0047042 | 2 | androsterone_dehydrogenase_(B-specific)_activity |
| GO:0047134 | 2 | protein-disulfide_reductase_activity |
| GO:0047150 | 2 | betaine-homocysteine_S-methyltransferase_activity |
| GO:0047192 | 2 | 1-alkylglycerophosphocholine_O-acetyltransferase_activity |
| GO:0047196 | 2 | long-chain-alcohol_O-fatty-acyltransferase_activity |
| GO:0047223 | 2 | beta-1,3-galactosyl-O-glycosyl-glycoprotein_beta-1,3-N-acetylglucosaminyltransferase_activity |
| GO:0047237 | 2 | glucuronylgalactosylproteoglycan_4-beta-N-acetylgalactosaminyltransferase_activity |
| GO:0047493 | 2 | ceramide_cholinephosphotransferase_activity |
| GO:0047522 | 2 | 15-oxoprostaglandin_13-oxidase_activity |
| GO:0047635 | 2 | alanine-oxo-acid_transaminase_activity |
| GO:0047661 | 2 | amino-acid_racemase_activity |
| GO:0047696 | 2 | beta-adrenergic_receptor_kinase_activity |
| GO:0047749 | 2 | cholestanetriol_26-monooxygenase_activity |
| GO:0047750 | 2 | cholestenol_delta-isomerase_activity |
| GO:0047787 | 2 | delta4-3-oxosteroid_5beta-reductase_activity |
| GO:0047804 | 2 | cysteine-S-conjugate_beta-lyase_activity |
| GO:0048007 | 2 | antigen_processing_and_presentation,_exogenous_lipid_antigen_via_MHC_class_Ib |
| GO:0048050 | 2 | post-embryonic_eye_morphogenesis |
| GO:0048069 | 2 | eye_pigmentation |
| GO:0048073 | 2 | regulation_of_eye_pigmentation |
| GO:0048101 | 2 | calcium-_and_calmodulin-regulated_3',5'-cyclic-GMP_phosphodiesterase_activity |
| GO:0048104 | 2 | establishment_of_body_hair_or_bristle_planar_orientation |
| GO:0048105 | 2 | establishment_of_body_hair_planar_orientation |
| GO:0048133 | 2 | male_germ-line_stem_cell_division |
| GO:0048199 | 2 | vesicle_targeting,_to,_from_or_within_Golgi |
| GO:0048227 | 2 | plasma_membrane_to_endosome_transport |
| GO:0048241 | 2 | epinephrine_transport |
| GO:0048257 | 2 | 3'-flap_endonuclease_activity |
| GO:0048296 | 2 | regulation_of_isotype_switching_to_IgA_isotypes |
| GO:0048298 | 2 | positive_regulation_of_isotype_switching_to_IgA_isotypes |
| GO:0048318 | 2 | axial_mesoderm_development |
| GO:0048319 | 2 | axial_mesoderm_morphogenesis |
| GO:0048320 | 2 | axial_mesoderm_formation |
| GO:0048340 | 2 | paraxial_mesoderm_morphogenesis |
| GO:0048368 | 2 | lateral_mesoderm_development |
| GO:0048378 | 2 | regulation_of_lateral_mesodermal_cell_fate_specification |
| GO:0048382 | 2 | mesendoderm_development |
| GO:0048408 | 2 | epidermal_growth_factor_binding |
| GO:0048483 | 2 | autonomic_nervous_system_development |
| GO:0048499 | 2 | synaptic_vesicle_membrane_organization |
| GO:0048549 | 2 | positive_regulation_of_pinocytosis |
| GO:0048550 | 2 | negative_regulation_of_pinocytosis |
| GO:0048560 | 2 | establishment_of_anatomical_structure_orientation |
| GO:0048627 | 2 | myoblast_development |
| GO:0048633 | 2 | positive_regulation_of_skeletal_muscle_tissue_growth |
| GO:0048683 | 2 | regulation_of_collateral_sprouting_of_intact_axon_in_response_to_injury |
| GO:0048686 | 2 | regulation_of_sprouting_of_injured_axon |
| GO:0048690 | 2 | regulation_of_axon_extension_involved_in_regeneration |
| GO:0048696 | 2 | regulation_of_collateral_sprouting_in_absence_of_injury |
| GO:0048697 | 2 | positive_regulation_of_collateral_sprouting_in_absence_of_injury |
| GO:0048743 | 2 | positive_regulation_of_skeletal_muscle_fiber_development |
| GO:0048822 | 2 | enucleate_erythrocyte_development |
| GO:0048840 | 2 | otolith_development |
| GO:0048842 | 2 | positive_regulation_of_axon_extension_involved_in_axon_guidance |
| GO:0048850 | 2 | hypophysis_morphogenesis |
| GO:0048866 | 2 | stem_cell_fate_specification |
| GO:0048936 | 2 | peripheral_nervous_system_neuron_axonogenesis |
| GO:0050062 | 2 | long-chain-fatty-acyl-CoA_reductase_activity |
| GO:0050144 | 2 | nucleoside_deoxyribosyltransferase_activity |
| GO:0050211 | 2 | procollagen_galactosyltransferase_activity |
| GO:0050262 | 2 | ribosylnicotinamide_kinase_activity |
| GO:0050347 | 2 | trans-octaprenyltranstransferase_activity |
| GO:0050428 | 2 | 3'-phosphoadenosine_5'-phosphosulfate_biosynthetic_process |
| GO:0050473 | 2 | arachidonate_15-lipoxygenase_activity |
| GO:0050509 | 2 | N-acetylglucosaminyl-proteoglycan_4-beta-glucuronosyltransferase_activity |
| GO:0050610 | 2 | methylarsonate_reductase_activity |
| GO:0050651 | 2 | dermatan_sulfate_proteoglycan_biosynthetic_process |
| GO:0050653 | 2 | chondroitin_sulfate_proteoglycan_biosynthetic_process,_polysaccharide_chain_biosynthetic_process |
| GO:0050666 | 2 | regulation_of_homocysteine_metabolic_process |
| GO:0050692 | 2 | DBD_domain_binding |
| GO:0050705 | 2 | regulation_of_interleukin-1_alpha_secretion |
| GO:0050717 | 2 | positive_regulation_of_interleukin-1_alpha_secretion |
| GO:0050755 | 2 | chemokine_metabolic_process |
| GO:0050774 | 2 | negative_regulation_of_dendrite_morphogenesis |
| GO:0050816 | 2 | phosphothreonine_binding |
| GO:0050882 | 2 | voluntary_musculoskeletal_movement |
| GO:0050883 | 2 | musculoskeletal_movement,_spinal_reflex_action |
| GO:0050902 | 2 | leukocyte_adhesive_activation |
| GO:0050913 | 2 | sensory_perception_of_bitter_taste |
| GO:0050925 | 2 | negative_regulation_of_negative_chemotaxis |
| GO:0050975 | 2 | sensory_perception_of_touch |
| GO:0050992 | 2 | dimethylallyl_diphosphate_biosynthetic_process |
| GO:0050993 | 2 | dimethylallyl_diphosphate_metabolic_process |
| GO:0051011 | 2 | microtubule_minus-end_binding |
| GO:0051036 | 2 | regulation_of_endosome_size |
| GO:0051066 | 2 | dihydrobiopterin_metabolic_process |
| GO:0051081 | 2 | nuclear_envelope_disassembly |
| GO:0051083 | 2 | 'de_novo'_cotranslational_protein_folding |
| GO:0051123 | 2 | RNA_polymerase_II_transcriptional_preinitiation_complex_assembly |
| GO:0051124 | 2 | synaptic_growth_at_neuromuscular_junction |
| GO:0051125 | 2 | regulation_of_actin_nucleation |
| GO:0051204 | 2 | protein_insertion_into_mitochondrial_membrane |
| GO:0051231 | 2 | spindle_elongation |
| GO:0051292 | 2 | nuclear_pore_complex_assembly |
| GO:0051295 | 2 | establishment_of_meiotic_spindle_localization |
| GO:0051306 | 2 | mitotic_sister_chromatid_separation |
| GO:0051315 | 2 | attachment_of_spindle_microtubules_to_kinetochore_involved_in_mitotic_sister_chromatid_segregation |
| GO:0051342 | 2 | regulation_of_cyclic-nucleotide_phosphodiesterase_activity |
| GO:0051344 | 2 | negative_regulation_of_cyclic-nucleotide_phosphodiesterase_activity |
| GO:0051365 | 2 | cellular_response_to_potassium_ion_starvation |
| GO:0051373 | 2 | FATZ_binding |
| GO:0051388 | 2 | positive_regulation_of_nerve_growth_factor_receptor_signaling_pathway |
| GO:0051410 | 2 | detoxification_of_nitrogen_compound |
| GO:0051424 | 2 | corticotropin-releasing_hormone_binding |
| GO:0051454 | 2 | intracellular_pH_elevation |
| GO:0051462 | 2 | regulation_of_cortisol_secretion |
| GO:0051464 | 2 | positive_regulation_of_cortisol_secretion |
| GO:0051466 | 2 | positive_regulation_of_corticotropin-releasing_hormone_secretion |
| GO:0051490 | 2 | negative_regulation_of_filopodium_assembly |
| GO:0051538 | 2 | 3_iron,_4_sulfur_cluster_binding |
| GO:0051541 | 2 | elastin_metabolic_process |
| GO:0051563 | 2 | smooth_endoplasmic_reticulum_calcium_ion_homeostasis |
| GO:0051572 | 2 | negative_regulation_of_histone_H3-K4_methylation |
| GO:0051610 | 2 | serotonin_uptake |
| GO:0051659 | 2 | maintenance_of_mitochondrion_location |
| GO:0051661 | 2 | maintenance_of_centrosome_location |
| GO:0051684 | 2 | maintenance_of_Golgi_location |
| GO:0051685 | 2 | maintenance_of_ER_location |
| GO:0051725 | 2 | protein_de-ADP-ribosylation |
| GO:0051747 | 2 | cytosine_C-5_DNA_demethylase_activity |
| GO:0051764 | 2 | actin_crosslink_formation |
| GO:0051765 | 2 | inositol_tetrakisphosphate_kinase_activity |
| GO:0051790 | 2 | short-chain_fatty_acid_biosynthetic_process |
| GO:0051794 | 2 | regulation_of_catagen |
| GO:0051795 | 2 | positive_regulation_of_catagen |
| GO:0051799 | 2 | negative_regulation_of_hair_follicle_development |
| GO:0051902 | 2 | negative_regulation_of_mitochondrial_depolarization |
| GO:0051984 | 2 | positive_regulation_of_chromosome_segregation |
| GO:0052314 | 2 | phytoalexin_metabolic_process |
| GO:0052551 | 2 | response_to_defense-related_nitric_oxide_production_by_other_organism_involved_in_symbiotic_interaction |
| GO:0052565 | 2 | response_to_defense-related_host_nitric_oxide_production |
| GO:0052593 | 2 | tryptamine:oxygen_oxidoreductase_(deaminating)_activity |
| GO:0052594 | 2 | aminoacetone:oxygen_oxidoreductase(deaminating)_activity |
| GO:0052595 | 2 | aliphatic-amine_oxidase_activity |
| GO:0052596 | 2 | phenethylamine:oxygen_oxidoreductase_(deaminating)_activity |
| GO:0052632 | 2 | citrate_hydro-lyase_(cis-aconitate-forming)_activity |
| GO:0052633 | 2 | isocitrate_hydro-lyase_(cis-aconitate-forming)_activity |
| GO:0052654 | 2 | L-leucine_transaminase_activity |
| GO:0052655 | 2 | L-valine_transaminase_activity |
| GO:0052656 | 2 | L-isoleucine_transaminase_activity |
| GO:0052741 | 2 | (R)-limonene_6-monooxygenase_activity |
| GO:0052803 | 2 | imidazole-containing_compound_metabolic_process |
| GO:0052852 | 2 | very-long-chain-(S)-2-hydroxy-acid_oxidase_activity |
| GO:0052853 | 2 | long-chain-(S)-2-hydroxy-long-chain-acid_oxidase_activity |
| GO:0052854 | 2 | medium-chain-(S)-2-hydroxy-acid_oxidase_activity |
| GO:0052870 | 2 | tocopherol_omega-hydroxylase_activity |
| GO:0052871 | 2 | alpha-tocopherol_omega-hydroxylase_activity |
| GO:0052901 | 2 | spermine:oxygen_oxidoreductase_(spermidine-forming)_activity |
| GO:0055026 | 2 | negative_regulation_of_cardiac_muscle_tissue_development |
| GO:0055073 | 2 | cadmium_ion_homeostasis |
| GO:0055090 | 2 | acylglycerol_homeostasis |
| GO:0055095 | 2 | lipoprotein_particle_mediated_signaling |
| GO:0055096 | 2 | low-density_lipoprotein_particle_mediated_signaling |
| GO:0055123 | 2 | digestive_system_development |
| GO:0060002 | 2 | plus-end_directed_microfilament_motor_activity |
| GO:0060003 | 2 | copper_ion_export |
| GO:0060012 | 2 | synaptic_transmission,_glycinergic |
| GO:0060020 | 2 | Bergmann_glial_cell_differentiation |
| GO:0060025 | 2 | regulation_of_synaptic_activity |
| GO:0060028 | 2 | convergent_extension_involved_in_axis_elongation |
| GO:0060032 | 2 | notochord_regression |
| GO:0060051 | 2 | negative_regulation_of_protein_glycosylation |
| GO:0060058 | 2 | positive_regulation_of_apoptotic_process_involved_in_mammary_gland_involution |
| GO:0060060 | 2 | post-embryonic_retina_morphogenesis_in_camera-type_eye |
| GO:0060067 | 2 | cervix_development |
| GO:0060072 | 2 | large_conductance_calcium-activated_potassium_channel_activity |
| GO:0060077 | 2 | inhibitory_synapse |
| GO:0060083 | 2 | smooth_muscle_contraction_involved_in_micturition |
| GO:0060120 | 2 | inner_ear_receptor_cell_fate_commitment |
| GO:0060128 | 2 | corticotropin_hormone_secreting_cell_differentiation |
| GO:0060142 | 2 | regulation_of_syncytium_formation_by_plasma_membrane_fusion |
| GO:0060143 | 2 | positive_regulation_of_syncytium_formation_by_plasma_membrane_fusion |
| GO:0060149 | 2 | negative_regulation_of_posttranscriptional_gene_silencing |
| GO:0060151 | 2 | peroxisome_localization |
| GO:0060152 | 2 | microtubule-based_peroxisome_localization |
| GO:0060160 | 2 | negative_regulation_of_dopamine_receptor_signaling_pathway |
| GO:0060167 | 2 | regulation_of_adenosine_receptor_signaling_pathway |
| GO:0060171 | 2 | stereocilium_membrane |
| GO:0060211 | 2 | regulation_of_nuclear-transcribed_mRNA_poly(A)_tail_shortening |
| GO:0060213 | 2 | positive_regulation_of_nuclear-transcribed_mRNA_poly(A)_tail_shortening |
| GO:0060221 | 2 | retinal_rod_cell_differentiation |
| GO:0060266 | 2 | negative_regulation_of_respiratory_burst_involved_in_inflammatory_response |
| GO:0060268 | 2 | negative_regulation_of_respiratory_burst |
| GO:0060279 | 2 | positive_regulation_of_ovulation |
| GO:0060282 | 2 | positive_regulation_of_oocyte_development |
| GO:0060300 | 2 | regulation_of_cytokine_activity |
| GO:0060319 | 2 | primitive_erythrocyte_differentiation |
| GO:0060353 | 2 | regulation_of_cell_adhesion_molecule_production |
| GO:0060369 | 2 | positive_regulation_of_Fc_receptor_mediated_stimulatory_signaling_pathway |
| GO:0060370 | 2 | susceptibility_to_T_cell_mediated_cytotoxicity |
| GO:0060374 | 2 | mast_cell_differentiation |
| GO:0060379 | 2 | cardiac_muscle_cell_myoblast_differentiation |
| GO:0060385 | 2 | axonogenesis_involved_in_innervation |
| GO:0060399 | 2 | positive_regulation_of_growth_hormone_receptor_signaling_pathway |
| GO:0060414 | 2 | aorta_smooth_muscle_tissue_morphogenesis |
| GO:0060431 | 2 | primary_lung_bud_formation |
| GO:0060433 | 2 | bronchus_development |
| GO:0060435 | 2 | bronchiole_development |
| GO:0060437 | 2 | lung_growth |
| GO:0060440 | 2 | trachea_formation |
| GO:0060447 | 2 | bud_outgrowth_involved_in_lung_branching |
| GO:0060466 | 2 | activation_of_meiosis_involved_in_egg_activation |
| GO:0060467 | 2 | negative_regulation_of_fertilization |
| GO:0060468 | 2 | prevention_of_polyspermy |
| GO:0060480 | 2 | lung_goblet_cell_differentiation |
| GO:0060488 | 2 | orthogonal_dichotomous_subdivision_of_terminal_units_involved_in_lung_branching_morphogenesis |
| GO:0060489 | 2 | planar_dichotomous_subdivision_of_terminal_units_involved_in_lung_branching_morphogenesis |
| GO:0060490 | 2 | lateral_sprouting_involved_in_lung_morphogenesis |
| GO:0060512 | 2 | prostate_gland_morphogenesis |
| GO:0060529 | 2 | squamous_basal_epithelial_stem_cell_differentiation_involved_in_prostate_gland_acinus_development |
| GO:0060557 | 2 | positive_regulation_of_vitamin_D_biosynthetic_process |
| GO:0060587 | 2 | regulation_of_lipoprotein_lipid_oxidation |
| GO:0060594 | 2 | mammary_gland_specification |
| GO:0060595 | 2 | fibroblast_growth_factor_receptor_signaling_pathway_involved_in_mammary_gland_specification |
| GO:0060599 | 2 | lateral_sprouting_involved_in_mammary_gland_duct_morphogenesis |
| GO:0060615 | 2 | mammary_gland_bud_formation |
| GO:0060621 | 2 | negative_regulation_of_cholesterol_import |
| GO:0060633 | 2 | negative_regulation_of_transcription_initiation_from_RNA_polymerase_II_promoter |
| GO:0060667 | 2 | branch_elongation_involved_in_salivary_gland_morphogenesis |
| GO:0060669 | 2 | embryonic_placenta_morphogenesis |
| GO:0060689 | 2 | cell_differentiation_involved_in_salivary_gland_development |
| GO:0060694 | 2 | regulation_of_cholesterol_transporter_activity |
| GO:0060697 | 2 | positive_regulation_of_phospholipid_catabolic_process |
| GO:0060713 | 2 | labyrinthine_layer_morphogenesis |
| GO:0060729 | 2 | intestinal_epithelial_structure_maintenance |
| GO:0060730 | 2 | regulation_of_intestinal_epithelial_structure_maintenance |
| GO:0060738 | 2 | epithelial-mesenchymal_signaling_involved_in_prostate_gland_development |
| GO:0060764 | 2 | cell-cell_signaling_involved_in_mammary_gland_development |
| GO:0060769 | 2 | positive_regulation_of_epithelial_cell_proliferation_involved_in_prostate_gland_development |
| GO:0060849 | 2 | regulation_of_transcription_involved_in_lymphatic_endothelial_cell_fate_commitment |
| GO:0060872 | 2 | semicircular_canal_development |
| GO:0060900 | 2 | embryonic_camera-type_eye_formation |
| GO:0060914 | 2 | heart_formation |
| GO:0060915 | 2 | mesenchymal_cell_differentiation_involved_in_lung_development |
| GO:0060926 | 2 | pacemaker_cell_development |
| GO:0060932 | 2 | His-Purkinje_system_cell_differentiation |
| GO:0060956 | 2 | endocardial_cell_differentiation |
| GO:0060965 | 2 | negative_regulation_of_gene_silencing_by_miRNA |
| GO:0060967 | 2 | negative_regulation_of_gene_silencing_by_RNA |
| GO:0060976 | 2 | coronary_vasculature_development |
| GO:0060982 | 2 | coronary_artery_morphogenesis |
| GO:0060995 | 2 | cell-cell_signaling_involved_in_kidney_development |
| GO:0061004 | 2 | pattern_specification_involved_in_kidney_development |
| GO:0061017 | 2 | hepatoblast_differentiation |
| GO:0061031 | 2 | endodermal_digestive_tract_morphogenesis |
| GO:0061032 | 2 | visceral_serous_pericardium_development |
| GO:0061033 | 2 | secretion_by_lung_epithelial_cell_involved_in_lung_growth |
| GO:0061034 | 2 | olfactory_bulb_mitral_cell_layer_development |
| GO:0061047 | 2 | positive_regulation_of_branching_involved_in_lung_morphogenesis |
| GO:0061050 | 2 | regulation_of_cell_growth_involved_in_cardiac_muscle_cell_development |
| GO:0061056 | 2 | sclerotome_development |
| GO:0061113 | 2 | pancreas_morphogenesis |
| GO:0061146 | 2 | Peyer's_patch_morphogenesis |
| GO:0061156 | 2 | pulmonary_artery_morphogenesis |
| GO:0061179 | 2 | negative_regulation_of_insulin_secretion_involved_in_cellular_response_to_glucose_stimulus |
| GO:0061187 | 2 | regulation_of_chromatin_silencing_at_rDNA |
| GO:0061188 | 2 | negative_regulation_of_chromatin_silencing_at_rDNA |
| GO:0061289 | 2 | Wnt_receptor_signaling_pathway_involved_in_kidney_development |
| GO:0061290 | 2 | canonical_Wnt_receptor_signaling_pathway_involved_in_metanephric_kidney_development |
| GO:0061302 | 2 | smooth_muscle_cell-matrix_adhesion |
| GO:0061309 | 2 | cardiac_neural_crest_cell_development_involved_in_outflow_tract_morphogenesis |
| GO:0061323 | 2 | cell_proliferation_involved_in_heart_morphogenesis |
| GO:0061324 | 2 | canonical_Wnt_receptor_signaling_pathway_involved_in_positive_regulation_of_cardiac_outflow_tract_cell_proliferation |
| GO:0061325 | 2 | cell_proliferation_involved_in_outflow_tract_morphogenesis |
| GO:0061338 | 2 | atrioventricular_node_impulse_conduction_delay |
| GO:0061386 | 2 | closure_of_optic_fissure |
| GO:0061428 | 2 | negative_regulation_of_transcription_from_RNA_polymerase_II_promoter_in_response_to_hypoxia |
| GO:0070034 | 2 | telomeric_RNA_binding |
| GO:0070051 | 2 | fibrinogen_binding |
| GO:0070070 | 2 | proton-transporting_V-type_ATPase_complex_assembly |
| GO:0070072 | 2 | vacuolar_proton-transporting_V-type_ATPase_complex_assembly |
| GO:0070075 | 2 | tear_secretion |
| GO:0070080 | 2 | titin_Z_domain_binding |
| GO:0070093 | 2 | negative_regulation_of_glucagon_secretion |
| GO:0070094 | 2 | positive_regulation_of_glucagon_secretion |
| GO:0070101 | 2 | positive_regulation_of_chemokine-mediated_signaling_pathway |
| GO:0070103 | 2 | regulation_of_interleukin-6-mediated_signaling_pathway |
| GO:0070125 | 2 | mitochondrial_translational_elongation |
| GO:0070163 | 2 | regulation_of_adiponectin_secretion |
| GO:0070171 | 2 | negative_regulation_of_tooth_mineralization |
| GO:0070173 | 2 | regulation_of_enamel_mineralization |
| GO:0070178 | 2 | D-serine_metabolic_process |
| GO:0070182 | 2 | DNA_polymerase_binding |
| GO:0070199 | 2 | establishment_of_protein_localization_to_chromosome |
| GO:0070213 | 2 | protein_auto-ADP-ribosylation |
| GO:0070256 | 2 | negative_regulation_of_mucus_secretion |
| GO:0070257 | 2 | positive_regulation_of_mucus_secretion |
| GO:0070259 | 2 | tyrosyl-DNA_phosphodiesterase_activity |
| GO:0070266 | 2 | necroptosis |
| GO:0070288 | 2 | ferritin_complex |
| GO:0070294 | 2 | renal_sodium_ion_absorption |
| GO:0070300 | 2 | phosphatidic_acid_binding |
| GO:0070314 | 2 | G1_to_G0_transition |
| GO:0070350 | 2 | regulation_of_white_fat_cell_proliferation |
| GO:0070358 | 2 | actin_polymerization-dependent_cell_motility |
| GO:0070363 | 2 | mitochondrial_light_strand_promoter_sense_binding |
| GO:0070368 | 2 | positive_regulation_of_hepatocyte_differentiation |
| GO:0070369 | 2 | beta-catenin-TCF7L2_complex |
| GO:0070404 | 2 | NADH_binding |
| GO:0070408 | 2 | carbamoyl_phosphate_metabolic_process |
| GO:0070409 | 2 | carbamoyl_phosphate_biosynthetic_process |
| GO:0070417 | 2 | cellular_response_to_cold |
| GO:0070427 | 2 | nucleotide-binding_oligomerization_domain_containing_1_signaling_pathway |
| GO:0070428 | 2 | regulation_of_nucleotide-binding_oligomerization_domain_containing_1_signaling_pathway |
| GO:0070432 | 2 | regulation_of_nucleotide-binding_oligomerization_domain_containing_2_signaling_pathway |
| GO:0070435 | 2 | Shc-EGFR_complex |
| GO:0070436 | 2 | Grb2-EGFR_complex |
| GO:0070438 | 2 | mTOR-FKBP12-rapamycin_complex |
| GO:0070445 | 2 | regulation_of_oligodendrocyte_progenitor_proliferation |
| GO:0070447 | 2 | positive_regulation_of_oligodendrocyte_progenitor_proliferation |
| GO:0070483 | 2 | detection_of_hypoxia |
| GO:0070492 | 2 | oligosaccharide_binding |
| GO:0070537 | 2 | histone_H2A_K63-linked_deubiquitination |
| GO:0070557 | 2 | PCNA-p21_complex |
| GO:0070561 | 2 | vitamin_D_receptor_signaling_pathway |
| GO:0070573 | 2 | metallodipeptidase_activity |
| GO:0070593 | 2 | dendrite_self-avoidance |
| GO:0070637 | 2 | pyridine_nucleoside_metabolic_process |
| GO:0070638 | 2 | pyridine_nucleoside_catabolic_process |
| GO:0070673 | 2 | response_to_interleukin-18 |
| GO:0070683 | 2 | mitotic_cell_cycle_G2/M_transition_decatenation_checkpoint |
| GO:0070694 | 2 | deoxyribonucleoside_5'-monophosphate_N-glycosidase_activity |
| GO:0070704 | 2 | sterol_desaturase_activity |
| GO:0070734 | 2 | histone_H3-K27_methylation |
| GO:0070736 | 2 | protein-glycine_ligase_activity,_initiating |
| GO:0070739 | 2 | protein-glutamic_acid_ligase_activity |
| GO:0070743 | 2 | interleukin-23_complex |
| GO:0070765 | 2 | gamma-secretase_complex |
| GO:0070781 | 2 | response_to_biotin |
| GO:0070813 | 2 | hydrogen_sulfide_metabolic_process |
| GO:0070814 | 2 | hydrogen_sulfide_biosynthetic_process |
| GO:0070816 | 2 | phosphorylation_of_RNA_polymerase_II_C-terminal_domain |
| GO:0070831 | 2 | basement_membrane_assembly |
| GO:0070836 | 2 | caveola_assembly |
| GO:0070843 | 2 | misfolded_protein_transport |
| GO:0070844 | 2 | polyubiquitinated_protein_transport |
| GO:0070845 | 2 | polyubiquitinated_misfolded_protein_transport |
| GO:0070858 | 2 | negative_regulation_of_bile_acid_biosynthetic_process |
| GO:0070863 | 2 | positive_regulation_of_protein_exit_from_endoplasmic_reticulum |
| GO:0070885 | 2 | negative_regulation_of_calcineurin-NFAT_signaling_cascade |
| GO:0070890 | 2 | sodium-dependent_L-ascorbate_transmembrane_transporter_activity |
| GO:0070904 | 2 | transepithelial_L-ascorbic_acid_transport |
| GO:0070905 | 2 | serine_binding |
| GO:0070922 | 2 | small_RNA_loading_onto_RISC |
| GO:0070970 | 2 | interleukin-2_secretion |
| GO:0070971 | 2 | endoplasmic_reticulum_exit_site |
| GO:0070977 | 2 | bone_maturation |
| GO:0070991 | 2 | medium-chain-acyl-CoA_dehydrogenase_activity |
| GO:0071029 | 2 | nuclear_ncRNA_surveillance |
| GO:0071035 | 2 | nuclear_polyadenylation-dependent_rRNA_catabolic_process |
| GO:0071046 | 2 | nuclear_polyadenylation-dependent_ncRNA_catabolic_process |
| GO:0071065 | 2 | alpha9-beta1_integrin-vascular_cell_adhesion_molecule-1_complex |
| GO:0071104 | 2 | response_to_interleukin-9 |
| GO:0071109 | 2 | superior_temporal_gyrus_development |
| GO:0071166 | 2 | ribonucleoprotein_complex_localization |
| GO:0071169 | 2 | establishment_of_protein_localization_to_chromatin |
| GO:0071205 | 2 | protein_localization_to_juxtaparanode_region_of_axon |
| GO:0071207 | 2 | histone_pre-mRNA_stem-loop_binding |
| GO:0071233 | 2 | cellular_response_to_leucine |
| GO:0071242 | 2 | cellular_response_to_ammonium_ion |
| GO:0071253 | 2 | connexin_binding |
| GO:0071256 | 2 | translocon_complex |
| GO:0071281 | 2 | cellular_response_to_iron_ion |
| GO:0071307 | 2 | cellular_response_to_vitamin_K |
| GO:0071338 | 2 | positive_regulation_of_hair_follicle_cell_proliferation |
| GO:0071344 | 2 | diphosphate_metabolic_process |
| GO:0071376 | 2 | cellular_response_to_corticotropin-releasing_hormone_stimulus |
| GO:0071394 | 2 | cellular_response_to_testosterone_stimulus |
| GO:0071404 | 2 | cellular_response_to_low-density_lipoprotein_particle_stimulus |
| GO:0071455 | 2 | cellular_response_to_hyperoxia |
| GO:0071483 | 2 | cellular_response_to_blue_light |
| GO:0071501 | 2 | cellular_response_to_sterol_depletion |
| GO:0071502 | 2 | cellular_response_to_temperature_stimulus |
| GO:0071505 | 2 | response_to_mycophenolic_acid |
| GO:0071506 | 2 | cellular_response_to_mycophenolic_acid |
| GO:0071535 | 2 | RING-like_zinc_finger_domain_binding |
| GO:0071539 | 2 | protein_localization_to_centrosome |
| GO:0071557 | 2 | histone_H3-K27_demethylation |
| GO:0071558 | 2 | histone_demethylase_activity_(H3-K27_specific) |
| GO:0071568 | 2 | UFM1_conjugating_enzyme_activity |
| GO:0071578 | 2 | zinc_ion_import |
| GO:0071585 | 2 | detoxification_of_cadmium_ion |
| GO:0071593 | 2 | lymphocyte_aggregation |
| GO:0071595 | 2 | Nem1-Spo7_phosphatase_complex |
| GO:0071621 | 2 | granulocyte_chemotaxis |
| GO:0071624 | 2 | positive_regulation_of_granulocyte_chemotaxis |
| GO:0071633 | 2 | dihydroceramidase_activity |
| GO:0071638 | 2 | negative_regulation_of_monocyte_chemotactic_protein-1_production |
| GO:0071672 | 2 | negative_regulation_of_smooth_muscle_cell_chemotaxis |
| GO:0071673 | 2 | positive_regulation_of_smooth_muscle_cell_chemotaxis |
| GO:0071676 | 2 | negative_regulation_of_mononuclear_cell_migration |
| GO:0071709 | 2 | membrane_assembly |
| GO:0071724 | 2 | response_to_diacyl_bacterial_lipopeptide |
| GO:0071725 | 2 | response_to_triacyl_bacterial_lipopeptide |
| GO:0071726 | 2 | cellular_response_to_diacyl_bacterial_lipopeptide |
| GO:0071727 | 2 | cellular_response_to_triacyl_bacterial_lipopeptide |
| GO:0071733 | 2 | transcriptional_activation_by_promoter-enhancer_looping |
| GO:0071763 | 2 | nuclear_membrane_organization |
| GO:0071791 | 2 | chemokine_(C-C_motif)_ligand_5_binding |
| GO:0071810 | 2 | regulation_of_fever_generation_by_regulation_of_prostaglandin_secretion |
| GO:0071812 | 2 | positive_regulation_of_fever_generation_by_positive_regulation_of_prostaglandin_secretion |
| GO:0071847 | 2 | TNFSF11-mediated_signaling_pathway |
| GO:0071848 | 2 | positive_regulation_of_ERK1_and_ERK2_cascade_via_TNFSF11-mediated_signaling |
| GO:0071886 | 2 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine_binding |
| GO:0071888 | 2 | macrophage_apoptotic_process |
| GO:0071896 | 2 | protein_localization_to_adherens_junction |
| GO:0071898 | 2 | regulation_of_estrogen_receptor_binding |
| GO:0071899 | 2 | negative_regulation_of_estrogen_receptor_binding |
| GO:0071918 | 2 | urea_transmembrane_transport |
| GO:0071920 | 2 | cleavage_body |
| GO:0071930 | 2 | negative_regulation_of_transcription_involved_in_G1/S_phase_of_mitotic_cell_cycle |
| GO:0072004 | 2 | kidney_field_specification |
| GO:0072006 | 2 | nephron_development |
| GO:0072023 | 2 | thick_ascending_limb_development |
| GO:0072033 | 2 | renal_vesicle_formation |
| GO:0072053 | 2 | renal_inner_medulla_development |
| GO:0072054 | 2 | renal_outer_medulla_development |
| GO:0072086 | 2 | specification_of_loop_of_Henle_identity |
| GO:0072092 | 2 | ureteric_bud_invasion |
| GO:0072098 | 2 | anterior/posterior_pattern_specification_involved_in_kidney_development |
| GO:0072126 | 2 | positive_regulation_of_glomerular_mesangial_cell_proliferation |
| GO:0072134 | 2 | nephrogenic_mesenchyme_morphogenesis |
| GO:0072139 | 2 | glomerular_parietal_epithelial_cell_differentiation |
| GO:0072141 | 2 | renal_interstitial_cell_development |
| GO:0072156 | 2 | distal_tubule_morphogenesis |
| GO:0072166 | 2 | posterior_mesonephric_tubule_development |
| GO:0072181 | 2 | mesonephric_duct_formation |
| GO:0072189 | 2 | ureter_development |
| GO:0072190 | 2 | ureter_urothelium_development |
| GO:0072194 | 2 | kidney_smooth_muscle_tissue_development |
| GO:0072197 | 2 | ureter_morphogenesis |
| GO:0072208 | 2 | metanephric_smooth_muscle_tissue_development |
| GO:0072210 | 2 | metanephric_nephron_development |
| GO:0072224 | 2 | metanephric_glomerulus_development |
| GO:0072233 | 2 | metanephric_thick_ascending_limb_development |
| GO:0072236 | 2 | metanephric_loop_of_Henle_development |
| GO:0072249 | 2 | metanephric_glomerular_visceral_epithelial_cell_development |
| GO:0072262 | 2 | metanephric_glomerular_mesangial_cell_proliferation_involved_in_metanephros_development |
| GO:0072278 | 2 | metanephric_comma-shaped_body_morphogenesis |
| GO:0072301 | 2 | regulation_of_metanephric_glomerular_mesangial_cell_proliferation |
| GO:0072304 | 2 | regulation_of_mesenchymal_stem_cell_apoptotic_process_involved_in_metanephric_nephron_morphogenesis |
| GO:0072305 | 2 | negative_regulation_of_mesenchymal_stem_cell_apoptotic_process_involved_in_metanephric_nephron_morphogenesis |
| GO:0072308 | 2 | negative_regulation_of_metanephric_nephron_tubule_epithelial_cell_differentiation |
| GO:0072313 | 2 | metanephric_glomerular_epithelial_cell_development |
| GO:0072334 | 2 | UDP-galactose_transmembrane_transport |
| GO:0072345 | 2 | NAADP-sensitive_calcium-release_channel_activity |
| GO:0072358 | 2 | cardiovascular_system_development |
| GO:0072361 | 2 | regulation_of_glycolysis_by_regulation_of_transcription_from_RNA_polymerase_II_promoter |
| GO:0072364 | 2 | regulation_of_cellular_ketone_metabolic_process_by_regulation_of_transcription_from_RNA_polymerase_II_promoter |
| GO:0072368 | 2 | regulation_of_lipid_transport_by_negative_regulation_of_transcription_from_RNA_polymerase_II_promoter |
| GO:0072369 | 2 | regulation_of_lipid_transport_by_positive_regulation_of_transcription_from_RNA_polymerase_II_promoter |
| GO:0072387 | 2 | flavin_adenine_dinucleotide_metabolic_process |
| GO:0072513 | 2 | positive_regulation_of_secondary_heart_field_cardioblast_proliferation |
| GO:0072536 | 2 | interleukin-23_receptor_complex |
| GO:0072545 | 2 | tyrosine_binding |
| GO:0072560 | 2 | type_B_pancreatic_cell_maturation |
| GO:0072562 | 2 | blood_microparticle |
| GO:0072563 | 2 | endothelial_microparticle |
| GO:0072602 | 2 | interleukin-4_secretion |
| GO:0072610 | 2 | interleukin-12_secretion |
| GO:0072672 | 2 | neutrophil_extravasation |
| GO:0080121 | 2 | AMP_transport |
| GO:0080122 | 2 | AMP_transmembrane_transporter_activity |
| GO:0080154 | 2 | regulation_of_fertilization |
| GO:0086014 | 2 | regulation_of_atrial_cardiac_muscle_cell_action_potential |
| GO:0086019 | 2 | cell-cell_signaling_involved_in_cardiac_conduction |
| GO:0086029 | 2 | Purkinje_myocyte_to_ventricular_cardiac_muscle_cell_signaling |
| GO:0086067 | 2 | AV_node_cell_to_bundle_of_His_cell_communication |
| GO:0086068 | 2 | Purkinje_myocyte_to_ventricular_cardiac_muscle_cell_communication |
| GO:0086100 | 2 | endothelin_receptor_signaling_pathway |
| GO:0090007 | 2 | regulation_of_mitotic_anaphase |
| GO:0090034 | 2 | regulation_of_chaperone-mediated_protein_complex_assembly |
| GO:0090035 | 2 | positive_regulation_of_chaperone-mediated_protein_complex_assembly |
| GO:0090074 | 2 | negative_regulation_of_protein_homodimerization_activity |
| GO:0090080 | 2 | positive_regulation_of_MAPKKK_cascade_by_fibroblast_growth_factor_receptor_signaling_pathway |
| GO:0090082 | 2 | positive_regulation_of_heart_induction_by_negative_regulation_of_canonical_Wnt_receptor_signaling_pathway |
| GO:0090086 | 2 | negative_regulation_of_protein_deubiquitination |
| GO:0090118 | 2 | receptor-mediated_endocytosis_of_low-density_lipoprotein_particle_involved_in_cholesterol_transport |
| GO:0090154 | 2 | positive_regulation_of_sphingolipid_biosynthetic_process |
| GO:0090166 | 2 | Golgi_disassembly |
| GO:0090169 | 2 | regulation_of_spindle_assembly |
| GO:0090177 | 2 | establishment_of_planar_polarity_involved_in_neural_tube_closure |
| GO:0090202 | 2 | gene_looping |
| GO:0090234 | 2 | regulation_of_kinetochore_assembly |
| GO:0090238 | 2 | positive_regulation_of_arachidonic_acid_secretion |
| GO:0090241 | 2 | negative_regulation_of_histone_H4_acetylation |
| GO:0090245 | 2 | axis_elongation_involved_in_somitogenesis |
| GO:0090259 | 2 | regulation_of_retinal_ganglion_cell_axon_guidance |
| GO:0090260 | 2 | negative_regulation_of_retinal_ganglion_cell_axon_guidance |
| GO:0090264 | 2 | regulation_of_immune_complex_clearance_by_monocytes_and_macrophages |
| GO:0090265 | 2 | positive_regulation_of_immune_complex_clearance_by_monocytes_and_macrophages |
| GO:0090270 | 2 | regulation_of_fibroblast_growth_factor_production |
| GO:0090274 | 2 | positive_regulation_of_somatostatin_secretion |
| GO:0090281 | 2 | negative_regulation_of_calcium_ion_import |
| GO:0090291 | 2 | negative_regulation_of_osteoclast_proliferation |
| GO:0090313 | 2 | regulation_of_protein_targeting_to_membrane |
| GO:0090335 | 2 | regulation_of_brown_fat_cell_differentiation |
| GO:0090336 | 2 | positive_regulation_of_brown_fat_cell_differentiation |
| GO:0090361 | 2 | regulation_of_platelet-derived_growth_factor_production |
| GO:0090402 | 2 | oncogene-induced_senescence |
| GO:0090427 | 2 | activation_of_meiosis |
| GO:0097016 | 2 | L27_domain_binding |
| GO:0097025 | 2 | MPP7-DLG1-LIN7_complex |
| GO:0097033 | 2 | mitochondrial_respiratory_chain_complex_III_biogenesis |
| GO:0097034 | 2 | mitochondrial_respiratory_chain_complex_IV_biogenesis |
| GO:0097052 | 2 | L-kynurenine_metabolic_process |
| GO:0097053 | 2 | L-kynurenine_catabolic_process |
| GO:0097055 | 2 | agmatine_biosynthetic_process |
| GO:0097059 | 2 | CNTFR-CLCF1_complex |
| GO:0097062 | 2 | dendritic_spine_maintenance |
| GO:0097106 | 2 | postsynaptic_density_organization |
| GO:0097107 | 2 | postsynaptic_density_assembly |
| GO:0097114 | 2 | N-methyl-D-aspartate_receptor_clustering |
| GO:0097117 | 2 | guanylate_kinase-associated_protein_clustering |
| GO:0097118 | 2 | neuroligin_clustering |
| GO:0097155 | 2 | fasciculation_of_sensory_neuron_axon |
| GO:0097156 | 2 | fasciculation_of_motor_neuron_axon |
| GO:0097195 | 2 | pilomotor_reflex |
| GO:0097196 | 2 | Shu_complex |
| GO:0097201 | 2 | negative_regulation_of_transcription_from_RNA_polymerase_II_promoter_in_response_to_stress |
| GO:0097241 | 2 | hematopoietic_stem_cell_migration_to_bone_marrow |
| GO:0097300 | 2 | programmed_necrotic_cell_death |
| GO:1900027 | 2 | regulation_of_ruffle_assembly |
| GO:1900040 | 2 | regulation_of_interleukin-2_secretion |
| GO:1900041 | 2 | negative_regulation_of_interleukin-2_secretion |
| GO:1900084 | 2 | regulation_of_peptidyl-tyrosine_autophosphorylation |
| GO:1900086 | 2 | positive_regulation_of_peptidyl-tyrosine_autophosphorylation |
| GO:1900087 | 2 | positive_regulation_of_G1/S_transition_of_mitotic_cell_cycle |
| GO:1900094 | 2 | regulation_of_transcription_from_RNA_polymerase_II_promoter_involved_in_determination_of_left/right_symmetry |
| GO:1900117 | 2 | regulation_of_execution_phase_of_apoptosis |
| GO:1900133 | 2 | regulation_of_renin_secretion_into_blood_stream |
| GO:1900151 | 2 | regulation_of_nuclear-transcribed_mRNA_catabolic_process,_deadenylation-dependent_decay |
| GO:1900164 | 2 | nodal_signaling_pathway_involved_in_determination_of_lateral_mesoderm_left/right_asymmetry |
| GO:1900211 | 2 | regulation_of_mesenchymal_cell_apoptotic_process_involved_in_metanephros_development |
| GO:1900212 | 2 | negative_regulation_of_mesenchymal_cell_apoptotic_process_involved_in_metanephros_development |
| GO:1900214 | 2 | regulation_of_apoptotic_process_involved_in_metanephric_collecting_duct_development |
| GO:1900215 | 2 | negative_regulation_of_apoptotic_process_involved_in_metanephric_collecting_duct_development |
| GO:1900217 | 2 | regulation_of_apoptotic_process_involved_in_metanephric_nephron_tubule_development |
| GO:1900218 | 2 | negative_regulation_of_apoptotic_process_involved_in_metanephric_nephron_tubule_development |
| GO:1900224 | 2 | positive_regulation_of_nodal_signaling_pathway_involved_in_determination_of_lateral_mesoderm_left/right_asymmetry |
| GO:1900245 | 2 | positive_regulation_of_MDA-5_signaling_pathway |
| GO:1900246 | 2 | positive_regulation_of_RIG-I_signaling_pathway |
| GO:1900271 | 2 | regulation_of_long-term_synaptic_potentiation |
| GO:1900620 | 2 | acetate_ester_biosynthetic_process |
| GO:1900623 | 2 | regulation_of_monocyte_aggregation |
| GO:1900625 | 2 | positive_regulation_of_monocyte_aggregation |
| GO:1900736 | 2 | regulation_of_phospholipase_C-activating_G-protein_coupled_receptor_signaling_pathway |
| GO:1900744 | 2 | regulation_of_p38MAPK_cascade |
| GO:1900745 | 2 | positive_regulation_of_p38MAPK_cascade |
| GO:1900746 | 2 | regulation_of_vascular_endothelial_growth_factor_signaling_pathway |
| GO:1900747 | 2 | negative_regulation_of_vascular_endothelial_growth_factor_signaling_pathway |
| GO:1901016 | 2 | regulation_of_potassium_ion_transmembrane_transporter_activity |
| GO:2000001 | 2 | regulation_of_DNA_damage_checkpoint |
| GO:2000035 | 2 | regulation_of_stem_cell_division |
| GO:2000077 | 2 | negative_regulation_of_type_B_pancreatic_cell_development |
| GO:2000079 | 2 | regulation_of_canonical_Wnt_receptor_signaling_pathway_involved_in_controlling_type_B_pancreatic_cell_proliferation |
| GO:2000098 | 2 | negative_regulation_of_smooth_muscle_cell-matrix_adhesion |
| GO:2000124 | 2 | regulation_of_endocannabinoid_signaling_pathway |
| GO:2000172 | 2 | regulation_of_branching_morphogenesis_of_a_nerve |
| GO:2000210 | 2 | positive_regulation_of_anoikis |
| GO:2000224 | 2 | regulation_of_testosterone_biosynthetic_process |
| GO:2000275 | 2 | regulation_of_oxidative_phosphorylation_uncoupler_activity |
| GO:2000297 | 2 | negative_regulation_of_synapse_maturation |
| GO:2000304 | 2 | positive_regulation_of_ceramide_biosynthetic_process |
| GO:2000307 | 2 | regulation_of_tumor_necrosis_factor_(ligand)_superfamily_member_11_production |
| GO:2000309 | 2 | positive_regulation_of_tumor_necrosis_factor_(ligand)_superfamily_member_11_production |
| GO:2000342 | 2 | negative_regulation_of_chemokine_(C-X-C_motif)_ligand_2_production |
| GO:2000348 | 2 | regulation_of_CD40_signaling_pathway |
| GO:2000370 | 2 | positive_regulation_of_clathrin-mediated_endocytosis |
| GO:2000371 | 2 | regulation_of_DNA_topoisomerase_(ATP-hydrolyzing)_activity |
| GO:2000373 | 2 | positive_regulation_of_DNA_topoisomerase_(ATP-hydrolyzing)_activity |
| GO:2000381 | 2 | negative_regulation_of_mesoderm_development |
| GO:2000383 | 2 | regulation_of_ectoderm_development |
| GO:2000410 | 2 | regulation_of_thymocyte_migration |
| GO:2000412 | 2 | positive_regulation_of_thymocyte_migration |
| GO:2000416 | 2 | regulation_of_eosinophil_migration |
| GO:2000427 | 2 | positive_regulation_of_apoptotic_cell_clearance |
| GO:2000437 | 2 | regulation_of_monocyte_extravasation |
| GO:2000471 | 2 | regulation_of_hematopoietic_stem_cell_migration |
| GO:2000473 | 2 | positive_regulation_of_hematopoietic_stem_cell_migration |
| GO:2000474 | 2 | regulation_of_opioid_receptor_signaling_pathway |
| GO:2000479 | 2 | regulation_of_cAMP-dependent_protein_kinase_activity |
| GO:2000481 | 2 | positive_regulation_of_cAMP-dependent_protein_kinase_activity |
| GO:2000523 | 2 | regulation_of_T_cell_costimulation |
| GO:2000525 | 2 | positive_regulation_of_T_cell_costimulation |
| GO:2000532 | 2 | regulation_of_renal_albumin_absorption |
| GO:2000537 | 2 | regulation_of_B_cell_chemotaxis |
| GO:2000538 | 2 | positive_regulation_of_B_cell_chemotaxis |
| GO:2000539 | 2 | regulation_of_protein_geranylgeranylation |
| GO:2000541 | 2 | positive_regulation_of_protein_geranylgeranylation |
| GO:2000552 | 2 | negative_regulation_of_T-helper_2_cell_cytokine_production |
| GO:2000564 | 2 | regulation_of_CD8-positive,_alpha-beta_T_cell_proliferation |
| GO:2000592 | 2 | regulation_of_metanephric_DCT_cell_differentiation |
| GO:2000594 | 2 | positive_regulation_of_metanephric_DCT_cell_differentiation |
| GO:2000615 | 2 | regulation_of_histone_H3-K9_acetylation |
| GO:2000645 | 2 | negative_regulation_of_receptor_catabolic_process |
| GO:2000650 | 2 | negative_regulation_of_sodium_ion_transmembrane_transporter_activity |
| GO:2000660 | 2 | negative_regulation_of_interleukin-1-mediated_signaling_pathway |
| GO:2000664 | 2 | positive_regulation_of_interleukin-5_secretion |
| GO:2000667 | 2 | positive_regulation_of_interleukin-13_secretion |
| GO:2000683 | 2 | regulation_of_cellular_response_to_X-ray |
| GO:2000705 | 2 | regulation_of_dense_core_granule_biogenesis |
| GO:2000722 | 2 | regulation_of_cardiac_vascular_smooth_muscle_cell_differentiation |
| GO:2000742 | 2 | regulation_of_anterior_head_development |
| GO:2000744 | 2 | positive_regulation_of_anterior_head_development |
| GO:2000781 | 2 | positive_regulation_of_double-strand_break_repair |
| GO:2000811 | 2 | negative_regulation_of_anoikis |
| GO:2000820 | 2 | negative_regulation_of_transcription_from_RNA_polymerase_II_promoter_involved_in_smooth_muscle_cell_differentiation |
| GO:2000821 | 2 | regulation_of_grooming_behavior |
| GO:2000823 | 2 | regulation_of_androgen_receptor_activity |
| GO:2000824 | 2 | negative_regulation_of_androgen_receptor_activity |
| GO:2000846 | 2 | regulation_of_corticosteroid_hormone_secretion |
| GO:2000848 | 2 | positive_regulation_of_corticosteroid_hormone_secretion |
| GO:2000849 | 2 | regulation_of_glucocorticoid_secretion |
| GO:2000851 | 2 | positive_regulation_of_glucocorticoid_secretion |
| GO:2000910 | 2 | negative_regulation_of_sterol_import |
| GO:2000977 | 2 | regulation_of_forebrain_neuron_differentiation |
| GO:2000987 | 2 | positive_regulation_of_behavioral_fear_response |
| GO:2001027 | 2 | negative_regulation_of_endothelial_cell_chemotaxis |
| GO:2001032 | 2 | regulation_of_double-strand_break_repair_via_nonhomologous_end_joining |
| GO:2001035 | 2 | regulation_of_tongue_muscle_cell_differentiation |
| GO:2001037 | 2 | positive_regulation_of_tongue_muscle_cell_differentiation |
| GO:2001044 | 2 | regulation_of_integrin-mediated_signaling_pathway |
| GO:2001047 | 2 | regulation_of_G1/S_transition_checkpoint |
| GO:2001074 | 2 | regulation_of_metanephric_ureteric_bud_development |
| GO:2001076 | 2 | positive_regulation_of_metanephric_ureteric_bud_development |
| GO:2001106 | 2 | regulation_of_Rho_guanyl-nucleotide_exchange_factor_activity |
| GO:2001160 | 2 | regulation_of_histone_H3-K79_methylation |
| GO:2001162 | 2 | positive_regulation_of_histone_H3-K79_methylation |
| GO:2001187 | 2 | positive_regulation_of_CD8-positive,_alpha-beta_T_cell_activation |
| GO:2001199 | 2 | negative_regulation_of_dendritic_cell_differentiation |
| GO:2001224 | 2 | positive_regulation_of_neuron_migration |
| GO:2001241 | 2 | positive_regulation_of_extrinsic_apoptotic_signaling_pathway_in_absence_of_ligand |
| GO:2001256 | 2 | regulation_of_store-operated_calcium_entry |
| GO:2001293 | 2 | malonyl-CoA_metabolic_process |
| GO:2001295 | 2 | malonyl-CoA_biosynthetic_process |
| GO:0000009 | 1 | alpha-1,6-mannosyltransferase_activity |
| GO:0000016 | 1 | lactase_activity |
| GO:0000023 | 1 | maltose_metabolic_process |
| GO:0000035 | 1 | acyl_binding |
| GO:0000036 | 1 | ACP_phosphopantetheine_attachment_site_binding_involved_in_fatty_acid_biosynthetic_process |
| GO:0000053 | 1 | argininosuccinate_metabolic_process |
| GO:0000054 | 1 | ribosomal_subunit_export_from_nucleus |
| GO:0000056 | 1 | ribosomal_small_subunit_export_from_nucleus |
| GO:0000073 | 1 | spindle_pole_body_separation |
| GO:0000093 | 1 | mitotic_telophase |
| GO:0000095 | 1 | S-adenosylmethionine_transmembrane_transporter_activity |
| GO:0000110 | 1 | nucleotide-excision_repair_factor_1_complex |
| GO:0000126 | 1 | transcription_factor_TFIIIB_complex |
| GO:0000133 | 1 | polarisome |
| GO:0000150 | 1 | recombinase_activity |
| GO:0000160 | 1 | two-component_signal_transduction_system_(phosphorelay) |
| GO:0000170 | 1 | sphingosine_hydroxylase_activity |
| GO:0000171 | 1 | ribonuclease_MRP_activity |
| GO:0000173 | 1 | inactivation_of_MAPK_activity_involved_in_osmosensory_signaling_pathway |
| GO:0000210 | 1 | NAD+_diphosphatase_activity |
| GO:0000215 | 1 | tRNA_2'-phosphotransferase_activity |
| GO:0000224 | 1 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine_amidase_activity |
| GO:0000225 | 1 | N-acetylglucosaminylphosphatidylinositol_deacetylase_activity |
| GO:0000229 | 1 | cytoplasmic_chromosome |
| GO:0000235 | 1 | astral_microtubule |
| GO:0000237 | 1 | leptotene |
| GO:0000238 | 1 | zygotene |
| GO:0000240 | 1 | diplotene |
| GO:0000250 | 1 | lanosterol_synthase_activity |
| GO:0000254 | 1 | C-4_methylsterol_oxidase_activity |
| GO:0000257 | 1 | nitrilase_activity |
| GO:0000262 | 1 | mitochondrial_chromosome |
| GO:0000285 | 1 | 1-phosphatidylinositol-3-phosphate_5-kinase_activity |
| GO:0000296 | 1 | spermine_transport |
| GO:0000334 | 1 | 3-hydroxyanthranilate_3,4-dioxygenase_activity |
| GO:0000379 | 1 | tRNA-type_intron_splice_site_recognition_and_cleavage |
| GO:0000386 | 1 | second_spliceosomal_transesterification_activity |
| GO:0000401 | 1 | open_form_four-way_junction_DNA_binding |
| GO:0000402 | 1 | crossed_form_four-way_junction_DNA_binding |
| GO:0000409 | 1 | regulation_of_transcription_by_galactose |
| GO:0000411 | 1 | positive_regulation_of_transcription_by_galactose |
| GO:0000415 | 1 | negative_regulation_of_histone_H3-K36_methylation |
| GO:0000416 | 1 | positive_regulation_of_histone_H3-K36_methylation |
| GO:0000431 | 1 | regulation_of_transcription_from_RNA_polymerase_II_promoter_by_galactose |
| GO:0000433 | 1 | negative_regulation_of_transcription_from_RNA_polymerase_II_promoter_by_glucose |
| GO:0000435 | 1 | positive_regulation_of_transcription_from_RNA_polymerase_II_promoter_by_galactose |
| GO:0000437 | 1 | carbon_catabolite_repression_of_transcription_from_RNA_polymerase_II_promoter |
| GO:0000439 | 1 | core_TFIIH_complex |
| GO:0000701 | 1 | purine-specific_mismatch_base_pair_DNA_N-glycosylase_activity |
| GO:0000702 | 1 | oxidized_base_lesion_DNA_N-glycosylase_activity |
| GO:0000717 | 1 | nucleotide-excision_repair,_DNA_duplex_unwinding |
| GO:0000719 | 1 | photoreactive_repair |
| GO:0000720 | 1 | pyrimidine_dimer_repair_by_nucleotide-excision_repair |
| GO:0000741 | 1 | karyogamy |
| GO:0000778 | 1 | condensed_nuclear_chromosome_kinetochore |
| GO:0000789 | 1 | cytoplasmic_chromatin |
| GO:0000797 | 1 | condensin_core_heterodimer |
| GO:0000799 | 1 | nuclear_condensin_complex |
| GO:0000820 | 1 | regulation_of_glutamine_family_amino_acid_metabolic_process |
| GO:0000822 | 1 | inositol_hexakisphosphate_binding |
| GO:0000823 | 1 | inositol-1,4,5-trisphosphate_6-kinase_activity |
| GO:0000824 | 1 | inositol_tetrakisphosphate_3-kinase_activity |
| GO:0000831 | 1 | inositol_hexakisphosphate_6-kinase_activity |
| GO:0000835 | 1 | ER_ubiquitin_ligase_complex |
| GO:0000836 | 1 | Hrd1p_ubiquitin_ligase_complex |
| GO:0000912 | 1 | assembly_of_actomyosin_apparatus_involved_in_cell_cycle_cytokinesis |
| GO:0000915 | 1 | cytokinesis,_actomyosin_contractile_ring_assembly |
| GO:0000920 | 1 | cytokinetic_cell_separation |
| GO:0000921 | 1 | septin_ring_assembly |
| GO:0000964 | 1 | mitochondrial_RNA_5'-end_processing |
| GO:0000972 | 1 | transcription-dependent_tethering_of_RNA_polymerase_II_gene_DNA_at_nuclear_periphery |
| GO:0001010 | 1 | sequence-specific_DNA_binding_transcription_factor_recruiting_transcription_factor_activity |
| GO:0001011 | 1 | sequence-specific_DNA_binding_RNA_polymerase_recruiting_transcription_factor_activity |
| GO:0001013 | 1 | RNA_polymerase_I_regulatory_region_DNA_binding |
| GO:0001042 | 1 | RNA_polymerase_I_core_binding |
| GO:0001079 | 1 | nitrogen_catabolite_regulation_of_transcription_from_RNA_polymerase_II_promoter |
| GO:0001080 | 1 | nitrogen_catabolite_activation_of_transcription_from_RNA_polymerase_II_promoter |
| GO:0001082 | 1 | RNA_polymerase_I_transcription_factor_binding_transcription_factor_activity |
| GO:0001083 | 1 | RNA_polymerase_II_basal_transcription_factor_binding_transcription_factor_activity |
| GO:0001087 | 1 | TFIIB-class_binding_transcription_factor_activity |
| GO:0001091 | 1 | RNA_polymerase_II_basal_transcription_factor_binding |
| GO:0001093 | 1 | TFIIB-class_transcription_factor_binding |
| GO:0001163 | 1 | RNA_polymerase_I_regulatory_region_sequence-specific_DNA_binding |
| GO:0001164 | 1 | RNA_polymerase_I_CORE_element_sequence-specific_DNA_binding |
| GO:0001167 | 1 | sequence-specific_DNA_binding_RNA_polymerase_I_transcription_factor_activity |
| GO:0001169 | 1 | RNA_polymerase_I_CORE_element_sequence-specific_DNA_binding_transcription_factor_activity |
| GO:0001186 | 1 | RNA_polymerase_I_transcription_factor_recruiting_transcription_factor_activity |
| GO:0001187 | 1 | RNA_polymerase_I_CORE_element_sequence-specific_DNA_binding_transcription_factor_recruiting_transcription_factor_activity |
| GO:0001188 | 1 | RNA_polymerase_I_transcriptional_preinitiation_complex_assembly |
| GO:0001189 | 1 | RNA_polymerase_I_transcriptional_preinitiation_complex_assembly_at_the_promoter_for_the_nuclear_large_rRNA_transcript |
| GO:0001205 | 1 | RNA_polymerase_II_distal_enhancer_sequence-specific_DNA_binding_transcription_factor_activity_involved_in_positive_regulation_of_transcription |
| GO:0001315 | 1 | age-dependent_response_to_reactive_oxygen_species |
| GO:0001401 | 1 | mitochondrial_sorting_and_assembly_machinery_complex |
| GO:0001512 | 1 | dihydronicotinamide_riboside_quinone_reductase_activity |
| GO:0001519 | 1 | peptide_amidation |
| GO:0001532 | 1 | interleukin-21_receptor_activity |
| GO:0001534 | 1 | radial_spoke |
| GO:0001544 | 1 | initiation_of_primordial_ovarian_follicle_growth |
| GO:0001552 | 1 | ovarian_follicle_atresia |
| GO:0001567 | 1 | cholesterol_25-hydroxylase_activity |
| GO:0001575 | 1 | globoside_metabolic_process |
| GO:0001579 | 1 | medium-chain_fatty_acid_transport |
| GO:0001594 | 1 | trace-amine_receptor_activity |
| GO:0001596 | 1 | angiotensin_type_I_receptor_activity |
| GO:0001604 | 1 | urotensin_II_receptor_activity |
| GO:0001616 | 1 | growth_hormone_secretagogue_receptor_activity |
| GO:0001621 | 1 | ADP_receptor_activity |
| GO:0001626 | 1 | nociceptin_receptor_activity |
| GO:0001631 | 1 | cysteinyl_leukotriene_receptor_activity |
| GO:0001634 | 1 | pituitary_adenylate_cyclase-activating_polypeptide_receptor_activity |
| GO:0001680 | 1 | tRNA_3'-terminal_CCA_addition |
| GO:0001681 | 1 | sialate_O-acetylesterase_activity |
| GO:0001682 | 1 | tRNA_5'-leader_removal |
| GO:0001716 | 1 | L-amino-acid_oxidase_activity |
| GO:0001731 | 1 | formation_of_translation_preinitiation_complex |
| GO:0001735 | 1 | prenylcysteine_oxidase_activity |
| GO:0001760 | 1 | aminocarboxymuconate-semialdehyde_decarboxylase_activity |
| GO:0001787 | 1 | natural_killer_cell_proliferation |
| GO:0001788 | 1 | antibody-dependent_cellular_cytotoxicity |
| GO:0001791 | 1 | IgM_binding |
| GO:0001794 | 1 | type_IIa_hypersensitivity |
| GO:0001809 | 1 | positive_regulation_of_type_IV_hypersensitivity |
| GO:0001828 | 1 | inner_cell_mass_cellular_morphogenesis |
| GO:0001830 | 1 | trophectodermal_cell_fate_commitment |
| GO:0001844 | 1 | protein_insertion_into_mitochondrial_membrane_involved_in_apoptotic_signaling_pathway |
| GO:0001850 | 1 | complement_component_C3a_binding |
| GO:0001851 | 1 | complement_component_C3b_binding |
| GO:0001855 | 1 | complement_component_C4b_binding |
| GO:0001861 | 1 | complement_component_C4b_receptor_activity |
| GO:0001873 | 1 | polysaccharide_receptor_activity |
| GO:0001874 | 1 | (1->3)-beta-D-glucan_receptor_activity |
| GO:0001879 | 1 | detection_of_yeast |
| GO:0001884 | 1 | pyrimidine_nucleoside_binding |
| GO:0001896 | 1 | autolysis |
| GO:0001918 | 1 | farnesylated_protein_binding |
| GO:0001923 | 1 | B-1_B_cell_differentiation |
| GO:0001928 | 1 | regulation_of_exocyst_assembly |
| GO:0001969 | 1 | regulation_of_activation_of_membrane_attack_complex |
| GO:0001970 | 1 | positive_regulation_of_activation_of_membrane_attack_complex |
| GO:0001980 | 1 | regulation_of_systemic_arterial_blood_pressure_by_ischemic_conditions |
| GO:0001984 | 1 | vasodilation_of_artery_involved_in_baroreceptor_response_to_increased_systemic_arterial_blood_pressure |
| GO:0001986 | 1 | negative_regulation_of_the_force_of_heart_contraction_involved_in_baroreceptor_response_to_increased_systemic_arterial_blood_pressure |
| GO:0001987 | 1 | vasoconstriction_of_artery_involved_in_baroreceptor_response_to_lowering_of_systemic_arterial_blood_pressure |
| GO:0001991 | 1 | regulation_of_systemic_arterial_blood_pressure_by_circulatory_renin-angiotensin |
| GO:0001997 | 1 | positive_regulation_of_the_force_of_heart_contraction_by_epinephrine-norepinephrine |
| GO:0001998 | 1 | angiotensin_mediated_vasoconstriction_involved_in_regulation_of_systemic_arterial_blood_pressure |
| GO:0001999 | 1 | renal_response_to_blood_flow_involved_in_circulatory_renin-angiotensin_regulation_of_systemic_arterial_blood_pressure |
| GO:0002001 | 1 | renin_secretion_into_blood_stream |
| GO:0002030 | 1 | inhibitory_G-protein_coupled_receptor_phosphorylation |
| GO:0002033 | 1 | vasodilation_by_angiotensin_involved_in_regulation_of_systemic_arterial_blood_pressure |
| GO:0002037 | 1 | negative_regulation_of_L-glutamate_transport |
| GO:0002041 | 1 | intussusceptive_angiogenesis |
| GO:0002055 | 1 | adenine_binding |
| GO:0002058 | 1 | uracil_binding |
| GO:0002059 | 1 | thymine_binding |
| GO:0002061 | 1 | pyrimidine_base_binding |
| GO:0002071 | 1 | glandular_epithelial_cell_maturation |
| GO:0002077 | 1 | acrosome_matrix_dispersal |
| GO:0002083 | 1 | 4-hydroxybenzoate_decaprenyltransferase_activity |
| GO:0002084 | 1 | protein_depalmitoylation |
| GO:0002085 | 1 | inhibition_of_neuroepithelial_cell_differentiation |
| GO:0002094 | 1 | polyprenyltransferase_activity |
| GO:0002113 | 1 | interleukin-33_binding |
| GO:0002114 | 1 | interleukin-33_receptor_activity |
| GO:0002121 | 1 | inter-male_aggressive_behavior |
| GO:0002135 | 1 | CTP_binding |
| GO:0002148 | 1 | hypochlorous_acid_metabolic_process |
| GO:0002149 | 1 | hypochlorous_acid_biosynthetic_process |
| GO:0002152 | 1 | bile_acid_conjugation |
| GO:0002154 | 1 | thyroid_hormone_mediated_signaling_pathway |
| GO:0002158 | 1 | osteoclast_proliferation |
| GO:0002159 | 1 | desmosome_assembly |
| GO:0002194 | 1 | hepatocyte_cell_migration |
| GO:0002215 | 1 | defense_response_to_nematode |
| GO:0002232 | 1 | leukocyte_chemotaxis_involved_in_inflammatory_response |
| GO:0002254 | 1 | kinin_cascade |
| GO:0002314 | 1 | germinal_center_B_cell_differentiation |
| GO:0002316 | 1 | follicular_B_cell_differentiation |
| GO:0002317 | 1 | plasma_cell_differentiation |
| GO:0002343 | 1 | peripheral_B_cell_selection |
| GO:0002344 | 1 | B_cell_affinity_maturation |
| GO:0002352 | 1 | B_cell_negative_selection |
| GO:0002353 | 1 | plasma_kallikrein-kinin_cascade |
| GO:0002355 | 1 | detection_of_tumor_cell |
| GO:0002358 | 1 | B_cell_homeostatic_proliferation |
| GO:0002368 | 1 | B_cell_cytokine_production |
| GO:0002371 | 1 | dendritic_cell_cytokine_production |
| GO:0002372 | 1 | myeloid_dendritic_cell_cytokine_production |
| GO:0002380 | 1 | immunoglobulin_secretion_involved_in_immune_response |
| GO:0002412 | 1 | antigen_transcytosis_by_M_cells_in_mucosal-associated_lymphoid_tissue |
| GO:0002426 | 1 | immunoglobulin_production_in_mucosal_tissue |
| GO:0002432 | 1 | granuloma_formation |
| GO:0002445 | 1 | type_II_hypersensitivity |
| GO:0002448 | 1 | mast_cell_mediated_immunity |
| GO:0002457 | 1 | T_cell_antigen_processing_and_presentation |
| GO:0002461 | 1 | tolerance_induction_dependent_upon_immune_response |
| GO:0002462 | 1 | tolerance_induction_to_nonself_antigen |
| GO:0002476 | 1 | antigen_processing_and_presentation_of_endogenous_peptide_antigen_via_MHC_class_Ib |
| GO:0002484 | 1 | antigen_processing_and_presentation_of_endogenous_peptide_antigen_via_MHC_class_I_via_ER_pathway |
| GO:0002485 | 1 | antigen_processing_and_presentation_of_endogenous_peptide_antigen_via_MHC_class_I_via_ER_pathway,_TAP-dependent |
| GO:0002488 | 1 | antigen_processing_and_presentation_of_endogenous_peptide_antigen_via_MHC_class_Ib_via_ER_pathway |
| GO:0002489 | 1 | antigen_processing_and_presentation_of_endogenous_peptide_antigen_via_MHC_class_Ib_via_ER_pathway,_TAP-dependent |
| GO:0002501 | 1 | peptide_antigen_assembly_with_MHC_protein_complex |
| GO:0002502 | 1 | peptide_antigen_assembly_with_MHC_class_I_protein_complex |
| GO:0002508 | 1 | central_tolerance_induction |
| GO:0002518 | 1 | lymphocyte_chemotaxis_across_high_endothelial_venule |
| GO:0002520 | 1 | immune_system_development |
| GO:0002524 | 1 | hypersensitivity |
| GO:0002525 | 1 | acute_inflammatory_response_to_non-antigenic_stimulus |
| GO:0002534 | 1 | cytokine_production_involved_in_inflammatory_response |
| GO:0002536 | 1 | respiratory_burst_involved_in_inflammatory_response |
| GO:0002541 | 1 | activation_of_plasma_proteins_involved_in_acute_inflammatory_response |
| GO:0002572 | 1 | pro-T_cell_differentiation |
| GO:0002575 | 1 | basophil_chemotaxis |
| GO:0002580 | 1 | regulation_of_antigen_processing_and_presentation_of_peptide_or_polysaccharide_antigen_via_MHC_class_II |
| GO:0002581 | 1 | negative_regulation_of_antigen_processing_and_presentation_of_peptide_or_polysaccharide_antigen_via_MHC_class_II |
| GO:0002583 | 1 | regulation_of_antigen_processing_and_presentation_of_peptide_antigen |
| GO:0002585 | 1 | positive_regulation_of_antigen_processing_and_presentation_of_peptide_antigen |
| GO:0002589 | 1 | regulation_of_antigen_processing_and_presentation_of_peptide_antigen_via_MHC_class_I |
| GO:0002591 | 1 | positive_regulation_of_antigen_processing_and_presentation_of_peptide_antigen_via_MHC_class_I |
| GO:0002605 | 1 | negative_regulation_of_dendritic_cell_antigen_processing_and_presentation |
| GO:0002640 | 1 | regulation_of_immunoglobulin_biosynthetic_process |
| GO:0002642 | 1 | positive_regulation_of_immunoglobulin_biosynthetic_process |
| GO:0002646 | 1 | regulation_of_central_tolerance_induction |
| GO:0002648 | 1 | positive_regulation_of_central_tolerance_induction |
| GO:0002649 | 1 | regulation_of_tolerance_induction_to_self_antigen |
| GO:0002651 | 1 | positive_regulation_of_tolerance_induction_to_self_antigen |
| GO:0002652 | 1 | regulation_of_tolerance_induction_dependent_upon_immune_response |
| GO:0002654 | 1 | positive_regulation_of_tolerance_induction_dependent_upon_immune_response |
| GO:0002658 | 1 | regulation_of_peripheral_tolerance_induction |
| GO:0002660 | 1 | positive_regulation_of_peripheral_tolerance_induction |
| GO:0002721 | 1 | regulation_of_B_cell_cytokine_production |
| GO:0002730 | 1 | regulation_of_dendritic_cell_cytokine_production |
| GO:0002731 | 1 | negative_regulation_of_dendritic_cell_cytokine_production |
| GO:0002769 | 1 | natural_killer_cell_inhibitory_signaling_pathway |
| GO:0002776 | 1 | antimicrobial_peptide_secretion |
| GO:0002779 | 1 | antibacterial_peptide_secretion |
| GO:0002840 | 1 | regulation_of_T_cell_mediated_immune_response_to_tumor_cell |
| GO:0002842 | 1 | positive_regulation_of_T_cell_mediated_immune_response_to_tumor_cell |
| GO:0002849 | 1 | regulation_of_peripheral_T_cell_tolerance_induction |
| GO:0002851 | 1 | positive_regulation_of_peripheral_T_cell_tolerance_induction |
| GO:0002875 | 1 | negative_regulation_of_chronic_inflammatory_response_to_antigenic_stimulus |
| GO:0002880 | 1 | regulation_of_chronic_inflammatory_response_to_non-antigenic_stimulus |
| GO:0002882 | 1 | positive_regulation_of_chronic_inflammatory_response_to_non-antigenic_stimulus |
| GO:0002895 | 1 | regulation_of_central_B_cell_tolerance_induction |
| GO:0002897 | 1 | positive_regulation_of_central_B_cell_tolerance_induction |
| GO:0002904 | 1 | positive_regulation_of_B_cell_apoptotic_process |
| GO:0003017 | 1 | lymph_circulation |
| GO:0003026 | 1 | regulation_of_systemic_arterial_blood_pressure_by_aortic_arch_baroreceptor_feedback |
| GO:0003050 | 1 | regulation_of_systemic_arterial_blood_pressure_by_atrial_natriuretic_peptide |
| GO:0003058 | 1 | hormonal_regulation_of_the_force_of_heart_contraction |
| GO:0003059 | 1 | positive_regulation_of_the_force_of_heart_contraction_by_epinephrine |
| GO:0003061 | 1 | positive_regulation_of_the_force_of_heart_contraction_by_norepinephrine |
| GO:0003065 | 1 | positive_regulation_of_heart_rate_by_epinephrine |
| GO:0003069 | 1 | vasodilation_by_acetylcholine_involved_in_regulation_of_systemic_arterial_blood_pressure |
| GO:0003083 | 1 | negative_regulation_of_renal_output_by_angiotensin |
| GO:0003106 | 1 | negative_regulation_of_glomerular_filtration_by_angiotensin |
| GO:0003108 | 1 | negative_regulation_of_the_force_of_heart_contraction_by_chemical_signal |
| GO:0003131 | 1 | mesodermal-endodermal_cell_signaling |
| GO:0003142 | 1 | cardiogenic_plate_morphogenesis |
| GO:0003144 | 1 | embryonic_heart_tube_formation |
| GO:0003146 | 1 | heart_jogging |
| GO:0003161 | 1 | cardiac_conduction_system_development |
| GO:0003163 | 1 | sinoatrial_node_development |
| GO:0003167 | 1 | atrioventricular_bundle_cell_differentiation |
| GO:0003168 | 1 | cardiac_Purkinje_fiber_cell_differentiation |
| GO:0003171 | 1 | atrioventricular_valve_development |
| GO:0003175 | 1 | tricuspid_valve_development |
| GO:0003184 | 1 | pulmonary_valve_morphogenesis |
| GO:0003192 | 1 | mitral_valve_formation |
| GO:0003193 | 1 | pulmonary_valve_formation |
| GO:0003205 | 1 | cardiac_chamber_development |
| GO:0003209 | 1 | cardiac_atrium_morphogenesis |
| GO:0003210 | 1 | cardiac_atrium_formation |
| GO:0003223 | 1 | ventricular_compact_myocardium_morphogenesis |
| GO:0003231 | 1 | cardiac_ventricle_development |
| GO:0003236 | 1 | sinus_venosus_morphogenesis |
| GO:0003241 | 1 | growth_involved_in_heart_morphogenesis |
| GO:0003259 | 1 | cardioblast_anterior-lateral_migration |
| GO:0003260 | 1 | cardioblast_migration |
| GO:0003270 | 1 | Notch_signaling_pathway_involved_in_regulation_of_secondary_heart_field_cardioblast_proliferation |
| GO:0003273 | 1 | cell_migration_involved_in_endocardial_cushion_formation |
| GO:0003285 | 1 | septum_secundum_development |
| GO:0003311 | 1 | pancreatic_D_cell_differentiation |
| GO:0003318 | 1 | cell_migration_to_the_midline_involved_in_heart_development |
| GO:0003322 | 1 | pancreatic_A_cell_development |
| GO:0003327 | 1 | type_B_pancreatic_cell_fate_commitment |
| GO:0003330 | 1 | regulation_of_extracellular_matrix_constituent_secretion |
| GO:0003331 | 1 | positive_regulation_of_extracellular_matrix_constituent_secretion |
| GO:0003342 | 1 | proepicardium_development |
| GO:0003356 | 1 | regulation_of_cilium_beat_frequency |
| GO:0003359 | 1 | noradrenergic_neuron_fate_commitment |
| GO:0003363 | 1 | lamellipodium_assembly_involved_in_ameboidal_cell_migration |
| GO:0003373 | 1 | dynamin_polymerization_involved_in_membrane_fission |
| GO:0003374 | 1 | dynamin_polymerization_involved_in_mitochondrial_fission |
| GO:0003383 | 1 | apical_constriction |
| GO:0003402 | 1 | planar_cell_polarity_pathway_involved_in_axis_elongation |
| GO:0003404 | 1 | optic_vesicle_morphogenesis |
| GO:0003409 | 1 | optic_cup_structural_organization |
| GO:0003414 | 1 | chondrocyte_morphogenesis_involved_in_endochondral_bone_morphogenesis |
| GO:0003415 | 1 | chondrocyte_hypertrophy |
| GO:0003418 | 1 | growth_plate_cartilage_chondrocyte_differentiation |
| GO:0003429 | 1 | growth_plate_cartilage_chondrocyte_morphogenesis |
| GO:0003692 | 1 | left-handed_Z-DNA_binding |
| GO:0003721 | 1 | telomeric_template_RNA_reverse_transcriptase_activity |
| GO:0003830 | 1 | beta-1,4-mannosylglycoprotein_4-beta-N-acetylglucosaminyltransferase_activity |
| GO:0003834 | 1 | beta-carotene_15,15'-monooxygenase_activity |
| GO:0003837 | 1 | beta-ureidopropionase_activity |
| GO:0003842 | 1 | 1-pyrroline-5-carboxylate_dehydrogenase_activity |
| GO:0003844 | 1 | 1,4-alpha-glucan_branching_enzyme_activity |
| GO:0003851 | 1 | 2-hydroxyacylsphingosine_1-beta-galactosyltransferase_activity |
| GO:0003860 | 1 | 3-hydroxyisobutyryl-CoA_hydrolase_activity |
| GO:0003867 | 1 | 4-aminobutyrate_transaminase_activity |
| GO:0003869 | 1 | 4-nitrophenylphosphatase_activity |
| GO:0003874 | 1 | 6-pyruvoyltetrahydropterin_synthase_activity |
| GO:0003881 | 1 | CDP-diacylglycerol-inositol_3-phosphatidyltransferase_activity |
| GO:0003882 | 1 | CDP-diacylglycerol-serine_O-phosphatidyltransferase_activity |
| GO:0003908 | 1 | methylated-DNA-[protein]-cysteine_S-methyltransferase_activity |
| GO:0003919 | 1 | FMN_adenylyltransferase_activity |
| GO:0003921 | 1 | GMP_synthase_activity |
| GO:0003922 | 1 | GMP_synthase_(glutamine-hydrolyzing)_activity |
| GO:0003933 | 1 | GTP_cyclohydrolase_activity |
| GO:0003934 | 1 | GTP_cyclohydrolase_I_activity |
| GO:0003937 | 1 | IMP_cyclohydrolase_activity |
| GO:0003939 | 1 | L-iditol_2-dehydrogenase_activity |
| GO:0003940 | 1 | L-iduronidase_activity |
| GO:0003944 | 1 | N-acetylglucosamine-1-phosphodiester_alpha-N-acetylglucosaminidase_activity |
| GO:0003947 | 1 | (N-acetylneuraminyl)-galactosylglucosylceramide_N-acetylgalactosaminyltransferase_activity |
| GO:0003952 | 1 | NAD+_synthase_(glutamine-hydrolyzing)_activity |
| GO:0003957 | 1 | NAD(P)+_transhydrogenase_(B-specific)_activity |
| GO:0003958 | 1 | NADPH-hemoprotein_reductase_activity |
| GO:0003968 | 1 | RNA-directed_RNA_polymerase_activity |
| GO:0003972 | 1 | RNA_ligase_(ATP)_activity |
| GO:0003975 | 1 | UDP-N-acetylglucosamine-dolichyl-phosphate_N-acetylglucosaminephosphotransferase_activity |
| GO:0003976 | 1 | UDP-N-acetylglucosamine-lysosomal-enzyme_N-acetylglucosaminephosphotransferase_activity |
| GO:0003977 | 1 | UDP-N-acetylglucosamine_diphosphorylase_activity |
| GO:0003978 | 1 | UDP-glucose_4-epimerase_activity |
| GO:0003979 | 1 | UDP-glucose_6-dehydrogenase_activity |
| GO:0003983 | 1 | UTP:glucose-1-phosphate_uridylyltransferase_activity |
| GO:0003996 | 1 | acyl-CoA_ligase_activity |
| GO:0003999 | 1 | adenine_phosphoribosyltransferase_activity |
| GO:0004001 | 1 | adenosine_kinase_activity |
| GO:0004014 | 1 | adenosylmethionine_decarboxylase_activity |
| GO:0004018 | 1 | N6-(1,2-dicarboxyethyl)AMP_AMP-lyase_(fumarate-forming)_activity |
| GO:0004034 | 1 | aldose_1-epimerase_activity |
| GO:0004037 | 1 | allantoicase_activity |
| GO:0004042 | 1 | acetyl-CoA:L-glutamate_N-acetyltransferase_activity |
| GO:0004043 | 1 | L-aminoadipate-semialdehyde_dehydrogenase_activity |
| GO:0004044 | 1 | amidophosphoribosyltransferase_activity |
| GO:0004055 | 1 | argininosuccinate_synthase_activity |
| GO:0004057 | 1 | arginyltransferase_activity |
| GO:0004058 | 1 | aromatic-L-amino-acid_decarboxylase_activity |
| GO:0004059 | 1 | aralkylamine_N-acetyltransferase_activity |
| GO:0004061 | 1 | arylformamidase_activity |
| GO:0004070 | 1 | aspartate_carbamoyltransferase_activity |
| GO:0004077 | 1 | biotin-[acetyl-CoA-carboxylase]_ligase_activity |
| GO:0004078 | 1 | biotin-[methylcrotonoyl-CoA-carboxylase]_ligase_activity |
| GO:0004079 | 1 | biotin-[methylmalonyl-CoA-carboxytransferase]_ligase_activity |
| GO:0004080 | 1 | biotin-[propionyl-CoA-carboxylase_(ATP-hydrolyzing)]_ligase_activity |
| GO:0004081 | 1 | bis(5'-nucleosyl)-tetraphosphatase_(asymmetrical)_activity |
| GO:0004085 | 1 | butyryl-CoA_dehydrogenase_activity |
| GO:0004087 | 1 | carbamoyl-phosphate_synthase_(ammonia)_activity |
| GO:0004088 | 1 | carbamoyl-phosphate_synthase_(glutamine-hydrolyzing)_activity |
| GO:0004092 | 1 | carnitine_O-acetyltransferase_activity |
| GO:0004096 | 1 | catalase_activity |
| GO:0004098 | 1 | cerebroside-sulfatase_activity |
| GO:0004102 | 1 | choline_O-acetyltransferase_activity |
| GO:0004108 | 1 | citrate_(Si)-synthase_activity |
| GO:0004113 | 1 | 2',3'-cyclic-nucleotide_3'-phosphodiesterase_activity |
| GO:0004121 | 1 | cystathionine_beta-lyase_activity |
| GO:0004122 | 1 | cystathionine_beta-synthase_activity |
| GO:0004123 | 1 | cystathionine_gamma-lyase_activity |
| GO:0004124 | 1 | cysteine_synthase_activity |
| GO:0004131 | 1 | cytosine_deaminase_activity |
| GO:0004132 | 1 | dCMP_deaminase_activity |
| GO:0004133 | 1 | glycogen_debranching_enzyme_activity |
| GO:0004134 | 1 | 4-alpha-glucanotransferase_activity |
| GO:0004135 | 1 | amylo-alpha-1,6-glucosidase_activity |
| GO:0004137 | 1 | deoxycytidine_kinase_activity |
| GO:0004138 | 1 | deoxyguanosine_kinase_activity |
| GO:0004139 | 1 | deoxyribose-phosphate_aldolase_activity |
| GO:0004148 | 1 | dihydrolipoyl_dehydrogenase_activity |
| GO:0004149 | 1 | dihydrolipoyllysine-residue_succinyltransferase_activity |
| GO:0004151 | 1 | dihydroorotase_activity |
| GO:0004155 | 1 | 6,7-dihydropteridine_reductase_activity |
| GO:0004163 | 1 | diphosphomevalonate_decarboxylase_activity |
| GO:0004164 | 1 | diphthine_synthase_activity |
| GO:0004168 | 1 | dolichol_kinase_activity |
| GO:0004170 | 1 | dUTP_diphosphatase_activity |
| GO:0004304 | 1 | estrone_sulfotransferase_activity |
| GO:0004306 | 1 | ethanolamine-phosphate_cytidylyltransferase_activity |
| GO:0004310 | 1 | farnesyl-diphosphate_farnesyltransferase_activity |
| GO:0004313 | 1 | [acyl-carrier-protein]_S-acetyltransferase_activity |
| GO:0004316 | 1 | 3-oxoacyl-[acyl-carrier-protein]_reductase_(NADPH)_activity |
| GO:0004317 | 1 | 3-hydroxypalmitoyl-[acyl-carrier-protein]_dehydratase_activity |
| GO:0004319 | 1 | enoyl-[acyl-carrier-protein]_reductase_(NADPH,_B-specific)_activity |
| GO:0004324 | 1 | ferredoxin-NADP+_reductase_activity |
| GO:0004325 | 1 | ferrochelatase_activity |
| GO:0004326 | 1 | tetrahydrofolylpolyglutamate_synthase_activity |
| GO:0004333 | 1 | fumarate_hydratase_activity |
| GO:0004336 | 1 | galactosylceramidase_activity |
| GO:0004339 | 1 | glucan_1,4-alpha-glucosidase_activity |
| GO:0004340 | 1 | glucokinase_activity |
| GO:0004341 | 1 | gluconolactonase_activity |
| GO:0004343 | 1 | glucosamine_6-phosphate_N-acetyltransferase_activity |
| GO:0004347 | 1 | glucose-6-phosphate_isomerase_activity |
| GO:0004349 | 1 | glutamate_5-kinase_activity |
| GO:0004350 | 1 | glutamate-5-semialdehyde_dehydrogenase_activity |
| GO:0004361 | 1 | glutaryl-CoA_dehydrogenase_activity |
| GO:0004362 | 1 | glutathione-disulfide_reductase_activity |
| GO:0004363 | 1 | glutathione_synthase_activity |
| GO:0004371 | 1 | glycerone_kinase_activity |
| GO:0004375 | 1 | glycine_dehydrogenase_(decarboxylating)_activity |
| GO:0004377 | 1 | GDP-Man:Man3GlcNAc2-PP-Dol_alpha-1,2-mannosyltransferase_activity |
| GO:0004378 | 1 | GDP-Man:Man1GlcNAc2-PP-Dol_alpha-1,3-mannosyltransferase_activity |
| GO:0004380 | 1 | glycoprotein-fucosylgalactoside_alpha-N-acetylgalactosaminyltransferase_activity |
| GO:0004381 | 1 | fucosylgalactoside_3-alpha-galactosyltransferase_activity |
| GO:0004382 | 1 | guanosine-diphosphatase_activity |
| GO:0004394 | 1 | heparan_sulfate_2-O-sulfotransferase_activity |
| GO:0004395 | 1 | hexaprenyldihydroxybenzoate_methyltransferase_activity |
| GO:0004397 | 1 | histidine_ammonia-lyase_activity |
| GO:0004398 | 1 | histidine_decarboxylase_activity |
| GO:0004408 | 1 | holocytochrome-c_synthase_activity |
| GO:0004411 | 1 | homogentisate_1,2-dioxygenase_activity |
| GO:0004418 | 1 | hydroxymethylbilane_synthase_activity |
| GO:0004420 | 1 | hydroxymethylglutaryl-CoA_reductase_(NADPH)_activity |
| GO:0004423 | 1 | iduronate-2-sulfatase_activity |
| GO:0004441 | 1 | inositol-1,4-bisphosphate_1-phosphatase_activity |
| GO:0004446 | 1 | inositol-hexakisphosphate_phosphatase_activity |
| GO:0004454 | 1 | ketohexokinase_activity |
| GO:0004458 | 1 | D-lactate_dehydrogenase_(cytochrome)_activity |
| GO:0004462 | 1 | lactoylglutathione_lyase_activity |
| GO:0004463 | 1 | leukotriene-A4_hydrolase_activity |
| GO:0004471 | 1 | malate_dehydrogenase_(decarboxylating)_activity |
| GO:0004479 | 1 | methionyl-tRNA_formyltransferase_activity |
| GO:0004482 | 1 | mRNA_(guanine-N7-)-methyltransferase_activity |
| GO:0004483 | 1 | mRNA_(nucleoside-2'-O-)-methyltransferase_activity |
| GO:0004484 | 1 | mRNA_guanylyltransferase_activity |
| GO:0004486 | 1 | methylenetetrahydrofolate_dehydrogenase_[NAD(P)+]_activity |
| GO:0004489 | 1 | methylenetetrahydrofolate_reductase_(NADPH)_activity |
| GO:0004490 | 1 | methylglutaconyl-CoA_hydratase_activity |
| GO:0004491 | 1 | methylmalonate-semialdehyde_dehydrogenase_(acylating)_activity |
| GO:0004492 | 1 | methylmalonyl-CoA_decarboxylase_activity |
| GO:0004493 | 1 | methylmalonyl-CoA_epimerase_activity |
| GO:0004494 | 1 | methylmalonyl-CoA_mutase_activity |
| GO:0004496 | 1 | mevalonate_kinase_activity |
| GO:0004498 | 1 | calcidiol_1-monooxygenase_activity |
| GO:0004502 | 1 | kynurenine_3-monooxygenase_activity |
| GO:0004503 | 1 | monophenol_monooxygenase_activity |
| GO:0004504 | 1 | peptidylglycine_monooxygenase_activity |
| GO:0004506 | 1 | squalene_monooxygenase_activity |
| GO:0004508 | 1 | steroid_17-alpha-monooxygenase_activity |
| GO:0004511 | 1 | tyrosine_3-monooxygenase_activity |
| GO:0004512 | 1 | inositol-3-phosphate_synthase_activity |
| GO:0004513 | 1 | neolactotetraosylceramide_alpha-2,3-sialyltransferase_activity |
| GO:0004530 | 1 | deoxyribonuclease_I_activity |
| GO:0004534 | 1 | 5'-3'_exoribonuclease_activity |
| GO:0004555 | 1 | alpha,alpha-trehalase_activity |
| GO:0004561 | 1 | alpha-N-acetylglucosaminidase_activity |
| GO:0004564 | 1 | beta-fructofuranosidase_activity |
| GO:0004567 | 1 | beta-mannosidase_activity |
| GO:0004573 | 1 | mannosyl-oligosaccharide_glucosidase_activity |
| GO:0004574 | 1 | oligo-1,6-glucosidase_activity |
| GO:0004575 | 1 | sucrose_alpha-glucosidase_activity |
| GO:0004577 | 1 | N-acetylglucosaminyldiphosphodolichol_N-acetylglucosaminyltransferase_activity |
| GO:0004578 | 1 | chitobiosyldiphosphodolichol_beta-mannosyltransferase_activity |
| GO:0004581 | 1 | dolichyl-phosphate_beta-glucosyltransferase_activity |
| GO:0004585 | 1 | ornithine_carbamoyltransferase_activity |
| GO:0004586 | 1 | ornithine_decarboxylase_activity |
| GO:0004587 | 1 | ornithine-oxo-acid_transaminase_activity |
| GO:0004588 | 1 | orotate_phosphoribosyltransferase_activity |
| GO:0004590 | 1 | orotidine-5'-phosphate_decarboxylase_activity |
| GO:0004595 | 1 | pantetheine-phosphate_adenylyltransferase_activity |
| GO:0004597 | 1 | peptide-aspartate_beta-dioxygenase_activity |
| GO:0004598 | 1 | peptidylamidoglycolate_lyase_activity |
| GO:0004603 | 1 | phenylethanolamine_N-methyltransferase_activity |
| GO:0004607 | 1 | phosphatidylcholine-sterol_O-acyltransferase_activity |
| GO:0004608 | 1 | phosphatidylethanolamine_N-methyltransferase_activity |
| GO:0004610 | 1 | phosphoacetylglucosamine_mutase_activity |
| GO:0004617 | 1 | phosphoglycerate_dehydrogenase_activity |
| GO:0004621 | 1 | glycosylphosphatidylinositol_phospholipase_D_activity |
| GO:0004631 | 1 | phosphomevalonate_kinase_activity |
| GO:0004632 | 1 | phosphopantothenate--cysteine_ligase_activity |
| GO:0004633 | 1 | phosphopantothenoylcysteine_decarboxylase_activity |
| GO:0004637 | 1 | phosphoribosylamine-glycine_ligase_activity |
| GO:0004638 | 1 | phosphoribosylaminoimidazole_carboxylase_activity |
| GO:0004639 | 1 | phosphoribosylaminoimidazolesuccinocarboxamide_synthase_activity |
| GO:0004640 | 1 | phosphoribosylanthranilate_isomerase_activity |
| GO:0004641 | 1 | phosphoribosylformylglycinamidine_cyclo-ligase_activity |
| GO:0004642 | 1 | phosphoribosylformylglycinamidine_synthase_activity |
| GO:0004643 | 1 | phosphoribosylaminoimidazolecarboxamide_formyltransferase_activity |
| GO:0004644 | 1 | phosphoribosylglycinamide_formyltransferase_activity |
| GO:0004648 | 1 | O-phospho-L-serine:2-oxoglutarate_aminotransferase_activity |
| GO:0004651 | 1 | polynucleotide_5'-phosphatase_activity |
| GO:0004654 | 1 | polyribonucleotide_nucleotidyltransferase_activity |
| GO:0004655 | 1 | porphobilinogen_synthase_activity |
| GO:0004671 | 1 | protein_C-terminal_S-isoprenylcysteine_carboxyl_O-methyltransferase_activity |
| GO:0004676 | 1 | 3-phosphoinositide-dependent_protein_kinase_activity |
| GO:0004686 | 1 | elongation_factor-2_kinase_activity |
| GO:0004698 | 1 | calcium-dependent_protein_kinase_C_activity |
| GO:0004728 | 1 | receptor_signaling_protein_tyrosine_phosphatase_activity |
| GO:0004729 | 1 | oxygen-dependent_protoporphyrinogen_oxidase_activity |
| GO:0004730 | 1 | pseudouridylate_synthase_activity |
| GO:0004731 | 1 | purine-nucleoside_phosphorylase_activity |
| GO:0004733 | 1 | pyridoxamine-phosphate_oxidase_activity |
| GO:0004736 | 1 | pyruvate_carboxylase_activity |
| GO:0004742 | 1 | dihydrolipoyllysine-residue_acetyltransferase_activity |
| GO:0004747 | 1 | ribokinase_activity |
| GO:0004750 | 1 | ribulose-phosphate_3-epimerase_activity |
| GO:0004751 | 1 | ribose-5-phosphate_isomerase_activity |
| GO:0004753 | 1 | saccharopine_dehydrogenase_activity |
| GO:0004757 | 1 | sepiapterin_reductase_activity |
| GO:0004760 | 1 | serine-pyruvate_transaminase_activity |
| GO:0004773 | 1 | steryl-sulfatase_activity |
| GO:0004777 | 1 | succinate-semialdehyde_dehydrogenase_(NAD+)_activity |
| GO:0004782 | 1 | sulfinoalanine_decarboxylase_activity |
| GO:0004788 | 1 | thiamine_diphosphokinase_activity |
| GO:0004796 | 1 | thromboxane-A_synthase_activity |
| GO:0004801 | 1 | sedoheptulose-7-phosphate:D-glyceraldehyde-3-phosphate_glyceronetransferase_activity |
| GO:0004807 | 1 | triose-phosphate_isomerase_activity |
| GO:0004808 | 1 | tRNA_(5-methylaminomethyl-2-thiouridylate)-methyltransferase_activity |
| GO:0004810 | 1 | tRNA_adenylyltransferase_activity |
| GO:0004819 | 1 | glutamine-tRNA_ligase_activity |
| GO:0004824 | 1 | lysine-tRNA_ligase_activity |
| GO:0004838 | 1 | L-tyrosine:2-oxoglutarate_aminotransferase_activity |
| GO:0004853 | 1 | uroporphyrinogen_decarboxylase_activity |
| GO:0004855 | 1 | xanthine_oxidase_activity |
| GO:0004856 | 1 | xylulokinase_activity |
| GO:0004874 | 1 | aryl_hydrocarbon_receptor_activity |
| GO:0004876 | 1 | complement_component_C3a_receptor_activity |
| GO:0004877 | 1 | complement_component_C3b_receptor_activity |
| GO:0004883 | 1 | glucocorticoid_receptor_activity |
| GO:0004900 | 1 | erythropoietin_receptor_activity |
| GO:0004903 | 1 | growth_hormone_receptor_activity |
| GO:0004919 | 1 | interleukin-9_receptor_activity |
| GO:0004921 | 1 | interleukin-11_receptor_activity |
| GO:0004925 | 1 | prolactin_receptor_activity |
| GO:0004940 | 1 | beta1-adrenergic_receptor_activity |
| GO:0004941 | 1 | beta2-adrenergic_receptor_activity |
| GO:0004943 | 1 | C3a_anaphylatoxin_receptor_activity |
| GO:0004960 | 1 | thromboxane_receptor_activity |
| GO:0004961 | 1 | thromboxane_A2_receptor_activity |
| GO:0004963 | 1 | follicle-stimulating_hormone_receptor_activity |
| GO:0004964 | 1 | luteinizing_hormone_receptor_activity |
| GO:0004978 | 1 | corticotropin_receptor_activity |
| GO:0004979 | 1 | beta-endorphin_receptor_activity |
| GO:0004990 | 1 | oxytocin_receptor_activity |
| GO:0004997 | 1 | thyrotropin-releasing_hormone_receptor_activity |
| GO:0005008 | 1 | hepatocyte_growth_factor-activated_receptor_activity |
| GO:0005009 | 1 | insulin-activated_receptor_activity |
| GO:0005018 | 1 | platelet-derived_growth_factor_alpha-receptor_activity |
| GO:0005020 | 1 | stem_cell_factor_receptor_activity |
| GO:0005034 | 1 | osmosensor_activity |
| GO:0005055 | 1 | laminin_receptor_activity |
| GO:0005098 | 1 | Ran_GTPase_activator_activity |
| GO:0005115 | 1 | receptor_tyrosine_kinase-like_orphan_receptor_binding |
| GO:0005121 | 1 | Toll_binding |
| GO:0005128 | 1 | erythropoietin_receptor_binding |
| GO:0005129 | 1 | granulocyte_macrophage_colony-stimulating_factor_receptor_binding |
| GO:0005130 | 1 | granulocyte_colony-stimulating_factor_receptor_binding |
| GO:0005135 | 1 | interleukin-3_receptor_binding |
| GO:0005136 | 1 | interleukin-4_receptor_binding |
| GO:0005139 | 1 | interleukin-7_receptor_binding |
| GO:0005140 | 1 | interleukin-9_receptor_binding |
| GO:0005141 | 1 | interleukin-10_receptor_binding |
| GO:0005142 | 1 | interleukin-11_receptor_binding |
| GO:0005144 | 1 | interleukin-13_receptor_binding |
| GO:0005147 | 1 | oncostatin-M_receptor_binding |
| GO:0005150 | 1 | interleukin-1,_Type_I_receptor_binding |
| GO:0005163 | 1 | nerve_growth_factor_receptor_binding |
| GO:0005171 | 1 | hepatocyte_growth_factor_receptor_binding |
| GO:0005175 | 1 | CD27_receptor_binding |
| GO:0005183 | 1 | gonadotropin_hormone-releasing_hormone_activity |
| GO:0005199 | 1 | structural_constituent_of_cell_wall |
| GO:0005224 | 1 | ATP-binding_and_phosphorylation-dependent_chloride_channel_activity |
| GO:0005252 | 1 | open_rectifier_potassium_channel_activity |
| GO:0005260 | 1 | channel-conductance-controlling_ATPase_activity |
| GO:0005287 | 1 | high_affinity_basic_amino_acid_transmembrane_transporter_activity |
| GO:0005289 | 1 | high_affinity_arginine_transmembrane_transporter_activity |
| GO:0005298 | 1 | proline:sodium_symporter_activity |
| GO:0005302 | 1 | L-tyrosine_transmembrane_transporter_activity |
| GO:0005307 | 1 | choline:sodium_symporter_activity |
| GO:0005308 | 1 | creatine_transporter_activity |
| GO:0005309 | 1 | creatine:sodium_symporter_activity |
| GO:0005316 | 1 | high_affinity_inorganic_phosphate:sodium_symporter_activity |
| GO:0005325 | 1 | peroxisomal_fatty-acyl-CoA_transporter_activity |
| GO:0005330 | 1 | dopamine:sodium_symporter_activity |
| GO:0005334 | 1 | norepinephrine:sodium_symporter_activity |
| GO:0005335 | 1 | serotonin:sodium_symporter_activity |
| GO:0005353 | 1 | fructose_transmembrane_transporter_activity |
| GO:0005362 | 1 | low-affinity_glucose:sodium_symporter_activity |
| GO:0005365 | 1 | myo-inositol_transmembrane_transporter_activity |
| GO:0005367 | 1 | myo-inositol:sodium_symporter_activity |
| GO:0005368 | 1 | taurine_transmembrane_transporter_activity |
| GO:0005369 | 1 | taurine:sodium_symporter_activity |
| GO:0005456 | 1 | CMP-N-acetylneuraminate_transmembrane_transporter_activity |
| GO:0005461 | 1 | UDP-glucuronic_acid_transmembrane_transporter_activity |
| GO:0005463 | 1 | UDP-N-acetylgalactosamine_transmembrane_transporter_activity |
| GO:0005464 | 1 | UDP-xylose_transmembrane_transporter_activity |
| GO:0005477 | 1 | pyruvate_secondary_active_transmembrane_transporter_activity |
| GO:0005503 | 1 | all-trans_retinal_binding |
| GO:0005519 | 1 | cytoskeletal_regulatory_protein_binding |
| GO:0005550 | 1 | pheromone_binding |
| GO:0005582 | 1 | collagen_type_XV |
| GO:0005585 | 1 | collagen_type_II |
| GO:0005586 | 1 | collagen_type_III |
| GO:0005590 | 1 | collagen_type_VII |
| GO:0005591 | 1 | collagen_type_VIII |
| GO:0005595 | 1 | collagen_type_XII |
| GO:0005596 | 1 | collagen_type_XIV |
| GO:0005597 | 1 | collagen_type_XVI |
| GO:0005598 | 1 | short-chain_collagen |
| GO:0005600 | 1 | collagen_type_XIII |
| GO:0005673 | 1 | transcription_factor_TFIIE_complex |
| GO:0005685 | 1 | U1_snRNP |
| GO:0005690 | 1 | U4atac_snRNP |
| GO:0005731 | 1 | nucleolus_organizer_region |
| GO:0005754 | 1 | mitochondrial_proton-transporting_ATP_synthase,_catalytic_core |
| GO:0005757 | 1 | mitochondrial_permeability_transition_pore_complex |
| GO:0005816 | 1 | spindle_pole_body |
| GO:0005854 | 1 | nascent_polypeptide-associated_complex |
| GO:0005879 | 1 | axonemal_microtubule |
| GO:0005889 | 1 | hydrogen:potassium-exchanging_ATPase_complex |
| GO:0005895 | 1 | interleukin-5_receptor_complex |
| GO:0005898 | 1 | interleukin-13_receptor_complex |
| GO:0005914 | 1 | spot_adherens_junction |
| GO:0005943 | 1 | 1-phosphatidylinositol-4-phosphate_3-kinase,_class_IA_complex |
| GO:0005953 | 1 | CAAX-protein_geranylgeranyltransferase_complex |
| GO:0005967 | 1 | mitochondrial_pyruvate_dehydrogenase_complex |
| GO:0005969 | 1 | serine-pyruvate_aminotransferase_complex |
| GO:0005971 | 1 | ribonucleoside-diphosphate_reductase_complex |
| GO:0005982 | 1 | starch_metabolic_process |
| GO:0005983 | 1 | starch_catabolic_process |
| GO:0005985 | 1 | sucrose_metabolic_process |
| GO:0005993 | 1 | trehalose_catabolic_process |
| GO:0005998 | 1 | xylulose_catabolic_process |
| GO:0006037 | 1 | cell_wall_chitin_metabolic_process |
| GO:0006046 | 1 | N-acetylglucosamine_catabolic_process |
| GO:0006049 | 1 | UDP-N-acetylglucosamine_catabolic_process |
| GO:0006050 | 1 | mannosamine_metabolic_process |
| GO:0006051 | 1 | N-acetylmannosamine_metabolic_process |
| GO:0006060 | 1 | sorbitol_metabolic_process |
| GO:0006062 | 1 | sorbitol_catabolic_process |
| GO:0006064 | 1 | glucuronate_catabolic_process |
| GO:0006117 | 1 | acetaldehyde_metabolic_process |
| GO:0006121 | 1 | mitochondrial_electron_transport,_succinate_to_ubiquinone |
| GO:0006147 | 1 | guanine_catabolic_process |
| GO:0006148 | 1 | inosine_catabolic_process |
| GO:0006157 | 1 | deoxyadenosine_catabolic_process |
| GO:0006173 | 1 | dADP_biosynthetic_process |
| GO:0006175 | 1 | dATP_biosynthetic_process |
| GO:0006193 | 1 | ITP_catabolic_process |
| GO:0006203 | 1 | dGTP_catabolic_process |
| GO:0006214 | 1 | thymidine_catabolic_process |
| GO:0006226 | 1 | dUMP_biosynthetic_process |
| GO:0006256 | 1 | UDP_catabolic_process |
| GO:0006258 | 1 | UDP-glucose_catabolic_process |
| GO:0006272 | 1 | leading_strand_elongation |
| GO:0006343 | 1 | establishment_of_chromatin_silencing |
| GO:0006393 | 1 | termination_of_mitochondrial_transcription |
| GO:0006404 | 1 | RNA_import_into_nucleus |
| GO:0006409 | 1 | tRNA_export_from_nucleus |
| GO:0006425 | 1 | glutaminyl-tRNA_aminoacylation |
| GO:0006430 | 1 | lysyl-tRNA_aminoacylation |
| GO:0006463 | 1 | steroid_hormone_receptor_complex_assembly |
| GO:0006480 | 1 | N-terminal_protein_amino_acid_methylation |
| GO:0006500 | 1 | N-terminal_protein_palmitoylation |
| GO:0006507 | 1 | GPI_anchor_release |
| GO:0006530 | 1 | asparagine_catabolic_process |
| GO:0006535 | 1 | cysteine_biosynthetic_process_from_serine |
| GO:0006550 | 1 | isoleucine_catabolic_process |
| GO:0006557 | 1 | S-adenosylmethioninamine_biosynthetic_process |
| GO:0006567 | 1 | threonine_catabolic_process |
| GO:0006571 | 1 | tyrosine_biosynthetic_process |
| GO:0006585 | 1 | dopamine_biosynthetic_process_from_tyrosine |
| GO:0006608 | 1 | snRNP_protein_import_into_nucleus |
| GO:0006616 | 1 | SRP-dependent_cotranslational_protein_targeting_to_membrane,_translocation |
| GO:0006617 | 1 | SRP-dependent_cotranslational_protein_targeting_to_membrane,_signal_sequence_recognition |
| GO:0006666 | 1 | 3-keto-sphinganine_metabolic_process |
| GO:0006669 | 1 | sphinganine-1-phosphate_biosynthetic_process |
| GO:0006671 | 1 | phytosphingosine_metabolic_process |
| GO:0006679 | 1 | glucosylceramide_biosynthetic_process |
| GO:0006683 | 1 | galactosylceramide_catabolic_process |
| GO:0006711 | 1 | estrogen_catabolic_process |
| GO:0006713 | 1 | glucocorticoid_catabolic_process |
| GO:0006741 | 1 | NADP_biosynthetic_process |
| GO:0006746 | 1 | FADH2_metabolic_process |
| GO:0006747 | 1 | FAD_biosynthetic_process |
| GO:0006756 | 1 | AMP_phosphorylation |
| GO:0006780 | 1 | uroporphyrinogen_III_biosynthetic_process |
| GO:0006781 | 1 | succinyl-CoA_pathway |
| GO:0006789 | 1 | bilirubin_conjugation |
| GO:0006797 | 1 | polyphosphate_metabolic_process |
| GO:0006842 | 1 | tricarboxylic_acid_transport |
| GO:0006843 | 1 | mitochondrial_citrate_transport |
| GO:0006848 | 1 | pyruvate_transport |
| GO:0006867 | 1 | asparagine_transport |
| GO:0006876 | 1 | cellular_cadmium_ion_homeostasis |
| GO:0006948 | 1 | induction_by_virus_of_host_cell-cell_fusion |
| GO:0006994 | 1 | positive_regulation_of_transcription_via_sterol_regulatory_element_binding_involved_in_ER-nuclear_sterol_response_pathway |
| GO:0007039 | 1 | vacuolar_protein_catabolic_process |
| GO:0007056 | 1 | spindle_assembly_involved_in_female_meiosis |
| GO:0007057 | 1 | spindle_assembly_involved_in_female_meiosis_I |
| GO:0007060 | 1 | male_meiosis_chromosome_segregation |
| GO:0007072 | 1 | positive_regulation_of_transcription_on_exit_from_mitosis |
| GO:0007108 | 1 | cytokinesis,_initiation_of_separation |
| GO:0007136 | 1 | meiotic_prophase_II |
| GO:0007137 | 1 | meiotic_metaphase_II |
| GO:0007174 | 1 | epidermal_growth_factor_catabolic_process |
| GO:0007192 | 1 | adenylate_cyclase-activating_serotonin_receptor_signaling_pathway |
| GO:0007198 | 1 | adenylate_cyclase-inhibiting_serotonin_receptor_signaling_pathway |
| GO:0007208 | 1 | phospholipase_C-activating_serotonin_receptor_signaling_pathway |
| GO:0007225 | 1 | patched_ligand_maturation |
| GO:0007231 | 1 | osmosensory_signaling_pathway |
| GO:0007285 | 1 | primary_spermatocyte_growth |
| GO:0007321 | 1 | sperm_displacement |
| GO:0007344 | 1 | pronuclear_fusion |
| GO:0007354 | 1 | zygotic_determination_of_anterior/posterior_axis,_embryo |
| GO:0007371 | 1 | ventral_midline_determination |
| GO:0007387 | 1 | anterior_compartment_pattern_formation |
| GO:0007388 | 1 | posterior_compartment_specification |
| GO:0007400 | 1 | neuroblast_fate_determination |
| GO:0007440 | 1 | foregut_morphogenesis |
| GO:0007443 | 1 | Malpighian_tubule_morphogenesis |
| GO:0007468 | 1 | regulation_of_rhodopsin_gene_expression |
| GO:0007495 | 1 | visceral_mesoderm-endoderm_interaction_involved_in_midgut_development |
| GO:0007499 | 1 | ectoderm_and_mesoderm_interaction |
| GO:0007502 | 1 | digestive_tract_mesoderm_development |
| GO:0007522 | 1 | visceral_muscle_development |
| GO:0007527 | 1 | adult_somatic_muscle_development |
| GO:0007576 | 1 | nucleolar_fragmentation |
| GO:0007624 | 1 | ultradian_rhythm |
| GO:0007634 | 1 | optokinetic_behavior |
| GO:0008024 | 1 | positive_transcription_elongation_factor_complex_b |
| GO:0008029 | 1 | pentraxin_receptor_activity |
| GO:0008039 | 1 | synaptic_target_recognition |
| GO:0008049 | 1 | male_courtship_behavior |
| GO:0008050 | 1 | female_courtship_behavior |
| GO:0008052 | 1 | sensory_organ_boundary_specification |
| GO:0008057 | 1 | eye_pigment_granule_organization |
| GO:0008111 | 1 | alpha-methylacyl-CoA_racemase_activity |
| GO:0008112 | 1 | nicotinamide_N-methyltransferase_activity |
| GO:0008115 | 1 | sarcosine_oxidase_activity |
| GO:0008116 | 1 | prostaglandin-I_synthase_activity |
| GO:0008117 | 1 | sphinganine-1-phosphate_aldolase_activity |
| GO:0008118 | 1 | N-acetyllactosaminide_alpha-2,3-sialyltransferase_activity |
| GO:0008120 | 1 | ceramide_glucosyltransferase_activity |
| GO:0008123 | 1 | cholesterol_7-alpha-monooxygenase_activity |
| GO:0008126 | 1 | acetylesterase_activity |
| GO:0008127 | 1 | quercetin_2,3-dioxygenase_activity |
| GO:0008169 | 1 | C-methyltransferase_activity |
| GO:0008208 | 1 | C21-steroid_hormone_catabolic_process |
| GO:0008251 | 1 | tRNA-specific_adenosine_deaminase_activity |
| GO:0008254 | 1 | 3'-nucleotidase_activity |
| GO:0008262 | 1 | importin-alpha_export_receptor_activity |
| GO:0008263 | 1 | pyrimidine-specific_mismatch_base_pair_DNA_N-glycosylase_activity |
| GO:0008265 | 1 | Mo-molybdopterin_cofactor_sulfurase_activity |
| GO:0008267 | 1 | poly-glutamine_tract_binding |
| GO:0008275 | 1 | gamma-tubulin_small_complex |
| GO:0008280 | 1 | cohesin_core_heterodimer |
| GO:0008303 | 1 | caspase_complex |
| GO:0008310 | 1 | single-stranded_DNA_specific_3'-5'_exodeoxyribonuclease_activity |
| GO:0008336 | 1 | gamma-butyrobetaine_dioxygenase_activity |
| GO:0008349 | 1 | MAP_kinase_kinase_kinase_kinase_activity |
| GO:0008352 | 1 | katanin_complex |
| GO:0008387 | 1 | steroid_7-alpha-hydroxylase_activity |
| GO:0008389 | 1 | coumarin_7-hydroxylase_activity |
| GO:0008390 | 1 | testosterone_16-alpha-hydroxylase_activity |
| GO:0008397 | 1 | sterol_12-alpha-hydroxylase_activity |
| GO:0008398 | 1 | sterol_14-demethylase_activity |
| GO:0008403 | 1 | 25-hydroxycholecalciferol-24-hydroxylase_activity |
| GO:0008404 | 1 | arachidonic_acid_14,15-epoxygenase_activity |
| GO:0008405 | 1 | arachidonic_acid_11,12-epoxygenase_activity |
| GO:0008413 | 1 | 8-oxo-7,8-dihydroguanosine_triphosphate_pyrophosphatase_activity |
| GO:0008418 | 1 | protein-N-terminal_asparagine_amidohydrolase_activity |
| GO:0008419 | 1 | RNA_lariat_debranching_enzyme_activity |
| GO:0008424 | 1 | glycoprotein_6-alpha-L-fucosyltransferase_activity |
| GO:0008425 | 1 | 2-polyprenyl-6-methoxy-1,4-benzoquinone_methyltransferase_activity |
| GO:0008427 | 1 | calcium-dependent_protein_kinase_inhibitor_activity |
| GO:0008434 | 1 | vitamin_D3_receptor_activity |
| GO:0008437 | 1 | thyrotropin-releasing_hormone_activity |
| GO:0008442 | 1 | 3-hydroxyisobutyrate_dehydrogenase_activity |
| GO:0008444 | 1 | CDP-diacylglycerol-glycerol-3-phosphate_3-phosphatidyltransferase_activity |
| GO:0008445 | 1 | D-aspartate_oxidase_activity |
| GO:0008446 | 1 | GDP-mannose_4,6-dehydratase_activity |
| GO:0008448 | 1 | N-acetylglucosamine-6-phosphate_deacetylase_activity |
| GO:0008452 | 1 | RNA_ligase_activity |
| GO:0008455 | 1 | alpha-1,6-mannosylglycoprotein_2-beta-N-acetylglucosaminyltransferase_activity |
| GO:0008456 | 1 | alpha-N-acetylgalactosaminidase_activity |
| GO:0008457 | 1 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide_beta-1,3-acetylglucosaminyltransferase_activity |
| GO:0008458 | 1 | carnitine_O-octanoyltransferase_activity |
| GO:0008460 | 1 | dTDP-glucose_4,6-dehydratase_activity |
| GO:0008465 | 1 | glycerate_dehydrogenase_activity |
| GO:0008470 | 1 | isovaleryl-CoA_dehydrogenase_activity |
| GO:0008478 | 1 | pyridoxal_kinase_activity |
| GO:0008480 | 1 | sarcosine_dehydrogenase_activity |
| GO:0008482 | 1 | sulfite_oxidase_activity |
| GO:0008488 | 1 | gamma-glutamyl_carboxylase_activity |
| GO:0008493 | 1 | tetracycline_transporter_activity |
| GO:0008495 | 1 | protoheme_IX_farnesyltransferase_activity |
| GO:0008507 | 1 | sodium:iodide_symporter_activity |
| GO:0008521 | 1 | acetyl-CoA_transporter_activity |
| GO:0008531 | 1 | riboflavin_kinase_activity |
| GO:0008532 | 1 | N-acetyllactosaminide_beta-1,3-N-acetylglucosaminyltransferase_activity |
| GO:0008534 | 1 | oxidized_purine_base_lesion_DNA_N-glycosylase_activity |
| GO:0008554 | 1 | sodium-exporting_ATPase_activity,_phosphorylative_mechanism |
| GO:0008555 | 1 | chloride-transporting_ATPase_activity |
| GO:0008579 | 1 | JUN_kinase_phosphatase_activity |
| GO:0008592 | 1 | regulation_of_Toll_signaling_pathway |
| GO:0008609 | 1 | alkylglycerone-phosphate_synthase_activity |
| GO:0008613 | 1 | diuretic_hormone_activity |
| GO:0008622 | 1 | epsilon_DNA_polymerase_complex |
| GO:0008627 | 1 | induction_of_apoptosis_by_ionic_changes |
| GO:0008636 | 1 | activation_of_caspase_activity_by_protein_phosphorylation |
| GO:0008650 | 1 | rRNA_(uridine-2'-O-)-methyltransferase_activity |
| GO:0008667 | 1 | 2,3-dihydro-2,3-dihydroxybenzoate_dehydrogenase_activity |
| GO:0008689 | 1 | 3-demethylubiquinone-9_3-O-methyltransferase_activity |
| GO:0008700 | 1 | 4-hydroxy-2-oxoglutarate_aldolase_activity |
| GO:0008705 | 1 | methionine_synthase_activity |
| GO:0008709 | 1 | cholate_7-alpha-dehydrogenase_activity |
| GO:0008721 | 1 | D-serine_ammonia-lyase_activity |
| GO:0008725 | 1 | DNA-3-methyladenine_glycosylase_activity |
| GO:0008747 | 1 | N-acetylneuraminate_lyase_activity |
| GO:0008750 | 1 | NAD(P)+_transhydrogenase_(AB-specific)_activity |
| GO:0008761 | 1 | UDP-N-acetylglucosamine_2-epimerase_activity |
| GO:0008768 | 1 | UDP-sugar_diphosphatase_activity |
| GO:0008783 | 1 | agmatinase_activity |
| GO:0008785 | 1 | alkyl_hydroperoxide_reductase_activity |
| GO:0008792 | 1 | arginine_decarboxylase_activity |
| GO:0008794 | 1 | arsenate_reductase_(glutaredoxin)_activity |
| GO:0008796 | 1 | bis(5'-nucleosyl)-tetraphosphatase_activity |
| GO:0008798 | 1 | beta-aspartyl-peptidase_activity |
| GO:0008802 | 1 | betaine-aldehyde_dehydrogenase_activity |
| GO:0008803 | 1 | bis(5'-nucleosyl)-tetraphosphatase_(symmetrical)_activity |
| GO:0008808 | 1 | cardiolipin_synthase_activity |
| GO:0008812 | 1 | choline_dehydrogenase_activity |
| GO:0008815 | 1 | citrate_(pro-3S)-lyase_activity |
| GO:0008817 | 1 | cob(I)yrinic_acid_a,c-diamide_adenosyltransferase_activity |
| GO:0008831 | 1 | dTDP-4-dehydrorhamnose_reductase_activity |
| GO:0008843 | 1 | endochitinase_activity |
| GO:0008859 | 1 | exoribonuclease_II_activity |
| GO:0008887 | 1 | glycerate_kinase_activity |
| GO:0008890 | 1 | glycine_C-acetyltransferase_activity |
| GO:0008891 | 1 | glycolate_oxidase_activity |
| GO:0008892 | 1 | guanine_deaminase_activity |
| GO:0008893 | 1 | guanosine-3',5'-bis(diphosphate)_3'-diphosphatase_activity |
| GO:0008894 | 1 | guanosine-5'-triphosphate,3'-diphosphate_diphosphatase_activity |
| GO:0008897 | 1 | holo-[acyl-carrier-protein]_synthase_activity |
| GO:0008903 | 1 | hydroxypyruvate_isomerase_activity |
| GO:0008917 | 1 | lipopolysaccharide_N-acetylglucosaminyltransferase_activity |
| GO:0008955 | 1 | peptidoglycan_glycosyltransferase_activity |
| GO:0008963 | 1 | phospho-N-acetylmuramoyl-pentapeptide-transferase_activity |
| GO:0008973 | 1 | phosphopentomutase_activity |
| GO:0008988 | 1 | rRNA_(adenine-N6-)-methyltransferase_activity |
| GO:0008995 | 1 | ribonuclease_E_activity |
| GO:0009000 | 1 | selenocysteine_lyase_activity |
| GO:0009019 | 1 | tRNA_(guanine-N1-)-methyltransferase_activity |
| GO:0009021 | 1 | tRNA_(uracil-5-)-methyltransferase_activity |
| GO:0009032 | 1 | thymidine_phosphorylase_activity |
| GO:0009041 | 1 | uridylate_kinase_activity |
| GO:0009073 | 1 | aromatic_amino_acid_family_biosynthetic_process |
| GO:0009095 | 1 | aromatic_amino_acid_family_biosynthetic_process,_prephenate_pathway |
| GO:0009098 | 1 | leucine_biosynthetic_process |
| GO:0009115 | 1 | xanthine_catabolic_process |
| GO:0009131 | 1 | pyrimidine_nucleoside_monophosphate_catabolic_process |
| GO:0009140 | 1 | pyrimidine_nucleoside_diphosphate_catabolic_process |
| GO:0009178 | 1 | pyrimidine_deoxyribonucleoside_monophosphate_catabolic_process |
| GO:0009182 | 1 | purine_deoxyribonucleoside_diphosphate_metabolic_process |
| GO:0009183 | 1 | purine_deoxyribonucleoside_diphosphate_biosynthetic_process |
| GO:0009193 | 1 | pyrimidine_ribonucleoside_diphosphate_metabolic_process |
| GO:0009195 | 1 | pyrimidine_ribonucleoside_diphosphate_catabolic_process |
| GO:0009216 | 1 | purine_deoxyribonucleoside_triphosphate_biosynthetic_process |
| GO:0009222 | 1 | pyrimidine_ribonucleotide_catabolic_process |
| GO:0009227 | 1 | nucleotide-sugar_catabolic_process |
| GO:0009229 | 1 | thiamine_diphosphate_biosynthetic_process |
| GO:0009231 | 1 | riboflavin_biosynthetic_process |
| GO:0009233 | 1 | menaquinone_metabolic_process |
| GO:0009234 | 1 | menaquinone_biosynthetic_process |
| GO:0009237 | 1 | siderophore_metabolic_process |
| GO:0009238 | 1 | enterobactin_metabolic_process |
| GO:0009239 | 1 | enterobactin_biosynthetic_process |
| GO:0009305 | 1 | protein_biotinylation |
| GO:0009317 | 1 | acetyl-CoA_carboxylase_complex |
| GO:0009330 | 1 | DNA_topoisomerase_complex_(ATP-hydrolyzing) |
| GO:0009348 | 1 | ornithine_carbamoyltransferase_complex |
| GO:0009360 | 1 | DNA_polymerase_III_complex |
| GO:0009368 | 1 | endopeptidase_Clp_complex |
| GO:0009372 | 1 | quorum_sensing |
| GO:0009386 | 1 | translational_attenuation |
| GO:0009398 | 1 | FMN_biosynthetic_process |
| GO:0009399 | 1 | nitrogen_fixation |
| GO:0009401 | 1 | phosphoenolpyruvate-dependent_sugar_phosphotransferase_system |
| GO:0009403 | 1 | toxin_biosynthetic_process |
| GO:0009438 | 1 | methylglyoxal_metabolic_process |
| GO:0009443 | 1 | pyridoxal_5'-phosphate_salvage |
| GO:0009449 | 1 | gamma-aminobutyric_acid_biosynthetic_process |
| GO:0009645 | 1 | response_to_low_light_intensity_stimulus |
| GO:0009673 | 1 | low_affinity_phosphate_transmembrane_transporter_activity |
| GO:0009720 | 1 | detection_of_hormone_stimulus |
| GO:0009757 | 1 | hexose_mediated_signaling |
| GO:0009787 | 1 | regulation_of_abscisic_acid_mediated_signaling_pathway |
| GO:0009789 | 1 | positive_regulation_of_abscisic_acid_mediated_signaling_pathway |
| GO:0009794 | 1 | regulation_of_mitotic_cell_cycle,_embryonic |
| GO:0009814 | 1 | defense_response,_incompatible_interaction |
| GO:0009817 | 1 | defense_response_to_fungus,_incompatible_interaction |
| GO:0009841 | 1 | mitochondrial_endopeptidase_Clp_complex |
| GO:0009917 | 1 | sterol_5-alpha_reductase_activity |
| GO:0009931 | 1 | calcium-dependent_protein_serine/threonine_kinase_activity |
| GO:0009946 | 1 | proximal/distal_axis_specification |
| GO:0009949 | 1 | polarity_specification_of_anterior/posterior_axis |
| GO:0009956 | 1 | radial_pattern_formation |
| GO:0009994 | 1 | oocyte_differentiation |
| GO:0010032 | 1 | meiotic_chromosome_condensation |
| GO:0010034 | 1 | response_to_acetate |
| GO:0010106 | 1 | cellular_response_to_iron_ion_starvation |
| GO:0010160 | 1 | formation_of_organ_boundary |
| GO:0010182 | 1 | sugar_mediated_signaling_pathway |
| GO:0010189 | 1 | vitamin_E_biosynthetic_process |
| GO:0010232 | 1 | vascular_transport |
| GO:0010255 | 1 | glucose_mediated_signaling_pathway |
| GO:0010260 | 1 | organ_senescence |
| GO:0010266 | 1 | response_to_vitamin_B1 |
| GO:0010273 | 1 | detoxification_of_copper_ion |
| GO:0010286 | 1 | heat_acclimation |
| GO:0010309 | 1 | acireductone_dioxygenase_[iron(II)-requiring]_activity |
| GO:0010333 | 1 | terpene_synthase_activity |
| GO:0010350 | 1 | cellular_response_to_magnesium_starvation |
| GO:0010370 | 1 | perinucleolar_chromocenter |
| GO:0010383 | 1 | cell_wall_polysaccharide_metabolic_process |
| GO:0010385 | 1 | double-stranded_methylated_DNA_binding |
| GO:0010387 | 1 | signalosome_assembly |
| GO:0010420 | 1 | polyprenyldihydroxybenzoate_methyltransferase_activity |
| GO:0010421 | 1 | hydrogen_peroxide-mediated_programmed_cell_death |
| GO:0010446 | 1 | response_to_alkalinity |
| GO:0010477 | 1 | response_to_sulfur_dioxide |
| GO:0010501 | 1 | RNA_secondary_structure_unwinding |
| GO:0010509 | 1 | polyamine_homeostasis |
| GO:0010513 | 1 | positive_regulation_of_phosphatidylinositol_biosynthetic_process |
| GO:0010571 | 1 | positive_regulation_of_DNA_replication_involved_in_S_phase |
| GO:0010573 | 1 | vascular_endothelial_growth_factor_production |
| GO:0010577 | 1 | metalloenzyme_activator_activity |
| GO:0010609 | 1 | posttranscriptional_regulation_of_gene_expression_by_mRNA_localization |
| GO:0010610 | 1 | regulation_of_mRNA_stability_involved_in_response_to_stress |
| GO:0010612 | 1 | regulation_of_cardiac_muscle_adaptation |
| GO:0010616 | 1 | negative_regulation_of_cardiac_muscle_adaptation |
| GO:0010625 | 1 | positive_regulation_of_Schwann_cell_proliferation |
| GO:0010644 | 1 | cell_communication_by_electrical_coupling |
| GO:0010649 | 1 | regulation_of_cell_communication_by_electrical_coupling |
| GO:0010663 | 1 | positive_regulation_of_striated_muscle_cell_apoptotic_process |
| GO:0010666 | 1 | positive_regulation_of_cardiac_muscle_cell_apoptotic_process |
| GO:0010702 | 1 | regulation_of_histolysis |
| GO:0010703 | 1 | negative_regulation_of_histolysis |
| GO:0010710 | 1 | regulation_of_collagen_catabolic_process |
| GO:0010711 | 1 | negative_regulation_of_collagen_catabolic_process |
| GO:0010722 | 1 | regulation_of_ferrochelatase_activity |
| GO:0010732 | 1 | regulation_of_protein_glutathionylation |
| GO:0010734 | 1 | negative_regulation_of_protein_glutathionylation |
| GO:0010747 | 1 | positive_regulation_of_plasma_membrane_long-chain_fatty_acid_transport |
| GO:0010749 | 1 | regulation_of_nitric_oxide_mediated_signal_transduction |
| GO:0010751 | 1 | negative_regulation_of_nitric_oxide_mediated_signal_transduction |
| GO:0010752 | 1 | regulation_of_cGMP-mediated_signaling |
| GO:0010754 | 1 | negative_regulation_of_cGMP-mediated_signaling |
| GO:0010760 | 1 | negative_regulation_of_macrophage_chemotaxis |
| GO:0010768 | 1 | negative_regulation_of_transcription_from_RNA_polymerase_II_promoter_in_response_to_UV-induced_DNA_damage |
| GO:0010793 | 1 | regulation_of_mRNA_export_from_nucleus |
| GO:0010814 | 1 | substance_P_catabolic_process |
| GO:0010816 | 1 | calcitonin_catabolic_process |
| GO:0010825 | 1 | positive_regulation_of_centrosome_duplication |
| GO:0010836 | 1 | negative_regulation_of_protein_ADP-ribosylation |
| GO:0010847 | 1 | regulation_of_chromatin_assembly |
| GO:0010852 | 1 | cyclase_inhibitor_activity |
| GO:0010854 | 1 | adenylate_cyclase_regulator_activity |
| GO:0010855 | 1 | adenylate_cyclase_inhibitor_activity |
| GO:0010858 | 1 | calcium-dependent_protein_kinase_regulator_activity |
| GO:0010859 | 1 | calcium-dependent_cysteine-type_endopeptidase_inhibitor_activity |
| GO:0010878 | 1 | cholesterol_storage |
| GO:0010900 | 1 | negative_regulation_of_phosphatidylcholine_catabolic_process |
| GO:0010904 | 1 | regulation_of_UDP-glucose_catabolic_process |
| GO:0010905 | 1 | negative_regulation_of_UDP-glucose_catabolic_process |
| GO:0010917 | 1 | negative_regulation_of_mitochondrial_membrane_potential |
| GO:0010918 | 1 | positive_regulation_of_mitochondrial_membrane_potential |
| GO:0010920 | 1 | negative_regulation_of_inositol_phosphate_biosynthetic_process |
| GO:0010924 | 1 | regulation_of_inositol-polyphosphate_5-phosphatase_activity |
| GO:0010925 | 1 | positive_regulation_of_inositol-polyphosphate_5-phosphatase_activity |
| GO:0010932 | 1 | regulation_of_macrophage_tolerance_induction |
| GO:0010933 | 1 | positive_regulation_of_macrophage_tolerance_induction |
| GO:0010934 | 1 | macrophage_cytokine_production |
| GO:0010987 | 1 | negative_regulation_of_high-density_lipoprotein_particle_clearance |
| GO:0012510 | 1 | trans-Golgi_network_transport_vesicle_membrane |
| GO:0014029 | 1 | neural_crest_formation |
| GO:0014042 | 1 | positive_regulation_of_neuron_maturation |
| GO:0014053 | 1 | negative_regulation_of_gamma-aminobutyric_acid_secretion |
| GO:0014057 | 1 | positive_regulation_of_acetylcholine_secretion |
| GO:0014705 | 1 | C_zone |
| GO:0014717 | 1 | regulation_of_satellite_cell_activation_involved_in_skeletal_muscle_regeneration |
| GO:0014718 | 1 | positive_regulation_of_satellite_cell_activation_involved_in_skeletal_muscle_regeneration |
| GO:0014739 | 1 | positive_regulation_of_muscle_hyperplasia |
| GO:0014744 | 1 | positive_regulation_of_muscle_adaptation |
| GO:0014804 | 1 | terminal_cisterna_lumen |
| GO:0014807 | 1 | regulation_of_somitogenesis |
| GO:0014809 | 1 | regulation_of_skeletal_muscle_contraction_by_regulation_of_release_of_sequestered_calcium_ion |
| GO:0014813 | 1 | satellite_cell_commitment |
| GO:0014822 | 1 | detection_of_wounding |
| GO:0014835 | 1 | myoblast_cell_differentiation_involved_in_skeletal_muscle_regeneration |
| GO:0014841 | 1 | satellite_cell_proliferation |
| GO:0014842 | 1 | regulation_of_satellite_cell_proliferation |
| GO:0014849 | 1 | ureter_smooth_muscle_contraction |
| GO:0014853 | 1 | regulation_of_excitatory_postsynaptic_membrane_potential_involved_in_skeletal_muscle_contraction |
| GO:0014857 | 1 | regulation_of_skeletal_muscle_cell_proliferation |
| GO:0014858 | 1 | positive_regulation_of_skeletal_muscle_cell_proliferation |
| GO:0014873 | 1 | response_to_muscle_activity_involved_in_regulation_of_muscle_adaptation |
| GO:0014878 | 1 | response_to_electrical_stimulus_involved_in_regulation_of_muscle_adaptation |
| GO:0014895 | 1 | smooth_muscle_hypertrophy |
| GO:0014904 | 1 | myotube_cell_development |
| GO:0015019 | 1 | heparan-alpha-glucosaminide_N-acetyltransferase_activity |
| GO:0015052 | 1 | beta3-adrenergic_receptor_activity |
| GO:0015054 | 1 | gastrin_receptor_activity |
| GO:0015055 | 1 | secretin_receptor_activity |
| GO:0015067 | 1 | amidinotransferase_activity |
| GO:0015068 | 1 | glycine_amidinotransferase_activity |
| GO:0015086 | 1 | cadmium_ion_transmembrane_transporter_activity |
| GO:0015087 | 1 | cobalt_ion_transmembrane_transporter_activity |
| GO:0015094 | 1 | lead_ion_transmembrane_transporter_activity |
| GO:0015099 | 1 | nickel_cation_transmembrane_transporter_activity |
| GO:0015100 | 1 | vanadium_ion_transmembrane_transporter_activity |
| GO:0015112 | 1 | nitrate_transmembrane_transporter_activity |
| GO:0015117 | 1 | thiosulfate_transmembrane_transporter_activity |
| GO:0015119 | 1 | hexose_phosphate_transmembrane_transporter_activity |
| GO:0015126 | 1 | canalicular_bile_acid_transmembrane_transporter_activity |
| GO:0015129 | 1 | lactate_transmembrane_transporter_activity |
| GO:0015130 | 1 | mevalonate_transmembrane_transporter_activity |
| GO:0015132 | 1 | prostaglandin_transmembrane_transporter_activity |
| GO:0015136 | 1 | sialic_acid_transmembrane_transporter_activity |
| GO:0015152 | 1 | glucose-6-phosphate_transmembrane_transporter_activity |
| GO:0015173 | 1 | aromatic_amino_acid_transmembrane_transporter_activity |
| GO:0015182 | 1 | L-asparagine_transmembrane_transporter_activity |
| GO:0015195 | 1 | L-threonine_transmembrane_transporter_activity |
| GO:0015227 | 1 | acyl_carnitine_transporter_activity |
| GO:0015230 | 1 | FAD_transmembrane_transporter_activity |
| GO:0015246 | 1 | fatty-acyl_group_transporter_activity |
| GO:0015319 | 1 | sodium:inorganic_phosphate_symporter_activity |
| GO:0015320 | 1 | phosphate_ion_carrier_activity |
| GO:0015326 | 1 | cationic_amino_acid_transmembrane_transporter_activity |
| GO:0015327 | 1 | cystine:glutamate_antiporter_activity |
| GO:0015334 | 1 | high_affinity_oligopeptide_transporter_activity |
| GO:0015350 | 1 | methotrexate_transporter_activity |
| GO:0015361 | 1 | low_affinity_sodium:dicarboxylate_symporter_activity |
| GO:0015362 | 1 | high_affinity_sodium:dicarboxylate_symporter_activity |
| GO:0015367 | 1 | oxoglutarate:malate_antiporter_activity |
| GO:0015373 | 1 | monovalent_anion:sodium_symporter_activity |
| GO:0015378 | 1 | sodium:chloride_symporter_activity |
| GO:0015390 | 1 | purine-specific_nucleoside:sodium_symporter_activity |
| GO:0015410 | 1 | manganese-transporting_ATPase_activity |
| GO:0015432 | 1 | bile_acid-exporting_ATPase_activity |
| GO:0015439 | 1 | heme-transporting_ATPase_activity |
| GO:0015495 | 1 | gamma-aminobutyric_acid:hydrogen_symporter_activity |
| GO:0015499 | 1 | formate_transmembrane_transporter_activity |
| GO:0015578 | 1 | mannose_transmembrane_transporter_activity |
| GO:0015607 | 1 | fatty-acyl-CoA_transporter_activity |
| GO:0015616 | 1 | DNA_translocase_activity |
| GO:0015633 | 1 | zinc_transporting_ATPase_activity |
| GO:0015675 | 1 | nickel_cation_transport |
| GO:0015676 | 1 | vanadium_ion_transport |
| GO:0015684 | 1 | ferrous_iron_transport |
| GO:0015692 | 1 | lead_ion_transport |
| GO:0015700 | 1 | arsenite_transport |
| GO:0015707 | 1 | nitrite_transport |
| GO:0015727 | 1 | lactate_transport |
| GO:0015728 | 1 | mevalonate_transport |
| GO:0015739 | 1 | sialic_acid_transport |
| GO:0015746 | 1 | citrate_transport |
| GO:0015782 | 1 | CMP-N-acetylneuraminate_transport |
| GO:0015787 | 1 | UDP-glucuronic_acid_transport |
| GO:0015789 | 1 | UDP-N-acetylgalactosamine_transport |
| GO:0015790 | 1 | UDP-xylose_transport |
| GO:0015798 | 1 | myo-inositol_transport |
| GO:0015805 | 1 | S-adenosylmethionine_transport |
| GO:0015817 | 1 | histidine_transport |
| GO:0015826 | 1 | threonine_transport |
| GO:0015828 | 1 | tyrosine_transport |
| GO:0015846 | 1 | polyamine_transport |
| GO:0015854 | 1 | guanine_transport |
| GO:0015874 | 1 | norepinephrine_transport |
| GO:0015876 | 1 | acetyl-CoA_transport |
| GO:0015881 | 1 | creatine_transport |
| GO:0015883 | 1 | FAD_transport |
| GO:0015888 | 1 | thiamine_transport |
| GO:0015904 | 1 | tetracycline_transport |
| GO:0015910 | 1 | peroxisomal_long-chain_fatty_acid_import |
| GO:0015919 | 1 | peroxisomal_membrane_transport |
| GO:0015922 | 1 | aspartate_oxidase_activity |
| GO:0015927 | 1 | trehalase_activity |
| GO:0015938 | 1 | coenzyme_A_catabolic_process |
| GO:0015957 | 1 | bis(5'-nucleosidyl)_oligophosphate_biosynthetic_process |
| GO:0015960 | 1 | diadenosine_polyphosphate_biosynthetic_process |
| GO:0015962 | 1 | diadenosine_triphosphate_metabolic_process |
| GO:0015964 | 1 | diadenosine_triphosphate_catabolic_process |
| GO:0015965 | 1 | diadenosine_tetraphosphate_metabolic_process |
| GO:0015966 | 1 | diadenosine_tetraphosphate_biosynthetic_process |
| GO:0015969 | 1 | guanosine_tetraphosphate_metabolic_process |
| GO:0016005 | 1 | phospholipase_A2_activator_activity |
| GO:0016011 | 1 | dystroglycan_complex |
| GO:0016028 | 1 | rhabdomere |
| GO:0016034 | 1 | maleylacetoacetate_isomerase_activity |
| GO:0016078 | 1 | tRNA_catabolic_process |
| GO:0016092 | 1 | prenol_catabolic_process |
| GO:0016095 | 1 | polyprenol_catabolic_process |
| GO:0016108 | 1 | tetraterpenoid_metabolic_process |
| GO:0016116 | 1 | carotenoid_metabolic_process |
| GO:0016119 | 1 | carotene_metabolic_process |
| GO:0016150 | 1 | translation_release_factor_activity,_codon_nonspecific |
| GO:0016153 | 1 | urocanate_hydratase_activity |
| GO:0016154 | 1 | pyrimidine-nucleoside_phosphorylase_activity |
| GO:0016189 | 1 | synaptic_vesicle_to_endosome_fusion |
| GO:0016191 | 1 | synaptic_vesicle_uncoating |
| GO:0016231 | 1 | beta-N-acetylglucosaminidase_activity |
| GO:0016232 | 1 | HNK-1_sulfotransferase_activity |
| GO:0016246 | 1 | RNA_interference |
| GO:0016250 | 1 | N-sulfoglucosamine_sulfohydrolase_activity |
| GO:0016259 | 1 | selenocysteine_metabolic_process |
| GO:0016260 | 1 | selenocysteine_biosynthetic_process |
| GO:0016263 | 1 | glycoprotein-N-acetylgalactosamine_3-beta-galactosyltransferase_activity |
| GO:0016269 | 1 | O-glycan_processing,_core_3 |
| GO:0016277 | 1 | [myelin_basic_protein]-arginine_N-methyltransferase_activity |
| GO:0016287 | 1 | glycerone-phosphate_O-acyltransferase_activity |
| GO:0016332 | 1 | establishment_or_maintenance_of_polarity_of_embryonic_epithelium |
| GO:0016344 | 1 | meiotic_chromosome_movement_towards_spindle_pole |
| GO:0016412 | 1 | serine_O-acyltransferase_activity |
| GO:0016414 | 1 | O-octanoyltransferase_activity |
| GO:0016427 | 1 | tRNA_(cytosine)_methyltransferase_activity |
| GO:0016428 | 1 | tRNA_(cytosine-5-)-methyltransferase_activity |
| GO:0016436 | 1 | rRNA_(uridine)_methyltransferase_activity |
| GO:0016495 | 1 | C-X3-C_chemokine_receptor_activity |
| GO:0016496 | 1 | substance_P_receptor_activity |
| GO:0016517 | 1 | interleukin-12_receptor_activity |
| GO:0016519 | 1 | gastric_inhibitory_peptide_receptor_activity |
| GO:0016521 | 1 | pituitary_adenylate_cyclase_activating_polypeptide_activity |
| GO:0016560 | 1 | protein_import_into_peroxisome_matrix,_docking |
| GO:0016590 | 1 | ACF_complex |
| GO:0016598 | 1 | protein_arginylation |
| GO:0016618 | 1 | hydroxypyruvate_reductase_activity |
| GO:0016642 | 1 | oxidoreductase_activity,_acting_on_the_CH-NH2_group_of_donors,_disulfide_as_acceptor |
| GO:0016711 | 1 | flavonoid_3'-monooxygenase_activity |
| GO:0016727 | 1 | oxidoreductase_activity,_acting_on_CH_or_CH2_groups,_oxygen_as_acceptor |
| GO:0016730 | 1 | oxidoreductase_activity,_acting_on_iron-sulfur_proteins_as_donors |
| GO:0016731 | 1 | oxidoreductase_activity,_acting_on_iron-sulfur_proteins_as_donors,_NAD_or_NADP_as_acceptor |
| GO:0016751 | 1 | S-succinyltransferase_activity |
| GO:0016768 | 1 | spermine_synthase_activity |
| GO:0016784 | 1 | 3-mercaptopyruvate_sulfurtransferase_activity |
| GO:0016785 | 1 | transferase_activity,_transferring_selenium-containing_groups |
| GO:0016794 | 1 | diphosphoric_monoester_hydrolase_activity |
| GO:0016815 | 1 | hydrolase_activity,_acting_on_carbon-nitrogen_(but_not_peptide)_bonds,_in_nitriles |
| GO:0016826 | 1 | hydrolase_activity,_acting_on_acid_sulfur-nitrogen_bonds |
| GO:0016872 | 1 | intramolecular_lyase_activity |
| GO:0016898 | 1 | oxidoreductase_activity,_acting_on_the_CH-OH_group_of_donors,_cytochrome_as_acceptor |
| GO:0016913 | 1 | follicle-stimulating_hormone_activity |
| GO:0016937 | 1 | short-branched-chain-acyl-CoA_dehydrogenase_activity |
| GO:0016938 | 1 | kinesin_I_complex |
| GO:0016971 | 1 | flavin-linked_sulfhydryl_oxidase_activity |
| GO:0016979 | 1 | lipoate-protein_ligase_activity |
| GO:0016992 | 1 | lipoate_synthase_activity |
| GO:0017005 | 1 | 3'-tyrosyl-DNA_phosphodiesterase_activity |
| GO:0017018 | 1 | myosin_phosphatase_activity |
| GO:0017020 | 1 | myosin_phosphatase_regulator_activity |
| GO:0017045 | 1 | corticotropin-releasing_hormone_activity |
| GO:0017061 | 1 | S-methyl-5-thioadenosine_phosphorylase_activity |
| GO:0017064 | 1 | fatty_acid_amide_hydrolase_activity |
| GO:0017065 | 1 | single-strand_selective_uracil_DNA_N-glycosylase_activity |
| GO:0017082 | 1 | mineralocorticoid_receptor_activity |
| GO:0017090 | 1 | meprin_A_complex |
| GO:0017096 | 1 | acetylserotonin_O-methyltransferase_activity |
| GO:0017113 | 1 | dihydropyrimidine_dehydrogenase_(NADP+)_activity |
| GO:0017125 | 1 | deoxycytidyl_transferase_activity |
| GO:0017126 | 1 | nucleologenesis |
| GO:0017130 | 1 | poly(C)_RNA_binding |
| GO:0017168 | 1 | 5-oxoprolinase_(ATP-hydrolyzing)_activity |
| GO:0017172 | 1 | cysteine_dioxygenase_activity |
| GO:0017174 | 1 | glycine_N-methyltransferase_activity |
| GO:0017185 | 1 | peptidyl-lysine_hydroxylation |
| GO:0017188 | 1 | aspartate_N-acetyltransferase_activity |
| GO:0017196 | 1 | N-terminal_peptidyl-methionine_acetylation |
| GO:0018009 | 1 | N-terminal_peptidyl-L-cysteine_N-palmitoylation |
| GO:0018011 | 1 | N-terminal_peptidyl-alanine_methylation |
| GO:0018012 | 1 | N-terminal_peptidyl-alanine_trimethylation |
| GO:0018016 | 1 | N-terminal_peptidyl-proline_dimethylation |
| GO:0018025 | 1 | calmodulin-lysine_N-methyltransferase_activity |
| GO:0018103 | 1 | protein_C-linked_glycosylation |
| GO:0018112 | 1 | proline_racemase_activity |
| GO:0018114 | 1 | threonine_racemase_activity |
| GO:0018117 | 1 | protein_adenylylation |
| GO:0018146 | 1 | keratan_sulfate_biosynthetic_process |
| GO:0018160 | 1 | peptidyl-pyrromethane_cofactor_linkage |
| GO:0018169 | 1 | ribosomal_S6-glutamic_acid_ligase_activity |
| GO:0018175 | 1 | protein_nucleotidylation |
| GO:0018184 | 1 | protein_polyamination |
| GO:0018190 | 1 | protein_octanoylation |
| GO:0018191 | 1 | peptidyl-serine_octanoylation |
| GO:0018192 | 1 | enzyme_active_site_formation_via_L-cysteine_persulfide |
| GO:0018194 | 1 | peptidyl-alanine_modification |
| GO:0018211 | 1 | peptidyl-tryptophan_modification |
| GO:0018271 | 1 | biotin-protein_ligase_activity |
| GO:0018272 | 1 | protein-pyridoxal-5-phosphate_linkage_via_peptidyl-N6-pyridoxal_phosphate-L-lysine |
| GO:0018293 | 1 | protein-FAD_linkage |
| GO:0018307 | 1 | enzyme_active_site_formation |
| GO:0018317 | 1 | protein_C-linked_glycosylation_via_tryptophan |
| GO:0018350 | 1 | protein_esterification |
| GO:0018364 | 1 | peptidyl-glutamine_methylation |
| GO:0018395 | 1 | peptidyl-lysine_hydroxylation_to_5-hydroxy-L-lysine |
| GO:0018406 | 1 | protein_C-linked_glycosylation_via_2'-alpha-mannosyl-L-tryptophan |
| GO:0018454 | 1 | acetoacetyl-CoA_reductase_activity |
| GO:0018455 | 1 | alcohol_dehydrogenase_[NAD(P)+]_activity |
| GO:0018467 | 1 | formaldehyde_dehydrogenase_activity |
| GO:0018478 | 1 | malonate-semialdehyde_dehydrogenase_(acetylating)_activity |
| GO:0018585 | 1 | fluorene_oxygenase_activity |
| GO:0018738 | 1 | S-formylglutathione_hydrolase_activity |
| GO:0018773 | 1 | acetylpyruvate_hydrolase_activity |
| GO:0018872 | 1 | arsonoacetate_metabolic_process |
| GO:0018916 | 1 | nitrobenzene_metabolic_process |
| GO:0018917 | 1 | fluorene_metabolic_process |
| GO:0018924 | 1 | mandelate_metabolic_process |
| GO:0018931 | 1 | naphthalene_metabolic_process |
| GO:0019035 | 1 | viral_integration_complex |
| GO:0019056 | 1 | modulation_by_virus_of_host_transcription |
| GO:0019061 | 1 | uncoating_of_virus |
| GO:0019066 | 1 | translocation_of_virus_into_host_cell |
| GO:0019088 | 1 | immortalization_of_host_cell_by_virus |
| GO:0019089 | 1 | transmission_of_virus |
| GO:0019101 | 1 | female_somatic_sex_determination |
| GO:0019102 | 1 | male_somatic_sex_determination |
| GO:0019115 | 1 | benzaldehyde_dehydrogenase_activity |
| GO:0019118 | 1 | phenanthrene-epoxide_hydrolase_activity |
| GO:0019119 | 1 | phenanthrene-9,10-epoxide_hydrolase_activity |
| GO:0019135 | 1 | deoxyhypusine_monooxygenase_activity |
| GO:0019145 | 1 | aminobutyraldehyde_dehydrogenase_activity |
| GO:0019153 | 1 | protein-disulfide_reductase_(glutathione)_activity |
| GO:0019171 | 1 | 3-hydroxyacyl-[acyl-carrier-protein]_dehydratase_activity |
| GO:0019184 | 1 | nonribosomal_peptide_biosynthetic_process |
| GO:0019185 | 1 | snRNA-activating_protein_complex |
| GO:0019187 | 1 | beta-1,4-mannosyltransferase_activity |
| GO:0019202 | 1 | amino_acid_kinase_activity |
| GO:0019244 | 1 | lactate_biosynthetic_process_from_pyruvate |
| GO:0019249 | 1 | lactate_biosynthetic_process |
| GO:0019262 | 1 | N-acetylneuraminate_catabolic_process |
| GO:0019290 | 1 | siderophore_biosynthetic_process |
| GO:0019310 | 1 | inositol_catabolic_process |
| GO:0019336 | 1 | phenol-containing_compound_catabolic_process |
| GO:0019341 | 1 | dibenzo-p-dioxin_catabolic_process |
| GO:0019343 | 1 | cysteine_biosynthetic_process_via_cystathionine |
| GO:0019357 | 1 | nicotinate_nucleotide_biosynthetic_process |
| GO:0019358 | 1 | nicotinate_nucleotide_salvage |
| GO:0019365 | 1 | pyridine_nucleotide_salvage |
| GO:0019389 | 1 | glucuronoside_metabolic_process |
| GO:0019391 | 1 | glucuronoside_catabolic_process |
| GO:0019402 | 1 | galactitol_metabolic_process |
| GO:0019407 | 1 | hexitol_catabolic_process |
| GO:0019413 | 1 | acetate_biosynthetic_process |
| GO:0019428 | 1 | allantoin_biosynthetic_process |
| GO:0019464 | 1 | glycine_decarboxylation_via_glycine_cleavage_system |
| GO:0019481 | 1 | L-alanine_catabolic_process,_by_transamination |
| GO:0019484 | 1 | beta-alanine_catabolic_process |
| GO:0019518 | 1 | L-threonine_catabolic_process_to_glycine |
| GO:0019519 | 1 | pentitol_metabolic_process |
| GO:0019520 | 1 | aldonic_acid_metabolic_process |
| GO:0019521 | 1 | D-gluconate_metabolic_process |
| GO:0019527 | 1 | pentitol_catabolic_process |
| GO:0019540 | 1 | siderophore_biosynthetic_process_from_catechol |
| GO:0019542 | 1 | propionate_biosynthetic_process |
| GO:0019543 | 1 | propionate_catabolic_process |
| GO:0019628 | 1 | urate_catabolic_process |
| GO:0019646 | 1 | aerobic_electron_transport_chain |
| GO:0019677 | 1 | NAD_catabolic_process |
| GO:0019719 | 1 | smooth_microsome |
| GO:0019731 | 1 | antibacterial_humoral_response |
| GO:0019770 | 1 | IgG_receptor_activity |
| GO:0019776 | 1 | Atg8_ligase_activity |
| GO:0019778 | 1 | APG12_activating_enzyme_activity |
| GO:0019780 | 1 | FAT10_activating_enzyme_activity |
| GO:0019782 | 1 | ISG15_activating_enzyme_activity |
| GO:0019799 | 1 | tubulin_N-acetyltransferase_activity |
| GO:0019807 | 1 | aspartoacylase_activity |
| GO:0019808 | 1 | polyamine_binding |
| GO:0019809 | 1 | spermidine_binding |
| GO:0019811 | 1 | cocaine_binding |
| GO:0019912 | 1 | cyclin-dependent_protein_kinase_activating_kinase_activity |
| GO:0019962 | 1 | type_I_interferon_binding |
| GO:0019964 | 1 | interferon-gamma_binding |
| GO:0019970 | 1 | interleukin-11_binding |
| GO:0020005 | 1 | symbiont-containing_vacuole_membrane |
| GO:0020012 | 1 | evasion_or_tolerance_of_host_immune_response |
| GO:0020021 | 1 | immortalization_of_host_cell |
| GO:0021501 | 1 | prechordal_plate_formation |
| GO:0021502 | 1 | neural_fold_elevation_formation |
| GO:0021509 | 1 | roof_plate_formation |
| GO:0021511 | 1 | spinal_cord_patterning |
| GO:0021526 | 1 | medial_motor_column_neuron_differentiation |
| GO:0021528 | 1 | commissural_neuron_differentiation_in_spinal_cord |
| GO:0021534 | 1 | cell_proliferation_in_hindbrain |
| GO:0021536 | 1 | diencephalon_development |
| GO:0021539 | 1 | subthalamus_development |
| GO:0021540 | 1 | corpus_callosum_morphogenesis |
| GO:0021541 | 1 | ammon_gyrus_development |
| GO:0021547 | 1 | midbrain-hindbrain_boundary_initiation |
| GO:0021548 | 1 | pons_development |
| GO:0021561 | 1 | facial_nerve_development |
| GO:0021572 | 1 | rhombomere_6_development |
| GO:0021586 | 1 | pons_maturation |
| GO:0021588 | 1 | cerebellum_formation |
| GO:0021589 | 1 | cerebellum_structural_organization |
| GO:0021590 | 1 | cerebellum_maturation |
| GO:0021591 | 1 | ventricular_system_development |
| GO:0021593 | 1 | rhombomere_morphogenesis |
| GO:0021594 | 1 | rhombomere_formation |
| GO:0021599 | 1 | abducens_nerve_formation |
| GO:0021623 | 1 | oculomotor_nerve_formation |
| GO:0021629 | 1 | olfactory_nerve_structural_organization |
| GO:0021633 | 1 | optic_nerve_structural_organization |
| GO:0021650 | 1 | vestibulocochlear_nerve_formation |
| GO:0021658 | 1 | rhombomere_3_morphogenesis |
| GO:0021660 | 1 | rhombomere_3_formation |
| GO:0021666 | 1 | rhombomere_5_formation |
| GO:0021679 | 1 | cerebellar_molecular_layer_development |
| GO:0021682 | 1 | nerve_maturation |
| GO:0021683 | 1 | cerebellar_granular_layer_morphogenesis |
| GO:0021697 | 1 | cerebellar_cortex_formation |
| GO:0021703 | 1 | locus_ceruleus_development |
| GO:0021740 | 1 | principal_sensory_nucleus_of_trigeminal_nerve_development |
| GO:0021762 | 1 | substantia_nigra_development |
| GO:0021764 | 1 | amygdala_development |
| GO:0021767 | 1 | mammillary_body_development |
| GO:0021768 | 1 | nucleus_accumbens_development |
| GO:0021769 | 1 | orbitofrontal_cortex_development |
| GO:0021771 | 1 | lateral_geniculate_nucleus_development |
| GO:0021779 | 1 | oligodendrocyte_cell_fate_commitment |
| GO:0021785 | 1 | branchiomotor_neuron_axon_guidance |
| GO:0021793 | 1 | chemorepulsion_of_branchiomotor_axon |
| GO:0021831 | 1 | embryonic_olfactory_bulb_interneuron_precursor_migration |
| GO:0021837 | 1 | motogenic_signaling_involved_in_postnatal_olfactory_bulb_interneuron_migration |
| GO:0021855 | 1 | hypothalamus_cell_migration |
| GO:0021858 | 1 | GABAergic_neuron_differentiation_in_basal_ganglia |
| GO:0021870 | 1 | Cajal-Retzius_cell_differentiation |
| GO:0021872 | 1 | forebrain_generation_of_neurons |
| GO:0021897 | 1 | forebrain_astrocyte_development |
| GO:0021902 | 1 | commitment_of_neuronal_cell_to_specific_neuron_type_in_forebrain |
| GO:0021905 | 1 | forebrain-midbrain_boundary_formation |
| GO:0021914 | 1 | negative_regulation_of_smoothened_signaling_pathway_involved_in_ventral_spinal_cord_patterning |
| GO:0021918 | 1 | regulation_of_transcription_from_RNA_polymerase_II_promoter_involved_in_somatic_motor_neuron_fate_commitment |
| GO:0021919 | 1 | BMP_signaling_pathway_involved_in_spinal_cord_dorsal/ventral_patterning |
| GO:0021920 | 1 | regulation_of_transcription_from_RNA_polymerase_II_promoter_involved_in_spinal_cord_association_neuron_specification |
| GO:0021924 | 1 | cell_proliferation_in_external_granule_layer |
| GO:0021930 | 1 | cerebellar_granule_cell_precursor_proliferation |
| GO:0021933 | 1 | radial_glia_guided_migration_of_cerebellar_granule_cell |
| GO:0021935 | 1 | cerebellar_granule_cell_precursor_tangential_migration |
| GO:0021941 | 1 | negative_regulation_of_cerebellar_granule_cell_precursor_proliferation |
| GO:0021942 | 1 | radial_glia_guided_migration_of_Purkinje_cell |
| GO:0021957 | 1 | corticospinal_tract_morphogenesis |
| GO:0021972 | 1 | corticospinal_neuron_axon_guidance_through_spinal_cord |
| GO:0021988 | 1 | olfactory_lobe_development |
| GO:0021990 | 1 | neural_plate_formation |
| GO:0021993 | 1 | initiation_of_neural_tube_closure |
| GO:0021997 | 1 | neural_plate_axis_specification |
| GO:0021998 | 1 | neural_plate_mediolateral_regionalization |
| GO:0022001 | 1 | negative_regulation_of_anterior_neural_cell_fate_commitment_of_the_neural_plate |
| GO:0022002 | 1 | negative_regulation_of_anterior_neural_cell_fate_commitment_of_the_neural_plate_by_Wnt_receptor_signaling_pathway |
| GO:0022004 | 1 | midbrain-hindbrain_boundary_maturation_during_brain_development |
| GO:0022012 | 1 | subpallium_cell_proliferation_in_forebrain |
| GO:0022018 | 1 | lateral_ganglionic_eminence_cell_proliferation |
| GO:0022037 | 1 | metencephalon_development |
| GO:0022400 | 1 | regulation_of_rhodopsin_mediated_signaling_pathway |
| GO:0022605 | 1 | oogenesis_stage |
| GO:0022840 | 1 | leak_channel_activity |
| GO:0022842 | 1 | narrow_pore_channel_activity |
| GO:0022851 | 1 | GABA-gated_chloride_ion_channel_activity |
| GO:0023029 | 1 | MHC_class_Ib_protein_binding |
| GO:0030007 | 1 | cellular_potassium_ion_homeostasis |
| GO:0030026 | 1 | cellular_manganese_ion_homeostasis |
| GO:0030037 | 1 | actin_filament_reorganization_involved_in_cell_cycle |
| GO:0030038 | 1 | contractile_actin_filament_bundle_assembly |
| GO:0030046 | 1 | parallel_actin_filament_bundle_assembly |
| GO:0030047 | 1 | actin_modification |
| GO:0030092 | 1 | regulation_of_flagellum_assembly |
| GO:0030103 | 1 | vasopressin_secretion |
| GO:0030107 | 1 | HLA-A_specific_inhibitory_MHC_class_I_receptor_activity |
| GO:0030108 | 1 | HLA-A_specific_activating_MHC_class_I_receptor_activity |
| GO:0030110 | 1 | HLA-C_specific_inhibitory_MHC_class_I_receptor_activity |
| GO:0030128 | 1 | clathrin_coat_of_endocytic_vesicle |
| GO:0030184 | 1 | nitric_oxide_transmembrane_transporter_activity |
| GO:0030226 | 1 | apolipoprotein_receptor_activity |
| GO:0030233 | 1 | deoxynucleotide_transmembrane_transporter_activity |
| GO:0030251 | 1 | guanylate_cyclase_inhibitor_activity |
| GO:0030267 | 1 | glyoxylate_reductase_(NADP)_activity |
| GO:0030273 | 1 | melanin-concentrating_hormone_receptor_activity |
| GO:0030290 | 1 | sphingolipid_activator_protein_activity |
| GO:0030298 | 1 | receptor_signaling_protein_tyrosine_kinase_activator_activity |
| GO:0030302 | 1 | deoxynucleotide_transport |
| GO:0030322 | 1 | stabilization_of_membrane_potential |
| GO:0030338 | 1 | CMP-N-acetylneuraminate_monooxygenase_activity |
| GO:0030342 | 1 | 1-alpha,25-dihydroxyvitamin_D3_24-hydroxylase_activity |
| GO:0030348 | 1 | syntaxin-3_binding |
| GO:0030351 | 1 | inositol-1,3,4,5,6-pentakisphosphate_3-phosphatase_activity |
| GO:0030352 | 1 | inositol-1,4,5,6-tetrakisphosphate_6-phosphatase_activity |
| GO:0030366 | 1 | Mo-molybdopterin_synthase_activity |
| GO:0030369 | 1 | ICAM-3_receptor_activity |
| GO:0030372 | 1 | high_molecular_weight_B_cell_growth_factor_receptor_binding |
| GO:0030377 | 1 | U-plasminogen_activator_receptor_activity |
| GO:0030378 | 1 | serine_racemase_activity |
| GO:0030379 | 1 | neurotensin_receptor_activity,_non-G-protein_coupled |
| GO:0030380 | 1 | interleukin-17E_receptor_binding |
| GO:0030387 | 1 | fructosamine-3-kinase_activity |
| GO:0030393 | 1 | fructoselysine_metabolic_process |
| GO:0030407 | 1 | formimidoyltransferase_activity |
| GO:0030409 | 1 | glutamate_formimidoyltransferase_activity |
| GO:0030412 | 1 | formimidoyltetrahydrofolate_cyclodeaminase_activity |
| GO:0030421 | 1 | defecation |
| GO:0030429 | 1 | kynureninase_activity |
| GO:0030450 | 1 | regulation_of_complement_activation,_classical_pathway |
| GO:0030451 | 1 | regulation_of_complement_activation,_alternative_pathway |
| GO:0030487 | 1 | inositol-4,5-bisphosphate_5-phosphatase_activity |
| GO:0030504 | 1 | inorganic_diphosphate_transmembrane_transporter_activity |
| GO:0030523 | 1 | dihydrolipoamide_S-acyltransferase_activity |
| GO:0030526 | 1 | granulocyte_macrophage_colony-stimulating_factor_receptor_complex |
| GO:0030549 | 1 | acetylcholine_receptor_activator_activity |
| GO:0030572 | 1 | phosphatidyltransferase_activity |
| GO:0030580 | 1 | quinone_cofactor_methyltransferase_activity |
| GO:0030586 | 1 | [methionine_synthase]_reductase_activity |
| GO:0030612 | 1 | arsenate_reductase_(thioredoxin)_activity |
| GO:0030616 | 1 | transforming_growth_factor_beta_receptor,_common-partner_cytoplasmic_mediator_activity |
| GO:0030619 | 1 | U1_snRNA_binding |
| GO:0030621 | 1 | U4_snRNA_binding |
| GO:0030682 | 1 | evasion_or_tolerance_of_host_defense_response |
| GO:0030683 | 1 | evasion_by_virus_of_host_immune_response |
| GO:0030690 | 1 | Noc1p-Noc2p_complex |
| GO:0030692 | 1 | Noc4p-Nop14p_complex |
| GO:0030729 | 1 | acetoacetate-CoA_ligase_activity |
| GO:0030731 | 1 | guanidinoacetate_N-methyltransferase_activity |
| GO:0030748 | 1 | amine_N-methyltransferase_activity |
| GO:0030791 | 1 | arsenite_methyltransferase_activity |
| GO:0030792 | 1 | methylarsonite_methyltransferase_activity |
| GO:0030813 | 1 | positive_regulation_of_nucleotide_catabolic_process |
| GO:0030824 | 1 | negative_regulation_of_cGMP_metabolic_process |
| GO:0030827 | 1 | negative_regulation_of_cGMP_biosynthetic_process |
| GO:0030842 | 1 | regulation_of_intermediate_filament_depolymerization |
| GO:0030844 | 1 | positive_regulation_of_intermediate_filament_depolymerization |
| GO:0030845 | 1 | phospholipase_C-inhibiting_G-protein_coupled_receptor_signaling_pathway |
| GO:0030874 | 1 | nucleolar_chromatin |
| GO:0030883 | 1 | endogenous_lipid_antigen_binding |
| GO:0030886 | 1 | negative_regulation_of_myeloid_dendritic_cell_activation |
| GO:0030887 | 1 | positive_regulation_of_myeloid_dendritic_cell_activation |
| GO:0030936 | 1 | transmembrane_collagen |
| GO:0030950 | 1 | establishment_or_maintenance_of_actin_cytoskeleton_polarity |
| GO:0030990 | 1 | intraflagellar_transport_particle |
| GO:0031021 | 1 | interphase_microtubule_organizing_center |
| GO:0031052 | 1 | chromosome_breakage |
| GO:0031071 | 1 | cysteine_desulfurase_activity |
| GO:0031073 | 1 | cholesterol_26-hydroxylase_activity |
| GO:0031081 | 1 | nuclear_pore_distribution |
| GO:0031087 | 1 | deadenylation-independent_decapping_of_nuclear-transcribed_mRNA |
| GO:0031118 | 1 | rRNA_pseudouridine_synthesis |
| GO:0031129 | 1 | inductive_cell-cell_signaling |
| GO:0031161 | 1 | phosphatidylinositol_catabolic_process |
| GO:0031167 | 1 | rRNA_methylation |
| GO:0031179 | 1 | peptide_modification |
| GO:0031208 | 1 | POZ_domain_binding |
| GO:0031209 | 1 | SCAR_complex |
| GO:0031240 | 1 | external_side_of_cell_outer_membrane |
| GO:0031257 | 1 | trailing_edge_membrane |
| GO:0031259 | 1 | uropod_membrane |
| GO:0031260 | 1 | pseudopodium_membrane |
| GO:0031275 | 1 | regulation_of_lateral_pseudopodium_assembly |
| GO:0031283 | 1 | negative_regulation_of_guanylate_cyclase_activity |
| GO:0031302 | 1 | intrinsic_to_endosome_membrane |
| GO:0031308 | 1 | intrinsic_to_nuclear_outer_membrane |
| GO:0031309 | 1 | integral_to_nuclear_outer_membrane |
| GO:0031337 | 1 | positive_regulation_of_sulfur_amino_acid_metabolic_process |
| GO:0031379 | 1 | RNA-directed_RNA_polymerase_complex |
| GO:0031380 | 1 | nuclear_RNA-directed_RNA_polymerase_complex |
| GO:0031393 | 1 | negative_regulation_of_prostaglandin_biosynthetic_process |
| GO:0031403 | 1 | lithium_ion_binding |
| GO:0031429 | 1 | box_H/ACA_snoRNP_complex |
| GO:0031433 | 1 | telethonin_binding |
| GO:0031441 | 1 | negative_regulation_of_mRNA_3'-end_processing |
| GO:0031468 | 1 | nuclear_envelope_reassembly |
| GO:0031493 | 1 | nucleosomal_histone_binding |
| GO:0031531 | 1 | thyrotropin-releasing_hormone_receptor_binding |
| GO:0031547 | 1 | brain-derived_neurotrophic_factor_receptor_signaling_pathway |
| GO:0031559 | 1 | oxidosqualene_cyclase_activity |
| GO:0031583 | 1 | phospholipase_D-activating_G-protein_coupled_receptor_signaling_pathway |
| GO:0031587 | 1 | positive_regulation_of_inositol_1,4,5-trisphosphate-sensitive_calcium-release_channel_activity |
| GO:0031590 | 1 | wybutosine_metabolic_process |
| GO:0031591 | 1 | wybutosine_biosynthetic_process |
| GO:0031592 | 1 | centrosomal_corona |
| GO:0031595 | 1 | nuclear_proteasome_complex |
| GO:0031619 | 1 | homologous_chromosome_orientation_involved_in_meiotic_metaphase_I_plate_congression |
| GO:0031626 | 1 | beta-endorphin_binding |
| GO:0031629 | 1 | synaptic_vesicle_fusion_to_presynaptic_membrane |
| GO:0031658 | 1 | negative_regulation_of_cyclin-dependent_protein_kinase_activity_involved_in_G1/S |
| GO:0031666 | 1 | positive_regulation_of_lipopolysaccharide-mediated_signaling_pathway |
| GO:0031680 | 1 | G-protein_beta/gamma-subunit_complex |
| GO:0031687 | 1 | A2A_adenosine_receptor_binding |
| GO:0031692 | 1 | alpha-1B_adrenergic_receptor_binding |
| GO:0031695 | 1 | alpha-2B_adrenergic_receptor_binding |
| GO:0031697 | 1 | beta-1_adrenergic_receptor_binding |
| GO:0031699 | 1 | beta-3_adrenergic_receptor_binding |
| GO:0031700 | 1 | adrenomedullin_receptor_binding |
| GO:0031703 | 1 | type_2_angiotensin_receptor_binding |
| GO:0031705 | 1 | bombesin_receptor_binding |
| GO:0031707 | 1 | endothelin_A_receptor_binding |
| GO:0031708 | 1 | endothelin_B_receptor_binding |
| GO:0031723 | 1 | CXCR4_chemokine_receptor_binding |
| GO:0031724 | 1 | CXCR5_chemokine_receptor_binding |
| GO:0031729 | 1 | CCR4_chemokine_receptor_binding |
| GO:0031739 | 1 | cholecystokinin_receptor_binding |
| GO:0031741 | 1 | type_B_gastrin/cholecystokinin_receptor_binding |
| GO:0031748 | 1 | D1_dopamine_receptor_binding |
| GO:0031749 | 1 | D2_dopamine_receptor_binding |
| GO:0031750 | 1 | D3_dopamine_receptor_binding |
| GO:0031768 | 1 | ghrelin_receptor_binding |
| GO:0031769 | 1 | glucagon_receptor_binding |
| GO:0031770 | 1 | growth_hormone-releasing_hormone_receptor_binding |
| GO:0031771 | 1 | type_1_hypocretin_receptor_binding |
| GO:0031772 | 1 | type_2_hypocretin_receptor_binding |
| GO:0031775 | 1 | lutropin-choriogonadotropic_hormone_receptor_binding |
| GO:0031776 | 1 | melanin-concentrating_hormone_receptor_binding |
| GO:0031777 | 1 | type_1_melanin-concentrating_hormone_receptor_binding |
| GO:0031802 | 1 | type_5_metabotropic_glutamate_receptor_binding |
| GO:0031811 | 1 | G-protein_coupled_nucleotide_receptor_binding |
| GO:0031812 | 1 | P2Y1_nucleotide_receptor_binding |
| GO:0031834 | 1 | neurokinin_receptor_binding |
| GO:0031835 | 1 | substance_P_receptor_binding |
| GO:0031841 | 1 | neuropeptide_Y_receptor_binding |
| GO:0031851 | 1 | kappa-type_opioid_receptor_binding |
| GO:0031854 | 1 | orexigenic_neuropeptide_QRFP_receptor_binding |
| GO:0031855 | 1 | oxytocin_receptor_binding |
| GO:0031856 | 1 | parathyroid_hormone_receptor_binding |
| GO:0031860 | 1 | telomeric_3'_overhang_formation |
| GO:0031861 | 1 | prolactin-releasing_peptide_receptor_binding |
| GO:0031867 | 1 | EP4_subtype_prostaglandin_E2_receptor_binding |
| GO:0031870 | 1 | thromboxane_A2_receptor_binding |
| GO:0031871 | 1 | proteinase_activated_receptor_binding |
| GO:0031877 | 1 | somatostatin_receptor_binding |
| GO:0031895 | 1 | V1B_vasopressin_receptor_binding |
| GO:0031945 | 1 | positive_regulation_of_glucocorticoid_metabolic_process |
| GO:0031948 | 1 | positive_regulation_of_glucocorticoid_biosynthetic_process |
| GO:0031959 | 1 | mineralocorticoid_receptor_signaling_pathway |
| GO:0031997 | 1 | N-terminal_myristoylation_domain_binding |
| GO:0032003 | 1 | interleukin-28_receptor_binding |
| GO:0032011 | 1 | ARF_protein_signal_transduction |
| GO:0032023 | 1 | trypsinogen_activation |
| GO:0032028 | 1 | myosin_head/neck_binding |
| GO:0032050 | 1 | clathrin_heavy_chain_binding |
| GO:0032051 | 1 | clathrin_light_chain_binding |
| GO:0032076 | 1 | negative_regulation_of_deoxyribonuclease_activity |
| GO:0032077 | 1 | positive_regulation_of_deoxyribonuclease_activity |
| GO:0032078 | 1 | negative_regulation_of_endodeoxyribonuclease_activity |
| GO:0032093 | 1 | SAM_domain_binding |
| GO:0032113 | 1 | regulation_of_carbohydrate_phosphatase_activity |
| GO:0032127 | 1 | dense_core_granule_membrane |
| GO:0032144 | 1 | 4-aminobutyrate_transaminase_complex |
| GO:0032145 | 1 | succinate-semialdehyde_dehydrogenase_binding |
| GO:0032184 | 1 | SUMO_polymer_binding |
| GO:0032185 | 1 | septin_cytoskeleton_organization |
| GO:0032223 | 1 | negative_regulation_of_synaptic_transmission,_cholinergic |
| GO:0032242 | 1 | regulation_of_nucleoside_transport |
| GO:0032244 | 1 | positive_regulation_of_nucleoside_transport |
| GO:0032280 | 1 | symmetric_synapse |
| GO:0032286 | 1 | central_nervous_system_myelin_maintenance |
| GO:0032290 | 1 | peripheral_nervous_system_myelin_formation |
| GO:0032316 | 1 | regulation_of_Ran_GTPase_activity |
| GO:0032377 | 1 | regulation_of_intracellular_lipid_transport |
| GO:0032380 | 1 | regulation_of_intracellular_sterol_transport |
| GO:0032383 | 1 | regulation_of_intracellular_cholesterol_transport |
| GO:0032394 | 1 | MHC_class_Ib_receptor_activity |
| GO:0032397 | 1 | activating_MHC_class_I_receptor_activity |
| GO:0032399 | 1 | HECT_domain_binding |
| GO:0032406 | 1 | MutLbeta_complex_binding |
| GO:0032408 | 1 | MutSbeta_complex_binding |
| GO:0032425 | 1 | positive_regulation_of_mismatch_repair |
| GO:0032427 | 1 | GBD_domain_binding |
| GO:0032439 | 1 | endosome_localization |
| GO:0032440 | 1 | 2-alkenal_reductase_[NAD(P)]_activity |
| GO:0032448 | 1 | DNA_hairpin_binding |
| GO:0032466 | 1 | negative_regulation_of_cytokinesis |
| GO:0032468 | 1 | Golgi_calcium_ion_homeostasis |
| GO:0032470 | 1 | elevation_of_endoplasmic_reticulum_calcium_ion_concentration |
| GO:0032472 | 1 | Golgi_calcium_ion_transport |
| GO:0032473 | 1 | external_side_of_mitochondrial_outer_membrane |
| GO:0032474 | 1 | otolith_morphogenesis |
| GO:0032483 | 1 | regulation_of_Rab_protein_signal_transduction |
| GO:0032498 | 1 | detection_of_muramyl_dipeptide |
| GO:0032499 | 1 | detection_of_peptidoglycan |
| GO:0032500 | 1 | muramyl_dipeptide_binding |
| GO:0032514 | 1 | positive_regulation_of_protein_phosphatase_type_2B_activity |
| GO:0032532 | 1 | regulation_of_microvillus_length |
| GO:0032541 | 1 | cortical_endoplasmic_reticulum |
| GO:0032542 | 1 | sulfiredoxin_activity |
| GO:0032546 | 1 | deoxyribonucleoside_binding |
| GO:0032548 | 1 | pyrimidine_deoxyribonucleoside_binding |
| GO:0032556 | 1 | pyrimidine_deoxyribonucleotide_binding |
| GO:0032574 | 1 | 5'-3'_RNA_helicase_activity |
| GO:0032575 | 1 | ATP-dependent_5'-3'_RNA_helicase_activity |
| GO:0032579 | 1 | apical_lamina_of_hyaline_layer |
| GO:0032581 | 1 | ER-dependent_peroxisome_organization |
| GO:0032595 | 1 | B_cell_receptor_transport_within_lipid_bilayer |
| GO:0032596 | 1 | protein_transport_into_membrane_raft |
| GO:0032597 | 1 | B_cell_receptor_transport_into_membrane_raft |
| GO:0032599 | 1 | protein_transport_out_of_membrane_raft |
| GO:0032600 | 1 | chemokine_receptor_transport_out_of_membrane_raft |
| GO:0032607 | 1 | interferon-alpha_production |
| GO:0032613 | 1 | interleukin-10_production |
| GO:0032615 | 1 | interleukin-12_production |
| GO:0032621 | 1 | interleukin-18_production |
| GO:0032623 | 1 | interleukin-2_production |
| GO:0032632 | 1 | interleukin-3_production |
| GO:0032637 | 1 | interleukin-8_production |
| GO:0032644 | 1 | regulation_of_fractalkine_production |
| GO:0032646 | 1 | regulation_of_hepatocyte_growth_factor_production |
| GO:0032658 | 1 | regulation_of_interleukin-15_production |
| GO:0032679 | 1 | regulation_of_TRAIL_production |
| GO:0032681 | 1 | regulation_of_lymphotoxin_A_production |
| GO:0032688 | 1 | negative_regulation_of_interferon-beta_production |
| GO:0032701 | 1 | negative_regulation_of_interleukin-18_production |
| GO:0032738 | 1 | positive_regulation_of_interleukin-15_production |
| GO:0032762 | 1 | mast_cell_cytokine_production |
| GO:0032782 | 1 | bile_acid_secretion |
| GO:0032788 | 1 | saturated_monocarboxylic_acid_metabolic_process |
| GO:0032789 | 1 | unsaturated_monocarboxylic_acid_metabolic_process |
| GO:0032791 | 1 | lead_ion_binding |
| GO:0032802 | 1 | low-density_lipoprotein_particle_receptor_catabolic_process |
| GO:0032804 | 1 | negative_regulation_of_low-density_lipoprotein_particle_receptor_catabolic_process |
| GO:0032805 | 1 | positive_regulation_of_low-density_lipoprotein_particle_receptor_catabolic_process |
| GO:0032812 | 1 | positive_regulation_of_epinephrine_secretion |
| GO:0032848 | 1 | negative_regulation_of_cellular_pH_reduction |
| GO:0032853 | 1 | positive_regulation_of_Ran_GTPase_activity |
| GO:0032903 | 1 | regulation_of_nerve_growth_factor_production |
| GO:0032904 | 1 | negative_regulation_of_nerve_growth_factor_production |
| GO:0032912 | 1 | negative_regulation_of_transforming_growth_factor_beta2_production |
| GO:0032913 | 1 | negative_regulation_of_transforming_growth_factor_beta3_production |
| GO:0032938 | 1 | negative_regulation_of_translation_in_response_to_oxidative_stress |
| GO:0032972 | 1 | regulation_of_muscle_filament_sliding_speed |
| GO:0032976 | 1 | release_of_matrix_enzymes_from_mitochondria |
| GO:0032980 | 1 | keratinocyte_activation |
| GO:0032996 | 1 | Bcl3-Bcl10_complex |
| GO:0033011 | 1 | perinuclear_theca |
| GO:0033029 | 1 | regulation_of_neutrophil_apoptotic_process |
| GO:0033031 | 1 | positive_regulation_of_neutrophil_apoptotic_process |
| GO:0033037 | 1 | polysaccharide_localization |
| GO:0033079 | 1 | immature_T_cell_proliferation |
| GO:0033080 | 1 | immature_T_cell_proliferation_in_thymus |
| GO:0033123 | 1 | positive_regulation_of_purine_nucleotide_catabolic_process |
| GO:0033126 | 1 | positive_regulation_of_GTP_catabolic_process |
| GO:0033142 | 1 | progesterone_receptor_binding |
| GO:0033165 | 1 | interphotoreceptor_matrix |
| GO:0033173 | 1 | calcineurin-NFAT_signaling_cascade |
| GO:0033182 | 1 | regulation_of_histone_ubiquitination |
| GO:0033193 | 1 | Lsd1/2_complex |
| GO:0033195 | 1 | response_to_alkyl_hydroperoxide |
| GO:0033218 | 1 | amide_binding |
| GO:0033234 | 1 | negative_regulation_of_protein_sumoylation |
| GO:0033274 | 1 | response_to_vitamin_B2 |
| GO:0033315 | 1 | meiotic_cell_cycle_DNA_replication_checkpoint |
| GO:0033319 | 1 | UDP-D-xylose_metabolic_process |
| GO:0033320 | 1 | UDP-D-xylose_biosynthetic_process |
| GO:0033328 | 1 | peroxisome_membrane_targeting_sequence_binding |
| GO:0033341 | 1 | regulation_of_collagen_binding |
| GO:0033342 | 1 | negative_regulation_of_collagen_binding |
| GO:0033345 | 1 | asparagine_catabolic_process_via_L-aspartate |
| GO:0033370 | 1 | maintenance_of_protein_location_in_mast_cell_secretory_granule |
| GO:0033371 | 1 | T_cell_secretory_granule_organization |
| GO:0033373 | 1 | maintenance_of_protease_location_in_mast_cell_secretory_granule |
| GO:0033377 | 1 | maintenance_of_protein_location_in_T_cell_secretory_granule |
| GO:0033379 | 1 | maintenance_of_protease_location_in_T_cell_secretory_granule |
| GO:0033382 | 1 | maintenance_of_granzyme_B_location_in_T_cell_secretory_granule |
| GO:0033385 | 1 | geranylgeranyl_diphosphate_metabolic_process |
| GO:0033386 | 1 | geranylgeranyl_diphosphate_biosynthetic_process |
| GO:0033387 | 1 | putrescine_biosynthetic_process_from_ornithine |
| GO:0033388 | 1 | putrescine_biosynthetic_process_from_arginine |
| GO:0033490 | 1 | cholesterol_biosynthetic_process_via_lathosterol |
| GO:0033505 | 1 | floor_plate_morphogenesis |
| GO:0033514 | 1 | L-lysine_catabolic_process_to_acetyl-CoA_via_L-pipecolate |
| GO:0033577 | 1 | protein_glycosylation_in_endoplasmic_reticulum |
| GO:0033588 | 1 | Elongator_holoenzyme_complex |
| GO:0033590 | 1 | response_to_cobalamin |
| GO:0033593 | 1 | BRCA2-MAGE-D1_complex |
| GO:0033594 | 1 | response_to_hydroxyisoflavone |
| GO:0033595 | 1 | response_to_genistein |
| GO:0033606 | 1 | chemokine_receptor_transport_within_lipid_bilayer |
| GO:0033622 | 1 | integrin_activation |
| GO:0033624 | 1 | negative_regulation_of_integrin_activation |
| GO:0033678 | 1 | 5'-3'_DNA/RNA_helicase_activity |
| GO:0033680 | 1 | ATP-dependent_DNA/RNA_helicase_activity |
| GO:0033682 | 1 | ATP-dependent_5'-3'_DNA/RNA_helicase_activity |
| GO:0033684 | 1 | regulation_of_luteinizing_hormone_secretion |
| GO:0033686 | 1 | positive_regulation_of_luteinizing_hormone_secretion |
| GO:0033693 | 1 | neurofilament_bundle_assembly |
| GO:0033699 | 1 | DNA_5'-adenosine_monophosphate_hydrolase_activity |
| GO:0033735 | 1 | aspartate_dehydrogenase_activity |
| GO:0033737 | 1 | 1-pyrroline_dehydrogenase_activity |
| GO:0033746 | 1 | histone_demethylase_activity_(H3-R2_specific) |
| GO:0033749 | 1 | histone_demethylase_activity_(H4-R3_specific) |
| GO:0033753 | 1 | establishment_of_ribosome_localization |
| GO:0033778 | 1 | 7alpha-hydroxycholest-4-en-3-one_12alpha-hydroxylase_activity |
| GO:0033780 | 1 | taurochenodeoxycholate_6alpha-hydroxylase_activity |
| GO:0033781 | 1 | cholesterol_24-hydroxylase_activity |
| GO:0033782 | 1 | 24-hydroxycholesterol_7alpha-hydroxylase_activity |
| GO:0033783 | 1 | 25-hydroxycholesterol_7alpha-hydroxylase_activity |
| GO:0033791 | 1 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholestanoyl-CoA_24-hydroxylase_activity |
| GO:0033798 | 1 | thyroxine_5-deiodinase_activity |
| GO:0033814 | 1 | propanoyl-CoA_C-acyltransferase_activity |
| GO:0033819 | 1 | lipoyl(octanoyl)_transferase_activity |
| GO:0033823 | 1 | procollagen_glucosyltransferase_activity |
| GO:0033858 | 1 | N-acetylgalactosamine_kinase_activity |
| GO:0033862 | 1 | UMP_kinase_activity |
| GO:0033867 | 1 | Fas-activated_serine/threonine_kinase_activity |
| GO:0033871 | 1 | [heparan_sulfate]-glucosamine_3-sulfotransferase_2_activity |
| GO:0033878 | 1 | hormone-sensitive_lipase_activity |
| GO:0033882 | 1 | choloyl-CoA_hydrolase_activity |
| GO:0033919 | 1 | glucan_1,3-alpha-glucosidase_activity |
| GO:0033925 | 1 | mannosyl-glycoprotein_endo-beta-N-acetylglucosaminidase_activity |
| GO:0033961 | 1 | cis-stilbene-oxide_hydrolase_activity |
| GO:0033979 | 1 | box_H/ACA_snoRNA_metabolic_process |
| GO:0033981 | 1 | D-dopachrome_decarboxylase_activity |
| GO:0033986 | 1 | response_to_methanol |
| GO:0033989 | 1 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA_hydratase_activity |
| GO:0034012 | 1 | FAD-AMP_lyase_(cyclizing)_activity |
| GO:0034021 | 1 | response_to_silicon_dioxide |
| GO:0034023 | 1 | 5-(carboxyamino)imidazole_ribonucleotide_mutase_activity |
| GO:0034038 | 1 | deoxyhypusine_synthase_activity |
| GO:0034040 | 1 | lipid-transporting_ATPase_activity |
| GO:0034041 | 1 | sterol-transporting_ATPase_activity |
| GO:0034118 | 1 | regulation_of_erythrocyte_aggregation |
| GO:0034120 | 1 | positive_regulation_of_erythrocyte_aggregation |
| GO:0034126 | 1 | positive_regulation_of_MyD88-dependent_toll-like_receptor_signaling_pathway |
| GO:0034127 | 1 | regulation_of_MyD88-independent_toll-like_receptor_signaling_pathway |
| GO:0034147 | 1 | regulation_of_toll-like_receptor_5_signaling_pathway |
| GO:0034148 | 1 | negative_regulation_of_toll-like_receptor_5_signaling_pathway |
| GO:0034155 | 1 | regulation_of_toll-like_receptor_7_signaling_pathway |
| GO:0034157 | 1 | positive_regulation_of_toll-like_receptor_7_signaling_pathway |
| GO:0034164 | 1 | negative_regulation_of_toll-like_receptor_9_signaling_pathway |
| GO:0034165 | 1 | positive_regulation_of_toll-like_receptor_9_signaling_pathway |
| GO:0034188 | 1 | apolipoprotein_A-I_receptor_activity |
| GO:0034189 | 1 | very-low-density_lipoprotein_particle_binding |
| GO:0034191 | 1 | apolipoprotein_A-I_receptor_binding |
| GO:0034196 | 1 | acylglycerol_transport |
| GO:0034197 | 1 | triglyceride_transport |
| GO:0034198 | 1 | cellular_response_to_amino_acid_starvation |
| GO:0034211 | 1 | GTP-dependent_protein_kinase_activity |
| GO:0034213 | 1 | quinolinate_catabolic_process |
| GO:0034230 | 1 | enkephalin_processing |
| GO:0034231 | 1 | islet_amyloid_polypeptide_processing |
| GO:0034255 | 1 | regulation_of_urea_metabolic_process |
| GO:0034263 | 1 | autophagy_in_response_to_ER_overload |
| GO:0034275 | 1 | kynurenic_acid_metabolic_process |
| GO:0034276 | 1 | kynurenic_acid_biosynthetic_process |
| GO:0034337 | 1 | RNA_folding |
| GO:0034338 | 1 | short-chain_carboxylesterase_activity |
| GO:0034346 | 1 | positive_regulation_of_type_III_interferon_production |
| GO:0034356 | 1 | NAD_biosynthesis_via_nicotinamide_riboside_salvage_pathway |
| GO:0034359 | 1 | mature_chylomicron |
| GO:0034360 | 1 | chylomicron_remnant |
| GO:0034365 | 1 | discoidal_high-density_lipoprotein_particle |
| GO:0034373 | 1 | intermediate-density_lipoprotein_particle_remodeling |
| GO:0034378 | 1 | chylomicron_assembly |
| GO:0034401 | 1 | regulation_of_transcription_by_chromatin_organization |
| GO:0034417 | 1 | bisphosphoglycerate_3-phosphatase_activity |
| GO:0034423 | 1 | autophagic_vacuole_lumen |
| GO:0034441 | 1 | plasma_lipoprotein_particle_oxidation |
| GO:0034458 | 1 | 3'-5'_RNA_helicase_activity |
| GO:0034466 | 1 | chromaffin_granule_lumen |
| GO:0034472 | 1 | snRNA_3'-end_processing |
| GO:0034474 | 1 | U2_snRNA_3'-end_processing |
| GO:0034508 | 1 | centromere_complex_assembly |
| GO:0034512 | 1 | box_C/D_snoRNA_binding |
| GO:0034513 | 1 | box_H/ACA_snoRNA_binding |
| GO:0034516 | 1 | response_to_vitamin_B6 |
| GO:0034545 | 1 | fumarylpyruvate_hydrolase_activity |
| GO:0034552 | 1 | respiratory_chain_complex_II_assembly |
| GO:0034553 | 1 | mitochondrial_respiratory_chain_complex_II_assembly |
| GO:0034589 | 1 | hydroxyproline_transport |
| GO:0034590 | 1 | L-hydroxyproline_transmembrane_transporter_activity |
| GO:0034602 | 1 | oxoglutarate_dehydrogenase_(NAD+)_activity |
| GO:0034632 | 1 | retinol_transporter_activity |
| GO:0034633 | 1 | retinol_transport |
| GO:0034647 | 1 | histone_demethylase_activity_(H3-trimethyl-K4_specific) |
| GO:0034666 | 1 | alpha2-beta1_integrin_complex |
| GO:0034667 | 1 | alpha3-beta1_integrin_complex |
| GO:0034676 | 1 | alpha6-beta4_integrin_complex |
| GO:0034696 | 1 | response_to_prostaglandin_F_stimulus |
| GO:0034699 | 1 | response_to_luteinizing_hormone_stimulus |
| GO:0034714 | 1 | type_III_transforming_growth_factor_beta_receptor_binding |
| GO:0034721 | 1 | histone_H3-K4_demethylation,_trimethyl-H3-K4-specific |
| GO:0034722 | 1 | gamma-glutamyl-peptidase_activity |
| GO:0034725 | 1 | DNA_replication-dependent_nucleosome_disassembly |
| GO:0034751 | 1 | aryl_hydrocarbon_receptor_complex |
| GO:0034752 | 1 | cytosolic_aryl_hydrocarbon_receptor_complex |
| GO:0034769 | 1 | basement_membrane_disassembly |
| GO:0034899 | 1 | trimethylamine_monooxygenase_activity |
| GO:0034959 | 1 | endothelin_maturation |
| GO:0034971 | 1 | histone_H3-R17_methylation |
| GO:0034986 | 1 | iron_chaperone_activity |
| GO:0035003 | 1 | subapical_complex |
| GO:0035026 | 1 | leading_edge_cell_differentiation |
| GO:0035034 | 1 | histone_acetyltransferase_regulator_activity |
| GO:0035037 | 1 | sperm_entry |
| GO:0035038 | 1 | female_pronucleus_assembly |
| GO:0035039 | 1 | male_pronucleus_assembly |
| GO:0035042 | 1 | fertilization,_exchange_of_chromosomal_proteins |
| GO:0035048 | 1 | splicing_factor_protein_import_into_nucleus |
| GO:0035063 | 1 | nuclear_speck_organization |
| GO:0035087 | 1 | siRNA_loading_onto_RISC_involved_in_RNA_interference |
| GO:0035105 | 1 | sterol_regulatory_element_binding_protein_import_into_nucleus |
| GO:0035226 | 1 | glutamate-cysteine_ligase_catalytic_subunit_binding |
| GO:0035227 | 1 | regulation_of_glutamate-cysteine_ligase_activity |
| GO:0035229 | 1 | positive_regulation_of_glutamate-cysteine_ligase_activity |
| GO:0035248 | 1 | alpha-1,4-N-acetylgalactosaminyltransferase_activity |
| GO:0035262 | 1 | gonad_morphogenesis |
| GO:0035272 | 1 | exocrine_system_development |
| GO:0035280 | 1 | miRNA_loading_onto_RISC_involved_in_gene_silencing_by_miRNA |
| GO:0035299 | 1 | inositol_pentakisphosphate_2-kinase_activity |
| GO:0035332 | 1 | positive_regulation_of_hippo_signaling_cascade |
| GO:0035344 | 1 | hypoxanthine_transport |
| GO:0035350 | 1 | FAD_transmembrane_transport |
| GO:0035364 | 1 | thymine_transport |
| GO:0035375 | 1 | zymogen_binding |
| GO:0035377 | 1 | transepithelial_water_transport |
| GO:0035378 | 1 | carbon_dioxide_transmembrane_transport |
| GO:0035379 | 1 | carbon_dioxide_transmembrane_transporter_activity |
| GO:0035400 | 1 | histone_tyrosine_kinase_activity |
| GO:0035401 | 1 | histone_kinase_activity_(H3-Y41_specific) |
| GO:0035406 | 1 | histone-tyrosine_phosphorylation |
| GO:0035409 | 1 | histone_H3-Y41_phosphorylation |
| GO:0035410 | 1 | dihydrotestosterone_17-beta-dehydrogenase_activity |
| GO:0035411 | 1 | catenin_import_into_nucleus |
| GO:0035415 | 1 | regulation_of_mitotic_prometaphase |
| GO:0035417 | 1 | negative_regulation_of_mitotic_prometaphase |
| GO:0035427 | 1 | purine_nucleoside_transmembrane_transport |
| GO:0035428 | 1 | hexose_transmembrane_transport |
| GO:0035444 | 1 | nickel_cation_transmembrane_transport |
| GO:0035445 | 1 | borate_transmembrane_transport |
| GO:0035472 | 1 | choriogonadotropin_hormone_receptor_activity |
| GO:0035478 | 1 | chylomicron_binding |
| GO:0035480 | 1 | regulation_of_Notch_signaling_pathway_involved_in_heart_induction |
| GO:0035481 | 1 | positive_regulation_of_Notch_signaling_pathway_involved_in_heart_induction |
| GO:0035490 | 1 | regulation_of_leukotriene_production_involved_in_inflammatory_response |
| GO:0035491 | 1 | positive_regulation_of_leukotriene_production_involved_in_inflammatory_response |
| GO:0035498 | 1 | carnosine_metabolic_process |
| GO:0035499 | 1 | carnosine_biosynthetic_process |
| GO:0035500 | 1 | MH2_domain_binding |
| GO:0035501 | 1 | MH1_domain_binding |
| GO:0035504 | 1 | regulation_of_myosin_light_chain_kinase_activity |
| GO:0035505 | 1 | positive_regulation_of_myosin_light_chain_kinase_activity |
| GO:0035509 | 1 | negative_regulation_of_myosin-light-chain-phosphatase_activity |
| GO:0035513 | 1 | oxidative_RNA_demethylation |
| GO:0035515 | 1 | oxidative_RNA_demethylase_activity |
| GO:0035516 | 1 | oxidative_DNA_demethylase_activity |
| GO:0035538 | 1 | carbohydrate_response_element_binding |
| GO:0035539 | 1 | 8-oxo-7,8-dihydrodeoxyguanosine_triphosphate_pyrophosphatase_activity |
| GO:0035545 | 1 | determination_of_left/right_asymmetry_in_nervous_system |
| GO:0035547 | 1 | regulation_of_interferon-beta_secretion |
| GO:0035548 | 1 | negative_regulation_of_interferon-beta_secretion |
| GO:0035553 | 1 | oxidative_single-stranded_RNA_demethylation |
| GO:0035562 | 1 | negative_regulation_of_chromatin_binding |
| GO:0035563 | 1 | positive_regulation_of_chromatin_binding |
| GO:0035568 | 1 | N-terminal_peptidyl-proline_methylation |
| GO:0035570 | 1 | N-terminal_peptidyl-serine_methylation |
| GO:0035572 | 1 | N-terminal_peptidyl-serine_dimethylation |
| GO:0035573 | 1 | N-terminal_peptidyl-serine_trimethylation |
| GO:0035589 | 1 | G-protein_coupled_purinergic_nucleotide_receptor_signaling_pathway |
| GO:0035594 | 1 | ganglioside_binding |
| GO:0035602 | 1 | fibroblast_growth_factor_receptor_signaling_pathway_involved_in_negative_regulation_of_apoptotic_process_in_bone_marrow |
| GO:0035603 | 1 | fibroblast_growth_factor_receptor_signaling_pathway_involved_in_hemopoiesis |
| GO:0035604 | 1 | fibroblast_growth_factor_receptor_signaling_pathway_involved_in_positive_regulation_of_cell_proliferation_in_bone_marrow |
| GO:0035605 | 1 | peptidyl-cysteine_S-nitrosylase_activity |
| GO:0035606 | 1 | peptidyl-cysteine_S-trans-nitrosylation |
| GO:0035611 | 1 | protein_branching_point_deglutamylation |
| GO:0035615 | 1 | clathrin_adaptor_activity |
| GO:0035621 | 1 | ER_to_Golgi_ceramide_transport |
| GO:0035622 | 1 | intrahepatic_bile_duct_development |
| GO:0035623 | 1 | renal_glucose_absorption |
| GO:0035627 | 1 | ceramide_transport |
| GO:0035638 | 1 | signal_maturation |
| GO:0035642 | 1 | histone_methyltransferase_activity_(H3-R17_specific) |
| GO:0035643 | 1 | L-DOPA_receptor_activity |
| GO:0035644 | 1 | phosphoanandamide_dephosphorylation |
| GO:0035645 | 1 | enteric_smooth_muscle_cell_differentiation |
| GO:0035646 | 1 | endosome_to_melanosome_transport |
| GO:0035659 | 1 | Wnt_receptor_signaling_pathway_involved_in_wound_healing,_spreading_of_epidermal_cells |
| GO:0035662 | 1 | Toll-like_receptor_4_binding |
| GO:0035663 | 1 | Toll-like_receptor_2_binding |
| GO:0035664 | 1 | TIRAP-dependent_toll-like_receptor_signaling_pathway |
| GO:0035665 | 1 | TIRAP-dependent_toll-like_receptor_4_signaling_pathway |
| GO:0035668 | 1 | TRAM-dependent_toll-like_receptor_signaling_pathway |
| GO:0035669 | 1 | TRAM-dependent_toll-like_receptor_4_signaling_pathway |
| GO:0035684 | 1 | helper_T_cell_extravasation |
| GO:0035686 | 1 | flagellar_fibrous_sheath |
| GO:0035691 | 1 | macrophage_migration_inhibitory_factor_signaling_pathway |
| GO:0035692 | 1 | macrophage_migration_inhibitory_factor_receptor_complex |
| GO:0035693 | 1 | NOS2-CD74_complex |
| GO:0035695 | 1 | mitochondrion_degradation_by_induced_vacuole_formation |
| GO:0035696 | 1 | monocyte_extravasation |
| GO:0035704 | 1 | helper_T_cell_chemotaxis |
| GO:0035705 | 1 | T-helper_17_cell_chemotaxis |
| GO:0035708 | 1 | interleukin-4-dependent_isotype_switching_to_IgE_isotypes |
| GO:0035709 | 1 | memory_T_cell_activation |
| GO:0035712 | 1 | T-helper_2_cell_activation |
| GO:0035715 | 1 | chemokine_(C-C_motif)_ligand_2_binding |
| GO:0035716 | 1 | chemokine_(C-C_motif)_ligand_12_binding |
| GO:0035718 | 1 | macrophage_migration_inhibitory_factor_binding |
| GO:0035721 | 1 | intraflagellar_retrograde_transport |
| GO:0035723 | 1 | interleukin-15-mediated_signaling_pathway |
| GO:0035724 | 1 | CD24_biosynthetic_process |
| GO:0035727 | 1 | lysophosphatidic_acid_binding |
| GO:0035730 | 1 | S-nitrosoglutathione_binding |
| GO:0035731 | 1 | dinitrosyl-iron_complex_binding |
| GO:0035732 | 1 | nitric_oxide_storage |
| GO:0035743 | 1 | CD4-positive,_alpha-beta_T_cell_cytokine_production |
| GO:0035745 | 1 | T-helper_2_cell_cytokine_production |
| GO:0035750 | 1 | protein_localization_to_myelin_sheath_abaxonal_region |
| GO:0035752 | 1 | lysosomal_lumen_pH_elevation |
| GO:0035755 | 1 | cardiolipin_hydrolase_activity |
| GO:0035757 | 1 | chemokine_(C-C_motif)_ligand_19_binding |
| GO:0035758 | 1 | chemokine_(C-C_motif)_ligand_21_binding |
| GO:0035766 | 1 | cell_chemotaxis_to_fibroblast_growth_factor |
| GO:0035768 | 1 | endothelial_cell_chemotaxis_to_fibroblast_growth_factor |
| GO:0035782 | 1 | mature_natural_killer_cell_chemotaxis |
| GO:0035786 | 1 | protein_complex_oligomerization |
| GO:0035787 | 1 | cell_migration_involved_in_kidney_development |
| GO:0035790 | 1 | platelet-derived_growth_factor_receptor-alpha_signaling_pathway |
| GO:0035844 | 1 | cloaca_development |
| GO:0035846 | 1 | oviduct_epithelium_development |
| GO:0035847 | 1 | uterine_epithelium_development |
| GO:0035849 | 1 | nephric_duct_elongation |
| GO:0035852 | 1 | horizontal_cell_localization |
| GO:0035861 | 1 | site_of_double-strand_break |
| GO:0035867 | 1 | alphav-beta3_integrin-IGF-1-IGF1R_complex |
| GO:0035870 | 1 | dITP_diphosphatase_activity |
| GO:0035871 | 1 | protein_K11-linked_deubiquitination |
| GO:0035873 | 1 | lactate_transmembrane_transport |
| GO:0035874 | 1 | cellular_response_to_copper_ion_starvation |
| GO:0035878 | 1 | nail_development |
| GO:0035883 | 1 | enteroendocrine_cell_differentiation |
| GO:0035898 | 1 | parathyroid_hormone_secretion |
| GO:0035907 | 1 | dorsal_aorta_development |
| GO:0035910 | 1 | ascending_aorta_morphogenesis |
| GO:0035921 | 1 | desmosome_disassembly |
| GO:0035922 | 1 | foramen_ovale_closure |
| GO:0035926 | 1 | chemokine_(C-C_motif)_ligand_2_secretion |
| GO:0035930 | 1 | corticosteroid_hormone_secretion |
| GO:0035933 | 1 | glucocorticoid_secretion |
| GO:0035945 | 1 | mitochondrial_ncRNA_surveillance |
| GO:0035946 | 1 | mitochondrial_mRNA_surveillance |
| GO:0035964 | 1 | COPI-coated_vesicle_budding |
| GO:0035971 | 1 | peptidyl-histidine_dephosphorylation |
| GO:0035978 | 1 | histone_H2A-S139_phosphorylation |
| GO:0035987 | 1 | endodermal_cell_differentiation |
| GO:0035989 | 1 | tendon_development |
| GO:0035992 | 1 | tendon_formation |
| GO:0035994 | 1 | response_to_muscle_stretch |
| GO:0035998 | 1 | 7,8-dihydroneopterin_3'-triphosphate_biosynthetic_process |
| GO:0036004 | 1 | GAF_domain_binding |
| GO:0036010 | 1 | protein_localization_in_endosome |
| GO:0036024 | 1 | protein_C_inhibitor-TMPRSS7_complex |
| GO:0036025 | 1 | protein_C_inhibitor-TMPRSS11E_complex |
| GO:0036026 | 1 | protein_C_inhibitor-PLAT_complex |
| GO:0036027 | 1 | protein_C_inhibitor-PLAU_complex |
| GO:0036028 | 1 | protein_C_inhibitor-thrombin_complex |
| GO:0036029 | 1 | protein_C_inhibitor-KLK3_complex |
| GO:0036030 | 1 | protein_C_inhibitor-plasma_kallikrein_complex |
| GO:0036031 | 1 | recruitment_of_mRNA_capping_enzyme_to_RNA_polymerase_II_holoenzyme_complex |
| GO:0036033 | 1 | mediator_complex_binding |
| GO:0036046 | 1 | protein_demalonylation |
| GO:0036047 | 1 | peptidyl-lysine_demalonylation |
| GO:0036048 | 1 | protein_desuccinylation |
| GO:0036049 | 1 | peptidyl-lysine_desuccinylation |
| GO:0036053 | 1 | glomerular_endothelium_fenestra |
| GO:0036054 | 1 | protein-malonyllysine_demalonylase_activity |
| GO:0036055 | 1 | protein-succinyllysine_desuccinylase_activity |
| GO:0036058 | 1 | filtration_diaphragm_assembly |
| GO:0036060 | 1 | slit_diaphragm_assembly |
| GO:0036066 | 1 | protein_O-linked_fucosylation |
| GO:0036071 | 1 | N-glycan_fucosylation |
| GO:0036088 | 1 | D-serine_catabolic_process |
| GO:0036091 | 1 | positive_regulation_of_transcription_from_RNA_polymerase_II_promoter_in_response_to_oxidative_stress |
| GO:0036105 | 1 | peroxisome_membrane_class-1_targeting_sequence_binding |
| GO:0036131 | 1 | prostaglandin_D2_11-ketoreductase_activity |
| GO:0036138 | 1 | peptidyl-histidine_hydroxylation |
| GO:0036139 | 1 | peptidyl-histidine_dioxygenase_activity |
| GO:0036140 | 1 | peptidyl-asparagine_3-dioxygenase_activity |
| GO:0036143 | 1 | kringle_domain_binding |
| GO:0036158 | 1 | outer_dynein_arm_assembly |
| GO:0036159 | 1 | inner_dynein_arm_assembly |
| GO:0036261 | 1 | 7-methylguanosine_cap_hypermethylation |
| GO:0038027 | 1 | apolipoprotein_A-I-mediated_signaling_pathway |
| GO:0038046 | 1 | enkephalin_receptor_activity |
| GO:0038047 | 1 | morphine_receptor_activity |
| GO:0038048 | 1 | dynorphin_receptor_activity |
| GO:0038052 | 1 | estrogen-activated_sequence-specific_DNA_binding_RNA_polymerase_II_transcription_factor_activity |
| GO:0038095 | 1 | Fc-epsilon_receptor_signaling_pathway |
| GO:0038100 | 1 | nodal_binding |
| GO:0038101 | 1 | sequestering_of_nodal_from_receptor_via_nodal_binding |
| GO:0038106 | 1 | choriogonadotropin_hormone_binding |
| GO:0038113 | 1 | interleukin-9-mediated_signaling_pathway |
| GO:0038114 | 1 | interleukin-21-mediated_signaling_pathway |
| GO:0038115 | 1 | chemokine_(C-C_motif)_ligand_19_signaling_pathway |
| GO:0038116 | 1 | chemokine_(C-C_motif)_ligand_21_signaling_pathway |
| GO:0038117 | 1 | C-C_motif_chemokine_19_receptor_activity |
| GO:0038121 | 1 | C-C_motif_chemokine_21_receptor_activity |
| GO:0038161 | 1 | prolactin_signaling_pathway |
| GO:0038162 | 1 | erythropoietin-mediated_signaling_pathway |
| GO:0039008 | 1 | pronephric_nephron_tubule_morphogenesis |
| GO:0039020 | 1 | pronephric_nephron_tubule_development |
| GO:0039023 | 1 | pronephric_duct_morphogenesis |
| GO:0039529 | 1 | RIG-I_signaling_pathway |
| GO:0039532 | 1 | negative_regulation_of_viral-induced_cytoplasmic_pattern_recognition_receptor_signaling_pathway |
| GO:0039534 | 1 | negative_regulation_of_MDA-5_signaling_pathway |
| GO:0039536 | 1 | negative_regulation_of_RIG-I_signaling_pathway |
| GO:0040030 | 1 | regulation_of_molecular_function,_epigenetic |
| GO:0040032 | 1 | post-embryonic_body_morphogenesis |
| GO:0040040 | 1 | thermosensory_behavior |
| GO:0042007 | 1 | interleukin-18_binding |
| GO:0042017 | 1 | interleukin-22_binding |
| GO:0042018 | 1 | interleukin-22_receptor_activity |
| GO:0042022 | 1 | interleukin-12_receptor_complex |
| GO:0042025 | 1 | host_cell_nucleus |
| GO:0042033 | 1 | chemokine_biosynthetic_process |
| GO:0042084 | 1 | 5-methyltetrahydrofolate-dependent_methyltransferase_activity |
| GO:0042134 | 1 | rRNA_primary_transcript_binding |
| GO:0042137 | 1 | sequestering_of_neurotransmitter |
| GO:0042160 | 1 | lipoprotein_modification |
| GO:0042161 | 1 | lipoprotein_oxidation |
| GO:0042163 | 1 | interleukin-12_beta_subunit_binding |
| GO:0042164 | 1 | interleukin-12_alpha_subunit_binding |
| GO:0042171 | 1 | lysophosphatidic_acid_acyltransferase_activity |
| GO:0042182 | 1 | ketone_catabolic_process |
| GO:0042222 | 1 | interleukin-1_biosynthetic_process |
| GO:0042228 | 1 | interleukin-8_biosynthetic_process |
| GO:0042231 | 1 | interleukin-13_biosynthetic_process |
| GO:0042253 | 1 | granulocyte_macrophage_colony-stimulating_factor_biosynthetic_process |
| GO:0042265 | 1 | peptidyl-asparagine_hydroxylation |
| GO:0042270 | 1 | protection_from_natural_killer_cell_mediated_cytotoxicity |
| GO:0042276 | 1 | error-prone_translesion_synthesis |
| GO:0042282 | 1 | hydroxymethylglutaryl-CoA_reductase_activity |
| GO:0042284 | 1 | sphingolipid_delta-4_desaturase_activity |
| GO:0042292 | 1 | URM1_activating_enzyme_activity |
| GO:0042309 | 1 | homoiothermy |
| GO:0042313 | 1 | protein_kinase_C_deactivation |
| GO:0042324 | 1 | hypocretin_receptor_binding |
| GO:0042340 | 1 | keratan_sulfate_catabolic_process |
| GO:0042356 | 1 | GDP-4-dehydro-D-rhamnose_reductase_activity |
| GO:0042357 | 1 | thiamine_diphosphate_metabolic_process |
| GO:0042368 | 1 | vitamin_D_biosynthetic_process |
| GO:0042374 | 1 | phylloquinone_metabolic_process |
| GO:0042376 | 1 | phylloquinone_catabolic_process |
| GO:0042377 | 1 | vitamin_K_catabolic_process |
| GO:0042378 | 1 | quinone_cofactor_catabolic_process |
| GO:0042404 | 1 | thyroid_hormone_catabolic_process |
| GO:0042406 | 1 | extrinsic_to_endoplasmic_reticulum_membrane |
| GO:0042413 | 1 | carnitine_catabolic_process |
| GO:0042443 | 1 | phenylethylamine_metabolic_process |
| GO:0042465 | 1 | kinesis |
| GO:0042466 | 1 | chemokinesis |
| GO:0042489 | 1 | negative_regulation_of_odontogenesis_of_dentin-containing_tooth |
| GO:0042497 | 1 | triacyl_lipopeptide_binding |
| GO:0042512 | 1 | negative_regulation_of_tyrosine_phosphorylation_of_Stat1_protein |
| GO:0042539 | 1 | hypotonic_salinity_response |
| GO:0042564 | 1 | NLS-dependent_protein_nuclear_import_complex |
| GO:0042568 | 1 | insulin-like_growth_factor_binary_complex |
| GO:0042585 | 1 | germinal_vesicle |
| GO:0042586 | 1 | peptide_deformylase_activity |
| GO:0042602 | 1 | riboflavin_reductase_(NADPH)_activity |
| GO:0042624 | 1 | ATPase_activity,_uncoupled |
| GO:0042628 | 1 | mating_plug_formation |
| GO:0042636 | 1 | negative_regulation_of_hair_cycle |
| GO:0042643 | 1 | actomyosin,_actin_part |
| GO:0042660 | 1 | positive_regulation_of_cell_fate_specification |
| GO:0042664 | 1 | negative_regulation_of_endodermal_cell_fate_specification |
| GO:0042665 | 1 | regulation_of_ectodermal_cell_fate_specification |
| GO:0042666 | 1 | negative_regulation_of_ectodermal_cell_fate_specification |
| GO:0042667 | 1 | auditory_receptor_cell_fate_specification |
| GO:0042694 | 1 | muscle_cell_fate_specification |
| GO:0042696 | 1 | menarche |
| GO:0042697 | 1 | menopause |
| GO:0042700 | 1 | luteinizing_hormone_signaling_pathway |
| GO:0042703 | 1 | menstruation |
| GO:0042709 | 1 | succinate-CoA_ligase_complex |
| GO:0042720 | 1 | mitochondrial_inner_membrane_peptidase_complex |
| GO:0042724 | 1 | thiamine-containing_compound_biosynthetic_process |
| GO:0042747 | 1 | circadian_sleep/wake_cycle,_REM_sleep |
| GO:0042766 | 1 | nucleosome_mobilization |
| GO:0042779 | 1 | tRNA_3'-trailer_cleavage |
| GO:0042795 | 1 | snRNA_transcription_from_RNA_polymerase_II_promoter |
| GO:0042796 | 1 | snRNA_transcription_from_RNA_polymerase_III_promoter |
| GO:0042801 | 1 | polo_kinase_kinase_activity |
| GO:0042827 | 1 | platelet_dense_granule |
| GO:0042839 | 1 | D-glucuronate_metabolic_process |
| GO:0042840 | 1 | D-glucuronate_catabolic_process |
| GO:0042842 | 1 | D-xylose_biosynthetic_process |
| GO:0042843 | 1 | D-xylose_catabolic_process |
| GO:0042892 | 1 | chloramphenicol_transport |
| GO:0042908 | 1 | xenobiotic_transport |
| GO:0042922 | 1 | neuromedin_U_receptor_binding |
| GO:0042924 | 1 | neuromedin_U_binding |
| GO:0042936 | 1 | dipeptide_transporter_activity |
| GO:0042941 | 1 | D-alanine_transport |
| GO:0042946 | 1 | glucoside_transport |
| GO:0042947 | 1 | glucoside_transmembrane_transporter_activity |
| GO:0042978 | 1 | ornithine_decarboxylase_activator_activity |
| GO:0042991 | 1 | transcription_factor_import_into_nucleus |
| GO:0042998 | 1 | positive_regulation_of_Golgi_to_plasma_membrane_protein_transport |
| GO:0042999 | 1 | regulation_of_Golgi_to_plasma_membrane_CFTR_protein_transport |
| GO:0043000 | 1 | Golgi_to_plasma_membrane_CFTR_protein_transport |
| GO:0043002 | 1 | negative_regulation_of_Golgi_to_plasma_membrane_CFTR_protein_transport |
| GO:0043004 | 1 | cytoplasmic_sequestering_of_CFTR_protein |
| GO:0043012 | 1 | regulation_of_fusion_of_sperm_to_egg_plasma_membrane |
| GO:0043016 | 1 | regulation_of_lymphotoxin_A_biosynthetic_process |
| GO:0043017 | 1 | positive_regulation_of_lymphotoxin_A_biosynthetic_process |
| GO:0043033 | 1 | isoamylase_complex |
| GO:0043035 | 1 | chromatin_insulator_sequence_binding |
| GO:0043048 | 1 | dolichyl_monophosphate_biosynthetic_process |
| GO:0043060 | 1 | meiotic_metaphase_I_plate_congression |
| GO:0043063 | 1 | intercellular_bridge_organization |
| GO:0043091 | 1 | L-arginine_import |
| GO:0043095 | 1 | regulation_of_GTP_cyclohydrolase_I_activity |
| GO:0043105 | 1 | negative_regulation_of_GTP_cyclohydrolase_I_activity |
| GO:0043111 | 1 | replication_fork_arrest |
| GO:0043126 | 1 | regulation_of_1-phosphatidylinositol_4-kinase_activity |
| GO:0043128 | 1 | positive_regulation_of_1-phosphatidylinositol_4-kinase_activity |
| GO:0043152 | 1 | induction_of_bacterial_agglutination |
| GO:0043177 | 1 | organic_acid_binding |
| GO:0043181 | 1 | vacuolar_sequestering |
| GO:0043207 | 1 | response_to_external_biotic_stimulus |
| GO:0043221 | 1 | SMC_protein_binding |
| GO:0043250 | 1 | sodium-dependent_organic_anion_transmembrane_transporter_activity |
| GO:0043251 | 1 | sodium-dependent_organic_anion_transport |
| GO:0043257 | 1 | laminin-8_complex |
| GO:0043265 | 1 | ectoplasm |
| GO:0043310 | 1 | negative_regulation_of_eosinophil_degranulation |
| GO:0043311 | 1 | positive_regulation_of_eosinophil_degranulation |
| GO:0043316 | 1 | cytotoxic_T_cell_degranulation |
| GO:0043375 | 1 | CD8-positive,_alpha-beta_T_cell_lineage_commitment |
| GO:0043376 | 1 | regulation_of_CD8-positive,_alpha-beta_T_cell_differentiation |
| GO:0043378 | 1 | positive_regulation_of_CD8-positive,_alpha-beta_T_cell_differentiation |
| GO:0043381 | 1 | negative_regulation_of_memory_T_cell_differentiation |
| GO:0043400 | 1 | cortisol_secretion |
| GO:0043402 | 1 | glucocorticoid_mediated_signaling_pathway |
| GO:0043404 | 1 | corticotropin-releasing_hormone_receptor_activity |
| GO:0043418 | 1 | homocysteine_catabolic_process |
| GO:0043420 | 1 | anthranilate_metabolic_process |
| GO:0043438 | 1 | acetoacetic_acid_metabolic_process |
| GO:0043455 | 1 | regulation_of_secondary_metabolic_process |
| GO:0043456 | 1 | regulation_of_pentose-phosphate_shunt |
| GO:0043480 | 1 | pigment_accumulation_in_tissues |
| GO:0043504 | 1 | mitochondrial_DNA_repair |
| GO:0043509 | 1 | activin_A_complex |
| GO:0043519 | 1 | regulation_of_myosin_II_filament_organization |
| GO:0043538 | 1 | regulation_of_actin_phosphorylation |
| GO:0043540 | 1 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase_1_complex |
| GO:0043544 | 1 | lipoamide_binding |
| GO:0043556 | 1 | regulation_of_translation_in_response_to_oxidative_stress |
| GO:0043602 | 1 | nitrate_catabolic_process |
| GO:0043605 | 1 | cellular_amide_catabolic_process |
| GO:0043622 | 1 | cortical_microtubule_organization |
| GO:0043626 | 1 | PCNA_complex |
| GO:0043651 | 1 | linoleic_acid_metabolic_process |
| GO:0043652 | 1 | engulfment_of_apoptotic_cell |
| GO:0043696 | 1 | dedifferentiation |
| GO:0043697 | 1 | cell_dedifferentiation |
| GO:0043715 | 1 | 2,3-diketo-5-methylthiopentyl-1-phosphate_enolase_activity |
| GO:0043716 | 1 | 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate_phosphatase_activity |
| GO:0043733 | 1 | DNA-3-methylbase_glycosylase_activity |
| GO:0043754 | 1 | dihydrolipoyllysine-residue_(2-methylpropanoyl)transferase_activity |
| GO:0043783 | 1 | oxidoreductase_activity,_oxidizing_metal_ions_with_flavin_as_acceptor |
| GO:0043795 | 1 | glyceraldehyde_oxidoreductase_activity |
| GO:0043843 | 1 | ADP-specific_glucokinase_activity |
| GO:0043874 | 1 | acireductone_synthase_activity |
| GO:0043890 | 1 | N-acetylgalactosamine-6-sulfatase_activity |
| GO:0043916 | 1 | DNA-7-methylguanine_glycosylase_activity |
| GO:0043924 | 1 | suramin_binding |
| GO:0043958 | 1 | acryloyl-CoA_reductase_activity |
| GO:0043973 | 1 | histone_H3-K4_acetylation |
| GO:0043990 | 1 | histone_H2A-S1_phosphorylation |
| GO:0043997 | 1 | histone_acetyltransferase_activity_(H4-K12_specific) |
| GO:0044007 | 1 | dissemination_or_transmission_of_symbiont_from_host |
| GO:0044090 | 1 | positive_regulation_of_vacuole_organization |
| GO:0044111 | 1 | development_involved_in_symbiotic_interaction |
| GO:0044139 | 1 | modulation_of_growth_of_symbiont_on_or_near_host |
| GO:0044140 | 1 | negative_regulation_of_growth_of_symbiont_on_or_near_host_surface |
| GO:0044214 | 1 | fully_spanning_plasma_membrane |
| GO:0044258 | 1 | intestinal_lipid_catabolic_process |
| GO:0044302 | 1 | dentate_gyrus_mossy_fiber |
| GO:0044316 | 1 | cone_cell_pedicle |
| GO:0044322 | 1 | endoplasmic_reticulum_quality_control_compartment |
| GO:0044327 | 1 | dendritic_spine_head |
| GO:0044328 | 1 | canonical_Wnt_receptor_signaling_pathway_involved_in_positive_regulation_of_endothelial_cell_migration |
| GO:0044329 | 1 | canonical_Wnt_receptor_signaling_pathway_involved_in_positive_regulation_of_cell-cell_adhesion |
| GO:0044330 | 1 | canonical_Wnt_receptor_signaling_pathway_involved_in_positive_regulation_of_wound_healing |
| GO:0044331 | 1 | cell-cell_adhesion_mediated_by_cadherin |
| GO:0044342 | 1 | type_B_pancreatic_cell_proliferation |
| GO:0044345 | 1 | stromal-epithelial_cell_signaling_involved_in_prostate_gland_development |
| GO:0044346 | 1 | fibroblast_apoptotic_process |
| GO:0044352 | 1 | pinosome |
| GO:0044354 | 1 | macropinosome |
| GO:0044355 | 1 | clearance_of_foreign_intracellular_DNA |
| GO:0044356 | 1 | clearance_of_foreign_intracellular_DNA_by_conversion_of_DNA_cytidine_to_uridine |
| GO:0044387 | 1 | negative_regulation_of_protein_kinase_activity_by_regulation_of_protein_phosphorylation |
| GO:0044501 | 1 | modulation_of_signal_transduction_in_other_organism |
| GO:0044524 | 1 | protein_sulfhydration |
| GO:0044531 | 1 | modulation_of_programmed_cell_death_in_other_organism |
| GO:0044532 | 1 | modulation_of_apoptotic_process_in_other_organism |
| GO:0044533 | 1 | positive_regulation_of_apoptotic_process_in_other_organism |
| GO:0044540 | 1 | L-cystine_L-cysteine-lyase_(deaminating) |
| GO:0044547 | 1 | DNA_topoisomerase_binding |
| GO:0044594 | 1 | 17-beta-hydroxysteroid_dehydrogenase_(NAD+)_activity |
| GO:0044620 | 1 | ACP_phosphopantetheine_attachment_site_binding |
| GO:0045032 | 1 | ADP-activated_nucleotide_receptor_activity |
| GO:0045062 | 1 | extrathymic_T_cell_selection |
| GO:0045065 | 1 | cytotoxic_T_cell_differentiation |
| GO:0045081 | 1 | negative_regulation_of_interleukin-10_biosynthetic_process |
| GO:0045083 | 1 | negative_regulation_of_interleukin-12_biosynthetic_process |
| GO:0045092 | 1 | interleukin-18_receptor_complex |
| GO:0045108 | 1 | regulation_of_intermediate_filament_polymerization_or_depolymerization |
| GO:0045112 | 1 | integrin_biosynthetic_process |
| GO:0045127 | 1 | N-acetylglucosamine_kinase_activity |
| GO:0045128 | 1 | negative_regulation_of_reciprocal_meiotic_recombination |
| GO:0045130 | 1 | keratan_sulfotransferase_activity |
| GO:0045145 | 1 | single-stranded_DNA_specific_5'-3'_exodeoxyribonuclease_activity |
| GO:0045160 | 1 | myosin_I_complex |
| GO:0045175 | 1 | basal_protein_localization |
| GO:0045189 | 1 | connective_tissue_growth_factor_biosynthetic_process |
| GO:0045196 | 1 | establishment_or_maintenance_of_neuroblast_polarity |
| GO:0045199 | 1 | maintenance_of_epithelial_cell_apical/basal_polarity |
| GO:0045200 | 1 | establishment_of_neuroblast_polarity |
| GO:0045204 | 1 | MAPK_export_from_nucleus |
| GO:0045208 | 1 | MAPK_phosphatase_export_from_nucleus |
| GO:0045209 | 1 | MAPK_phosphatase_export_from_nucleus,_leptomycin_B_sensitive |
| GO:0045210 | 1 | FasL_biosynthetic_process |
| GO:0045212 | 1 | neurotransmitter_receptor_biosynthetic_process |
| GO:0045219 | 1 | regulation_of_FasL_biosynthetic_process |
| GO:0045221 | 1 | negative_regulation_of_FasL_biosynthetic_process |
| GO:0045226 | 1 | extracellular_polysaccharide_biosynthetic_process |
| GO:0045234 | 1 | protein_palmitoleylation |
| GO:0045244 | 1 | succinate-CoA_ligase_complex_(GDP-forming) |
| GO:0045267 | 1 | proton-transporting_ATP_synthase,_catalytic_core |
| GO:0045273 | 1 | respiratory_chain_complex_II |
| GO:0045297 | 1 | post-mating_behavior |
| GO:0045299 | 1 | otolith_mineralization |
| GO:0045324 | 1 | late_endosome_to_vacuole_transport |
| GO:0045342 | 1 | MHC_class_II_biosynthetic_process |
| GO:0045352 | 1 | interleukin-1_Type_I_receptor_antagonist_activity |
| GO:0045353 | 1 | interleukin-1_Type_II_receptor_antagonist_activity |
| GO:0045355 | 1 | negative_regulation_of_interferon-alpha_biosynthetic_process |
| GO:0045358 | 1 | negative_regulation_of_interferon-beta_biosynthetic_process |
| GO:0045360 | 1 | regulation_of_interleukin-1_biosynthetic_process |
| GO:0045362 | 1 | positive_regulation_of_interleukin-1_biosynthetic_process |
| GO:0045366 | 1 | regulation_of_interleukin-13_biosynthetic_process |
| GO:0045368 | 1 | positive_regulation_of_interleukin-13_biosynthetic_process |
| GO:0045381 | 1 | regulation_of_interleukin-18_biosynthetic_process |
| GO:0045403 | 1 | negative_regulation_of_interleukin-4_biosynthetic_process |
| GO:0045405 | 1 | regulation_of_interleukin-5_biosynthetic_process |
| GO:0045407 | 1 | positive_regulation_of_interleukin-5_biosynthetic_process |
| GO:0045513 | 1 | interleukin-27_binding |
| GO:0045517 | 1 | interleukin-20_receptor_binding |
| GO:0045518 | 1 | interleukin-22_receptor_binding |
| GO:0045554 | 1 | regulation_of_TRAIL_biosynthetic_process |
| GO:0045556 | 1 | positive_regulation_of_TRAIL_biosynthetic_process |
| GO:0045575 | 1 | basophil_activation |
| GO:0045583 | 1 | regulation_of_cytotoxic_T_cell_differentiation |
| GO:0045585 | 1 | positive_regulation_of_cytotoxic_T_cell_differentiation |
| GO:0045609 | 1 | positive_regulation_of_auditory_receptor_cell_differentiation |
| GO:0045633 | 1 | positive_regulation_of_mechanoreceptor_differentiation |
| GO:0045643 | 1 | regulation_of_eosinophil_differentiation |
| GO:0045645 | 1 | positive_regulation_of_eosinophil_differentiation |
| GO:0045659 | 1 | negative_regulation_of_neutrophil_differentiation |
| GO:0045660 | 1 | positive_regulation_of_neutrophil_differentiation |
| GO:0045703 | 1 | ketoreductase_activity |
| GO:0045713 | 1 | low-density_lipoprotein_particle_receptor_biosynthetic_process |
| GO:0045720 | 1 | negative_regulation_of_integrin_biosynthetic_process |
| GO:0045724 | 1 | positive_regulation_of_flagellum_assembly |
| GO:0045728 | 1 | respiratory_burst_after_phagocytosis |
| GO:0045735 | 1 | nutrient_reservoir_activity |
| GO:0045751 | 1 | negative_regulation_of_Toll_signaling_pathway |
| GO:0045769 | 1 | negative_regulation_of_asymmetric_cell_division |
| GO:0045799 | 1 | positive_regulation_of_chromatin_assembly_or_disassembly |
| GO:0045827 | 1 | negative_regulation_of_isoprenoid_metabolic_process |
| GO:0045870 | 1 | positive_regulation_of_retroviral_genome_replication |
| GO:0045872 | 1 | positive_regulation_of_rhodopsin_gene_expression |
| GO:0045914 | 1 | negative_regulation_of_catecholamine_metabolic_process |
| GO:0045917 | 1 | positive_regulation_of_complement_activation |
| GO:0045925 | 1 | positive_regulation_of_female_receptivity |
| GO:0045957 | 1 | negative_regulation_of_complement_activation,_alternative_pathway |
| GO:0045959 | 1 | negative_regulation_of_complement_activation,_classical_pathway |
| GO:0045963 | 1 | negative_regulation_of_dopamine_metabolic_process |
| GO:0045993 | 1 | negative_regulation_of_translational_initiation_by_iron |
| GO:0045994 | 1 | positive_regulation_of_translational_initiation_by_iron |
| GO:0046032 | 1 | ADP_catabolic_process |
| GO:0046048 | 1 | UDP_metabolic_process |
| GO:0046056 | 1 | dADP_metabolic_process |
| GO:0046061 | 1 | dATP_catabolic_process |
| GO:0046079 | 1 | dUMP_catabolic_process |
| GO:0046086 | 1 | adenosine_biosynthetic_process |
| GO:0046090 | 1 | deoxyadenosine_metabolic_process |
| GO:0046092 | 1 | deoxycytidine_metabolic_process |
| GO:0046105 | 1 | thymidine_biosynthetic_process |
| GO:0046111 | 1 | xanthine_biosynthetic_process |
| GO:0046120 | 1 | deoxyribonucleoside_biosynthetic_process |
| GO:0046124 | 1 | purine_deoxyribonucleoside_catabolic_process |
| GO:0046126 | 1 | pyrimidine_deoxyribonucleoside_biosynthetic_process |
| GO:0046127 | 1 | pyrimidine_deoxyribonucleoside_catabolic_process |
| GO:0046144 | 1 | D-alanine_family_amino_acid_metabolic_process |
| GO:0046166 | 1 | glyceraldehyde-3-phosphate_biosynthetic_process |
| GO:0046167 | 1 | glycerol-3-phosphate_biosynthetic_process |
| GO:0046176 | 1 | aldonic_acid_catabolic_process |
| GO:0046177 | 1 | D-gluconate_catabolic_process |
| GO:0046203 | 1 | spermidine_catabolic_process |
| GO:0046208 | 1 | spermine_catabolic_process |
| GO:0046210 | 1 | nitric_oxide_catabolic_process |
| GO:0046226 | 1 | coumarin_catabolic_process |
| GO:0046271 | 1 | phenylpropanoid_catabolic_process |
| GO:0046293 | 1 | formaldehyde_biosynthetic_process |
| GO:0046294 | 1 | formaldehyde_catabolic_process |
| GO:0046296 | 1 | glycolate_catabolic_process |
| GO:0046316 | 1 | gluconokinase_activity |
| GO:0046317 | 1 | regulation_of_glucosylceramide_biosynthetic_process |
| GO:0046318 | 1 | negative_regulation_of_glucosylceramide_biosynthetic_process |
| GO:0046327 | 1 | glycerol_biosynthetic_process_from_pyruvate |
| GO:0046331 | 1 | lateral_inhibition |
| GO:0046333 | 1 | octopamine_metabolic_process |
| GO:0046352 | 1 | disaccharide_catabolic_process |
| GO:0046358 | 1 | butyrate_biosynthetic_process |
| GO:0046359 | 1 | butyrate_catabolic_process |
| GO:0046360 | 1 | 2-oxobutyrate_biosynthetic_process |
| GO:0046361 | 1 | 2-oxobutyrate_metabolic_process |
| GO:0046370 | 1 | fructose_biosynthetic_process |
| GO:0046379 | 1 | extracellular_polysaccharide_metabolic_process |
| GO:0046380 | 1 | N-acetylneuraminate_biosynthetic_process |
| GO:0046417 | 1 | chorismate_metabolic_process |
| GO:0046436 | 1 | D-alanine_metabolic_process |
| GO:0046437 | 1 | D-amino_acid_biosynthetic_process |
| GO:0046443 | 1 | FAD_metabolic_process |
| GO:0046444 | 1 | FMN_metabolic_process |
| GO:0046448 | 1 | tropane_alkaloid_metabolic_process |
| GO:0046491 | 1 | L-methylmalonyl-CoA_metabolic_process |
| GO:0046497 | 1 | nicotinate_nucleotide_metabolic_process |
| GO:0046525 | 1 | xylosylprotein_4-beta-galactosyltransferase_activity |
| GO:0046534 | 1 | positive_regulation_of_photoreceptor_cell_differentiation |
| GO:0046538 | 1 | 2,3-bisphosphoglycerate-dependent_phosphoglycerate_mutase_activity |
| GO:0046539 | 1 | histamine_N-methyltransferase_activity |
| GO:0046554 | 1 | malate_dehydrogenase_(NADP+)_activity |
| GO:0046570 | 1 | methylthioribulose_1-phosphate_dehydratase_activity |
| GO:0046587 | 1 | positive_regulation_of_calcium-dependent_cell-cell_adhesion |
| GO:0046588 | 1 | negative_regulation_of_calcium-dependent_cell-cell_adhesion |
| GO:0046597 | 1 | negative_regulation_of_viral_entry_into_host_cell |
| GO:0046601 | 1 | positive_regulation_of_centriole_replication |
| GO:0046604 | 1 | positive_regulation_of_mitotic_centrosome_separation |
| GO:0046618 | 1 | drug_export |
| GO:0046674 | 1 | induction_of_retinal_programmed_cell_death |
| GO:0046680 | 1 | response_to_DDT |
| GO:0046690 | 1 | response_to_tellurium_ion |
| GO:0046692 | 1 | sperm_competition |
| GO:0046707 | 1 | IDP_metabolic_process |
| GO:0046709 | 1 | IDP_catabolic_process |
| GO:0046713 | 1 | borate_transport |
| GO:0046715 | 1 | borate_transmembrane_transporter_activity |
| GO:0046724 | 1 | oxalic_acid_secretion |
| GO:0046811 | 1 | histone_deacetylase_inhibitor_activity |
| GO:0046814 | 1 | virion_attachment,_binding_of_host_cell_surface_coreceptor |
| GO:0046817 | 1 | chemokine_receptor_antagonist_activity |
| GO:0046833 | 1 | positive_regulation_of_RNA_export_from_nucleus |
| GO:0046864 | 1 | isoprenoid_transport |
| GO:0046865 | 1 | terpenoid_transport |
| GO:0046900 | 1 | tetrahydrofolylpolyglutamate_metabolic_process |
| GO:0046901 | 1 | tetrahydrofolylpolyglutamate_biosynthetic_process |
| GO:0046904 | 1 | calcium_oxalate_binding |
| GO:0046911 | 1 | metal_chelating_activity |
| GO:0046916 | 1 | cellular_transition_metal_ion_homeostasis |
| GO:0046921 | 1 | alpha-(1->6)-fucosyltransferase_activity |
| GO:0046946 | 1 | hydroxylysine_metabolic_process |
| GO:0046947 | 1 | hydroxylysine_biosynthetic_process |
| GO:0046963 | 1 | 3'-phosphoadenosine_5'-phosphosulfate_transport |
| GO:0046964 | 1 | 3'-phosphoadenosine_5'-phosphosulfate_transmembrane_transporter_activity |
| GO:0046986 | 1 | negative_regulation_of_hemoglobin_biosynthetic_process |
| GO:0046996 | 1 | oxidoreductase_activity,_acting_on_paired_donors,_with_incorporation_or_reduction_of_molecular_oxygen,_with_NADH_or_NADPH_as_one_donor,_and_the_other_dehydrogenated |
| GO:0047012 | 1 | sterol-4-alpha-carboxylate_3-dehydrogenase_(decarboxylating)_activity |
| GO:0047015 | 1 | 3-hydroxy-2-methylbutyryl-CoA_dehydrogenase_activity |
| GO:0047016 | 1 | cholest-5-ene-3-beta,7-alpha-diol_3-beta-dehydrogenase_activity |
| GO:0047017 | 1 | prostaglandin-F_synthase_activity |
| GO:0047020 | 1 | 15-hydroxyprostaglandin-D_dehydrogenase_(NADP+)_activity |
| GO:0047021 | 1 | 15-hydroxyprostaglandin_dehydrogenase_(NADP+)_activity |
| GO:0047035 | 1 | testosterone_dehydrogenase_(NAD+)_activity |
| GO:0047058 | 1 | vitamin-K-epoxide_reductase_(warfarin-insensitive)_activity |
| GO:0047066 | 1 | phospholipid-hydroperoxide_glutathione_peroxidase_activity |
| GO:0047105 | 1 | 4-trimethylammoniobutyraldehyde_dehydrogenase_activity |
| GO:0047127 | 1 | thiomorpholine-carboxylate_dehydrogenase_activity |
| GO:0047130 | 1 | saccharopine_dehydrogenase_(NADP+,_L-lysine-forming)_activity |
| GO:0047131 | 1 | saccharopine_dehydrogenase_(NAD+,_L-glutamate-forming)_activity |
| GO:0047173 | 1 | phosphatidylcholine-retinol_O-acyltransferase_activity |
| GO:0047220 | 1 | galactosylxylosylprotein_3-beta-galactosyltransferase_activity |
| GO:0047225 | 1 | acetylgalactosaminyl-O-glycosyl-glycoprotein_beta-1,6-N-acetylglucosaminyltransferase_activity |
| GO:0047256 | 1 | lactosylceramide_1,3-N-acetyl-beta-D-glucosaminyltransferase_activity |
| GO:0047273 | 1 | galactosylgalactosylglucosylceramide_beta-D-acetylgalactosaminyltransferase_activity |
| GO:0047277 | 1 | globoside_alpha-N-acetylgalactosaminyltransferase_activity |
| GO:0047280 | 1 | nicotinamide_phosphoribosyltransferase_activity |
| GO:0047288 | 1 | monosialoganglioside_sialyltransferase_activity |
| GO:0047290 | 1 | (alpha-N-acetylneuraminyl-2,3-beta-galactosyl-1,3)-N-acetyl-galactosaminide_6-alpha-sialyltransferase_activity |
| GO:0047291 | 1 | lactosylceramide_alpha-2,3-sialyltransferase_activity |
| GO:0047293 | 1 | 4-hydroxybenzoate_nonaprenyltransferase_activity |
| GO:0047298 | 1 | (S)-3-amino-2-methylpropionate_transaminase_activity |
| GO:0047305 | 1 | (R)-3-amino-2-methylpropionate-pyruvate_transaminase_activity |
| GO:0047312 | 1 | L-phenylalanine:pyruvate_aminotransferase_activity |
| GO:0047315 | 1 | kynurenine-glyoxylate_transaminase_activity |
| GO:0047316 | 1 | glutamine-phenylpyruvate_transaminase_activity |
| GO:0047323 | 1 | [3-methyl-2-oxobutanoate_dehydrogenase_(acetyl-transferring)]_kinase_activity |
| GO:0047325 | 1 | inositol_tetrakisphosphate_1-kinase_activity |
| GO:0047341 | 1 | fucose-1-phosphate_guanylyltransferase_activity |
| GO:0047389 | 1 | glycerophosphocholine_phosphodiesterase_activity |
| GO:0047391 | 1 | alkylglycerophosphoethanolamine_phosphodiesterase_activity |
| GO:0047394 | 1 | glycerophosphoinositol_inositolphosphodiesterase_activity |
| GO:0047395 | 1 | glycerophosphoinositol_glycerophosphodiesterase_activity |
| GO:0047408 | 1 | alkenylglycerophosphocholine_hydrolase_activity |
| GO:0047409 | 1 | alkenylglycerophosphoethanolamine_hydrolase_activity |
| GO:0047444 | 1 | N-acylneuraminate-9-phosphate_synthase_activity |
| GO:0047453 | 1 | ATP-dependent_NAD(P)H-hydrate_dehydratase_activity |
| GO:0047464 | 1 | heparosan-N-sulfate-glucuronate_5-epimerase_activity |
| GO:0047536 | 1 | 2-aminoadipate_transaminase_activity |
| GO:0047545 | 1 | 2-hydroxyglutarate_dehydrogenase_activity |
| GO:0047547 | 1 | 2-methylcitrate_dehydratase_activity |
| GO:0047560 | 1 | 3-dehydrosphinganine_reductase_activity |
| GO:0047568 | 1 | 3-oxo-5-beta-steroid_4-dehydrogenase_activity |
| GO:0047598 | 1 | 7-dehydrocholesterol_reductase_activity |
| GO:0047620 | 1 | acylglycerol_kinase_activity |
| GO:0047621 | 1 | acylpyruvate_hydrolase_activity |
| GO:0047638 | 1 | albendazole_monooxygenase_activity |
| GO:0047676 | 1 | arachidonate-CoA_ligase_activity |
| GO:0047704 | 1 | bile-salt_sulfotransferase_activity |
| GO:0047708 | 1 | biotinidase_activity |
| GO:0047710 | 1 | bis(5'-adenosyl)-triphosphatase_activity |
| GO:0047714 | 1 | galactolipase_activity |
| GO:0047726 | 1 | iron-cytochrome-c_reductase_activity |
| GO:0047730 | 1 | carnosine_synthase_activity |
| GO:0047734 | 1 | CDP-glycerol_diphosphatase_activity |
| GO:0047743 | 1 | chlordecone_reductase_activity |
| GO:0047747 | 1 | cholate-CoA_ligase_activity |
| GO:0047757 | 1 | chondroitin-glucuronate_5-epimerase_activity |
| GO:0047783 | 1 | corticosterone_18-monooxygenase_activity |
| GO:0047800 | 1 | cysteamine_dioxygenase_activity |
| GO:0047837 | 1 | D-xylose_1-dehydrogenase_(NADP+)_activity |
| GO:0047840 | 1 | dCTP_diphosphatase_activity |
| GO:0047844 | 1 | deoxycytidine_deaminase_activity |
| GO:0047865 | 1 | dimethylglycine_dehydrogenase_activity |
| GO:0047874 | 1 | dolichyldiphosphatase_activity |
| GO:0047915 | 1 | ganglioside_galactosyltransferase_activity |
| GO:0047933 | 1 | glucose-1,6-bisphosphate_synthase_activity |
| GO:0047936 | 1 | glucose_1-dehydrogenase_[NAD(P)]_activity |
| GO:0047939 | 1 | L-glucuronate_reductase_activity |
| GO:0047945 | 1 | L-glutamine:pyruvate_aminotransferase_activity |
| GO:0047962 | 1 | glycine_N-benzoyltransferase_activity |
| GO:0047963 | 1 | glycine_N-choloyltransferase_activity |
| GO:0047969 | 1 | glyoxylate_oxidase_activity |
| GO:0047977 | 1 | hepoxilin-epoxide_hydrolase_activity |
| GO:0047982 | 1 | homocysteine_desulfhydrase_activity |
| GO:0047988 | 1 | hydroxyacid-oxoacid_transhydrogenase_activity |
| GO:0048006 | 1 | antigen_processing_and_presentation,_endogenous_lipid_antigen_via_MHC_class_Ib |
| GO:0048033 | 1 | heme_o_metabolic_process |
| GO:0048034 | 1 | heme_O_biosynthetic_process |
| GO:0048040 | 1 | UDP-glucuronate_decarboxylase_activity |
| GO:0048074 | 1 | negative_regulation_of_eye_pigmentation |
| GO:0048075 | 1 | positive_regulation_of_eye_pigmentation |
| GO:0048086 | 1 | negative_regulation_of_developmental_pigmentation |
| GO:0048160 | 1 | primary_follicle_stage |
| GO:0048176 | 1 | regulation_of_hepatocyte_growth_factor_biosynthetic_process |
| GO:0048178 | 1 | negative_regulation_of_hepatocyte_growth_factor_biosynthetic_process |
| GO:0048180 | 1 | activin_complex |
| GO:0048194 | 1 | Golgi_vesicle_budding |
| GO:0048203 | 1 | vesicle_targeting,_trans-Golgi_to_endosome |
| GO:0048210 | 1 | Golgi_vesicle_fusion_to_target_membrane |
| GO:0048213 | 1 | Golgi_vesicle_prefusion_complex_stabilization |
| GO:0048237 | 1 | rough_endoplasmic_reticulum_lumen |
| GO:0048239 | 1 | negative_regulation_of_DNA_recombination_at_telomere |
| GO:0048242 | 1 | epinephrine_secretion |
| GO:0048243 | 1 | norepinephrine_secretion |
| GO:0048244 | 1 | phytanoyl-CoA_dioxygenase_activity |
| GO:0048248 | 1 | CXCR3_chemokine_receptor_binding |
| GO:0048262 | 1 | determination_of_dorsal/ventral_asymmetry |
| GO:0048269 | 1 | methionine_adenosyltransferase_complex |
| GO:0048270 | 1 | methionine_adenosyltransferase_regulator_activity |
| GO:0048289 | 1 | isotype_switching_to_IgE_isotypes |
| GO:0048291 | 1 | isotype_switching_to_IgG_isotypes |
| GO:0048308 | 1 | organelle_inheritance |
| GO:0048313 | 1 | Golgi_inheritance |
| GO:0048327 | 1 | axial_mesodermal_cell_fate_specification |
| GO:0048338 | 1 | mesoderm_structural_organization |
| GO:0048352 | 1 | paraxial_mesoderm_structural_organization |
| GO:0048370 | 1 | lateral_mesoderm_formation |
| GO:0048371 | 1 | lateral_mesodermal_cell_differentiation |
| GO:0048389 | 1 | intermediate_mesoderm_development |
| GO:0048392 | 1 | intermediate_mesodermal_cell_differentiation |
| GO:0048403 | 1 | brain-derived_neurotrophic_factor_binding |
| GO:0048476 | 1 | Holliday_junction_resolvase_complex |
| GO:0048486 | 1 | parasympathetic_nervous_system_development |
| GO:0048553 | 1 | negative_regulation_of_metalloenzyme_activity |
| GO:0048561 | 1 | establishment_of_organ_orientation |
| GO:0048570 | 1 | notochord_morphogenesis |
| GO:0048597 | 1 | post-embryonic_camera-type_eye_morphogenesis |
| GO:0048613 | 1 | embryonic_ectodermal_digestive_tract_morphogenesis |
| GO:0048621 | 1 | post-embryonic_digestive_tract_morphogenesis |
| GO:0048632 | 1 | negative_regulation_of_skeletal_muscle_tissue_growth |
| GO:0048669 | 1 | collateral_sprouting_in_absence_of_injury |
| GO:0048677 | 1 | axon_extension_involved_in_regeneration |
| GO:0048682 | 1 | sprouting_of_injured_axon |
| GO:0048685 | 1 | negative_regulation_of_collateral_sprouting_of_intact_axon_in_response_to_injury |
| GO:0048687 | 1 | positive_regulation_of_sprouting_of_injured_axon |
| GO:0048688 | 1 | negative_regulation_of_sprouting_of_injured_axon |
| GO:0048691 | 1 | positive_regulation_of_axon_extension_involved_in_regeneration |
| GO:0048692 | 1 | negative_regulation_of_axon_extension_involved_in_regeneration |
| GO:0048749 | 1 | compound_eye_development |
| GO:0048763 | 1 | calcium-induced_calcium_release_activity |
| GO:0048773 | 1 | erythrophore_differentiation |
| GO:0048789 | 1 | cytoskeletal_matrix_organization_at_active_zone |
| GO:0048790 | 1 | maintenance_of_presynaptic_active_zone_structure |
| GO:0048820 | 1 | hair_follicle_maturation |
| GO:0048855 | 1 | adenohypophysis_morphogenesis |
| GO:0048865 | 1 | stem_cell_fate_commitment |
| GO:0048874 | 1 | homeostasis_of_number_of_cells_in_a_free-living_population |
| GO:0048894 | 1 | efferent_axon_development_in_a_lateral_line_nerve |
| GO:0050031 | 1 | L-pipecolate_oxidase_activity |
| GO:0050038 | 1 | L-xylulose_reductase_(NADP+)_activity |
| GO:0050046 | 1 | lathosterol_oxidase_activity |
| GO:0050080 | 1 | malonyl-CoA_decarboxylase_activity |
| GO:0050104 | 1 | L-gulonate_3-dehydrogenase_activity |
| GO:0050113 | 1 | inositol_oxygenase_activity |
| GO:0050115 | 1 | myosin-light-chain-phosphatase_activity |
| GO:0050121 | 1 | N-acylglucosamine_2-epimerase_activity |
| GO:0050124 | 1 | N-acylneuraminate-9-phosphatase_activity |
| GO:0050145 | 1 | nucleoside_phosphate_kinase_activity |
| GO:0050152 | 1 | omega-amidase_activity |
| GO:0050178 | 1 | phenylpyruvate_tautomerase_activity |
| GO:0050197 | 1 | phytanate-CoA_ligase_activity |
| GO:0050201 | 1 | fucokinase_activity |
| GO:0050221 | 1 | prostaglandin-E2_9-reductase_activity |
| GO:0050277 | 1 | sedoheptulokinase_activity |
| GO:0050290 | 1 | sphingomyelin_phosphodiesterase_D_activity |
| GO:0050313 | 1 | sulfur_dioxygenase_activity |
| GO:0050333 | 1 | thiamin-triphosphatase_activity |
| GO:0050353 | 1 | trimethyllysine_dioxygenase_activity |
| GO:0050379 | 1 | UDP-glucuronate_5'-epimerase_activity |
| GO:0050429 | 1 | calcium-dependent_phospholipase_C_activity |
| GO:0050462 | 1 | N-acetylneuraminate_synthase_activity |
| GO:0050479 | 1 | glyceryl-ether_monooxygenase_activity |
| GO:0050480 | 1 | imidazolonepropionase_activity |
| GO:0050512 | 1 | lactosylceramide_4-alpha-galactosyltransferase_activity |
| GO:0050528 | 1 | acyloxyacyl_hydrolase_activity |
| GO:0050560 | 1 | aspartate-tRNA(Asn)_ligase_activity |
| GO:0050561 | 1 | glutamate-tRNA(Gln)_ligase_activity |
| GO:0050571 | 1 | 1,5-anhydro-D-fructose_reductase_activity |
| GO:0050577 | 1 | GDP-L-fucose_synthase_activity |
| GO:0050591 | 1 | quinine_3-monooxygenase_activity |
| GO:0050613 | 1 | delta14-sterol_reductase_activity |
| GO:0050614 | 1 | delta24-sterol_reductase_activity |
| GO:0050646 | 1 | 5-oxo-6E,8Z,11Z,14Z-icosatetraenoic_acid_binding |
| GO:0050647 | 1 | 5-hydroxy-6E,8Z,11Z,14Z-icosatetraenoic_acid_binding |
| GO:0050648 | 1 | 5(S)-hydroxyperoxy-6E,8Z,11Z,14Z-icosatetraenoic_acid_binding |
| GO:0050649 | 1 | testosterone_6-beta-hydroxylase_activity |
| GO:0050652 | 1 | dermatan_sulfate_proteoglycan_biosynthetic_process,_polysaccharide_chain_biosynthetic_process |
| GO:0050659 | 1 | N-acetylgalactosamine_4-sulfate_6-O-sulfotransferase_activity |
| GO:0050668 | 1 | positive_regulation_of_homocysteine_metabolic_process |
| GO:0050674 | 1 | urothelial_cell_proliferation |
| GO:0050675 | 1 | regulation_of_urothelial_cell_proliferation |
| GO:0050677 | 1 | positive_regulation_of_urothelial_cell_proliferation |
| GO:0050683 | 1 | AF-1_domain_binding |
| GO:0050703 | 1 | interleukin-1_alpha_secretion |
| GO:0050713 | 1 | negative_regulation_of_interleukin-1_beta_secretion |
| GO:0050720 | 1 | interleukin-1_beta_biosynthetic_process |
| GO:0050722 | 1 | regulation_of_interleukin-1_beta_biosynthetic_process |
| GO:0050725 | 1 | positive_regulation_of_interleukin-1_beta_biosynthetic_process |
| GO:0050747 | 1 | positive_regulation_of_lipoprotein_metabolic_process |
| GO:0050752 | 1 | regulation_of_fractalkine_biosynthetic_process |
| GO:0050754 | 1 | positive_regulation_of_fractalkine_biosynthetic_process |
| GO:0050756 | 1 | fractalkine_metabolic_process |
| GO:0050758 | 1 | regulation_of_thymidylate_synthase_biosynthetic_process |
| GO:0050760 | 1 | negative_regulation_of_thymidylate_synthase_biosynthetic_process |
| GO:0050783 | 1 | cocaine_metabolic_process |
| GO:0050785 | 1 | advanced_glycation_end-product_receptor_activity |
| GO:0050809 | 1 | diazepam_binding |
| GO:0050812 | 1 | regulation_of_acyl-CoA_biosynthetic_process |
| GO:0050822 | 1 | peptide_stabilization |
| GO:0050823 | 1 | peptide_antigen_stabilization |
| GO:0050827 | 1 | toxin_receptor_binding |
| GO:0050831 | 1 | male-specific_defense_response_to_bacterium |
| GO:0050833 | 1 | pyruvate_transmembrane_transporter_activity |
| GO:0050859 | 1 | negative_regulation_of_B_cell_receptor_signaling_pathway |
| GO:0050894 | 1 | determination_of_affect |
| GO:0050924 | 1 | positive_regulation_of_negative_chemotaxis |
| GO:0050928 | 1 | negative_regulation_of_positive_chemotaxis |
| GO:0050929 | 1 | induction_of_negative_chemotaxis |
| GO:0050968 | 1 | detection_of_chemical_stimulus_involved_in_sensory_perception_of_pain |
| GO:0050973 | 1 | detection_of_mechanical_stimulus_involved_in_equilibrioception |
| GO:0050983 | 1 | deoxyhypusine_biosynthetic_process_from_spermidine |
| GO:0051025 | 1 | negative_regulation_of_immunoglobulin_secretion |
| GO:0051031 | 1 | tRNA_transport |
| GO:0051040 | 1 | regulation_of_calcium-independent_cell-cell_adhesion |
| GO:0051041 | 1 | positive_regulation_of_calcium-independent_cell-cell_adhesion |
| GO:0051068 | 1 | dihydrolipoamide_metabolic_process |
| GO:0051089 | 1 | constitutive_protein_ectodomain_proteolysis |
| GO:0051102 | 1 | DNA_ligation_involved_in_DNA_recombination |
| GO:0051105 | 1 | regulation_of_DNA_ligation |
| GO:0051106 | 1 | positive_regulation_of_DNA_ligation |
| GO:0051126 | 1 | negative_regulation_of_actin_nucleation |
| GO:0051139 | 1 | metal_ion:hydrogen_antiporter_activity |
| GO:0051160 | 1 | L-xylitol_catabolic_process |
| GO:0051164 | 1 | L-xylitol_metabolic_process |
| GO:0051192 | 1 | prosthetic_group_binding |
| GO:0051257 | 1 | spindle_midzone_assembly_involved_in_meiosis |
| GO:0051264 | 1 | mono-olein_transacylation_activity |
| GO:0051265 | 1 | diolein_transacylation_activity |
| GO:0051267 | 1 | CP2_mannose-ethanolamine_phosphotransferase_activity |
| GO:0051280 | 1 | negative_regulation_of_release_of_sequestered_calcium_ion_into_cytosol |
| GO:0051311 | 1 | meiotic_metaphase_plate_congression |
| GO:0051317 | 1 | attachment_of_spindle_microtubules_to_meiotic_chromosome |
| GO:0051326 | 1 | telophase |
| GO:0051343 | 1 | positive_regulation_of_cyclic-nucleotide_phosphodiesterase_activity |
| GO:0051355 | 1 | proprioception_involved_in_equilibrioception |
| GO:0051377 | 1 | mannose-ethanolamine_phosphotransferase_activity |
| GO:0051381 | 1 | histamine_binding |
| GO:0051389 | 1 | inactivation_of_MAPKK_activity |
| GO:0051430 | 1 | corticotropin-releasing_hormone_receptor_1_binding |
| GO:0051435 | 1 | BH4_domain_binding |
| GO:0051447 | 1 | negative_regulation_of_meiotic_cell_cycle |
| GO:0051460 | 1 | negative_regulation_of_corticotropin_secretion |
| GO:0051531 | 1 | NFAT_protein_import_into_nucleus |
| GO:0051542 | 1 | elastin_biosynthetic_process |
| GO:0051543 | 1 | regulation_of_elastin_biosynthetic_process |
| GO:0051545 | 1 | negative_regulation_of_elastin_biosynthetic_process |
| GO:0051585 | 1 | negative_regulation_of_dopamine_uptake_involved_in_synaptic_transmission |
| GO:0051595 | 1 | response_to_methylglyoxal |
| GO:0051598 | 1 | meiotic_recombination_checkpoint |
| GO:0051615 | 1 | histamine_uptake |
| GO:0051621 | 1 | regulation_of_norepinephrine_uptake |
| GO:0051622 | 1 | negative_regulation_of_norepinephrine_uptake |
| GO:0051643 | 1 | endoplasmic_reticulum_localization |
| GO:0051658 | 1 | maintenance_of_nucleus_location |
| GO:0051660 | 1 | establishment_of_centrosome_localization |
| GO:0051664 | 1 | nuclear_pore_localization |
| GO:0051674 | 1 | localization_of_cell |
| GO:0051717 | 1 | inositol-1,3,4,5-tetrakisphosphate_3-phosphatase_activity |
| GO:0051718 | 1 | DNA_(cytosine-5-)-methyltransferase_activity,_acting_on_CpG_substrates |
| GO:0051722 | 1 | protein_C-terminal_methylesterase_activity |
| GO:0051723 | 1 | protein_methylesterase_activity |
| GO:0051724 | 1 | NAD_transporter_activity |
| GO:0051732 | 1 | polyribonucleotide_kinase_activity |
| GO:0051736 | 1 | ATP-dependent_polyribonucleotide_5'-hydroxyl-kinase_activity |
| GO:0051748 | 1 | UTP-monosaccharide-1-phosphate_uridylyltransferase_activity |
| GO:0051754 | 1 | meiotic_sister_chromatid_cohesion,_centromeric |
| GO:0051786 | 1 | all-trans-retinol_13,14-reductase_activity |
| GO:0051792 | 1 | medium-chain_fatty_acid_biosynthetic_process |
| GO:0051793 | 1 | medium-chain_fatty_acid_catabolic_process |
| GO:0051800 | 1 | phosphatidylinositol-3,4-bisphosphate_3-phosphatase_activity |
| GO:0051805 | 1 | evasion_or_tolerance_of_immune_response_of_other_organism_involved_in_symbiotic_interaction |
| GO:0051807 | 1 | evasion_or_tolerance_of_defense_response_of_other_organism_involved_in_symbiotic_interaction |
| GO:0051821 | 1 | dissemination_or_transmission_of_organism_from_other_organism_involved_in_symbiotic_interaction |
| GO:0051882 | 1 | mitochondrial_depolarization |
| GO:0051884 | 1 | regulation_of_anagen |
| GO:0051885 | 1 | positive_regulation_of_anagen |
| GO:0051892 | 1 | negative_regulation_of_cardioblast_differentiation |
| GO:0051903 | 1 | S-(hydroxymethyl)glutathione_dehydrogenase_activity |
| GO:0051908 | 1 | double-stranded_DNA_specific_5'-3'_exodeoxyribonuclease_activity |
| GO:0051913 | 1 | regulation_of_synaptic_plasticity_by_chemical_substance |
| GO:0051914 | 1 | positive_regulation_of_synaptic_plasticity_by_chemical_substance |
| GO:0051915 | 1 | induction_of_synaptic_plasticity_by_chemical_substance |
| GO:0051939 | 1 | gamma-aminobutyric_acid_import |
| GO:0051945 | 1 | negative_regulation_of_catecholamine_uptake_involved_in_synaptic_transmission |
| GO:0051958 | 1 | methotrexate_transport |
| GO:0051977 | 1 | lysophospholipid_transport |
| GO:0051987 | 1 | positive_regulation_of_attachment_of_spindle_microtubules_to_kinetochore |
| GO:0051990 | 1 | (R)-2-hydroxyglutarate_dehydrogenase_activity |
| GO:0051996 | 1 | squalene_synthase_activity |
| GO:0052026 | 1 | modulation_by_symbiont_of_host_transcription |
| GO:0052027 | 1 | modulation_by_symbiont_of_host_signal_transduction_pathway |
| GO:0052040 | 1 | modulation_by_symbiont_of_host_programmed_cell_death |
| GO:0052042 | 1 | positive_regulation_by_symbiont_of_host_programmed_cell_death |
| GO:0052047 | 1 | interaction_with_other_organism_via_secreted_substance_involved_in_symbiotic_interaction |
| GO:0052063 | 1 | induction_by_symbiont_of_defense-related_host_nitric_oxide_production |
| GO:0052066 | 1 | entry_of_symbiont_into_host_cell_by_promotion_of_host_phagocytosis |
| GO:0052097 | 1 | interspecies_quorum_sensing |
| GO:0052106 | 1 | quorum_sensing_involved_in_interaction_with_host |
| GO:0052150 | 1 | modulation_by_symbiont_of_host_apoptotic_process |
| GO:0052151 | 1 | positive_regulation_by_symbiont_of_host_apoptotic_process |
| GO:0052163 | 1 | modulation_by_symbiont_of_defense-related_host_nitric_oxide_production |
| GO:0052182 | 1 | modification_by_host_of_symbiont_morphology_or_physiology_via_secreted_substance |
| GO:0052190 | 1 | modulation_by_symbiont_of_host_phagocytosis |
| GO:0052191 | 1 | positive_regulation_by_symbiont_of_host_phagocytosis |
| GO:0052212 | 1 | modification_of_morphology_or_physiology_of_other_organism_via_secreted_substance_involved_in_symbiotic_interaction |
| GO:0052213 | 1 | interaction_with_symbiont_via_secreted_substance_involved_in_symbiotic_interaction |
| GO:0052231 | 1 | modulation_of_phagocytosis_in_other_organism_involved_in_symbiotic_interaction |
| GO:0052248 | 1 | modulation_of_programmed_cell_death_in_other_organism_involved_in_symbiotic_interaction |
| GO:0052250 | 1 | modulation_of_signal_transduction_in_other_organism_involved_in_symbiotic_interaction |
| GO:0052263 | 1 | induction_by_organism_of_defense-related_nitric_oxide_production_in_other_organism_involved_in_symbiotic_interaction |
| GO:0052302 | 1 | modulation_by_organism_of_defense-related_nitric_oxide_production_in_other_organism_involved_in_symbiotic_interaction |
| GO:0052330 | 1 | positive_regulation_by_organism_of_programmed_cell_death_in_other_organism_involved_in_symbiotic_interaction |
| GO:0052345 | 1 | positive_regulation_by_organism_of_defense-related_nitric_oxide_production_in_other_organism_involved_in_symbiotic_interaction |
| GO:0052347 | 1 | positive_regulation_by_symbiont_of_defense-related_host_nitric_oxide_production |
| GO:0052370 | 1 | entry_of_organism_into_cell_of_other_organism_by_promotion_of_phagocytosis_in_other_organism_involved_in_symbiotic_interaction |
| GO:0052381 | 1 | tRNA_dimethylallyltransferase_activity |
| GO:0052433 | 1 | modulation_by_organism_of_apoptotic_process_in_other_organism_involved_in_symbiotic_interaction |
| GO:0052501 | 1 | positive_regulation_by_organism_of_apoptotic_process_in_other_organism_involved_in_symbiotic_interaction |
| GO:0052522 | 1 | positive_regulation_by_organism_of_phagocytosis_in_other_organism_involved_in_symbiotic_interaction |
| GO:0052558 | 1 | induction_by_organism_of_immune_response_of_other_organism_involved_in_symbiotic_interaction |
| GO:0052559 | 1 | induction_by_symbiont_of_host_immune_response |
| GO:0052590 | 1 | sn-glycerol-3-phosphate:ubiquinone_oxidoreductase_activity |
| GO:0052591 | 1 | sn-glycerol-3-phosphate:ubiquinone-8_oxidoreductase_activity |
| GO:0052597 | 1 | diamine_oxidase_activity |
| GO:0052598 | 1 | histamine_oxidase_activity |
| GO:0052599 | 1 | methylputrescine_oxidase_activity |
| GO:0052600 | 1 | propane-1,3-diamine_oxidase_activity |
| GO:0052629 | 1 | phosphatidylinositol-3,5-bisphosphate_3-phosphatase_activity |
| GO:0052657 | 1 | guanine_phosphoribosyltransferase_activity |
| GO:0052692 | 1 | raffinose_alpha-galactosidase_activity |
| GO:0052725 | 1 | inositol-1,3,4-trisphosphate_6-kinase_activity |
| GO:0052726 | 1 | inositol-1,3,4-trisphosphate_5-kinase_activity |
| GO:0052731 | 1 | phosphocholine_phosphatase_activity |
| GO:0052732 | 1 | phosphoethanolamine_phosphatase_activity |
| GO:0052739 | 1 | phosphatidylserine_1-acylhydrolase_activity |
| GO:0052740 | 1 | 1-acyl-2-lysophosphatidylserine_acylhydrolase_activity |
| GO:0052746 | 1 | inositol_phosphorylation |
| GO:0052798 | 1 | beta-galactoside_alpha-2,3-sialyltransferase_activity |
| GO:0052821 | 1 | DNA-7-methyladenine_glycosylase_activity |
| GO:0052822 | 1 | DNA-3-methylguanine_glycosylase_activity |
| GO:0052826 | 1 | inositol_hexakisphosphate_2-phosphatase_activity |
| GO:0052827 | 1 | inositol_pentakisphosphate_phosphatase_activity |
| GO:0052872 | 1 | tocotrienol_omega-hydroxylase_activity |
| GO:0052884 | 1 | all-trans-retinyl-palmitate_hydrolase,_11-cis_retinol_forming_activity |
| GO:0052885 | 1 | all-trans-retinyl-ester_hydrolase,_11-cis_retinol_forming_activity |
| GO:0052890 | 1 | oxidoreductase_activity,_acting_on_the_CH-CH_group_of_donors,_with_a_flavin_as_acceptor |
| GO:0052894 | 1 | norspermine:oxygen_oxidoreductase_activity |
| GO:0052895 | 1 | N1-acetylspermine:oxygen_oxidoreductase_(N1-acetylspermidine-forming)_activity |
| GO:0052899 | 1 | N(1),N(12)-diacetylspermine:oxygen_oxidoreductase_(3-acetamidopropanal-forming)_activity |
| GO:0052902 | 1 | spermidine:oxygen_oxidoreductase_(3-aminopropanal-forming)_activity |
| GO:0052903 | 1 | N1-acetylspermine:oxygen_oxidoreductase_(3-acetamidopropanal-forming)_activity |
| GO:0052904 | 1 | N1-acetylspermidine:oxygen_oxidoreductase_(3-acetamidopropanal-forming)_activity |
| GO:0052906 | 1 | tRNA_(guanine(37)-N(1))-methyltransferase_activity |
| GO:0052909 | 1 | 18S_rRNA_(adenine(1779)-N(6)/adenine(1780)-N(6))-dimethyltransferase_activity |
| GO:0052917 | 1 | dol-P-Man:Man(7)GlcNAc(2)-PP-Dol_alpha-1,6-mannosyltransferase_activity |
| GO:0052918 | 1 | dol-P-Man:Man(8)GlcNAc(2)-PP-Dol_alpha-1,2-mannosyltransferase_activity |
| GO:0052925 | 1 | dol-P-Man:Man(5)GlcNAc(2)-PP-Dol_alpha-1,3-mannosyltransferase_activity |
| GO:0052926 | 1 | dol-P-Man:Man(6)GlcNAc(2)-PP-Dol_alpha-1,2-mannosyltransferase_activity |
| GO:0052927 | 1 | CTP:tRNA_cytidylyltransferase_activity |
| GO:0052928 | 1 | CTP:3'-cytidine-tRNA_cytidylyltransferase_activity |
| GO:0052929 | 1 | ATP:3'-cytidine-cytidine-tRNA_adenylyltransferase_activity |
| GO:0055018 | 1 | regulation_of_cardiac_muscle_fiber_development |
| GO:0055020 | 1 | positive_regulation_of_cardiac_muscle_fiber_development |
| GO:0055022 | 1 | negative_regulation_of_cardiac_muscle_tissue_growth |
| GO:0055023 | 1 | positive_regulation_of_cardiac_muscle_tissue_growth |
| GO:0055071 | 1 | manganese_ion_homeostasis |
| GO:0055076 | 1 | transition_metal_ion_homeostasis |
| GO:0055100 | 1 | adiponectin_binding |
| GO:0055104 | 1 | ligase_inhibitor_activity |
| GO:0055105 | 1 | ubiquitin-protein_ligase_inhibitor_activity |
| GO:0055107 | 1 | Golgi_to_secretory_granule_transport |
| GO:0055108 | 1 | Golgi_to_transport_vesicle_transport |
| GO:0055130 | 1 | D-alanine_catabolic_process |
| GO:0055131 | 1 | C3HC4-type_RING_finger_domain_binding |
| GO:0060001 | 1 | minus-end_directed_microfilament_motor_activity |
| GO:0060005 | 1 | vestibular_reflex |
| GO:0060010 | 1 | Sertoli_cell_fate_commitment |
| GO:0060016 | 1 | granulosa_cell_development |
| GO:0060035 | 1 | notochord_cell_development |
| GO:0060050 | 1 | positive_regulation_of_protein_glycosylation |
| GO:0060064 | 1 | Spemann_organizer_formation_at_the_anterior_end_of_the_primitive_streak |
| GO:0060073 | 1 | micturition |
| GO:0060086 | 1 | circadian_temperature_homeostasis |
| GO:0060091 | 1 | kinocilium |
| GO:0060096 | 1 | serotonin_secretion,_neurotransmission |
| GO:0060112 | 1 | generation_of_ovulation_cycle_rhythm |
| GO:0060118 | 1 | vestibular_receptor_cell_development |
| GO:0060127 | 1 | prolactin_secreting_cell_differentiation |
| GO:0060133 | 1 | somatotropin_secreting_cell_development |
| GO:0060139 | 1 | positive_regulation_of_apoptotic_process_by_virus |
| GO:0060163 | 1 | subpallium_neuron_fate_commitment |
| GO:0060165 | 1 | regulation_of_timing_of_subpallium_neuron_differentiation |
| GO:0060168 | 1 | positive_regulation_of_adenosine_receptor_signaling_pathway |
| GO:0060169 | 1 | negative_regulation_of_adenosine_receptor_signaling_pathway |
| GO:0060175 | 1 | brain-derived_neurotrophic_factor-activated_receptor_activity |
| GO:0060178 | 1 | regulation_of_exocyst_localization |
| GO:0060209 | 1 | estrus |
| GO:0060210 | 1 | metestrus |
| GO:0060217 | 1 | hemangioblast_cell_differentiation |
| GO:0060220 | 1 | camera-type_eye_photoreceptor_cell_fate_commitment |
| GO:0060244 | 1 | negative_regulation_of_cell_proliferation_involved_in_contact_inhibition |
| GO:0060245 | 1 | detection_of_cell_density |
| GO:0060246 | 1 | detection_of_cell_density_by_contact_stimulus |
| GO:0060248 | 1 | detection_of_cell_density_by_contact_stimulus_involved_in_contact_inhibition |
| GO:0060254 | 1 | regulation_of_N-terminal_protein_palmitoylation |
| GO:0060262 | 1 | negative_regulation_of_N-terminal_protein_palmitoylation |
| GO:0060265 | 1 | positive_regulation_of_respiratory_burst_involved_in_inflammatory_response |
| GO:0060280 | 1 | negative_regulation_of_ovulation |
| GO:0060283 | 1 | negative_regulation_of_oocyte_development |
| GO:0060299 | 1 | negative_regulation_of_sarcomere_organization |
| GO:0060301 | 1 | positive_regulation_of_cytokine_activity |
| GO:0060302 | 1 | negative_regulation_of_cytokine_activity |
| GO:0060304 | 1 | regulation_of_phosphatidylinositol_dephosphorylation |
| GO:0060305 | 1 | regulation_of_cell_diameter |
| GO:0060310 | 1 | regulation_of_elastin_catabolic_process |
| GO:0060311 | 1 | negative_regulation_of_elastin_catabolic_process |
| GO:0060313 | 1 | negative_regulation_of_blood_vessel_remodeling |
| GO:0060327 | 1 | cytoplasmic_actin-based_contraction_involved_in_cell_motility |
| GO:0060339 | 1 | negative_regulation_of_type_I_interferon-mediated_signaling_pathway |
| GO:0060342 | 1 | photoreceptor_inner_segment_membrane |
| GO:0060345 | 1 | spleen_trabecula_formation |
| GO:0060350 | 1 | endochondral_bone_morphogenesis |
| GO:0060354 | 1 | negative_regulation_of_cell_adhesion_molecule_production |
| GO:0060355 | 1 | positive_regulation_of_cell_adhesion_molecule_production |
| GO:0060356 | 1 | leucine_import |
| GO:0060366 | 1 | lambdoid_suture_morphogenesis |
| GO:0060367 | 1 | sagittal_suture_morphogenesis |
| GO:0060372 | 1 | regulation_of_atrial_cardiomyocyte_membrane_repolarization |
| GO:0060376 | 1 | positive_regulation_of_mast_cell_differentiation |
| GO:0060377 | 1 | negative_regulation_of_mast_cell_differentiation |
| GO:0060380 | 1 | regulation_of_single-stranded_telomeric_DNA_binding |
| GO:0060381 | 1 | positive_regulation_of_single-stranded_telomeric_DNA_binding |
| GO:0060382 | 1 | regulation_of_DNA_strand_elongation |
| GO:0060383 | 1 | positive_regulation_of_DNA_strand_elongation |
| GO:0060400 | 1 | negative_regulation_of_growth_hormone_receptor_signaling_pathway |
| GO:0060407 | 1 | negative_regulation_of_penile_erection |
| GO:0060408 | 1 | regulation_of_acetylcholine_metabolic_process |
| GO:0060409 | 1 | positive_regulation_of_acetylcholine_metabolic_process |
| GO:0060422 | 1 | peptidyl-dipeptidase_inhibitor_activity |
| GO:0060427 | 1 | lung_connective_tissue_development |
| GO:0060432 | 1 | lung_pattern_specification_process |
| GO:0060434 | 1 | bronchus_morphogenesis |
| GO:0060436 | 1 | bronchiole_morphogenesis |
| GO:0060439 | 1 | trachea_morphogenesis |
| GO:0060446 | 1 | branching_involved_in_open_tracheal_system_development |
| GO:0060450 | 1 | positive_regulation_of_hindgut_contraction |
| GO:0060458 | 1 | right_lung_development |
| GO:0060459 | 1 | left_lung_development |
| GO:0060460 | 1 | left_lung_morphogenesis |
| GO:0060461 | 1 | right_lung_morphogenesis |
| GO:0060464 | 1 | lung_lobe_formation |
| GO:0060465 | 1 | pharynx_development |
| GO:0060473 | 1 | cortical_granule |
| GO:0060482 | 1 | lobar_bronchus_development |
| GO:0060495 | 1 | cell-cell_signaling_involved_in_lung_development |
| GO:0060496 | 1 | mesenchymal-epithelial_cell_signaling_involved_in_lung_development |
| GO:0060502 | 1 | epithelial_cell_proliferation_involved_in_lung_morphogenesis |
| GO:0060503 | 1 | bud_dilation_involved_in_lung_branching |
| GO:0060516 | 1 | primary_prostatic_bud_elongation |
| GO:0060517 | 1 | epithelial_cell_proliferation_involved_in_prostatic_bud_elongation |
| GO:0060520 | 1 | activation_of_prostate_induction_by_androgen_receptor_signaling_pathway |
| GO:0060546 | 1 | negative_regulation_of_necroptosis |
| GO:0060549 | 1 | regulation_of_fructose_1,6-bisphosphate_1-phosphatase_activity |
| GO:0060550 | 1 | positive_regulation_of_fructose_1,6-bisphosphate_1-phosphatase_activity |
| GO:0060551 | 1 | regulation_of_fructose_1,6-bisphosphate_metabolic_process |
| GO:0060552 | 1 | positive_regulation_of_fructose_1,6-bisphosphate_metabolic_process |
| GO:0060554 | 1 | induction_of_necroptosis_of_activated-T_cells |
| GO:0060565 | 1 | inhibition_of_mitotic_anaphase-promoting_complex_activity |
| GO:0060577 | 1 | pulmonary_vein_morphogenesis |
| GO:0060580 | 1 | ventral_spinal_cord_interneuron_fate_determination |
| GO:0060588 | 1 | negative_regulation_of_lipoprotein_lipid_oxidation |
| GO:0060598 | 1 | dichotomous_subdivision_of_terminal_units_involved_in_mammary_gland_duct_morphogenesis |
| GO:0060611 | 1 | mammary_gland_fat_development |
| GO:0060623 | 1 | regulation_of_chromosome_condensation |
| GO:0060629 | 1 | regulation_of_homologous_chromosome_segregation |
| GO:0060649 | 1 | mammary_gland_bud_elongation |
| GO:0060658 | 1 | nipple_morphogenesis |
| GO:0060659 | 1 | nipple_sheath_formation |
| GO:0060661 | 1 | submandibular_salivary_gland_formation |
| GO:0060668 | 1 | regulation_of_branching_involved_in_salivary_gland_morphogenesis_by_extracellular_matrix-epithelial_cell_signaling |
| GO:0060681 | 1 | branch_elongation_involved_in_ureteric_bud_branching |
| GO:0060683 | 1 | regulation_of_branching_involved_in_salivary_gland_morphogenesis_by_epithelial-mesenchymal_signaling |
| GO:0060690 | 1 | epithelial_cell_differentiation_involved_in_salivary_gland_development |
| GO:0060691 | 1 | epithelial_cell_maturation_involved_in_salivary_gland_development |
| GO:0060695 | 1 | negative_regulation_of_cholesterol_transporter_activity |
| GO:0060699 | 1 | regulation_of_endoribonuclease_activity |
| GO:0060709 | 1 | glycogen_cell_differentiation_involved_in_embryonic_placenta_development |
| GO:0060721 | 1 | regulation_of_spongiotrophoblast_cell_proliferation |
| GO:0060723 | 1 | regulation_of_cell_proliferation_involved_in_embryonic_placenta_development |
| GO:0060731 | 1 | positive_regulation_of_intestinal_epithelial_structure_maintenance |
| GO:0060739 | 1 | mesenchymal-epithelial_cell_signaling_involved_in_prostate_gland_development |
| GO:0060751 | 1 | branch_elongation_involved_in_mammary_gland_duct_branching |
| GO:0060767 | 1 | epithelial_cell_proliferation_involved_in_prostate_gland_development |
| GO:0060782 | 1 | regulation_of_mesenchymal_cell_proliferation_involved_in_prostate_gland_development |
| GO:0060783 | 1 | mesenchymal_smoothened_signaling_pathway_involved_in_prostate_gland_development |
| GO:0060784 | 1 | regulation_of_cell_proliferation_involved_in_tissue_homeostasis |
| GO:0060785 | 1 | regulation_of_apoptosis_involved_in_tissue_homeostasis |
| GO:0060796 | 1 | regulation_of_transcription_involved_in_primary_germ_layer_cell_fate_commitment |
| GO:0060800 | 1 | regulation_of_cell_differentiation_involved_in_embryonic_placenta_development |
| GO:0060802 | 1 | epiblast_cell-extraembryonic_ectoderm_cell_signaling_involved_in_anterior/posterior_axis_specification |
| GO:0060803 | 1 | BMP_signaling_pathway_involved_in_mesodermal_cell_fate_specification |
| GO:0060804 | 1 | positive_regulation_of_Wnt_receptor_signaling_pathway_by_BMP_signaling_pathway |
| GO:0060806 | 1 | negative_regulation_of_cell_differentiation_involved_in_embryonic_placenta_development |
| GO:0060807 | 1 | regulation_of_transcription_from_RNA_polymerase_II_promoter_involved_in_definitive_endodermal_cell_fate_specification |
| GO:0060809 | 1 | mesodermal_to_mesenchymal_transition_involved_in_gastrulation |
| GO:0060816 | 1 | random_inactivation_of_X_chromosome |
| GO:0060821 | 1 | inactivation_of_X_chromosome_by_DNA_methylation |
| GO:0060823 | 1 | canonical_Wnt_receptor_signaling_pathway_involved_in_neural_plate_anterior/posterior_pattern_formation |
| GO:0060825 | 1 | fibroblast_growth_factor_receptor_signaling_pathway_involved_in_neural_plate_anterior/posterior_pattern_formation |
| GO:0060827 | 1 | regulation_of_canonical_Wnt_receptor_signaling_pathway_involved_in_neural_plate_anterior/posterior_pattern_formation |
| GO:0060829 | 1 | negative_regulation_of_canonical_Wnt_receptor_signaling_pathway_involved_in_neural_plate_anterior/posterior_pattern_formation |
| GO:0060830 | 1 | ciliary_receptor_clustering_involved_in_smoothened_signaling_pathway |
| GO:0060837 | 1 | blood_vessel_endothelial_cell_differentiation |
| GO:0060847 | 1 | endothelial_cell_fate_specification |
| GO:0060873 | 1 | anterior_semicircular_canal_development |
| GO:0060875 | 1 | lateral_semicircular_canal_development |
| GO:0060876 | 1 | semicircular_canal_formation |
| GO:0060879 | 1 | semicircular_canal_fusion |
| GO:0060904 | 1 | regulation_of_protein_folding_in_endoplasmic_reticulum |
| GO:0060920 | 1 | pacemaker_cell_differentiation |
| GO:0060921 | 1 | sinoatrial_node_cell_differentiation |
| GO:0060927 | 1 | pacemaker_cell_fate_commitment |
| GO:0060928 | 1 | atrioventricular_node_cell_development |
| GO:0060929 | 1 | atrioventricular_node_cell_fate_commitment |
| GO:0060931 | 1 | sinoatrial_node_cell_development |
| GO:0060948 | 1 | cardiac_vascular_smooth_muscle_cell_development |
| GO:0060961 | 1 | phospholipase_D_inhibitor_activity |
| GO:0060974 | 1 | cell_migration_involved_in_heart_formation |
| GO:0060975 | 1 | cardioblast_migration_to_the_midline_involved_in_heart_field_formation |
| GO:0060980 | 1 | cell_migration_involved_in_coronary_vasculogenesis |
| GO:0060981 | 1 | cell_migration_involved_in_coronary_angiogenesis |
| GO:0060988 | 1 | lipid_tube_assembly |
| GO:0061011 | 1 | hepatic_duct_development |
| GO:0061015 | 1 | snRNA_import_into_nucleus |
| GO:0061026 | 1 | cardiac_muscle_tissue_regeneration |
| GO:0061044 | 1 | negative_regulation_of_vascular_wound_healing |
| GO:0061048 | 1 | negative_regulation_of_branching_involved_in_lung_morphogenesis |
| GO:0061049 | 1 | cell_growth_involved_in_cardiac_muscle_cell_development |
| GO:0061051 | 1 | positive_regulation_of_cell_growth_involved_in_cardiac_muscle_cell_development |
| GO:0061052 | 1 | negative_regulation_of_cell_growth_involved_in_cardiac_muscle_cell_development |
| GO:0061090 | 1 | positive_regulation_of_sequestering_of_zinc_ion |
| GO:0061100 | 1 | lung_neuroendocrine_cell_differentiation |
| GO:0061102 | 1 | stomach_neuroendocrine_cell_differentiation |
| GO:0061103 | 1 | carotid_body_glomus_cell_differentiation |
| GO:0061104 | 1 | adrenal_chromaffin_cell_differentiation |
| GO:0061105 | 1 | regulation_of_stomach_neuroendocrine_cell_differentiation |
| GO:0061106 | 1 | negative_regulation_of_stomach_neuroendocrine_cell_differentiation |
| GO:0061108 | 1 | seminal_vesicle_epithelium_development |
| GO:0061110 | 1 | dense_core_granule_biogenesis |
| GO:0061115 | 1 | lung_proximal/distal_axis_specification |
| GO:0061117 | 1 | negative_regulation_of_heart_growth |
| GO:0061141 | 1 | lung_ciliated_cell_differentiation |
| GO:0061143 | 1 | alveolar_primary_septum_development |
| GO:0061144 | 1 | alveolar_secondary_septum_development |
| GO:0061149 | 1 | BMP_signaling_pathway_involved_in_ureter_morphogenesis |
| GO:0061151 | 1 | BMP_signaling_pathway_involved_in_renal_system_segmentation |
| GO:0061155 | 1 | pulmonary_artery_endothelial_tube_morphogenesis |
| GO:0061157 | 1 | mRNA_destabilization |
| GO:0061181 | 1 | regulation_of_chondrocyte_development |
| GO:0061185 | 1 | negative_regulation_of_dermatome_development |
| GO:0061189 | 1 | positive_regulation_of_sclerotome_development |
| GO:0061190 | 1 | regulation_of_sclerotome_development |
| GO:0061196 | 1 | fungiform_papilla_development |
| GO:0061217 | 1 | regulation_of_mesonephros_development |
| GO:0061301 | 1 | cerebellum_vasculature_morphogenesis |
| GO:0061310 | 1 | canonical_Wnt_receptor_signaling_pathway_involved_in_cardiac_neural_crest_cell_differentiation_involved_in_heart_development |
| GO:0061314 | 1 | Notch_signaling_involved_in_heart_development |
| GO:0061341 | 1 | non-canonical_Wnt_receptor_signaling_pathway_involved_in_heart_development |
| GO:0061346 | 1 | planar_cell_polarity_pathway_involved_in_heart_morphogenesis |
| GO:0061347 | 1 | planar_cell_polarity_pathway_involved_in_outflow_tract_morphogenesis |
| GO:0061348 | 1 | planar_cell_polarity_pathway_involved_in_ventricular_septum_morphogenesis |
| GO:0061349 | 1 | planar_cell_polarity_pathway_involved_in_cardiac_right_atrium_morphogenesis |
| GO:0061350 | 1 | planar_cell_polarity_pathway_involved_in_cardiac_muscle_tissue_morphogenesis |
| GO:0061354 | 1 | planar_cell_polarity_pathway_involved_in_pericardium_morphogenesis |
| GO:0061356 | 1 | regulation_of_Wnt_protein_secretion |
| GO:0061358 | 1 | negative_regulation_of_Wnt_protein_secretion |
| GO:0061360 | 1 | optic_chiasma_development |
| GO:0061369 | 1 | negative_regulation_of_testicular_blood_vessel_morphogenesis |
| GO:0061374 | 1 | mammillothalamic_axonal_tract_development |
| GO:0061377 | 1 | mammary_gland_lobule_development |
| GO:0061379 | 1 | inferior_colliculus_development |
| GO:0061381 | 1 | cell_migration_in_diencephalon |
| GO:0061419 | 1 | positive_regulation_of_transcription_from_RNA_polymerase_II_promoter_in_response_to_hypoxia |
| GO:0061441 | 1 | renal_artery_morphogenesis |
| GO:0070002 | 1 | glutamic-type_peptidase_activity |
| GO:0070012 | 1 | oligopeptidase_activity |
| GO:0070026 | 1 | nitric_oxide_binding |
| GO:0070037 | 1 | rRNA_(pseudouridine)_methyltransferase_activity |
| GO:0070060 | 1 | 'de_novo'_actin_filament_nucleation |
| GO:0070069 | 1 | cytochrome_complex |
| GO:0070077 | 1 | histone_arginine_demethylation |
| GO:0070078 | 1 | histone_H3-R2_demethylation |
| GO:0070079 | 1 | histone_H4-R3_demethylation |
| GO:0070086 | 1 | ubiquitin-dependent_endocytosis |
| GO:0070091 | 1 | glucagon_secretion |
| GO:0070104 | 1 | negative_regulation_of_interleukin-6-mediated_signaling_pathway |
| GO:0070105 | 1 | positive_regulation_of_interleukin-6-mediated_signaling_pathway |
| GO:0070106 | 1 | interleukin-27-mediated_signaling_pathway |
| GO:0070110 | 1 | ciliary_neurotrophic_factor_receptor_complex |
| GO:0070119 | 1 | ciliary_neurotrophic_factor_binding |
| GO:0070121 | 1 | Kupffer's_vesicle_development |
| GO:0070123 | 1 | transforming_growth_factor_beta_receptor_activity,_type_III |
| GO:0070126 | 1 | mitochondrial_translational_termination |
| GO:0070143 | 1 | mitochondrial_alanyl-tRNA_aminoacylation |
| GO:0070145 | 1 | mitochondrial_asparaginyl-tRNA_aminoacylation |
| GO:0070159 | 1 | mitochondrial_threonyl-tRNA_aminoacylation |
| GO:0070164 | 1 | negative_regulation_of_adiponectin_secretion |
| GO:0070165 | 1 | positive_regulation_of_adiponectin_secretion |
| GO:0070179 | 1 | D-serine_biosynthetic_process |
| GO:0070181 | 1 | SSU_rRNA_binding |
| GO:0070184 | 1 | mitochondrial_tyrosyl-tRNA_aminoacylation |
| GO:0070191 | 1 | methionine-R-sulfoxide_reductase_activity |
| GO:0070195 | 1 | growth_hormone_receptor_complex |
| GO:0070224 | 1 | sulfide:quinone_oxidoreductase_activity |
| GO:0070235 | 1 | regulation_of_activation-induced_cell_death_of_T_cells |
| GO:0070236 | 1 | negative_regulation_of_activation-induced_cell_death_of_T_cells |
| GO:0070242 | 1 | thymocyte_apoptotic_process |
| GO:0070247 | 1 | regulation_of_natural_killer_cell_apoptotic_process |
| GO:0070251 | 1 | pristanate-CoA_ligase_activity |
| GO:0070253 | 1 | somatostatin_secretion |
| GO:0070254 | 1 | mucus_secretion |
| GO:0070260 | 1 | 5'-tyrosyl-DNA_phosphodiesterase_activity |
| GO:0070262 | 1 | peptidyl-serine_dephosphorylation |
| GO:0070267 | 1 | oncosis |
| GO:0070274 | 1 | RES_complex |
| GO:0070278 | 1 | extracellular_matrix_constituent_secretion |
| GO:0070283 | 1 | radical_SAM_enzyme_activity |
| GO:0070287 | 1 | ferritin_receptor_activity |
| GO:0070315 | 1 | G1_to_G0_transition_involved_in_cell_differentiation |
| GO:0070317 | 1 | negative_regulation_of_G0_to_G1_transition |
| GO:0070327 | 1 | thyroid_hormone_transport |
| GO:0070335 | 1 | aspartate_binding |
| GO:0070345 | 1 | negative_regulation_of_fat_cell_proliferation |
| GO:0070347 | 1 | regulation_of_brown_fat_cell_proliferation |
| GO:0070349 | 1 | positive_regulation_of_brown_fat_cell_proliferation |
| GO:0070352 | 1 | positive_regulation_of_white_fat_cell_proliferation |
| GO:0070361 | 1 | mitochondrial_light_strand_promoter_anti-sense_binding |
| GO:0070362 | 1 | mitochondrial_heavy_strand_promoter_anti-sense_binding |
| GO:0070364 | 1 | mitochondrial_heavy_strand_promoter_sense_binding |
| GO:0070367 | 1 | negative_regulation_of_hepatocyte_differentiation |
| GO:0070370 | 1 | cellular_heat_acclimation |
| GO:0070384 | 1 | Harderian_gland_development |
| GO:0070401 | 1 | NADP+_binding |
| GO:0070407 | 1 | oxidation-dependent_protein_catabolic_process |
| GO:0070425 | 1 | negative_regulation_of_nucleotide-binding_oligomerization_domain_containing_signaling_pathway |
| GO:0070426 | 1 | positive_regulation_of_nucleotide-binding_oligomerization_domain_containing_signaling_pathway |
| GO:0070429 | 1 | negative_regulation_of_nucleotide-binding_oligomerization_domain_containing_1_signaling_pathway |
| GO:0070430 | 1 | positive_regulation_of_nucleotide-binding_oligomerization_domain_containing_1_signaling_pathway |
| GO:0070433 | 1 | negative_regulation_of_nucleotide-binding_oligomerization_domain_containing_2_signaling_pathway |
| GO:0070434 | 1 | positive_regulation_of_nucleotide-binding_oligomerization_domain_containing_2_signaling_pathway |
| GO:0070463 | 1 | tubulin-dependent_ATPase_activity |
| GO:0070489 | 1 | T_cell_aggregation |
| GO:0070494 | 1 | regulation_of_thrombin_receptor_signaling_pathway |
| GO:0070495 | 1 | negative_regulation_of_thrombin_receptor_signaling_pathway |
| GO:0070506 | 1 | high-density_lipoprotein_particle_receptor_activity |
| GO:0070510 | 1 | regulation_of_histone_H4-K20_methylation |
| GO:0070512 | 1 | positive_regulation_of_histone_H4-K20_methylation |
| GO:0070522 | 1 | ERCC4-ERCC1_complex |
| GO:0070524 | 1 | 11-beta-hydroxysteroid_dehydrogenase_(NADP+)_activity |
| GO:0070535 | 1 | histone_H2A_K63-linked_ubiquitination |
| GO:0070539 | 1 | linoleic_acid_binding |
| GO:0070541 | 1 | response_to_platinum_ion |
| GO:0070543 | 1 | response_to_linoleic_acid |
| GO:0070547 | 1 | L-tyrosine_aminotransferase_activity |
| GO:0070551 | 1 | endoribonuclease_activity,_cleaving_siRNA-paired_mRNA |
| GO:0070553 | 1 | nicotinic_acid_receptor_activity |
| GO:0070560 | 1 | protein_secretion_by_platelet |
| GO:0070563 | 1 | negative_regulation_of_vitamin_D_receptor_signaling_pathway |
| GO:0070564 | 1 | positive_regulation_of_vitamin_D_receptor_signaling_pathway |
| GO:0070585 | 1 | protein_localization_in_mitochondrion |
| GO:0070601 | 1 | centromeric_sister_chromatid_cohesion |
| GO:0070611 | 1 | histone_methyltransferase_activity_(H3-R2_specific) |
| GO:0070612 | 1 | histone_methyltransferase_activity_(H2A-R3_specific) |
| GO:0070625 | 1 | zymogen_granule_exocytosis |
| GO:0070626 | 1 | (S)-2-(5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamido)succinate_AMP-lyase_(fumarate-forming)_activity |
| GO:0070627 | 1 | ferrous_iron_import |
| GO:0070662 | 1 | mast_cell_proliferation |
| GO:0070667 | 1 | negative_regulation_of_mast_cell_proliferation |
| GO:0070684 | 1 | seminal_clot_liquefaction |
| GO:0070685 | 1 | macropinocytic_cup |
| GO:0070698 | 1 | type_I_activin_receptor_binding |
| GO:0070701 | 1 | mucus_layer |
| GO:0070702 | 1 | inner_mucus_layer |
| GO:0070703 | 1 | outer_mucus_layer |
| GO:0070715 | 1 | sodium-dependent_organic_cation_transport |
| GO:0070722 | 1 | Tle3-Aes_complex |
| GO:0070733 | 1 | protein_adenylyltransferase_activity |
| GO:0070740 | 1 | tubulin-glutamic_acid_ligase_activity |
| GO:0070773 | 1 | protein-N-terminal_glutamine_amidohydrolase_activity |
| GO:0070774 | 1 | phytoceramidase_activity |
| GO:0070818 | 1 | protoporphyrinogen_oxidase_activity |
| GO:0070821 | 1 | tertiary_granule_membrane |
| GO:0070826 | 1 | paraferritin_complex |
| GO:0070827 | 1 | chromatin_maintenance |
| GO:0070835 | 1 | chromium_ion_transmembrane_transporter_activity |
| GO:0070837 | 1 | dehydroascorbic_acid_transport |
| GO:0070839 | 1 | divalent_metal_ion_export |
| GO:0070840 | 1 | dynein_complex_binding |
| GO:0070846 | 1 | Hsp90_deacetylation |
| GO:0070853 | 1 | myosin_VI_binding |
| GO:0070860 | 1 | RNA_polymerase_I_core_factor_complex |
| GO:0070868 | 1 | heterochromatin_organization_involved_in_chromatin_silencing |
| GO:0070869 | 1 | heterochromatin_assembly_involved_in_chromatin_silencing |
| GO:0070889 | 1 | platelet_alpha_granule_organization |
| GO:0070892 | 1 | lipoteichoic_acid_receptor_activity |
| GO:0070926 | 1 | regulation_of_ATP:ADP_antiporter_activity |
| GO:0070948 | 1 | regulation_of_neutrophil_mediated_cytotoxicity |
| GO:0070949 | 1 | regulation_of_neutrophil_mediated_killing_of_symbiont_cell |
| GO:0070950 | 1 | regulation_of_neutrophil_mediated_killing_of_bacterium |
| GO:0070951 | 1 | regulation_of_neutrophil_mediated_killing_of_gram-negative_bacterium |
| GO:0070960 | 1 | positive_regulation_of_neutrophil_mediated_cytotoxicity |
| GO:0070961 | 1 | positive_regulation_of_neutrophil_mediated_killing_of_symbiont_cell |
| GO:0070962 | 1 | positive_regulation_of_neutrophil_mediated_killing_of_bacterium |
| GO:0070963 | 1 | positive_regulation_of_neutrophil_mediated_killing_of_gram-negative_bacterium |
| GO:0070974 | 1 | POU_domain_binding |
| GO:0070975 | 1 | FHA_domain_binding |
| GO:0070976 | 1 | TIR_domain_binding |
| GO:0070980 | 1 | biphenyl_catabolic_process |
| GO:0070981 | 1 | L-asparagine_biosynthetic_process |
| GO:0070982 | 1 | L-asparagine_metabolic_process |
| GO:0070990 | 1 | snRNP_binding |
| GO:0070994 | 1 | detection_of_oxidative_stress |
| GO:0071000 | 1 | response_to_magnetism |
| GO:0071033 | 1 | nuclear_retention_of_pre-mRNA_at_the_site_of_transcription |
| GO:0071042 | 1 | nuclear_polyadenylation-dependent_mRNA_catabolic_process |
| GO:0071047 | 1 | polyadenylation-dependent_mRNA_catabolic_process |
| GO:0071048 | 1 | nuclear_retention_of_unspliced_pre-mRNA_at_the_site_of_transcription |
| GO:0071073 | 1 | positive_regulation_of_phospholipid_biosynthetic_process |
| GO:0071105 | 1 | response_to_interleukin-11 |
| GO:0071110 | 1 | histone_biotinylation |
| GO:0071133 | 1 | alpha9-beta1_integrin-ADAM8_complex |
| GO:0071164 | 1 | RNA_trimethylguanosine_synthase_activity |
| GO:0071167 | 1 | ribonucleoprotein_complex_import_into_nucleus |
| GO:0071206 | 1 | establishment_of_protein_localization_to_juxtaparanode_region_of_axon |
| GO:0071209 | 1 | U7_snRNA_binding |
| GO:0071217 | 1 | cellular_response_to_external_biotic_stimulus |
| GO:0071226 | 1 | cellular_response_to_molecule_of_fungal_origin |
| GO:0071228 | 1 | cellular_response_to_tumor_cell |
| GO:0071247 | 1 | cellular_response_to_chromate |
| GO:0071258 | 1 | cellular_response_to_gravity |
| GO:0071259 | 1 | cellular_response_to_magnetism |
| GO:0071279 | 1 | cellular_response_to_cobalt_ion |
| GO:0071283 | 1 | cellular_response_to_iron(III)_ion |
| GO:0071284 | 1 | cellular_response_to_lead_ion |
| GO:0071298 | 1 | cellular_response_to_L-ascorbic_acid |
| GO:0071314 | 1 | cellular_response_to_cocaine |
| GO:0071324 | 1 | cellular_response_to_disaccharide_stimulus |
| GO:0071329 | 1 | cellular_response_to_sucrose_stimulus |
| GO:0071335 | 1 | hair_follicle_cell_proliferation |
| GO:0071351 | 1 | cellular_response_to_interleukin-18 |
| GO:0071352 | 1 | cellular_response_to_interleukin-2 |
| GO:0071393 | 1 | cellular_response_to_progesterone_stimulus |
| GO:0071401 | 1 | cellular_response_to_triglyceride |
| GO:0071403 | 1 | cellular_response_to_high_density_lipoprotein_particle_stimulus |
| GO:0071416 | 1 | cellular_response_to_tropane |
| GO:0071426 | 1 | ribonucleoprotein_complex_export_from_nucleus |
| GO:0071428 | 1 | rRNA-containing_ribonucleoprotein_complex_export_from_nucleus |
| GO:0071439 | 1 | clathrin_complex |
| GO:0071448 | 1 | cellular_response_to_alkyl_hydroperoxide |
| GO:0071449 | 1 | cellular_response_to_lipid_hydroperoxide |
| GO:0071466 | 1 | cellular_response_to_xenobiotic_stimulus |
| GO:0071469 | 1 | cellular_response_to_alkalinity |
| GO:0071477 | 1 | cellular_hypotonic_salinity_response |
| GO:0071492 | 1 | cellular_response_to_UV-A |
| GO:0071529 | 1 | cementum_mineralization |
| GO:0071543 | 1 | diphosphoinositol_polyphosphate_metabolic_process |
| GO:0071544 | 1 | diphosphoinositol_polyphosphate_catabolic_process |
| GO:0071566 | 1 | UFM1_activating_enzyme_activity |
| GO:0071567 | 1 | UFM1_hydrolase_activity |
| GO:0071579 | 1 | regulation_of_zinc_ion_transport |
| GO:0071580 | 1 | regulation_of_zinc_ion_transmembrane_transport |
| GO:0071581 | 1 | regulation_of_zinc_ion_import |
| GO:0071582 | 1 | negative_regulation_of_zinc_ion_transport |
| GO:0071583 | 1 | negative_regulation_of_zinc_ion_transmembrane_transport |
| GO:0071584 | 1 | negative_regulation_of_zinc_ion_import |
| GO:0071598 | 1 | neuronal_ribonucleoprotein_granule |
| GO:0071600 | 1 | otic_vesicle_morphogenesis |
| GO:0071602 | 1 | phytosphingosine_biosynthetic_process |
| GO:0071608 | 1 | macrophage_inflammatory_protein-1_alpha_production |
| GO:0071614 | 1 | linoleic_acid_epoxygenase_activity |
| GO:0071615 | 1 | oxidative_deethylation |
| GO:0071617 | 1 | lysophospholipid_acyltransferase_activity |
| GO:0071640 | 1 | regulation_of_macrophage_inflammatory_protein_1_alpha_production |
| GO:0071641 | 1 | negative_regulation_of_macrophage_inflammatory_protein_1_alpha_production |
| GO:0071649 | 1 | regulation_of_chemokine_(C-C_motif)_ligand_5_production |
| GO:0071651 | 1 | positive_regulation_of_chemokine_(C-C_motif)_ligand_5_production |
| GO:0071655 | 1 | regulation_of_granulocyte_colony-stimulating_factor_production |
| GO:0071657 | 1 | positive_regulation_of_granulocyte_colony-stimulating_factor_production |
| GO:0071658 | 1 | regulation_of_IP-10_production |
| GO:0071660 | 1 | positive_regulation_of_IP-10_production |
| GO:0071661 | 1 | regulation_of_granzyme_B_production |
| GO:0071663 | 1 | positive_regulation_of_granzyme_B_production |
| GO:0071665 | 1 | gamma-catenin-TCF7L2_complex |
| GO:0071670 | 1 | smooth_muscle_cell_chemotaxis |
| GO:0071677 | 1 | positive_regulation_of_mononuclear_cell_migration |
| GO:0071691 | 1 | cardiac_muscle_thin_filament_assembly |
| GO:0071712 | 1 | ER-associated_misfolded_protein_catabolic_process |
| GO:0071714 | 1 | icosanoid_transmembrane_transporter_activity |
| GO:0071722 | 1 | detoxification_of_arsenic-containing_substance |
| GO:0071782 | 1 | endoplasmic_reticulum_tubular_network |
| GO:0071796 | 1 | K6-linked_polyubiquitin_binding |
| GO:0071798 | 1 | response_to_prostaglandin_D_stimulus |
| GO:0071799 | 1 | cellular_response_to_prostaglandin_D_stimulus |
| GO:0071800 | 1 | podosome_assembly |
| GO:0071820 | 1 | N-box_binding |
| GO:0071862 | 1 | protein_phosphatase_type_1_activator_activity |
| GO:0071865 | 1 | regulation_of_apoptotic_process_in_bone_marrow |
| GO:0071866 | 1 | negative_regulation_of_apoptotic_process_in_bone_marrow |
| GO:0071873 | 1 | response_to_norepinephrine_stimulus |
| GO:0071874 | 1 | cellular_response_to_norepinephrine_stimulus |
| GO:0071878 | 1 | negative_regulation_of_adrenergic_receptor_signaling_pathway |
| GO:0071879 | 1 | positive_regulation_of_adrenergic_receptor_signaling_pathway |
| GO:0071881 | 1 | adenylate_cyclase-inhibiting_adrenergic_receptor_signaling_pathway |
| GO:0071882 | 1 | phospholipase_C-activating_adrenergic_receptor_signaling_pathway |
| GO:0071884 | 1 | vitamin_D_receptor_activator_activity |
| GO:0071893 | 1 | BMP_signaling_pathway_involved_in_nephric_duct_formation |
| GO:0071894 | 1 | histone_H2B_conserved_C-terminal_lysine_ubiquitination |
| GO:0071895 | 1 | odontoblast_differentiation |
| GO:0071914 | 1 | prominosome |
| GO:0071931 | 1 | positive_regulation_of_transcription_involved_in_G1/S_phase_of_mitotic_cell_cycle |
| GO:0071934 | 1 | thiamine_transmembrane_transport |
| GO:0071936 | 1 | coreceptor_activity_involved_in_Wnt_receptor_signaling_pathway |
| GO:0071938 | 1 | vitamin_A_transport |
| GO:0071947 | 1 | protein_deubiquitination_involved_in_ubiquitin-dependent_protein_catabolic_process |
| GO:0071953 | 1 | elastic_fiber |
| GO:0071954 | 1 | chemokine_(C-C_motif)_ligand_11_production |
| GO:0071955 | 1 | recycling_endosome_to_Golgi_transport |
| GO:0071971 | 1 | extracellular_vesicular_exosome_assembly |
| GO:0071987 | 1 | WD40-repeat_domain_binding |
| GO:0072003 | 1 | kidney_rudiment_formation |
| GO:0072007 | 1 | mesangial_cell_differentiation |
| GO:0072008 | 1 | glomerular_mesangial_cell_differentiation |
| GO:0072019 | 1 | proximal_convoluted_tubule_development |
| GO:0072024 | 1 | macula_densa_development |
| GO:0072027 | 1 | connecting_tubule_development |
| GO:0072028 | 1 | nephron_morphogenesis |
| GO:0072036 | 1 | mesenchymal_to_epithelial_transition_involved_in_renal_vesicle_formation |
| GO:0072046 | 1 | establishment_of_planar_polarity_involved_in_nephron_morphogenesis |
| GO:0072055 | 1 | renal_cortex_development |
| GO:0072059 | 1 | cortical_collecting_duct_development |
| GO:0072060 | 1 | outer_medullary_collecting_duct_development |
| GO:0072061 | 1 | inner_medullary_collecting_duct_development |
| GO:0072069 | 1 | DCT_cell_differentiation |
| GO:0072077 | 1 | renal_vesicle_morphogenesis |
| GO:0072096 | 1 | negative_regulation_of_branch_elongation_involved_in_ureteric_bud_branching |
| GO:0072097 | 1 | negative_regulation_of_branch_elongation_involved_in_ureteric_bud_branching_by_BMP_signaling_pathway |
| GO:0072099 | 1 | anterior/posterior_pattern_specification_involved_in_ureteric_bud_development |
| GO:0072100 | 1 | specification_of_ureteric_bud_anterior/posterior_symmetry |
| GO:0072101 | 1 | specification_of_ureteric_bud_anterior/posterior_symmetry_by_BMP_signaling_pathway |
| GO:0072105 | 1 | ureteric_peristalsis |
| GO:0072127 | 1 | renal_capsule_development |
| GO:0072130 | 1 | renal_capsule_specification |
| GO:0072148 | 1 | epithelial_cell_fate_commitment |
| GO:0072167 | 1 | specification_of_mesonephric_tubule_identity |
| GO:0072168 | 1 | specification_of_anterior_mesonephric_tubule_identity |
| GO:0072169 | 1 | specification_of_posterior_mesonephric_tubule_identity |
| GO:0072184 | 1 | renal_vesicle_progenitor_cell_differentiation |
| GO:0072192 | 1 | ureter_epithelial_cell_differentiation |
| GO:0072195 | 1 | kidney_smooth_muscle_cell_differentiation |
| GO:0072198 | 1 | mesenchymal_cell_proliferation_involved_in_ureter_development |
| GO:0072200 | 1 | negative_regulation_of_mesenchymal_cell_proliferation_involved_in_ureter_development |
| GO:0072209 | 1 | metanephric_mesangial_cell_differentiation |
| GO:0072213 | 1 | metanephric_capsule_development |
| GO:0072214 | 1 | metanephric_cortex_development |
| GO:0072219 | 1 | metanephric_cortical_collecting_duct_development |
| GO:0072227 | 1 | metanephric_macula_densa_development |
| GO:0072229 | 1 | metanephric_proximal_convoluted_tubule_development |
| GO:0072235 | 1 | metanephric_distal_tubule_development |
| GO:0072237 | 1 | metanephric_proximal_tubule_development |
| GO:0072239 | 1 | metanephric_glomerulus_vasculature_development |
| GO:0072240 | 1 | metanephric_DCT_cell_differentiation |
| GO:0072254 | 1 | metanephric_glomerular_mesangial_cell_differentiation |
| GO:0072255 | 1 | metanephric_glomerular_mesangial_cell_development |
| GO:0072259 | 1 | metanephric_interstitial_cell_development |
| GO:0072264 | 1 | metanephric_glomerular_endothelium_development |
| GO:0072267 | 1 | metanephric_capsule_specification |
| GO:0072268 | 1 | pattern_specification_involved_in_metanephros_development |
| GO:0072273 | 1 | metanephric_nephron_morphogenesis |
| GO:0072274 | 1 | metanephric_glomerular_basement_membrane_development |
| GO:0072283 | 1 | metanephric_renal_vesicle_morphogenesis |
| GO:0072285 | 1 | mesenchymal_to_epithelial_transition_involved_in_metanephric_renal_vesicle_formation |
| GO:0072286 | 1 | metanephric_connecting_tubule_development |
| GO:0072287 | 1 | metanephric_distal_tubule_morphogenesis |
| GO:0072302 | 1 | negative_regulation_of_metanephric_glomerular_mesangial_cell_proliferation |
| GO:0072303 | 1 | positive_regulation_of_glomerular_metanephric_mesangial_cell_proliferation |
| GO:0072319 | 1 | vesicle_uncoating |
| GO:0072333 | 1 | signal_transduction_by_p53_class_mediator_resulting_in_induction_of_anoikis |
| GO:0072340 | 1 | cellular_lactam_catabolic_process |
| GO:0072354 | 1 | histone_kinase_activity_(H3-T3_specific) |
| GO:0072355 | 1 | histone_H3-T3_phosphorylation |
| GO:0072362 | 1 | regulation_of_glycolysis_by_negative_regulation_of_transcription_from_RNA_polymerase_II_promoter |
| GO:0072363 | 1 | regulation_of_glycolysis_by_positive_regulation_of_transcription_from_RNA_polymerase_II_promoter |
| GO:0072365 | 1 | regulation_of_cellular_ketone_metabolic_process_by_negative_regulation_of_transcription_from_RNA_polymerase_II_promoter |
| GO:0072366 | 1 | regulation_of_cellular_ketone_metabolic_process_by_positive_regulation_of_transcription_from_RNA_polymerase_II_promoter |
| GO:0072388 | 1 | flavin_adenine_dinucleotide_biosynthetic_process |
| GO:0072428 | 1 | signal_transduction_involved_in_intra-S_DNA_damage_checkpoint |
| GO:0072517 | 1 | host_cell_viral_assembly_compartment |
| GO:0072539 | 1 | T-helper_17_cell_differentiation |
| GO:0072544 | 1 | L-DOPA_binding |
| GO:0072558 | 1 | NLRP1_inflammasome_complex |
| GO:0072570 | 1 | ADP-D-ribose_binding |
| GO:0072572 | 1 | poly-ADP-D-ribose_binding |
| GO:0072573 | 1 | tolerance_induction_to_lipopolysaccharide |
| GO:0072574 | 1 | hepatocyte_proliferation |
| GO:0072575 | 1 | epithelial_cell_proliferation_involved_in_liver_morphogenesis |
| GO:0072588 | 1 | box_H/ACA_RNP_complex |
| GO:0072606 | 1 | interleukin-8_secretion |
| GO:0072608 | 1 | interleukin-10_secretion |
| GO:0072658 | 1 | maintenance_of_protein_location_in_membrane |
| GO:0072660 | 1 | maintenance_of_protein_location_in_plasma_membrane |
| GO:0072674 | 1 | multinuclear_osteoclast_differentiation |
| GO:0072675 | 1 | osteoclast_fusion |
| GO:0072683 | 1 | T_cell_extravasation |
| GO:0072695 | 1 | regulation_of_DNA_recombination_at_telomere |
| GO:0080009 | 1 | mRNA_methylation |
| GO:0080048 | 1 | GDP-D-glucose_phosphorylase_activity |
| GO:0080125 | 1 | multicellular_structure_septum_development |
| GO:0080146 | 1 | L-cysteine_desulfhydrase_activity |
| GO:0080164 | 1 | regulation_of_nitric_oxide_metabolic_process |
| GO:0085018 | 1 | maintenance_of_symbiont-containing_vacuole_via_substance_secreted_by_host |
| GO:0085032 | 1 | modulation_by_symbiont_of_host_I-kappaB_kinase/NF-kappaB_cascade |
| GO:0086004 | 1 | regulation_of_cardiac_muscle_cell_contraction |
| GO:0086007 | 1 | voltage-gated_calcium_channel_activity_involved_in_regulation_of_cardiac_muscle_cell_action_potential |
| GO:0086015 | 1 | regulation_of_SA_node_cardiac_muscle_cell_action_potential |
| GO:0086023 | 1 | adrenergic_receptor_signaling_pathway_involved_in_heart_process |
| GO:0086046 | 1 | membrane_depolarization_involved_in_regulation_of_SA_node_cardiac_muscle_cell_action_potential |
| GO:0086047 | 1 | membrane_depolarization_involved_in_regulation_of_Purkinje_myocyte_cell_action_potential |
| GO:0086062 | 1 | voltage-gated_sodium_channel_activity_involved_in_regulation_of_Purkinje_myocyte_action_potential |
| GO:0086064 | 1 | cell_communication_by_electrical_coupling_involved_in_cardiac_conduction |
| GO:0086066 | 1 | atrial_cardiomyocyte_to_AV_node_cell_communication |
| GO:0086069 | 1 | bundle_of_His_cardiac_muscle_cell_to_Purkinje_myocyte_communication |
| GO:0086094 | 1 | positive_regulation_of_ryanodine-sensitive_calcium-release_channel_activity_by_adrenergic_receptor_signaling_pathway_involved_in_positive_regulation_of_cardiac_muscle_contraction |
| GO:0086097 | 1 | phospholipase_C-activating_angiotensin-mediated_signaling_pathway |
| GO:0086103 | 1 | G-protein_coupled_receptor_signaling_pathway_involved_in_heart_process |
| GO:0090010 | 1 | transforming_growth_factor_beta_receptor_signaling_pathway_involved_in_primitive_streak_formation |
| GO:0090024 | 1 | negative_regulation_of_neutrophil_chemotaxis |
| GO:0090042 | 1 | tubulin_deacetylation |
| GO:0090043 | 1 | regulation_of_tubulin_deacetylation |
| GO:0090044 | 1 | positive_regulation_of_tubulin_deacetylation |
| GO:0090069 | 1 | regulation_of_ribosome_biogenesis |
| GO:0090076 | 1 | relaxation_of_skeletal_muscle |
| GO:0090091 | 1 | positive_regulation_of_extracellular_matrix_disassembly |
| GO:0090094 | 1 | metanephric_cap_mesenchymal_cell_proliferation_involved_in_metanephros_development |
| GO:0090095 | 1 | regulation_of_metanephric_cap_mesenchymal_cell_proliferation |
| GO:0090096 | 1 | positive_regulation_of_metanephric_cap_mesenchymal_cell_proliferation |
| GO:0090104 | 1 | pancreatic_E_cell_differentiation |
| GO:0090107 | 1 | regulation_of_high-density_lipoprotein_particle_assembly |
| GO:0090131 | 1 | mesenchyme_migration |
| GO:0090138 | 1 | regulation_of_actin_cytoskeleton_organization_by_cell-cell_adhesion |
| GO:0090148 | 1 | membrane_fission |
| GO:0090149 | 1 | membrane_fission_involved_in_mitochondrial_fission |
| GO:0090155 | 1 | negative_regulation_of_sphingolipid_biosynthetic_process |
| GO:0090168 | 1 | Golgi_reassembly |
| GO:0090171 | 1 | chondrocyte_morphogenesis |
| GO:0090182 | 1 | regulation_of_secretion_of_lysosomal_enzymes |
| GO:0090194 | 1 | negative_regulation_of_glomerulus_development |
| GO:0090195 | 1 | chemokine_secretion |
| GO:0090198 | 1 | negative_regulation_of_chemokine_secretion |
| GO:0090210 | 1 | regulation_of_establishment_of_blood-brain_barrier |
| GO:0090212 | 1 | negative_regulation_of_establishment_of_blood-brain_barrier |
| GO:0090219 | 1 | negative_regulation_of_lipid_kinase_activity |
| GO:0090222 | 1 | centrosome-templated_microtubule_nucleation |
| GO:0090251 | 1 | protein_localization_involved_in_establishment_of_planar_polarity |
| GO:0090271 | 1 | positive_regulation_of_fibroblast_growth_factor_production |
| GO:0090272 | 1 | negative_regulation_of_fibroblast_growth_factor_production |
| GO:0090275 | 1 | negative_regulation_of_somatostatin_secretion |
| GO:0090283 | 1 | regulation_of_protein_glycosylation_in_Golgi |
| GO:0090285 | 1 | negative_regulation_of_protein_glycosylation_in_Golgi |
| GO:0090290 | 1 | positive_regulation_of_osteoclast_proliferation |
| GO:0090293 | 1 | nitrogen_catabolite_regulation_of_transcription |
| GO:0090294 | 1 | nitrogen_catabolite_activation_of_transcription |
| GO:0090296 | 1 | regulation_of_mitochondrial_DNA_replication |
| GO:0090298 | 1 | negative_regulation_of_mitochondrial_DNA_replication |
| GO:0090306 | 1 | spindle_assembly_involved_in_meiosis |
| GO:0090314 | 1 | positive_regulation_of_protein_targeting_to_membrane |
| GO:0090315 | 1 | negative_regulation_of_protein_targeting_to_membrane |
| GO:0090320 | 1 | regulation_of_chylomicron_remnant_clearance |
| GO:0090321 | 1 | positive_regulation_of_chylomicron_remnant_clearance |
| GO:0090324 | 1 | negative_regulation_of_oxidative_phosphorylation |
| GO:0090341 | 1 | negative_regulation_of_secretion_of_lysosomal_enzymes |
| GO:0090345 | 1 | cellular_organohalogen_metabolic_process |
| GO:0090346 | 1 | cellular_organofluorine_metabolic_process |
| GO:0090347 | 1 | regulation_of_cellular_organohalogen_metabolic_process |
| GO:0090348 | 1 | regulation_of_cellular_organofluorine_metabolic_process |
| GO:0090349 | 1 | negative_regulation_of_cellular_organohalogen_metabolic_process |
| GO:0090350 | 1 | negative_regulation_of_cellular_organofluorine_metabolic_process |
| GO:0090362 | 1 | positive_regulation_of_platelet-derived_growth_factor_production |
| GO:0090382 | 1 | phagosome_maturation |
| GO:0090409 | 1 | malonyl-CoA_synthetase_activity |
| GO:0090410 | 1 | malonate_catabolic_process |
| GO:0090420 | 1 | naphthalene-containing_compound_metabolic_process |
| GO:0097001 | 1 | ceramide_binding |
| GO:0097006 | 1 | regulation_of_plasma_lipoprotein_particle_levels |
| GO:0097009 | 1 | energy_homeostasis |
| GO:0097022 | 1 | lymphocyte_migration_into_lymph_node |
| GO:0097026 | 1 | dendritic_cell_dendrite_assembly |
| GO:0097032 | 1 | mitochondrial_respiratory_chain_complex_II_biogenesis |
| GO:0097037 | 1 | heme_export |
| GO:0097045 | 1 | phosphatidylserine_exposure_on_blood_platelet |
| GO:0097057 | 1 | TRAF2-GSTP1_complex |
| GO:0097089 | 1 | methyl-branched_fatty_acid_metabolic_process |
| GO:0097100 | 1 | supercoiled_DNA_binding |
| GO:0097109 | 1 | neuroligin_family_protein_binding |
| GO:0097112 | 1 | gamma-aminobutyric_acid_receptor_clustering |
| GO:0097115 | 1 | neurexin_clustering |
| GO:0097140 | 1 | BIM-BCL-xl_complex |
| GO:0097141 | 1 | BIM-BCL-2_complex |
| GO:0097151 | 1 | positive_regulation_of_inhibitory_postsynaptic_membrane_potential |
| GO:0097152 | 1 | mesenchymal_cell_apoptotic_process |
| GO:0097154 | 1 | GABAergic_neuron_differentiation |
| GO:0097157 | 1 | pre-mRNA_intronic_binding |
| GO:0097159 | 1 | organic_cyclic_compound_binding |
| GO:0097161 | 1 | DH_domain_binding |
| GO:0097162 | 1 | MADS_box_domain_binding |
| GO:0097179 | 1 | protease_inhibitor_complex |
| GO:0097180 | 1 | serine_protease_inhibitor_complex |
| GO:0097181 | 1 | protein_C_inhibitor-coagulation_factor_V_complex |
| GO:0097182 | 1 | protein_C_inhibitor-coagulation_factor_Xa_complex |
| GO:0097183 | 1 | protein_C_inhibitor-coagulation_factor_XI_complex |
| GO:0097184 | 1 | response_to_azide |
| GO:0097185 | 1 | cellular_response_to_azide |
| GO:0097186 | 1 | amelogenesis |
| GO:0097197 | 1 | tetraspanin-enriched_microdomain |
| GO:0097208 | 1 | alveolar_lamellar_body |
| GO:0097212 | 1 | lysosomal_membrane_organization |
| GO:0097213 | 1 | regulation_of_lysosomal_membrane_permeability |
| GO:0097214 | 1 | positive_regulation_of_lysosomal_membrane_permeability |
| GO:0097223 | 1 | sperm_part |
| GO:0097227 | 1 | sperm_annulus |
| GO:0097254 | 1 | renal_tubular_secretion |
| GO:0097264 | 1 | self_proteolysis |
| GO:0097272 | 1 | ammonia_homeostasis |
| GO:0097273 | 1 | creatinine_homeostasis |
| GO:0097274 | 1 | urea_homeostasis |
| GO:0097275 | 1 | cellular_ammonia_homeostasis |
| GO:0097276 | 1 | cellular_creatinine_homeostasis |
| GO:0097277 | 1 | cellular_urea_homeostasis |
| GO:0097286 | 1 | iron_ion_import |
| GO:0097291 | 1 | renal_phosphate_ion_absorption |
| GO:0097320 | 1 | membrane_tubulation |
| GO:1900003 | 1 | regulation_of_serine-type_endopeptidase_activity |
| GO:1900004 | 1 | negative_regulation_of_serine-type_endopeptidase_activity |
| GO:1900005 | 1 | positive_regulation_of_serine-type_endopeptidase_activity |
| GO:1900010 | 1 | regulation_of_corticotropin-releasing_hormone_receptor_activity |
| GO:1900011 | 1 | negative_regulation_of_corticotropin-releasing_hormone_receptor_activity |
| GO:1900015 | 1 | regulation_of_cytokine_production_involved_in_inflammatory_response |
| GO:1900016 | 1 | negative_regulation_of_cytokine_production_involved_in_inflammatory_response |
| GO:1900024 | 1 | regulation_of_substrate_adhesion-dependent_cell_spreading |
| GO:1900025 | 1 | negative_regulation_of_substrate_adhesion-dependent_cell_spreading |
| GO:1900028 | 1 | negative_regulation_of_ruffle_assembly |
| GO:1900029 | 1 | positive_regulation_of_ruffle_assembly |
| GO:1900034 | 1 | regulation_of_cellular_response_to_heat |
| GO:1900035 | 1 | negative_regulation_of_cellular_response_to_heat |
| GO:1900038 | 1 | negative_regulation_of_cellular_response_to_hypoxia |
| GO:1900052 | 1 | regulation_of_retinoic_acid_biosynthetic_process |
| GO:1900053 | 1 | negative_regulation_of_retinoic_acid_biosynthetic_process |
| GO:1900060 | 1 | negative_regulation_of_ceramide_biosynthetic_process |
| GO:1900102 | 1 | negative_regulation_of_endoplasmic_reticulum_unfolded_protein_response |
| GO:1900108 | 1 | negative_regulation_of_nodal_signaling_pathway |
| GO:1900122 | 1 | positive_regulation_of_receptor_binding |
| GO:1900126 | 1 | negative_regulation_of_hyaluronan_biosynthetic_process |
| GO:1900130 | 1 | regulation_of_lipid_binding |
| GO:1900131 | 1 | negative_regulation_of_lipid_binding |
| GO:1900134 | 1 | negative_regulation_of_renin_secretion_into_blood_stream |
| GO:1900135 | 1 | positive_regulation_of_renin_secretion_into_blood_stream |
| GO:1900138 | 1 | negative_regulation_of_phospholipase_A2_activity |
| GO:1900139 | 1 | negative_regulation_of_arachidonic_acid_secretion |
| GO:1900141 | 1 | regulation_of_oligodendrocyte_apoptotic_process |
| GO:1900142 | 1 | negative_regulation_of_oligodendrocyte_apoptotic_process |
| GO:1900146 | 1 | negative_regulation_of_nodal_signaling_pathway_involved_in_determination_of_left/right_asymmetry |
| GO:1900154 | 1 | regulation_of_bone_trabecula_formation |
| GO:1900155 | 1 | negative_regulation_of_bone_trabecula_formation |
| GO:1900157 | 1 | regulation_of_bone_mineralization_involved_in_bone_maturation |
| GO:1900158 | 1 | negative_regulation_of_bone_mineralization_involved_in_bone_maturation |
| GO:1900161 | 1 | regulation_of_phospholipid_scramblase_activity |
| GO:1900163 | 1 | positive_regulation_of_phospholipid_scramblase_activity |
| GO:1900165 | 1 | negative_regulation_of_interleukin-6_secretion |
| GO:1900166 | 1 | regulation_of_glial_cell_line-derived_neurotrophic_factor_secretion |
| GO:1900168 | 1 | positive_regulation_of_glial_cell_line-derived_neurotrophic_factor_secretion |
| GO:1900242 | 1 | regulation_of_synaptic_vesicle_endocytosis |
| GO:1900244 | 1 | positive_regulation_of_synaptic_vesicle_endocytosis |
| GO:1900273 | 1 | positive_regulation_of_long-term_synaptic_potentiation |
| GO:1900275 | 1 | negative_regulation_of_phospholipase_C_activity |
| GO:1900276 | 1 | regulation_of_proteinase_activated_receptor_activity |
| GO:1900279 | 1 | regulation_of_CD4-positive,_alpha_beta_T_cell_costimulation |
| GO:1900281 | 1 | positive_regulation_of_CD4-positive,_alpha_beta_T_cell_costimulation |
| GO:1900408 | 1 | negative_regulation_of_cellular_response_to_oxidative_stress |
| GO:1900451 | 1 | positive_regulation_of_glutamate_receptor_signaling_pathway |
| GO:1900452 | 1 | regulation_of_long_term_synaptic_depression |
| GO:1900737 | 1 | negative_regulation_of_phospholipase_C-activating_G-protein_coupled_receptor_signaling_pathway |
| GO:1900738 | 1 | positive_regulation_of_phospholipase_C-activating_G-protein_coupled_receptor_signaling_pathway |
| GO:1900739 | 1 | regulation_of_protein_insertion_into_mitochondrial_membrane_involved_in_apoptotic_signaling_pathway |
| GO:1900740 | 1 | positive_regulation_of_protein_insertion_into_mitochondrial_membrane_involved_in_apoptotic_signaling_pathway |
| GO:1900757 | 1 | regulation_of_D-amino-acid_oxidase_activity |
| GO:1900758 | 1 | negative_regulation_of_D-amino-acid_oxidase_activity |
| GO:1900825 | 1 | regulation_of_membrane_depolarization_involved_in_regulation_of_cardiac_muscle_cell_action_potential |
| GO:1900827 | 1 | positive_regulation_of_membrane_depolarization_involved_in_regulation_of_cardiac_muscle_cell_action_potential |
| GO:1901017 | 1 | negative_regulation_of_potassium_ion_transmembrane_transporter_activity |
| GO:1901018 | 1 | positive_regulation_of_potassium_ion_transmembrane_transporter_activity |
| GO:1901028 | 1 | regulation_of_mitochondrial_outer_membrane_permeabilization |
| GO:1901032 | 1 | negative_regulation_of_response_to_reactive_oxygen_species |
| GO:2000002 | 1 | negative_regulation_of_DNA_damage_checkpoint |
| GO:2000004 | 1 | regulation_of_metanephric_S-shaped_body_morphogenesis |
| GO:2000005 | 1 | negative_regulation_of_metanephric_S-shaped_body_morphogenesis |
| GO:2000006 | 1 | regulation_of_metanephric_comma-shaped_body_morphogenesis |
| GO:2000007 | 1 | negative_regulation_of_metanephric_comma-shaped_body_morphogenesis |
| GO:2000016 | 1 | negative_regulation_of_determination_of_dorsal_identity |
| GO:2000019 | 1 | negative_regulation_of_male_gonad_development |
| GO:2000042 | 1 | negative_regulation_of_double-strand_break_repair_via_homologous_recombination |
| GO:2000046 | 1 | regulation_of_G2_phase_of_mitotic_cell_cycle |
| GO:2000048 | 1 | negative_regulation_of_cell-cell_adhesion_mediated_by_cadherin |
| GO:2000051 | 1 | negative_regulation_of_non-canonical_Wnt_receptor_signaling_pathway |
| GO:2000055 | 1 | positive_regulation_of_Wnt_receptor_signaling_pathway_involved_in_dorsal/ventral_axis_specification |
| GO:2000056 | 1 | regulation_of_Wnt_receptor_signaling_pathway_involved_in_digestive_tract_morphogenesis |
| GO:2000057 | 1 | negative_regulation_of_Wnt_receptor_signaling_pathway_involved_in_digestive_tract_morphogenesis |
| GO:2000061 | 1 | regulation_of_ureter_smooth_muscle_cell_differentiation |
| GO:2000062 | 1 | negative_regulation_of_ureter_smooth_muscle_cell_differentiation |
| GO:2000063 | 1 | positive_regulation_of_ureter_smooth_muscle_cell_differentiation |
| GO:2000066 | 1 | positive_regulation_of_cortisol_biosynthetic_process |
| GO:2000080 | 1 | negative_regulation_of_canonical_Wnt_receptor_signaling_pathway_involved_in_controlling_type_B_pancreatic_cell_proliferation |
| GO:2000081 | 1 | positive_regulation_of_canonical_Wnt_receptor_signaling_pathway_involved_in_controlling_type_B_pancreatic_cell_proliferation |
| GO:2000118 | 1 | regulation_of_sodium-dependent_phosphate_transport |
| GO:2000119 | 1 | negative_regulation_of_sodium-dependent_phosphate_transport |
| GO:2000137 | 1 | negative_regulation_of_cell_proliferation_involved_in_heart_morphogenesis |
| GO:2000138 | 1 | positive_regulation_of_cell_proliferation_involved_in_heart_morphogenesis |
| GO:2000148 | 1 | regulation_of_planar_cell_polarity_pathway_involved_in_ventricular_septum_morphogenesis |
| GO:2000149 | 1 | negative_regulation_of_planar_cell_polarity_pathway_involved_in_ventricular_septum_morphogenesis |
| GO:2000150 | 1 | regulation_of_planar_cell_polarity_pathway_involved_in_cardiac_muscle_tissue_morphogenesis |
| GO:2000151 | 1 | negative_regulation_of_planar_cell_polarity_pathway_involved_in_cardiac_muscle_tissue_morphogenesis |
| GO:2000159 | 1 | regulation_of_planar_cell_polarity_pathway_involved_in_heart_morphogenesis |
| GO:2000160 | 1 | negative_regulation_of_planar_cell_polarity_pathway_involved_in_heart_morphogenesis |
| GO:2000161 | 1 | regulation_of_planar_cell_polarity_pathway_involved_in_cardiac_right_atrium_morphogenesis |
| GO:2000162 | 1 | negative_regulation_of_planar_cell_polarity_pathway_involved_in_cardiac_right_atrium_morphogenesis |
| GO:2000163 | 1 | regulation_of_planar_cell_polarity_pathway_involved_in_outflow_tract_morphogenesis |
| GO:2000164 | 1 | negative_regulation_of_planar_cell_polarity_pathway_involved_in_outflow_tract_morphogenesis |
| GO:2000165 | 1 | regulation_of_planar_cell_polarity_pathway_involved_in_pericardium_morphogenesis |
| GO:2000166 | 1 | negative_regulation_of_planar_cell_polarity_pathway_involved_in_pericardium_morphogenesis |
| GO:2000167 | 1 | regulation_of_planar_cell_polarity_pathway_involved_in_neural_tube_closure |
| GO:2000168 | 1 | negative_regulation_of_planar_cell_polarity_pathway_involved_in_neural_tube_closure |
| GO:2000173 | 1 | negative_regulation_of_branching_morphogenesis_of_a_nerve |
| GO:2000180 | 1 | negative_regulation_of_androgen_biosynthetic_process |
| GO:2000181 | 1 | negative_regulation_of_blood_vessel_morphogenesis |
| GO:2000211 | 1 | regulation_of_glutamate_metabolic_process |
| GO:2000213 | 1 | positive_regulation_of_glutamate_metabolic_process |
| GO:2000225 | 1 | negative_regulation_of_testosterone_biosynthetic_process |
| GO:2000226 | 1 | regulation_of_pancreatic_A_cell_differentiation |
| GO:2000227 | 1 | negative_regulation_of_pancreatic_A_cell_differentiation |
| GO:2000232 | 1 | regulation_of_rRNA_processing |
| GO:2000233 | 1 | negative_regulation_of_rRNA_processing |
| GO:2000252 | 1 | negative_regulation_of_feeding_behavior |
| GO:2000253 | 1 | positive_regulation_of_feeding_behavior |
| GO:2000254 | 1 | regulation_of_male_germ_cell_proliferation |
| GO:2000256 | 1 | positive_regulation_of_male_germ_cell_proliferation |
| GO:2000259 | 1 | positive_regulation_of_protein_activation_cascade |
| GO:2000271 | 1 | positive_regulation_of_fibroblast_apoptotic_process |
| GO:2000276 | 1 | negative_regulation_of_oxidative_phosphorylation_uncoupler_activity |
| GO:2000277 | 1 | positive_regulation_of_oxidative_phosphorylation_uncoupler_activity |
| GO:2000295 | 1 | regulation_of_hydrogen_peroxide_catabolic_process |
| GO:2000296 | 1 | negative_regulation_of_hydrogen_peroxide_catabolic_process |
| GO:2000298 | 1 | regulation_of_Rho-dependent_protein_serine/threonine_kinase_activity |
| GO:2000299 | 1 | negative_regulation_of_Rho-dependent_protein_serine/threonine_kinase_activity |
| GO:2000302 | 1 | positive_regulation_of_synaptic_vesicle_exocytosis |
| GO:2000313 | 1 | regulation_of_fibroblast_growth_factor_receptor_signaling_pathway_involved_in_neural_plate_anterior/posterior_pattern_formation |
| GO:2000320 | 1 | negative_regulation_of_T-helper_17_cell_differentiation |
| GO:2000322 | 1 | regulation_of_glucocorticoid_receptor_signaling_pathway |
| GO:2000323 | 1 | negative_regulation_of_glucocorticoid_receptor_signaling_pathway |
| GO:2000325 | 1 | regulation_of_ligand-dependent_nuclear_receptor_transcription_coactivator_activity |
| GO:2000326 | 1 | negative_regulation_of_ligand-dependent_nuclear_receptor_transcription_coactivator_activity |
| GO:2000332 | 1 | regulation_of_blood_microparticle_formation |
| GO:2000334 | 1 | positive_regulation_of_blood_microparticle_formation |
| GO:2000338 | 1 | regulation_of_chemokine_(C-X-C_motif)_ligand_1_production |
| GO:2000340 | 1 | positive_regulation_of_chemokine_(C-X-C_motif)_ligand_1_production |
| GO:2000346 | 1 | negative_regulation_of_hepatocyte_proliferation |
| GO:2000349 | 1 | negative_regulation_of_CD40_signaling_pathway |
| GO:2000354 | 1 | regulation_of_ovarian_follicle_development |
| GO:2000356 | 1 | regulation_of_kidney_smooth_muscle_cell_differentiation |
| GO:2000357 | 1 | negative_regulation_of_kidney_smooth_muscle_cell_differentiation |
| GO:2000358 | 1 | positive_regulation_of_kidney_smooth_muscle_cell_differentiation |
| GO:2000361 | 1 | regulation_of_prostaglandin-E_synthase_activity |
| GO:2000363 | 1 | positive_regulation_of_prostaglandin-E_synthase_activity |
| GO:2000367 | 1 | regulation_of_acrosomal_vesicle_exocytosis |
| GO:2000368 | 1 | positive_regulation_of_acrosomal_vesicle_exocytosis |
| GO:2000374 | 1 | regulation_of_oxygen_metabolic_process |
| GO:2000376 | 1 | positive_regulation_of_oxygen_metabolic_process |
| GO:2000382 | 1 | positive_regulation_of_mesoderm_development |
| GO:2000384 | 1 | negative_regulation_of_ectoderm_development |
| GO:2000386 | 1 | positive_regulation_of_ovarian_follicle_development |
| GO:2000387 | 1 | regulation_of_antral_ovarian_follicle_growth |
| GO:2000388 | 1 | positive_regulation_of_antral_ovarian_follicle_growth |
| GO:2000389 | 1 | regulation_of_neutrophil_extravasation |
| GO:2000391 | 1 | positive_regulation_of_neutrophil_extravasation |
| GO:2000392 | 1 | regulation_of_lamellipodium_morphogenesis |
| GO:2000405 | 1 | negative_regulation_of_T_cell_migration |
| GO:2000407 | 1 | regulation_of_T_cell_extravasation |
| GO:2000409 | 1 | positive_regulation_of_T_cell_extravasation |
| GO:2000413 | 1 | regulation_of_fibronectin-dependent_thymocyte_migration |
| GO:2000415 | 1 | positive_regulation_of_fibronectin-dependent_thymocyte_migration |
| GO:2000417 | 1 | negative_regulation_of_eosinophil_migration |
| GO:2000418 | 1 | positive_regulation_of_eosinophil_migration |
| GO:2000419 | 1 | regulation_of_eosinophil_extravasation |
| GO:2000420 | 1 | negative_regulation_of_eosinophil_extravasation |
| GO:2000438 | 1 | negative_regulation_of_monocyte_extravasation |
| GO:2000439 | 1 | positive_regulation_of_monocyte_extravasation |
| GO:2000449 | 1 | regulation_of_CD8-positive,_alpha-beta_T_cell_extravasation |
| GO:2000451 | 1 | positive_regulation_of_CD8-positive,_alpha-beta_T_cell_extravasation |
| GO:2000460 | 1 | regulation_of_eukaryotic_cell_surface_binding |
| GO:2000461 | 1 | negative_regulation_of_eukaryotic_cell_surface_binding |
| GO:2000477 | 1 | regulation_of_metanephric_glomerular_visceral_epithelial_cell_development |
| GO:2000478 | 1 | positive_regulation_of_metanephric_glomerular_visceral_epithelial_cell_development |
| GO:2000480 | 1 | negative_regulation_of_cAMP-dependent_protein_kinase_activity |
| GO:2000489 | 1 | regulation_of_hepatic_stellate_cell_activation |
| GO:2000491 | 1 | positive_regulation_of_hepatic_stellate_cell_activation |
| GO:2000498 | 1 | regulation_of_induction_of_apoptosis_in_response_to_chemical_stimulus |
| GO:2000500 | 1 | positive_regulation_of_induction_of_apoptosis_in_response_to_chemical_stimulus |
| GO:2000502 | 1 | negative_regulation_of_natural_killer_cell_chemotaxis |
| GO:2000504 | 1 | positive_regulation_of_blood_vessel_remodeling |
| GO:2000507 | 1 | positive_regulation_of_energy_homeostasis |
| GO:2000511 | 1 | regulation_of_granzyme_A_production |
| GO:2000513 | 1 | positive_regulation_of_granzyme_A_production |
| GO:2000517 | 1 | regulation_of_T-helper_1_cell_activation |
| GO:2000518 | 1 | negative_regulation_of_T-helper_1_cell_activation |
| GO:2000520 | 1 | regulation_of_immunological_synapse_formation |
| GO:2000522 | 1 | positive_regulation_of_immunological_synapse_formation |
| GO:2000526 | 1 | positive_regulation_of_glycoprotein_biosynthetic_process_involved_in_immunological_synapse_formation |
| GO:2000527 | 1 | regulation_of_myeloid_dendritic_cell_chemotaxis |
| GO:2000529 | 1 | positive_regulation_of_myeloid_dendritic_cell_chemotaxis |
| GO:2000533 | 1 | negative_regulation_of_renal_albumin_absorption |
| GO:2000534 | 1 | positive_regulation_of_renal_albumin_absorption |
| GO:2000535 | 1 | regulation_of_entry_of_bacterium_into_host_cell |
| GO:2000536 | 1 | negative_regulation_of_entry_of_bacterium_into_host_cell |
| GO:2000544 | 1 | regulation_of_endothelial_cell_chemotaxis_to_fibroblast_growth_factor |
| GO:2000545 | 1 | negative_regulation_of_endothelial_cell_chemotaxis_to_fibroblast_growth_factor |
| GO:2000548 | 1 | negative_regulation_of_dendritic_cell_dendrite_assembly |
| GO:2000549 | 1 | positive_regulation_of_dendritic_cell_dendrite_assembly |
| GO:2000554 | 1 | regulation_of_T-helper_1_cell_cytokine_production |
| GO:2000556 | 1 | positive_regulation_of_T-helper_1_cell_cytokine_production |
| GO:2000557 | 1 | regulation_of_immunoglobulin_production_in_mucosal_tissue |
| GO:2000558 | 1 | positive_regulation_of_immunoglobulin_production_in_mucosal_tissue |
| GO:2000559 | 1 | regulation_of_CD24_biosynthetic_process |
| GO:2000560 | 1 | positive_regulation_of_CD24_biosynthetic_process |
| GO:2000561 | 1 | regulation_of_CD4-positive,_alpha-beta_T_cell_proliferation |
| GO:2000562 | 1 | negative_regulation_of_CD4-positive,_alpha-beta_T_cell_proliferation |
| GO:2000563 | 1 | positive_regulation_of_CD4-positive,_alpha-beta_T_cell_proliferation |
| GO:2000566 | 1 | positive_regulation_of_CD8-positive,_alpha-beta_T_cell_proliferation |
| GO:2000569 | 1 | regulation_of_T-helper_2_cell_activation |
| GO:2000570 | 1 | positive_regulation_of_T-helper_2_cell_activation |
| GO:2000583 | 1 | regulation_of_platelet-derived_growth_factor_receptor-alpha_signaling_pathway |
| GO:2000584 | 1 | negative_regulation_of_platelet-derived_growth_factor_receptor-alpha_signaling_pathway |
| GO:2000586 | 1 | regulation_of_platelet-derived_growth_factor_receptor-beta_signaling_pathway |
| GO:2000587 | 1 | negative_regulation_of_platelet-derived_growth_factor_receptor-beta_signaling_pathway |
| GO:2000590 | 1 | negative_regulation_of_metanephric_mesenchymal_cell_migration |
| GO:2000595 | 1 | regulation_of_optic_nerve_formation |
| GO:2000597 | 1 | positive_regulation_of_optic_nerve_formation |
| GO:2000606 | 1 | regulation_of_cell_proliferation_involved_in_mesonephros_development |
| GO:2000607 | 1 | negative_regulation_of_cell_proliferation_involved_in_mesonephros_development |
| GO:2000612 | 1 | regulation_of_thyroid-stimulating_hormone_secretion |
| GO:2000617 | 1 | positive_regulation_of_histone_H3-K9_acetylation |
| GO:2000618 | 1 | regulation_of_histone_H4-K16_acetylation |
| GO:2000620 | 1 | positive_regulation_of_histone_H4-K16_acetylation |
| GO:2000622 | 1 | regulation_of_nuclear-transcribed_mRNA_catabolic_process,_nonsense-mediated_decay |
| GO:2000623 | 1 | negative_regulation_of_nuclear-transcribed_mRNA_catabolic_process,_nonsense-mediated_decay |
| GO:2000625 | 1 | regulation_of_miRNA_catabolic_process |
| GO:2000627 | 1 | positive_regulation_of_miRNA_catabolic_process |
| GO:2000628 | 1 | regulation_of_miRNA_metabolic_process |
| GO:2000630 | 1 | positive_regulation_of_miRNA_metabolic_process |
| GO:2000638 | 1 | regulation_of_SREBP-mediated_signaling_pathway |
| GO:2000639 | 1 | negative_regulation_of_SREBP-mediated_signaling_pathway |
| GO:2000643 | 1 | positive_regulation_of_early_endosome_to_late_endosome_transport |
| GO:2000646 | 1 | positive_regulation_of_receptor_catabolic_process |
| GO:2000647 | 1 | negative_regulation_of_stem_cell_proliferation |
| GO:2000653 | 1 | regulation_of_genetic_imprinting |
| GO:2000654 | 1 | regulation_of_cellular_response_to_testosterone_stimulus |
| GO:2000655 | 1 | negative_regulation_of_cellular_response_to_testosterone_stimulus |
| GO:2000656 | 1 | regulation_of_apolipoprotein_binding |
| GO:2000657 | 1 | negative_regulation_of_apolipoprotein_binding |
| GO:2000663 | 1 | negative_regulation_of_interleukin-5_secretion |
| GO:2000666 | 1 | negative_regulation_of_interleukin-13_secretion |
| GO:2000670 | 1 | positive_regulation_of_dendritic_cell_apoptotic_process |
| GO:2000680 | 1 | regulation_of_rubidium_ion_transport |
| GO:2000682 | 1 | positive_regulation_of_rubidium_ion_transport |
| GO:2000685 | 1 | positive_regulation_of_cellular_response_to_X-ray |
| GO:2000686 | 1 | regulation_of_rubidium_ion_transmembrane_transporter_activity |
| GO:2000688 | 1 | positive_regulation_of_rubidium_ion_transmembrane_transporter_activity |
| GO:2000690 | 1 | regulation_of_cardiac_muscle_cell_myoblast_differentiation |
| GO:2000691 | 1 | negative_regulation_of_cardiac_muscle_cell_myoblast_differentiation |
| GO:2000702 | 1 | regulation_of_fibroblast_growth_factor_receptor_signaling_pathway_involved_in_ureteric_bud_formation |
| GO:2000703 | 1 | negative_regulation_of_fibroblast_growth_factor_receptor_signaling_pathway_involved_in_ureteric_bud_formation |
| GO:2000706 | 1 | negative_regulation_of_dense_core_granule_biogenesis |
| GO:2000707 | 1 | positive_regulation_of_dense_core_granule_biogenesis |
| GO:2000723 | 1 | negative_regulation_of_cardiac_vascular_smooth_muscle_cell_differentiation |
| GO:2000724 | 1 | positive_regulation_of_cardiac_vascular_smooth_muscle_cell_differentiation |
| GO:2000726 | 1 | negative_regulation_of_cardiac_muscle_cell_differentiation |
| GO:2000733 | 1 | regulation_of_glial_cell-derived_neurotrophic_factor_receptor_signaling_pathway_involved_in_ureteric_bud_formation |
| GO:2000734 | 1 | negative_regulation_of_glial_cell-derived_neurotrophic_factor_receptor_signaling_pathway_involved_in_ureteric_bud_formation |
| GO:2000740 | 1 | negative_regulation_of_mesenchymal_stem_cell_differentiation |
| GO:2000752 | 1 | regulation_of_glucosylceramide_catabolic_process |
| GO:2000753 | 1 | positive_regulation_of_glucosylceramide_catabolic_process |
| GO:2000754 | 1 | regulation_of_sphingomyelin_catabolic_process |
| GO:2000755 | 1 | positive_regulation_of_sphingomyelin_catabolic_process |
| GO:2000759 | 1 | regulation_of_N-terminal_peptidyl-lysine_acetylation |
| GO:2000761 | 1 | positive_regulation_of_N-terminal_peptidyl-lysine_acetylation |
| GO:2000763 | 1 | positive_regulation_of_transcription_from_RNA_polymerase_II_promoter_involved_in_norepinephrine_biosynthetic_process |
| GO:2000764 | 1 | positive_regulation_of_semaphorin-plexin_signaling_pathway_involved_in_outflow_tract_morphogenesis |
| GO:2000777 | 1 | positive_regulation_of_proteasomal_ubiquitin-dependent_protein_catabolic_process_involved_in_cellular_response_to_hypoxia |
| GO:2000785 | 1 | regulation_of_autophagic_vacuole_assembly |
| GO:2000786 | 1 | positive_regulation_of_autophagic_vacuole_assembly |
| GO:2000790 | 1 | regulation_of_mesenchymal_cell_proliferation_involved_in_lung_development |
| GO:2000791 | 1 | negative_regulation_of_mesenchymal_cell_proliferation_involved_in_lung_development |
| GO:2000793 | 1 | cell_proliferation_involved_in_heart_valve_development |
| GO:2000795 | 1 | negative_regulation_of_epithelial_cell_proliferation_involved_in_lung_morphogenesis |
| GO:2000797 | 1 | regulation_of_amniotic_stem_cell_differentiation |
| GO:2000798 | 1 | negative_regulation_of_amniotic_stem_cell_differentiation |
| GO:2000800 | 1 | regulation_of_endocardial_cushion_to_mesenchymal_transition_involved_in_heart_valve_formation |
| GO:2000802 | 1 | positive_regulation_of_endocardial_cushion_to_mesenchymal_transition_involved_in_heart_valve_formation |
| GO:2000807 | 1 | regulation_of_synaptic_vesicle_clustering |
| GO:2000808 | 1 | negative_regulation_of_synaptic_vesicle_clustering |
| GO:2000812 | 1 | regulation_of_barbed-end_actin_filament_capping |
| GO:2000827 | 1 | mitochondrial_RNA_surveillance |
| GO:2000834 | 1 | regulation_of_androgen_secretion |
| GO:2000836 | 1 | positive_regulation_of_androgen_secretion |
| GO:2000870 | 1 | regulation_of_progesterone_secretion |
| GO:2000970 | 1 | regulation_of_detection_of_glucose |
| GO:2000971 | 1 | negative_regulation_of_detection_of_glucose |
| GO:2000976 | 1 | regulation_of_transcription_from_RNA_polymerase_II_promoter_involved_in_detection_of_glucose |
| GO:2000978 | 1 | negative_regulation_of_forebrain_neuron_differentiation |
| GO:2000979 | 1 | positive_regulation_of_forebrain_neuron_differentiation |
| GO:2000982 | 1 | positive_regulation_of_inner_ear_receptor_cell_differentiation |
| GO:2000983 | 1 | regulation_of_ATP_citrate_synthase_activity |
| GO:2000984 | 1 | negative_regulation_of_ATP_citrate_synthase_activity |
| GO:2001025 | 1 | positive_regulation_of_response_to_drug |
| GO:2001029 | 1 | regulation_of_cellular_glucuronidation |
| GO:2001031 | 1 | positive_regulation_of_cellular_glucuronidation |
| GO:2001033 | 1 | negative_regulation_of_double-strand_break_repair_via_nonhomologous_end_joining |
| GO:2001034 | 1 | positive_regulation_of_double-strand_break_repair_via_nonhomologous_end_joining |
| GO:2001040 | 1 | positive_regulation_of_cellular_response_to_drug |
| GO:2001045 | 1 | negative_regulation_of_integrin-mediated_signaling_pathway |
| GO:2001049 | 1 | regulation_of_tendon_cell_differentiation |
| GO:2001051 | 1 | positive_regulation_of_tendon_cell_differentiation |
| GO:2001052 | 1 | positive_regulation_of_G1/S_transition_checkpoint |
| GO:2001069 | 1 | glycogen_binding |
| GO:2001108 | 1 | positive_regulation_of_Rho_guanyl-nucleotide_exchange_factor_activity |
| GO:2001109 | 1 | regulation_of_lens_epithelial_cell_proliferation |
| GO:2001111 | 1 | positive_regulation_of_lens_epithelial_cell_proliferation |
| GO:2001138 | 1 | regulation_of_phospholipid_transport |
| GO:2001144 | 1 | regulation_of_phosphatidylinositol-3,4,5-trisphosphate_5-phosphatase_activity |
| GO:2001145 | 1 | negative_regulation_of_phosphatidylinositol-3,4,5-trisphosphate_5-phosphatase_activity |
| GO:2001151 | 1 | regulation_of_renal_water_transport |
| GO:2001153 | 1 | positive_regulation_of_renal_water_transport |
| GO:2001169 | 1 | regulation_of_ATP_biosynthetic_process |
| GO:2001170 | 1 | negative_regulation_of_ATP_biosynthetic_process |
| GO:2001171 | 1 | positive_regulation_of_ATP_biosynthetic_process |
| GO:2001176 | 1 | regulation_of_mediator_complex_assembly |
| GO:2001178 | 1 | positive_regulation_of_mediator_complex_assembly |
| GO:2001180 | 1 | negative_regulation_of_interleukin-10_secretion |
| GO:2001181 | 1 | positive_regulation_of_interleukin-10_secretion |
| GO:2001182 | 1 | regulation_of_interleukin-12_secretion |
| GO:2001183 | 1 | negative_regulation_of_interleukin-12_secretion |
| GO:2001186 | 1 | negative_regulation_of_CD8-positive,_alpha-beta_T_cell_activation |
| GO:2001188 | 1 | regulation_of_T_cell_activation_via_T_cell_receptor_contact_with_antigen_bound_to_MHC_molecule_on_antigen_presenting_cell |
| GO:2001189 | 1 | negative_regulation_of_T_cell_activation_via_T_cell_receptor_contact_with_antigen_bound_to_MHC_molecule_on_antigen_presenting_cell |
| GO:2001191 | 1 | regulation_of_gamma-delta_T_cell_activation_involved_in_immune_response |
| GO:2001193 | 1 | positive_regulation_of_gamma-delta_T_cell_activation_involved_in_immune_response |
| GO:2001201 | 1 | regulation_of_transforming_growth_factor-beta_secretion |
| GO:2001202 | 1 | negative_regulation_of_transforming_growth_factor-beta_secretion |
| GO:2001204 | 1 | regulation_of_osteoclast_development |
| GO:2001205 | 1 | negative_regulation_of_osteoclast_development |
| GO:2001213 | 1 | negative_regulation_of_vasculogenesis |
| GO:2001220 | 1 | negative_regulation_of_G2_phase_of_mitotic_cell_cycle |
| GO:2001223 | 1 | negative_regulation_of_neuron_migration |
| GO:2001245 | 1 | regulation_of_phosphatidylcholine_biosynthetic_process |
| GO:2001246 | 1 | negative_regulation_of_phosphatidylcholine_biosynthetic_process |
| GO:2001260 | 1 | regulation_of_semaphorin-plexin_signaling_pathway |
| GO:2001262 | 1 | positive_regulation_of_semaphorin-plexin_signaling_pathway |
| GO:2001263 | 1 | regulation_of_C-C_chemokine_binding |
| GO:2001264 | 1 | negative_regulation_of_C-C_chemokine_binding |
| GO:2001270 | 1 | regulation_of_cysteine-type_endopeptidase_activity_involved_in_execution_phase_of_apoptosis |
| GO:2001271 | 1 | negative_regulation_of_cysteine-type_endopeptidase_activity_involved_in_execution_phase_of_apoptosis |
| GO:2001273 | 1 | regulation_of_glucose_import_in_response_to_insulin_stimulus |
| GO:2001274 | 1 | negative_regulation_of_glucose_import_in_response_to_insulin_stimulus |
Format: map
Web label: GO Terms
Web label: key| (|count|): |desc
| Filename | Size | md5 |
|---|---|---|
| GO-map.txt | 11.29 MB | 7d8283e927db6444ec0586a05ea9cf0a |
| GO-mapnames.txt | 790.16 KB | dd871ffe242113f9e37a666a737ec5a9 |
| genomic_resource.yaml | 312.0 B | cda1767a1f9ded2807d993219444ea7a |
| statistics/ |