| Id: | gene_properties/gene_scores/LOEUF |
| Type: | gene_score |
| Version: | 0 |
| Summary: |
Degree of intolerance to predicted Loss-of-Function (pLoF) variation |
| Description: |
IntroductionDegree of intolerance to pLoF variation in each gene is assessed using the continuous metric of the observed/expected ratio. A confidence interval is estimated around the ratio. The loss-of-function observed/expected upper bound fraction (LOEUF) is the 90% upper bound of this confidence interval. Score definitionsLow scores or ranks indicate intolerance to loss of function mutations. Processing DetailsSupplementary Data, Table 11 downloaded on 2024 May 5 from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7334197/#MOESM1. DataPrep.py utilizes only scores for canonical transcripts (column canonical = TRUE). The 'oe_lof_upper' column in the downloaded file is used for LOEUF scores. LOEUF_ranks are calculated based on LOEUF scores. Resulting file LOEUF_scores.csv.gz has 3 columns: gene, LOEUF score, LOEUF rank. |
| Labels: |
| ID | Type | Description | Histograms | Range |
|---|---|---|---|---|
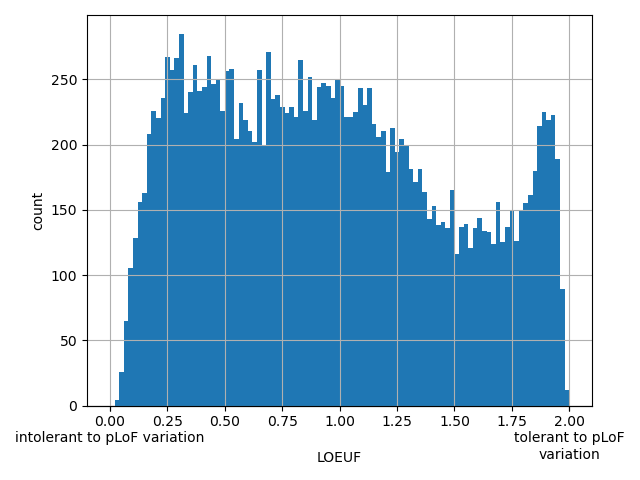
| LOEUF |
90% upper bound for the confidence interval for observed/expected pLoF variation ratio
Small values desc: intolerant to pLoF variation
Large values desc: tolerant to pLoF variation
|

|
[0.03, 2] | |
| LOEUF_rank |
Gene ranks after sorting by LOEUF scores
Small values desc: intolerant to pLoF variation
Large values desc: tolerant to pLoF variation
|

|
[1, 1.92e+04] |
| Filename | Size | md5 |
|---|---|---|
| DataPrep.py | 1.09 KB | ec139d6ccbaa42ca8aa8c303b497155e |
| LOEUF_scores.csv.gz | 129.89 KB | 2de3bb891f944af4d9cc3ac642294e99 |
| genomic_resource.yaml | 1.79 KB | b3855f4060b82193109415a205a64f47 |
| supplementary_dataset_11_full_constraint_metrics.tsv.gz | 13.02 MB | 338a3ed667290813cafbb72eabcba2e0 |
| statistics/ |