| Id: | gene_properties/gene_scores/new/pNull |
| Type: | gene_score |
| Version: | 0 |
| Summary: |
Probability of LoF tolerance |
| Description: |
The probability of being null (pNull) reflects a gene’s tolerance to loss-of-function (LoF) variants in both heterozygous and homozygous states. Genes with high pNull values show LoF variants at expected frequencies, indicating little or no selective constraint. Such genes are considered LoF-tolerant, meaning that complete loss of function is typically compatible with normal human health. The pNull score is computed alongside pLI and pRec using population-level LoF data from ExAC and gnomAD. Lek et al., Analysis of protein-coding genetic variation in 60,706 humans, Nature 2016 Downloaded on: 10/24/25 Dataprep.py takes the pNull column from Supplementary Table 13 "Gene Constraint" sheet and adds a pNull_rank column. |
| Labels: |
| ID | Type | Description | Histograms | Range |
|---|---|---|---|---|
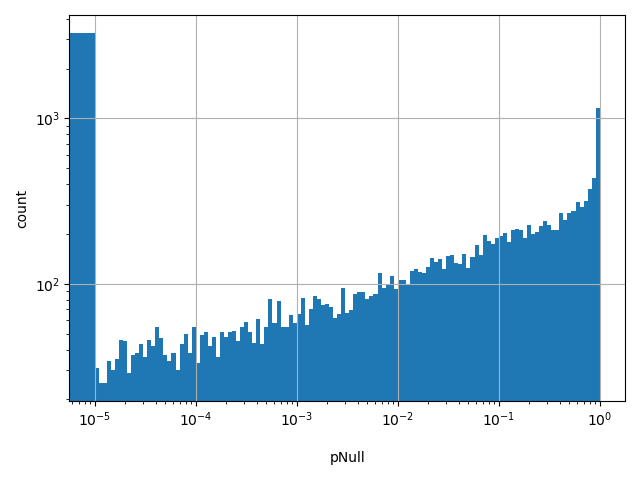
| pNull |
Probability of biallelic loss-of-function intolerance
|

|
[5.79e-127, 1] | |
| pNull_rank |
Gene rank after sorting by pNull score
|

|
[1, 1.82e+04] |
| Filename | Size | md5 |
|---|---|---|
| dataprep.py | 536.0 B | f4bd4dfbf7d124d8225c41758cfb962a |
| genomic_resource.yaml | 1.38 KB | d36b5d328ef55e887c707aeefdfba8ff |
| nature19057-SI Table 13.xlsx | 5.55 MB | 183617a934bf63b9df992104391bbecc |
| pNull.csv | 589.27 KB | 1e953bae62d1bf5545b5e7cc23f6a48e |
| statistics/ |